Abstract
Post-translational modification with one of the isoforms of the small ubiquitin-like modifier (SUMO) affects thousands of proteins in the human proteome. The binding of SUMO to SUMO interacting motifs (SIMs) can translate the SUMOylation event into functional consequences. The E3 ubiquitin ligase RNF4 contains multiple SIMs and connects SUMOylation to the ubiquitin pathway. SIM2 and SIM3 of RNF4 were shown to be the most important motifs to recognize SUMO chains. However, the study of SIM-SUMO complexes is complicated by their typically low affinity and variable binding of the SIMs in parallel and antiparallel orientations. We investigated properties of complexes formed by SUMO3 with peptides containing either SIM2 or SIM3 using molecular dynamics simulations. The affinities of the complexes were determined using a state-of-the-art free energy protocol and were found to be in good agreement with experimental data, thus corroborating our method. Long unrestrained simulations allowed a new interpretation of experimental results regarding the structure of the SIM-SUMO interface. We show that both SIM2 and SIM3 bind SUMO3 in parallel and antiparallel orientations and identified main interaction sites for acidic residues flanking the SIM. We noticed unusual SIM-SUMO interfaces in a previously reported NMR structure (PDB: 2mp2) of a complex formed by a SUMO3 dimer with the bivalent SIM2-SIM3 peptide. Computational determination of the individual SIM-SUMO affinities based on these structural arrangements yielded significantly higher dissociation constants. To our knowledge, our approach adds new opportunities to characterize individual SIM-SUMO complexes and suggests that further studies will be necessary to understand these interactions when occurring in multivalent form.
Significance
SUMOylation, the conjugation of the small ubiquitin-like modifier (SUMO), marks the SUMOylated protein for various cellular processes. The noncovalent interaction of SUMO and the SUMO-targeted ubiquitin ligase RNF4, for example, targets SUMOylated proteins for ubiquitination. We investigate the interaction of SUMO and the two most important SUMO interacting motifs (SIMs) of RNF4 by molecular dynamics simulations, including detailed free energy calculations. The numerical results for the affinities of these complex systems agree well with the experiment. Furthermore, the structures we sampled show important characteristics of the SIM-SUMO interface. This indicates that simulations are a valuable tool to understand SIM-SUMO complexes. Regarding the structures of RNF4-SIM-SUMO complexes, our results challenge some previous experimental deductions and point toward difficulties in understanding multivalent SIM-SUMO interactions.
Introduction
SUMOylation, the conjugation of the small ubiquitin-like modifier (SUMO) to a target protein, is a post-translational modification that affects thousands of proteins in the human proteome (1). The isoforms SUMO2 and SUMO3 are nearly identical and are also referred to as SUMO2/3. They share 50% sequence identity with the isoform SUMO1 (2).
The most prevalent type of noncovalent interaction between SUMO binding proteins and SUMO is mediated by SUMO interacting motifs (SIMs) (3). SIMs are peptide stretches that typically consist of four amino acids (4). Many SIMs, but not all, fall into one of three categories: (hhhh) SIMs, (hhXh) SIMs, and (hXhh) SIMs, where h is a hydrophobic and X is an acidic residue (5). SIMs are disordered in solution (6,7); however, upon binding the second β-sheet of SUMO, they adopt a β-sheet configuration themselves that either aligns in parallel or antiparallel orientation with respect to SUMO. Beside this backbone-mediated interaction, the SIM-SUMO interface is stabilized by interaction between side chains of the SIM and the hydrophobic groove between the second β-sheet and α-helix of SUMO (3).
Frequently, the SIMs are flanked by acidic residues or residues with a phosphorylation site (8). The interaction of negative charges near the SIM and positive charges on the SUMO surface is assumed to be a requirement for high-affinity binding as well as a determinant of the bound orientation (9).
SUMO2 and SUMO3 form covalent chains, typically via an isopeptide bond between the C-terminal glycine and Lys11. Proteins with multiple SIMs are predestined to bind polySUMO chains. The interaction of multiple SIMs in the E3 ubiquitin ligase RING finger protein 4 (RNF4) and polySUMO chains connects the latter to the ubiquitin pathway. Multivalent SIM-SUMO interaction in general and the interaction of multiple RNF4-SIMs and SUMO chains in particular have been the subject of multiple studies in past years (5,6,9,10). Direct structural information on these complexes is scarce, however. The structure of the complex formed by the SIM2-SIM3 peptide of RNF4 and diSUMO3, which has been solved with NMR by Xu et al., is the only available structure of a multivalent SIM-SUMO complex involving a SUMO chain in the UniProt database (PDB: 2mp2) (9). Note that the SUMO isoform nomenclature is used inconsistently in literature. Here, we stick to the names used in the UniProt database, in which the SUMO proteins investigated by Xu et al. are referred to as SUMO3, whereas Xu et al. called them SUMO2 in their publication.
Of the four SIMs in RNF4, SIM2 and SIM3 have been identified as the most important (6,9). Results regarding the interfaces of these SIMs in complex with SUMO are, however, somewhat puzzling. For example, Xu et al. found that the overall orientation of the bivalent binding of the SIM2-SIM3 peptide can flip relative to the diSUMO minimal chain, with each SIM switching binding between the proximal and the distal SUMO. Nevertheless, at the individual SIM-SUMO interfaces, SIM2 always adopted a parallel orientation, and SIM3 always adopted an antiparallel orientation (9). On the other hand, Kung et al., who used indirect NMR experiments, found that individual SIM2 and SIM3 peptides bind SUMO2 in parallel orientation and deduced that this would also be the case in multivalent interaction of RNF4-SIMs and SUMO chains (6).
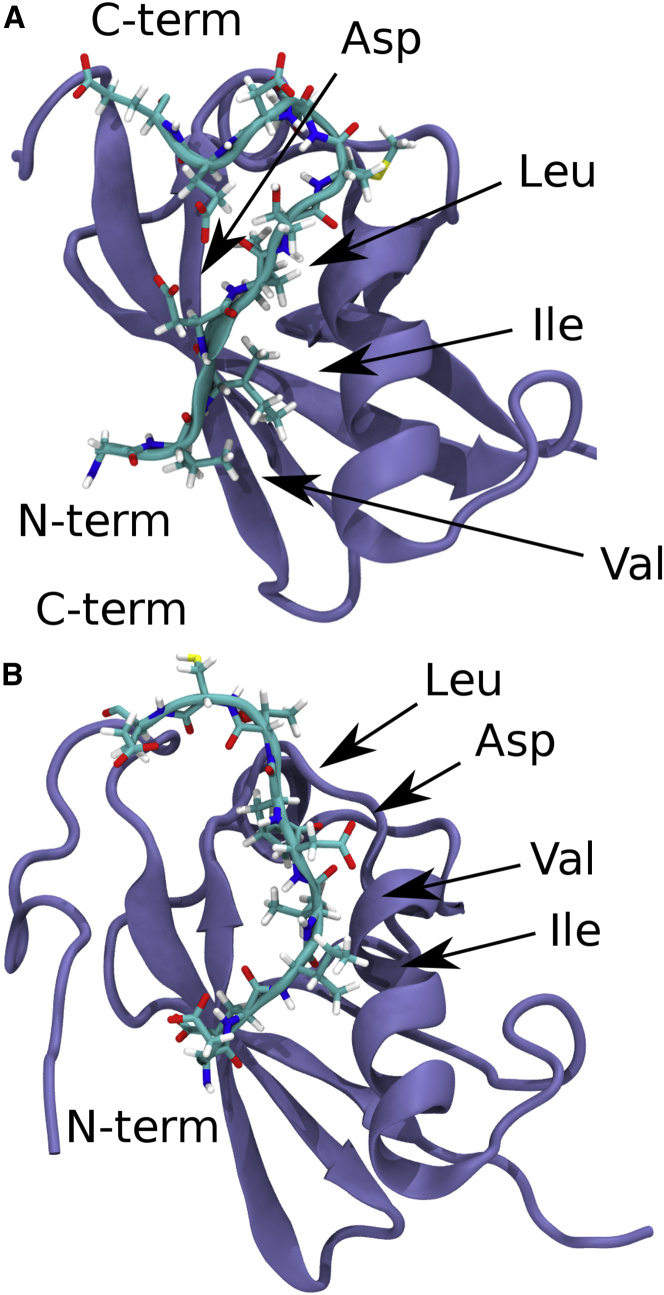
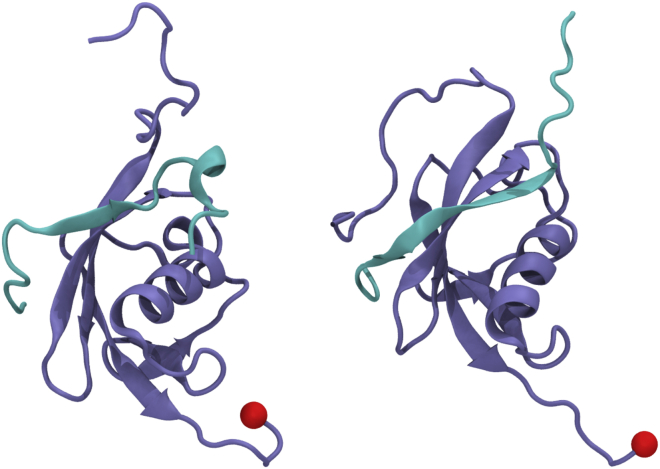
Furthermore, the individual SIM-SUMO interfaces in the SIM2-SIM3-diSUMO structure of Xu et al. show some remarkable deviations from typical examples of SIM-SUMO interfaces in the literature. Fig. 1 shows a comparison of the SIM peptide of MCAF1 (VIDL) (11) in complex with SUMO3 and the SIM2-SUMO3 interface from Xu et al. The SIM peptide of MCAF1 forms a β-sheet that aligns in parallel orientation with the second β-sheet of SUMO. The side chains of isoleucine and leucine on the second and fourth position of the MCAF1-SIM occupy the hydrophobic groove of SUMO. These structural features are characteristic for the SIM-SUMO interface (3,4) and can be found in all structures of a SIM-SUMO2/3 interface available on the UniProt database (June 2020, see PDB: 2d07 (12), PDB: 2n9e (13), PDB: 3uin (14), PDB: 5d2m (15), and PDB: 6k5r (16)). In contrast, the SIM2-SUMO3 interface from Xu et al. deviates from this picture in that only valine on the second position of SIM2 (IVDL) is turned toward the SIM binding groove, at a similar location where leucine from the fourth position of the MCAF1-SIM binds. Furthermore, the peptide forms no β-sheet and does not align with the second β-sheet of SUMO; thus, no stabilizing hydrogen bonds are formed. Similar deviations from the prototypical SIM-SUMO interface can also be found in the case of the individual SIM3-SUMO3 interface in the bivalent structure of Xu et al.
Figure 1.
Structures of SUMO3 in complex with SIM peptides. (A) Shown is a peptide containing the MCAF1-SIM (VIDL) taken from PDB: 2rpq. (B) The RNF4-SIM2 peptide taken from PDB: 2mp2 is shown. We show the same peptide that we use for the simulations, i.e., the SIM2 peptide found in Table 1. To see this figure in color, go online.
We aimed to understand this discrepancy by computer simulations of the SIM-SUMO complex. We used atomistic molecular dynamics (MD) simulations to investigate structural properties of complexes formed by peptides containing SIM2 or SIM3 of RNF4 and SUMO3 (see Table 1). We used the method of Woo and Roux (17) to estimate the affinities of the SIM2 and SIM3 peptides to SUMO, which also directly allowed us to quantify the contribution of either binding orientation to the overall affinity (18).
Table 1.
Sequences of the Considered Peptides (SIM2 and SIM3) and the SIM2-SIM3 Peptide Considered by Xu et al
| Peptide | Sequence |
|---|---|
| SIM2 | (rETAG)DEIVDLTCE(SLEPrw) |
| SIM3 | (TCESL)EPVVVDLTHND(Sw) |
| SIM2-SIM3 | TVGDEIVDLTCESLEPVVVDLTHND |
The SIMs themselves are underlined. The residues in parenthesis are cut off in our model but present in experiments by Kung et al. Small letters indicate residues that are not part of RNF4 but have been added by Kung et al. Note that in our smaller peptides, the termini were amidated/acetylated.
Our simulation results suggest that the monovalent SIM-SUMO interfaces excised from the NMR structure of Xu et al. show significantly lower affinities than is typically observed for the SIM-SUMO interaction. In contrast, SIM-SUMO interfaces found by molecular docking that are structurally similar to the usual SIM-SUMO interface bind in the expected affinity range. Our simulation results furthermore indicate that SIM2 and SIM3 peptides bind SUMO2/3 both in parallel and antiparallel orientation.
Methods
We used the CHARMM Gui for system construction (19). We solvated the protein-peptide complexes that were either obtained from PDB: 2mp2 or molecular docking using the HPEPDOC web server (20) in TIP3P water (21) such that the distance between complex and box edges was at least 1 nm. We added sodium or chloride ions to render the system charge neutral. The protonation state of amino acids was checked with propka (22), integrated in the playmolecule web server (23). We used the CHARMM 36m force field (24) and GROMACS version 2018 (25) for all simulations. The simulations were performed at 303.15 K using Nosé-Hoover temperature coupling (26) and at atmospheric pressure using Parrinello-Rahman pressure coupling (27). For more details on the optimal simulation parameters for the CHARMM force field in GROMACS, we refer to (28). Umbrella sampling was carried out using the PLUMED (29) patch for GROMACS, version 2.3.
We applied the method of Woo and Roux (17) to calculate the affinities of the SIM peptides to SUMO. The affinity of protein and peptide is characterized by the binding constant or equivalently its inverse, the dissociation constant . Here, the terms in square brackets are concentrations of protein-ligand complex, free protein, and free ligand. The binding constant is related to the free energy of binding with respect to a standard concentration by the equation . Typically, is set to 1 M. Here, R and T are gas constant and temperature.
Woo and Roux write in terms of the equilibrium constant in a system in which configuration (c), orientation (o), and angular part of the position (a) of the ligand are restrained, which we denote by :
| (1) |
The factors on the right hand side, except for , may be interpreted as correction factors that account for applying and releasing restraints on configuration (index c), angular part of the position (index a), and orientation (index o), whereas is the binding constant in the restrained system. We define orientation and angular part of the position of the ligand with respect to the protein using three angles and two angles , respectively. The configuration of the ligand is defined as the root mean-square deviation (RMSD) from a reference position after optimal alignment. Using a harmonic potential, we restrain the collective variables to reference values taken from the structures after an unrestrained 100-ns MD run. The force constants for the restraining potentials are 1200 kJ/mol for the angles and 4200 kJ/mol for the RMSD.
In total, we have six correction factors in the bound and six correction factors in the free state (the correction factors for the angular and orientational restraints comprise two and three factors, e.g., ). The underlined factors in Eq. 1 are accessible by numerical integration. All other contributions require the estimation of the probability density of the restrained collective variable in the case of the correction factors and of the distance of protein and peptide in the case of . For this, we used replica exchange umbrella sampling (REMD-US) (28,30) (exchanges between neighboring umbrella windows were attempted every 1000 steps) and the weighted histogram analysis method (WHAM) (31), as implemented by Grossfield (32).
To determine the contribution of parallel and antiparallel binding mode to the affinity, we treat the two binding modes as separate systems and add up the results: . See also our previous work (18) for more details on the method.
For the umbrella sampling parameters used in this study, we refer to the respective section in the Supporting Material. We prepared figures and manipulated structures with VMD (33).
Results
The basis for our simulations is the NMR structure of the SIM2-SIM3-diSUMO complex solved by Xu et al. (PDB: 2mp2). The SUMO3 dimer in PDB: 2mp2 consists of truncated SUMO3 monomers—namely, SUMO3-N11 and SUMO3-GG. To have identical proteins in all simulations, we therefore used truncated SUMO3 proteins with the 11 N-terminal residues cut off and the C-terminal two glycines cut off in all simulations.
To limit the necessary computation time, especially for the simulations needed for the estimation of the affinities, we cut off the peptides after the nearest acidic neighbors of the SIMs (see Table 1). It is a common procedure to investigate the properties of SIM-SUMO complexes using SIM peptides that only contain two to four residues flanking the SIM (11,34,35). To further test the validity of our model, we performed additional simulations using the full-length SIM2 peptide of Kung et al. in complex with the full-length SUMO3. Fig. S1 shows the fluctuations of the positions of the SIM2 peptide residues. Although the SIM itself and close neighbors are highly restrained by binding SUMO, the positions of remote residues fluctuate strongly, indicating that they are not involved in binding SUMO.
As pointed out in the Introduction, the individual SIM-SUMO interfaces in PDB: 2mp2 lack key stabilizing characteristics of the standard SIM-SUMO interface. Because no other structures of RNF4-SIM-SUMO interfaces were available, it was unclear whether the unusual interfaces are a result of the bivalent binding or typical for these SIMs. Although MD simulations are in principle able to sample the whole configuration space of the SIM-SUMO complexes considered here, there is no guarantee that this can be achieved in the available computation time.
As a first step toward clarifying whether SIM2 and SIM3 may also bind in the usual configurations, we thus used the HPEPDOC web server (20) to dock the SIM2 and SIM3 peptides to an individual SUMO structure excised from PDB: 2mp2. Both for SIM2 and SIM3 and in both orientations, the docking tool suggested structures that were close to the typical SIM-SUMO interface. Based on this, we decided to perform MD simulations starting in six different initial structures: the SIM2-SUMO3 complex in parallel orientation and the SIM3-SUMO3 complex in antiparallel orientation excised from the NMR structure of Xu et al., the docked SIM2-SUMO3 complexes in parallel and antiparallel orientation, and the docked SIM3-SUMO3 complexes in parallel and antiparallel orientation.
Estimation of affinities
We estimate the affinities using the protocol of Woo and Roux (17). This protocol involves multiple enhanced sampling simulations (here: umbrella sampling, i.e., restrained MD simulations; for more details, see Methods). We found that during all six sets of these enhanced sampling simulations, the regions of the configuration space visited by the individual trajectories did not overlap between these sets. This means that the following results have to be interpreted as affinities associated with the configuration space in the neighborhood of the starting structures.
The dissociation constants from the simulations starting in the structures excised from PDB: 2mp2 are mM for the SIM2 peptide and mM for the SIM3 peptide. These are 600- and 1800-fold higher, respectively, than the experimentally determined dissociation constants (Kung et al. measured mM for SIM2 and mM for SIM3) (6). The results of the simulations and experimental values from Kung et al. are summarized in Table 2. In contrast to the results obtained from the 2mp2 initial structures, these latter results are in excellent agreement with experimental measurements by Kung et al. We remark, however, that it is likely that our results benefit from cancelation of errors. We find that SIM2 binds SUMO in parallel and antiparallel orientation with similar affinities, whereas SIM3 shows higher affinity for the parallel orientation. The difference between these affinities is, however, relatively small, such that we would expect that both orientations could be seen in experiments.
Table 2.
Dissociation Constants: Results from Our Simulations and Reference Experimental Results
| SIM2 | SIM3 | |
|---|---|---|
| (mM) | 26.18 | – |
| (mM) | – | 163.47 |
| (mM) | 0.06 | 0.10 |
| (mM) | 0.10 | 0.30 |
| (mM) | 0.04 | 0.08 |
| (mM) |
and are dissociation constants estimated from simulations starting in the 2mp2 structures in parallel and antiparallel orientation, respectively.
and are dissociation constants estimated from simulations starting in the docked complexes in parallel and antiparallel orientation, respectively. is the resulting total dissociation constant from the docked simulations. is the reference experimental result from Kung et al.
We provide a detailed discussion of the individual contributions to the estimate of the binding free energy and their convergence in the Supporting Material. This seems especially necessary because the good agreement between experimental measurement and computational estimation of the free energy is one important indication that supports the assumption that the structures we sampled are close to experimentally relevant structures.
We have to remark again, however, that during the simulations starting with the NMR structures, the systems did not visit the apparently far more stable docked structures, despite microsecond-long sampling of the bound configurations (see also Supporting Material). Therefore, we should only consider our results locally converged.
Large-scale studies on pharmaceutically relevant ligand-receptor pairs indicate that free energy estimates from MD simulation exhibit systematic errors in the range of 1 kcal/mol (36). The statistical error found for affinity estimates with the method used in this study is typically much lower, even when shorter simulation times were used (18,37). This suggests that statistical errors can indicate higher accuracy than we actually have.
Structural properties of the complexes
To investigate structural properties of the monovalent SIM2- and SIM3-SUMO3 complexes, we performed unrestrained 1-μs-long MD simulations. The estimates of the affinities strongly suggest that, when considering monovalent complexes, the individual SIM-SUMO interfaces found in the NMR structure of the bivalent complex (PDB: 2mp2) are not relevant. In the following, we will therefore mostly focus on the simulations starting in the docked structures. We remark that also during these long unrestrained simulations, the trajectories starting in the NMR structures did not visit the more stable docked structures, nor did the trajectories starting in the docked structures visit the interfaces found in PDB: 2mp2.
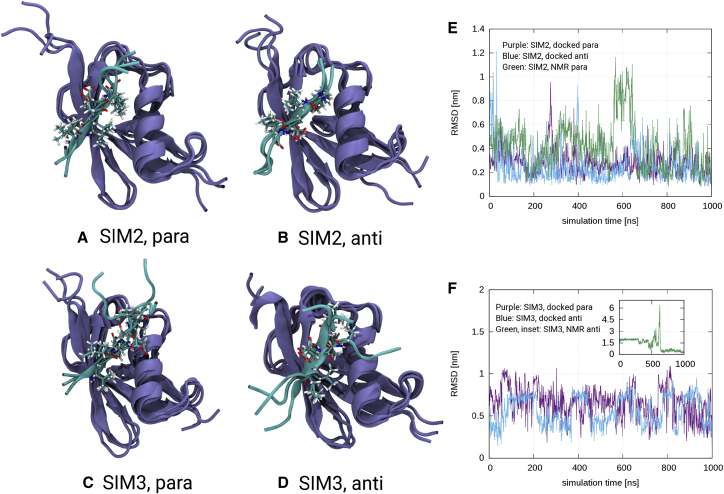
Fig. 2, A–D show snapshots of the simulations starting in the docked structures. Fig. 2, E and F show the RMSD of the SIM peptide backbone atoms with respect to the structure after the full simulation time. Note that here, the RMSD has been calculated after optimal alignment of the SUMO structure. Both for SIM2 and SIM3, the RMSD over the course of the simulation is smaller for the simulations starting in the docked structure compared with the simulations starting in the 2mp2 interfaces. This effect is particularly strong in the case of the SIM3 peptide: the system leaves the initial SIM-SUMO interface and changes the bound orientation during simulation that starts in the NMR structure (see also Figs. S4 and S5 for snapshots of the simulations starting in the NMR structures). Furthermore, Fig. 2, E and F show that in both orientations the SIM2 peptide is stronger restrained by binding than the SIM3 peptide.
Figure 2.
(A–D) Snapshots of the 1-μs simulations starting in the docked structures after 100, 500 ns, and 1 μs. SIMs are shown in atomic detail. For the SIM3 peptide in parallel orientation, the residue Val3 is additionally highlighted because it occupies the SUMO groove. (E and F) Shown is the RMSD of the backbone atoms of the SIM peptides with respect to the structure after full simulation time. To see this figure in color, go online.
Fig. 2, A–D also show that in the simulations starting in the docked structures, the SIM peptide forms a β-sheet that aligns either in parallel or antiparallel orientation with the second β-sheet of SUMO. Furthermore, in each case, multiple stable hydrogen bonds with the β-sheet of SUMO are formed (see Fig. S6). The groove of SUMO is occupied by the side chains of Val4 and Leu6 (SIM2 in parallel orientation), Ile3 and Asp5 (SIM2 in antiparallel orientation), Val3 and Val5 (SIM3 in parallel orientation), and VAL5 and Leu7 (SIM3 in antiparallel orientation). See also Figs. S7 and S8 for a comparison of the RNF4-SIM-SUMO interfaces found in our simulations and typical examples from literature.
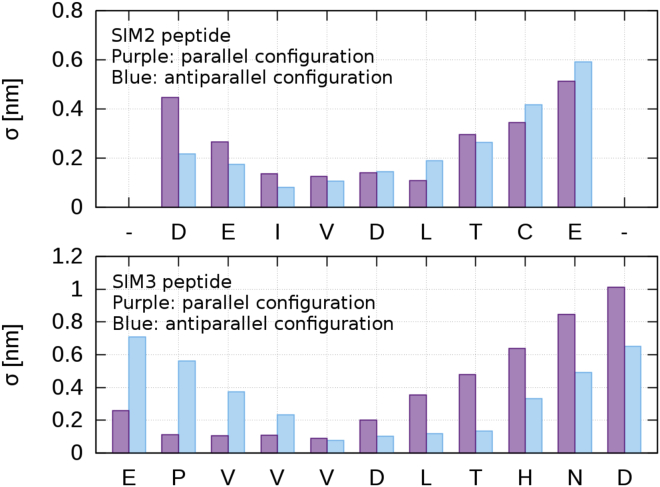
To investigate how much the SIM peptides are restricted in their motion by binding SUMO, we consider the standard deviation σ of the geometric centers of the SIM peptides residues. Because we are only interested in the motion of the SIM peptides relative to the SUMO, we first aligned 200 equally spaced structures from the 1-μs simulations using the SUMO structure as reference and then calculated the standard deviations from these aligned structures. The results are shown in Fig. 3.
Figure 3.
Standard deviations of the centers of the SIM peptide residues from their average position in complex with SUMO. We used 200 configurations of the simulation starting in the docked structures, written out every 5 ns. Top: SIM2; bottom: SIM3; purple: parallel orientation; blue: antiparallel orientation. To see this figure in color, go online.
The standard deviations indicate that the SIM cores that are involved in the β-sheet and hydrophobic interaction to SUMO are strongly restrained in their motion. This could certainly be expected. Still, this observation is important to note because it suggests that the structural properties of the SIM-SUMO interfaces found in Fig. 2, C–F are actually representative of the whole trajectory.
Role of flanking acidic residues
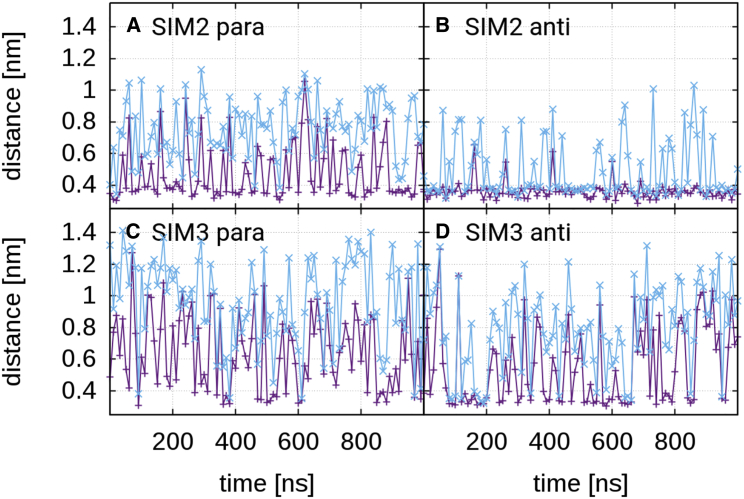
We then investigated the impact of the negatively charged amino acids that typically flank the core SIM sequence. Fig. 4 shows the two lowest distances between a negatively charged side chain flanking the SIM and a positively charged SUMO side chain against the simulation time (see Fig. S9 for structures of the complexes in which the charged side chains are highlighted and the Supporting Material for a precise definition of the observable). In the case of SIM2 in antiparallel orientation, the distances are constantly low. The reason for this is the formation of stable salt bridges between Asp1(SIM2 peptide) and Glu2(SIM2 peptide) and Lys34(SUMO3) and Lys41(SUMO3). In the case of SIM2 in parallel orientation and SIM3 in antiparallel orientation, less stable salt bridges form and break from time to time. These salt bridges form between Glu9(SIM2 peptide) and Lys34(SUMO3) and Lys41(SUMO3) in the case of SIM2 and between Glu1(SIM3 peptide) and Lys34(SUMO3) and Lys41(SUMO3) in the case of SIM3. Note also that for SIM3 in antiparallel orientation, salt bridges form only very infrequently during the second half of the simulation. Between flanking acidic residues of the SIM3 peptide in parallel orientation and SUMO3, no stable salt bridges form at all. The averages of the two lowest distances are 0.47 and 0.76 nm (SIM2 parallel), 0.36 and 0.50 nm (SIM2 antiparallel), 0.64 and 0.96 nm (SIM3 parallel), and 0.55 and 0.78 nm (SIM3 antiparallel).
Figure 4.
The two lowest distances between a negatively charged side chain of SIM peptide and a positively charged side chain of SUMO. (A) Shown are SIM2 in parallel orientation, (B) SIM2 in antiparallel orientation, (C) SIM3 in parallel orientation, (D) and SIM3 in antiparallel orientation. To see this figure in color, go online.
Together, our observation indicated that charged residues of the SIM2 and SIM3 peptides form salt bridges to Lys34 and Lys41, which are placed in the region between second β-sheet and α-helix of SUMO3. The equivalent residues in SUMO2 are Lys35 and Lys42. Consistent with our findings, these lysine residues have been identified as main interaction sites for negatively charged side chains of similar SIM peptides in multiple experimental studies (13,38, 39, 40).
As pointed out in the section above, the SIM2 peptide in antiparallel orientation shows the smallest average number of hydrogen bonds to SUMO (see Table S3), and only one hydrophobic side chain of the SIM2 peptide occupies the groove (see also Fig. 2 B). This indicates that the contribution of hydrogen bonds and hydrophobic interactions to the affinity between SIM peptide and SUMO is lower than in the other sampled structures (see also Fig. 2, A, C, and D).
The formation of stable salt bridges between Asp1(SIM2 peptide) and Glu2(SIM2 peptide) and Lys35(SUMO) and Lys42(SUMO) might be the reason for the high affinity of SIM2 and SUMO in antiparallel orientation, as estimated from our simulations. To further quantify the stabilizing effect of the salt bridges on the SIM-SUMO complex, we set up systems in which the acidic groups of Asp1, Glu2, and Glu9 in the case of the SIM2 peptide and Glu1 and Asp11 in the case of the SIM3 peptide were protonated. Following the same protocol as before, we calculated the affinities for the protonated systems (Table 3). Whereas the SIM2 peptide in parallel orientation was affected 25-fold by the protonation, the same peptide in antiparallel orientation showed a 120-fold lower affinity. In contrast, for the SIM3 peptide, the effect of the protonation was very limited in both orientations. These results together with the data shown in Fig. 4 support the interpretation that the ionic interactions between Asp1(SIM2 peptide) and Glu2(SIM2 peptide) and Lys34(SUMO3) and Lys41(SUMO3) are of particular importance for the stability of the SIM2 complex in antiparallel configuration.
Table 3.
Dissociation Constants for the Protonated Systems
| SIM2 | SIM3 | |
|---|---|---|
| (mM) | 1.53 | 0.16 |
| (mM) | 11.97 | 0.52 |
| (mM) | 1.36 | 0.12 |
Dissociation constants from our simulations for the parallel () and antiparallel () complexes and resulting total dissociation constants () with the flanking acidic residues protonated.
In parentheses, we give the results for the unprotonated systems.
Finally, we note that, except for SIM2 in antiparallel orientation, no stable salt bridges form between the central acidic residue (i.e., Asp5 of the SIM2 peptide or Asp6 of the SIM3 peptide) and positively charged side chains of SUMO (data not shown). In the case of SIM2 in antiparallel orientation, Asp5(SIM2 peptide) forms a salt bridge to Arg49(SUMO3). Note that Asp5(SIM2 peptide) at the central position has not been protonated by us. The affinities reported in Table 3 thus indicate that a hydrophobic residue on this position in the binding groove, as in the case of SIM3, results in a much higher affinity between the SIM peptide and SUMO and that a salt bridge between an acidic amino acid in the SIM and Arg49 has no strong stabilizing effect on the SIM-SUMO interface. Consistent with this observation, it has been found that the interaction of SIM peptides and SUMO1 as well as SUMO2 is largely unaffected by mutation of Arg54(SUMO1) and Arg50(SUMO2) (41,42). Arg(54) is the SUMO1 equivalent of Arg50(SUMO2) and Arg49(SUMO3).
Comparison with experimental results on the bound orientation
As pointed out in the introduction, Kung et al. found that individual SIM2 and SIM3 peptides bind SUMO2 exclusively in parallel orientation (6). This is in contrast to our results, which indicate that the peptides bind SUMO3 in both orientations with similar affinities. Kung et al. used two different experimental setups for the determination of the bound orientation.
In the case of SIM3, they attached the spin label MTSL at the N-terminus of the peptide and measured the loss in NMR signal intensity of the SUMO2 residues. From this experiment, they inferred that the SIM3 peptide binds SUMO2 in antiparallel orientation. Fig. S11 shows the structure of SUMO2, with the residues that exhibited the greatest loss in intensity highlighted. Our re-evaluation of the data by Kung et al. does not seem to be consistent with a model in which the SIM3 peptide binds only in parallel orientation, because this would not explain the great loss of intensity in the region around the N-terminal part of the α-helix. Instead, the data would be consistent with the SIM3 peptide binding in parallel and antiparallel orientation, which is also what our simulations suggest.
To investigate the orientation of SIM2, Kung et al. coordinated a paramagnetic ion at the C-terminus of SUMO2 such that it would broaden the resonances of SIM residues in close proximity and thus decrease their intensity. From the recorded spectra, they concluded that SIM2 binds in parallel orientation. Critical for this conclusion was the assumption that the C-terminus of SUMO2 resided near the N-terminal side of the SUMO’s second β-sheet and that the SIM peptide lies close to this β-sheet. Kung et al., however, did not provide or cite structural evidence for this assumption. To clarify this point, we consulted Spyracopoulos et al., who provided an NMR structure of SUMO2 including the full C-terminal part (PDB: 2n9e) in complex with a SIM peptide (13). Judging from this structure, it seems that the C-terminal part of SUMO2 is disordered, shows no nonbonded interactions with other parts of SUMO2, and is thus able to move around freely. To investigate the potential proximity of the C-terminal part of SUMO2 with any regions neighboring the SIM of the binding partner, we performed additional simulations starting from a structure in which the full-length SIM2 peptide used by Kung et al. (see also Table 1) was docked to the SUMO2 structure of Spyracopoulos et al. Fig. 5 shows snapshots of these simulations, and Fig. S12 shows the distance between the C-terminus of SUMO and the main SIM interacting regions of SUMO. We assumed that if the C-terminus of SUMO is close to one of the residues of these regions, it is also close to residues of the SIM peptide.
Figure 5.
Structures of docked SIM2 peptide and SUMO complexes in parallel and antiparallel orientation. These complexes resemble the experimental system by Kung et al. The C-terminus of SUMO, to which the paramagnetic ion has been attached, is highlighted in red. To see this figure in color, go online.
Fig. S12 shows that the C-terminus of SUMO is at a very similar distance to the N-terminus of the second β-sheet and to the N-terminus of the α-helix, which represent the opposite ends of the SIM binding groove. This finding suggests that the experimental setup of Kung et al. is not able to discriminate between the two possible orientations of the bound SIM peptide, because a decreased peak intensity would be expected on both sides of the peptide.
Discussion
Computational aspects
Woo and Roux specifically designed their free energy method for flexible ligands (17). For such ligands, convergence of estimates of the probability density of the distance between protein and peptide () from MD simulations is typically very slow. The key idea of the method is to restrict the flexibility of the ligand, which is expected to accelerate the convergence of the estimate of .
Here, we applied this method to investigate complexes formed by RNF4-SIM2 and RNF4-SIM3 peptides with SUMO3. We used umbrella sampling in combination with replica exchange of neighboring windows, and the simulation times for the critical collective variables distance and RMSD regularly exceeded 100 ns per window. First, monovalent SIM2-SUMO and SIM3-SUMO complexes were excised from the bivalent SIM2-SIM3-diSUMO NMR structure from PDB: 2mp2 (9) and used as initial configurations. Kung et al. provided reference values, which were determined by NMR, for the affinities of similar monovalent SIM2- and SIM3-SUMO complexes. We found the dissociation constants of SIM2 and SIM3 peptides in complex with SUMO3 to be 600- to 1800-fold higher than those experimentally determined for similar systems by Kung et al. and thus in a range much higher than typical SIM-SUMO interactions. This discrepancy indicated that both the initial equilibration and the systematic exploration of the configuration space using umbrella sampling of the RMSD failed to find high-affinity structures.
We performed a second set of simulations using docked initial configurations of the complexes. The resulting estimates of the affinities from simulation are in good agreement with experimental values by Kung et al., suggesting that the SIM-SUMO interfaces sampled would be representative of the real binding structures.
In the method of Woo and Roux (17), a series of correction factors has to be computed to account for the restraints on the ligand’s configuration. Among others, this requires umbrella sampling of the RMSD of the ligand in the bound state. Intuitively this may result in a further benefit of this method—namely, a greater independence of the results and convergence speed from the starting structure—because all possible bound configurations of the system would be sampled systematically. However, our results show that, in general, this is not true. Keeping in mind that the configurational restraints essentially turn the peptide into a rigid body, our results furthermore highlight the need of long simulation times to get a converged estimate for .
Properties of the SIM-SUMO complexes
We sampled the structures of the SIM2 and SIM3 peptides in parallel and antiparallel orientation with respect to SUMO3. In the simulations starting in the docked complexes, we found the SIM-SUMO interfaces to be very similar to known structures in the UniProt database (see also Table 4). These contain in all but one structure (PDB: 2d07 (12)) at least three negatively charged residues flanking the SIM. Furthermore, we observe an imbalance of the charges between the N-terminal and C-terminal side of the SIM. All SIMs flanked C-terminally by negative charges bind in parallel orientation, whereas the SIMs flanked N-terminally by negative charges bind in antiparallel orientation. The correlation between the position of acidic residues flanking the SIM and the orientation of the SIM to SUMO has been discussed previously (7,43). It suggests the presence of interactions between the negatively charged residues flanking the SIM and positively charged residues on the SUMO surface, specifically between the C-terminal side of the SUMO’s second β-sheet and the N-terminal part of the following α-helix. Indeed, several studies identified lysines 34 and 41 as important for SIM binding (13,38, 39, 40). Our simulations further corroborate this model and indicate that these are also the most important interaction sites for acidic residues of the SIM2 and SIM3 peptides of RNF4.
Table 4.
Seven SIM Peptide Sequences, for which Structural Data of Complexes with SUMO2/3 Are Available on UniProt
| PDB File | Protein | SIM | Orientation | Neg |
|---|---|---|---|---|
| 2d07 | TDG | RNMDVQEVQYTF | anti | 1 |
| 2n9e | RAP80 | EDAFIVIpSDpSD | para | −2 |
| 2rpq | MCAF1 | GVIDLTMDDEE | para | −4 |
| 3uin | RanBP2 | DDDVLIVYEL | anti | 2 |
| 5d2m | ZNF451 | EDDIQFVSEGP | anti | 2 |
| 5d2m | ZNF451 | VLEYIDLVSSDDEE | para | −3 |
| 6k5r | HCMV IE2 | DTAGCIVIpSDpSEEE | para | −5 |
The column protein gives the protein that contains the SIM. The SIMs are underlined, and phosphorylated serines are denoted by pS. The column Neg gives the difference between negatively charged residues flanking the SIM N-terminally and C-terminally.
The affinities determined in our simulations for SIM3 binding in parallel and antiparallel orientations differ by only 0.66 kcal/mol. This is relatively low compared with the typical error range of affinity estimates from MD simulations that stem from multiple issues like the use of empirical force fields and insufficient simulation time. Although some of these errors might have canceled each other in our case, we do not conclude here that SIM3 binds SUMO3 with higher affinity in parallel orientation.
From our simulations, we thus conclude that both SIM2 and SIM3 bind SUMO3 with similar affinities in both orientations, in contrast to the idea that (hhXh) SIMs bind SUMO2/3 exclusively in parallel orientation (5) and to the conclusions by Kung et al., who suggested that SIM2 and SIM3 bind SUMO2 in parallel orientation (6). After revisiting the experimental data that led to the latter proposal and considering our additional simulations regarding the localization of SUMO’s C-terminus relative to the SIM binding groove, we believe that the data by Kung et al. can be reconciled with our model of SIM3 binding in both orientations.
The SIM regions SIM-C and SIM-N of the scaffold protein DAXX provides an interesting comparison to the SIM2 and SIM3 regions of RNF4. Both SIM-C and SIM-N consist of the residues IIVL. Similar to the SIM2 and SIM3 regions of RNF4, the difference in negative charges N- and C-terminally of the DAXX SIM-C is only 1, and this SIM was found to bind SUMO1 and SUMO2 both in parallel and antiparallel orientation (7), in agreement with our conclusions for the RNF4 SIMs. On the other hand, SIM-N is flanked by a stretch of seven negatively charged residues on the C-terminal side and lacks such residues on the opposite side. Accordingly, Escobar-Cabrera et al. hypothesized that SIM-N would bind SUMO1 and SUMO2 only in parallel orientation and experimentally showed this to be true for SUMO1 (no investigations were reported for SUMO2) (7). Together, these findings and considerations support the idea that SIMs themselves are able to bind SUMO both in parallel and antiparallel orientation, in addition to the steering effects of flanking acidic residues.
We further ran simulations with protonated acidic residues neighboring SIM2 and SIM3. A large drop in binding free energy was observed for the SIM2 peptide in antiparallel orientation. This indicated the importance of the ionic interactions with Lys34 and Lys41. In general, our findings indicate that ionic interactions are more important for the affinity of SIM2 than SIM3 when binding to SUMO. This idea is consistent with the additional flanking acidic residue found in the SIM2 peptide and the extension of the hydrophobic patch found in the SIM3 peptide (by residue Pro2 and Val3), which likely result in stronger hydrophobic interactions of the latter with the SIM binding groove. However, despite the importance of ionic interactions for the binding orientation, we do not expect a complete loss of antiparallel binding of the SIM2 peptide if Asp1, Glu2, and Glu9 were mutated, because also for this SIM peptide, an antiparallel binding mode corresponding to the one observed for SIM3 would appear reasonable. In this configuration Val4 and Leu6 would occupy the SIM binding groove of SUMO instead of Ile3 and Asp5.
Finally, our simulations support the notion that the individual SIM-SUMO interfaces in the bivalent structure of SIM2-SIM3 with diSUMO3 by Xu et al. (PDB: 2mp2) (9) are deviating from typical architectures of SIM-SUMO interactions. They are lacking the typical β-sheet contact and hydrophobic interactions, which is consistent with very poor binding affinities in the medium to high millimolar range for the individual SIM-SUMO pairs as predicted by our simulations and calculations of the free energies of binding. Experimentally determined dissociation constants for the individual SIM-SUMO interactions were reported to be dramatically lower in the medium micromolar range (approximately 600- to 1800-fold lower (6)), which is the expected range to give a dissociation constant of 8.5 μM as the experimentally determined affinity of the bivalent complex (5). The bivalent binding mode and the possible SUMO chain topology surely add additional parameters important for the entire interaction interface that were not considered in this work. Nevertheless, the enormous deviation of the calculated individual SIM-SUMO affinities seems incompatible with this model and suggests that more research will be necessary in the future to understand the full complexity of multivalent SIM-SUMO interactions.
Author Contributions
All authors designed the research. A.K. performed the simulations and data analysis. All authors discussed the results and wrote the manuscript.
Acknowledgments
The authors thank the Deutsche Forschungsgemeinschaft (DFG) for funding via SFB 858 and SFB 1348. The simulations have been performed on the computing cluster PALMA2 at the University of Münster.
Editor: Alan Grossfield.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.09.003.
Contributor Information
Alex Kötter, Email: andheuer@wwu.de.
Andreas Heuer, Email: a_koet04@wwu.de.
Supporting Material
References
- 1.Hendriks I.A., Vertegaal A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 2.Gareau J.R., Lima C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cappadocia L., Lima C.D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2018;118:889–918. doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keusekotten K., Bade V.N., Praefcke G.J. Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem. J. 2014;457:207–214. doi: 10.1042/BJ20130753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kung C.C., Naik M.T., Huang T.-H. Structural analysis of poly-SUMO chain recognition by the RNF4-SIMs domain. Biochem. J. 2014;462:53–65. doi: 10.1042/BJ20140521. [DOI] [PubMed] [Google Scholar]
- 7.Escobar-Cabrera E., Okon M., McIntosh L.P. Characterizing the N- and C-terminal Small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J. Biol. Chem. 2011;286:19816–19829. doi: 10.1074/jbc.M111.231647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J., Durrin L.K., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y., Plechanovová A., Matthews S.J. Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4. Nat. Commun. 2014;5:4217. doi: 10.1038/ncomms5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kost L.J., Mootz H.D. A FRET sensor to monitor bivalent SUMO-SIM interactions in SUMO chain binding. ChemBioChem. 2018;19:177–184. doi: 10.1002/cbic.201700507. [DOI] [PubMed] [Google Scholar]
- 11.Sekiyama N., Ikegami T., Shirakawa M. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 12.Baba D., Maita N., Shirakawa M. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J. Mol. Biol. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Anamika, Spyracopoulos L. Molecular basis for phosphorylation-dependent SUMO recognition by the DNA repair protein RAP80. J. Biol. Chem. 2016;291:4417–4428. doi: 10.1074/jbc.M115.705061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gareau J.R., Reverter D., Lima C.D. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J. Biol. Chem. 2012;287:4740–4751. doi: 10.1074/jbc.M111.321141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cappadocia L., Pichler A., Lima C.D. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 2015;22:968–975. doi: 10.1038/nsmb.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi V., Chatterjee K.S., Das R. Casein kinase-2-mediated phosphorylation increases the SUMO-dependent activity of the cytomegalovirus transactivator IE2. J. Biol. Chem. 2019;294:14546–14561. doi: 10.1074/jbc.RA119.009601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo H.-J., Roux B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA. 2005;102:6825–6830. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kötter A., Mootz H.D., Heuer A. Standard binding free energy of a SIM-SUMO complex. J. Chem. Theory Comput. 2019;15:6403–6410. doi: 10.1021/acs.jctc.9b00428. [DOI] [PubMed] [Google Scholar]
- 19.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P., Jin B., Huang S.Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018;46:W443–W450. doi: 10.1093/nar/gky357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 22.Søndergaard C.R., Olsson M.H., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Rosell G., Giorgino T., De Fabritiis G. PlayMolecule ProteinPrepare: a web application for protein preparation for molecular dynamics simulations. J. Chem. Inf. Model. 2017;57:1511–1516. doi: 10.1021/acs.jcim.7b00190. [DOI] [PubMed] [Google Scholar]
- 24.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 26.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 27.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 28.Sugita Y., Okamoto Y. Replica exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 29.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 30.Bussi G. Hamiltonian replica exchange in GROMACS: a flexible implementation. Mol. Phys. 2014;112:379–384. [Google Scholar]
- 31.Kumar S., Bouzida D., Rosenberg J.M. The weighted histogram analysis method for free energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 32.Grossfield A. WHAM: the weighted histogram analysis method, version 2.0.9. http://membrane.urmc.rochester.edu
- 33.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 34.Namanja A.T., Li Y.J., Chen Y. Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J. Biol. Chem. 2012;287:3231–3240. doi: 10.1074/jbc.M111.293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J., Zhang Z., Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 36.Aldeghi M., Heifetz A., Biggin P.C. Predictions of ligand selectivity from absolute binding free energy calculations. J. Am. Chem. Soc. 2017;139:946–957. doi: 10.1021/jacs.6b11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu H., Cai W., Chipot C. New coarse variables for the accurate determination of standard binding free energies. J. Chem. Theory Comput. 2017;13:5173–5178. doi: 10.1021/acs.jctc.7b00791. [DOI] [PubMed] [Google Scholar]
- 38.Stehmeier P., Müller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Chang C.-C., Naik M.T., Shih H.M. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell. 2011;42:62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 40.Droescher M., Chaugule V.K., Pichler A. SUMO rules: regulatory concepts and their implication in neurologic functions. Neuromolecular Med. 2013;15:639–660. doi: 10.1007/s12017-013-8258-6. [DOI] [PubMed] [Google Scholar]
- 41.Taupitz K.F., Dörner W., Mootz H.D. Covalent capturing of transient SUMO-SIM interactions using unnatural amino acid mutagenesis and photocrosslinking. Chem. Europ. J. 2017;23:5978–5982. doi: 10.1002/chem.201605619. [DOI] [PubMed] [Google Scholar]
- 42.Brüninghoff K., Aust A., Mootz H.D. Identification of SUMO binding proteins enriched after covalent photo-cross-linking. ACS Chem. Biol. 2020;15:2406–2414. doi: 10.1021/acschembio.0c00609. [DOI] [PubMed] [Google Scholar]
- 43.Cappadocia L., Mascle X.H., Omichinski J.G. Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure. 2015;23:126–138. doi: 10.1016/j.str.2014.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.