Abstract
The α7 nicotinic acetylcholine receptor is a homopentameric ion channel from the Cys-loop receptor superfamily targeted for psychiatric indications and inflammatory pain. Molecular dynamics studies of the receptor have focused on residue mobility and global conformational changes to address receptor function. However, a comparative analysis of α7 with its homologs that cannot trigger channel opening has not been made so far. To identify the residues involved in α7 activation, we ran triplicate 500-ns molecular dynamics simulations with an α7 extracellular domain homology model and two acetylcholine-binding protein homologs. We tested the effect of ligand binding and amino acid sequence on the structure and dynamics of the three proteins. We found that mobile regions identified based on root mean-square deviation and root mean-square fluctuation values are not always consistent among the individual α7 extracellular domain simulations. Comparison of the replica-average properties of the three proteins based on dynamic cross-correlation maps showed that ligand binding affects the coupling between the C-loop and the Cys-loop, vestibular loop, and β1–β2 loops. In addition, the main-immunogenic-region-like domain of α7 went through correlated motions with multiple domains of the receptor. These correlated motions were absent or diminished in α7 homologs, suggesting a unique role in α7 activation.
Significance
The α7 nicotinic acetylcholine receptor is a key multifunctional ion channel found throughout the body but has no high-resolution experimental structure, a typical case for membrane proteins. Studies of the mechanisms of α7 nicotinic acetylcholine receptor function rely heavily on in silico models and various molecular-dynamic-based techniques. Our work reveals the limitations of basic metrics used in molecular dynamics simulations, such as root mean-square deviation and root mean-square fluctuation, in measuring the structural and dynamical properties of the α7 receptor. Our analyses that compare the behavior of α7 and its homologs identified the ligand-induced-correlated motions unique to α7. In addition, our results underline the importance of adequate sampling of the α7 conformational space in terms of simulation times and the use of replica simulations.
Introduction
α7 nicotinic acetylcholine receptor function and properties
The α7 nicotinic acetylcholine receptor (nAChR) is a member of the Cys-loop receptor superfamily. It is a homopentamer and is further distinguished from heteromeric nAChR by its high calcium permeability (1), fast desensitization kinetics (2), and five potential binding sites (3). Although the α7 nAChR has five potential binding sites, the binding of one or two acetylcholine molecules is sufficient to activate the receptor, and higher agonist concentrations promote receptor desensitization (4,5).
α7 nAChR is targeted for the treatment of diseases including the cognitive symptoms of schizophrenia (6,7), Alzheimer’s disease (8), and depression (9,10). In addition to these conditions, α7 nAChR is plays a role in the cholinergic antiinflammatory pathway (11,12), and some ligands targeting α7 were found to have antiinflammatory properties (13).
There is no high-resolution structure available for the α7 nAChR. Much of our structural understanding of the α7 nAChR extracellular domain (ECD) comes from the crystal structures of two α7 homologs: acetylcholine-binding protein (AChBP) and its humanized chimeric form α7-AChBP. Of the two proteins, AChBP has ∼24% and α7-AChBP has ∼64% homology with the human α7 ECD.
α7 nAChR and its homologs show subunit asymmetry during molecular dynamics simulations
Because of the challenges associated with the experimental analysis of α7 nAChR dynamics, molecular dynamics (MD) simulations have been used to understand the mechanism of the α7 ECD and transmembrane domain (TMD) motion using homology models, AChBP, or α7-AChBP in multiple studies (14). Simulations from some of these studies displayed subunit asymmetry in agonist- and antagonist-bound and apo (i.e. no ligand bound) states of the receptor (15, 16, 17, 18, 19, 20). In the context of this work, we broadly define asymmetry as significant nonidentical backbone configurations for subunits or nonequivalent subunit interfaces within the pentamer. The conclusions reached in most studies came from single MD simulations shorter than 100 ns, and the residues putatively involved in asymmetry or structural mobility have not been systematically investigated. Specifically, it is not certain whether the asymmetric behavior observed in these simulations represent the natural behavior of the system, for example, are caused by the stochastic motion of subunits, are computational artifacts, or are some combination of these factors.
Analysis of root mean-square deviation, root mean-square fluctuation, and amino acid composition can shed light on the mechanisms of α7 nAChR asymmetry
We investigated whether the observed dynamical and structural differences among α7 simulations reflect the genuine behavior of α7 and, if so, whether these mobile residues are unique to α7 nAChR versus AChBP and α7-AChBP proteins. AChBP and α7-AChBP proteins are not coupled with an ion channel in their native structures and cannot activate ECD-TMD chimeric receptors with 5-HT3A receptor TMD (21). Note that as one progresses from AChBP to the chimeric α7-AChBP and finally the α7 nAChR, the models become increasingly “α7 like” in terms of their amino acid content. Therefore, a comparison of the three systems can potentially shed light on the identity of the α7 ECD residues directly related to channel opening. To achieve this goal, we ran three independent 500-ns MD simulations and analyzed the root mean-square deviation (RMSD) and root mean-square fluctuation (RMSF) changes associated with these systems.
As additional metrics to measure the varying motion among the subunits and among the replica simulations, we calculated average per-residue RMSD-values. Variations in the per-residue RMSD-values among the replica simulations or between the epibatidine-bound and unbound simulations are suggestive of asymmetric motions at the tertiary level. We also analyzed the effect of epibatidine binding to each protein by comparing the results of fivefold epibatidine-bound and unbound MD simulations. Next, we calculated and plotted dynamic cross-correlation maps (DCCMs) to identify correlated motions between the different domains of these proteins. Finally, we used the data calculated for each system to find the relation between the α7 residue content and the structural and dynamical features of AChBP, α7-AChBP, and α7 ECD.
Materials and Methods
Ligand parametrization
The structure of epibatidine was drawn with MolView and was then subjected to a two-step geometry optimization procedure, with an initial HF/6-31G∗ optimization followed by a B3LYP/6-31G∗∗ optimization. All quantum mechanical calculations were run with Gaussian09 (22). The electrostatic potentials of the optimized structures were calculated by single-point calculations at the HF/6-31G∗ level. The atomic charges were generated via the RESP method using the antechamber module of AmberTools16 (23,24) with a net charge of +1 on the epibatidine molecule. The ligand parameters were generated using the tleap module with GAFF (25) as the force field.
Homology modeling of the α7 ECD
The α7 nAChR homology model was built using the Prime module of the Schrödinger 2014-2 Suite (26,27). The epibatidine-bound α7-AChBP chimera protein structure (Protein Data Bank, PDB: 3SQ6) (28) was used as the homology model template, and the ECD of the mature human α7 subunit sequence was used as the target sequence (Uniprot: P36544). The conserved residues in the template were kept as is, and nonconserved residues were optimized based on energy minimization. Loops smaller than five residues, including the tip regions of the β8–β9 loop, β1–β2 loop, Cys-loop, and C-loop, were refined by the Prime module, and larger loops including the vestibular loop (95–105) were refined during the MD simulations. Subunit symmetry was not enforced while generating the model, which yielded a protein model with slight asymmetry at the quaternary level. The mature protein α7 numbering was used to number the α7 ECD residues, and the five subunits were named A, B, C, D, and E in a counterclockwise fashion when viewing the receptor from above on the extracellular side. The PDB model starts from the canonical amino acid number 4, and all the corresponding residue numbers were adjusted to canonical numbering.
Assessment of the structure quality
The quality of the AChBP, α7-AChBP crystal structures and the α7 ECD homology model, was compared by measuring the Ramachandran outliers for all subunits of each structure. A comparison of the average Ramachandran scores of individual subunits of the three proteins showed that the AChBP structure has the lowest percentage of outliers before the MD simulations (1.1%), followed by the homology model of α7 ECD (2.2%) and the α7-AChBP (4.1%). The final structures were subjected to the contact analysis module of the UCSF Chimera software (29), and no clashes were found between residues.
MD simulations
All MD simulations were run with Amber16 (24). Epibatidine-bound Aplysia californica AChBP (30) and epibatidine-bound, humanized, chimeric Lymnaea stagnalis AChBP (28) (α7-AChBP) were taken from PDB (PDB: 2BYQ and 3SQ6, respectively). Homology modeling of the α7 ECD was done as described above. The unbound models for all three proteins were generated by removing the bound epibatidine molecules from all five binding sites of each protein before the simulations. The protein residues were parametrized with the ff14SB force field (31), and an octahedral TIP3P water box was added with sodium ions to neutralize the system. This system was minimized first with heavy restraints on all nonhydrogen atoms and then without restraints. Next, the minimized system was gradually heated to 300 K in an NVT ensemble with 5-kcal/mol-Å2 restraints on the backbone atoms over 2 ns, then equilibrated in an NPT ensemble at 1 atm at 300 K over 8 ns. The equilibrated structure was used as the starting point for three independent 500-ns NPT production runs. The outputs of the production runs were used to calculate the RMSD- and RMSF-values for each system.
Structural analyses of the MD trajectories
RMSD- and RMSF-values were calculated separately for the individual chains of each model with cpptraj (32), and the average chain RMSD- and RMSF-values were used as the RMSD- and RMSF-values for each simulation. The replica-averaged RMSD- and RMSF-values were calculated as the average of the three replica simulations. The reported RMSD-values were calculated as the average RMSD-value for the last 10 snapshots (10 ns) of each simulation. The two-dimensional (2D) RMSD plots were calculated for the whole protein over 500 frames of 1-ns time steps.
Comparison of the AChBP, α7-AChBP, and α7 ECD sequences
An arbitrary residue numbering based on the homologous domains of the three proteins was created for easier comparison of the three proteins because length differences, gaps, and insertions prevent the use of canonical numbering schemes for a meaningful comparison (Fig. S4). Alignment of the protein sequences were done with Clustal Omega (33). The AChBP sequence was used as the basis for the numbering because it has the largest number of amino acids.
Jensen-Shannon divergence calculations
All Jensen-Shannon divergence (JSD) values were calculated with the ENCORE application of the MDAnalysis software (34,35). The clustering ensemble similarity method was used for the divergence calculations. K-means clustering with five clusters, 10 initial cluster searches, and 300 maximal iterations were run for the clustering step. The divergence values were calculated for the replica simulations of the same protein in the same ligation state.
DCCM calculations
All DCCM calculations were run with Amber cpptraj. Aligned trajectories of each set of replicas for the epibatidine-bound and unbound simulations with AChBP, α7-AChBP, and α7 ECD were combined for the DCCM calculations. The matrix keyword was used to calculate the covariances of the individual residues, and the correlation matrix was calculated based on these values. The resulting dynamic cross-correlation (DCC) values calculated for the five chains were then averaged to obtain the final DCCMs. The results were plotted as a heatmap using the Python package matplotlib. The difference matrix was calculated as the difference between the DCC-values of the epibatidine-bound and unbound simulations.
Per-residue RMSD calculations
For each simulation, the trajectories of the individual chains were generated with cpptraj, and all five trajectories were aligned to the first frame of chain A. The average per-residue RMSD-values were calculated over 500 ns for the individual simulations separately as the average of the per-residue RMSD-values of the five chains of the same simulation. For the comparisons between the epibatidine-bound and unbound simulations, the average per-residue RMSD-values were calculated as the average of the three replica simulations for both states.
Results
The RMSD-values of AChBP replica simulations converge over 500 ns
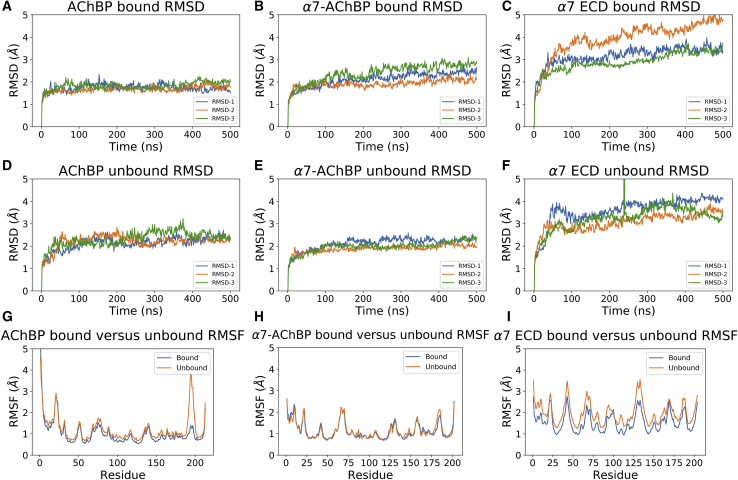
The average RMSD-values of the AChBP replica simulations had no large variations in the epibatidine-bound model over 500 ns (Fig. 1 A). For the unbound AChBP model, RMSD variations up to 1 Å were observed between the replica simulations during the first 350 ns, but the values started to converge after this point (Fig. 1 D). The plateaued, replica-averaged RMSD-values for the three replica simulations were 1.8 and 2.3 Å for the epibatidine-bound and unbound models, respectively.
Figure 1.
Chain-averaged RMSD-values calculated for the (A) epibatidine-bound AChBP, (B) epibatidine-bound α7-AChBP, (C) epibatidine-bound α7 ECD, (D) unbound AChBP, (E) unbound α7-AChBP, (F) unbound , α7 ECD, and comparison of the RMSF values calculated for epibatidine-bound and unbound (G) AChBP, (H) α7-AChBP, and (I) α7 ECD in three replicate 500-ns MD simulations. To see this figure in color, go online.
Pairwise RMSD-values comparing the RMSD-value of each frame with respect to all the other frames in the trajectory were calculated (2D RMSD) to better understand the conformational changes over time. The epibatidine-bound simulations had a uniform RMSD distribution suggesting little change in conformation throughout the trajectory (Fig. S1 A). For the unbound simulations, there was a relatively larger variation between the earlier and later frames, but all three trajectories showed consistently low RMSD-values after 250 ns (Fig. S1 B).
JSDs suggest a larger similarity among epibatidine-bound calculations
Next, we calculated JSDs to measure the similarity of the replica simulations with respect to each other. JSD is a metric of measuring similarity between two probability distributions that can be used to measure the similarity of the ensembles or MD trajectories based on a variety of structural parameters. The clustering method selected for the JSD calculations was a clustering-based ensemble similarity. In this method, the trajectory is split into individual frames at each time point, and clusters are formed based on structural similarity. Specifically, the pairwise RMSD matrix is calculated as a measure of structural similarity, and the closest structures by this metric are assigned into a given total number of clusters. Once the clusters are obtained, the JSDs are calculated by comparing the variations among these clusters in terms of their distance from each other. The lowest JSD-value of 0 implies completely similar trajectories, and the highest value of ln2 (0.693) implies completely different trajectories. This approach was previously used to compare the MD trajectories produced through different protein force fields (36).
The resulting JSD-values from our calculations can be seen in Table S1. The differences in these values suggest that the epibatidine-bound replica simulations are more similar to each other where simulations 1 and 2 had particularly low JSDs, whereas the unbound simulations had consistently large values.
AChBP RMSF and per-residue RMSD-values only differ at the C-loop
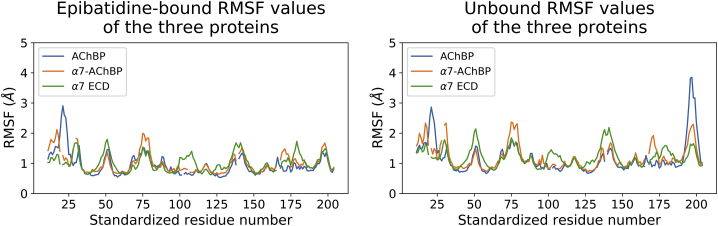
The activation mechanism of the α7 nAChR is considered to involve multiple regions that may have different mobilities in the presence or absence of a bound ligand. On the other hand, AChBP has not evolved as an ion channel whereby ligand binding triggers movements at the parts of the protein in contact with a TMD. Because of this functional difference, the only domain that needs to be mobile is the C-loop of the protein to facilitate binding of the ligand acetylcholine. Consistent with this idea, the replica-averaged RMSF plots of the epibatidine-bound and unbound AChBP simulations were nearly identical with the exception of the C-loop region (191–200) (Fig. 2). This region had a ∼2.5 Å larger RMSF at the peak point of the unbound simulations (Fig. 1 G). Further, a comparison of the RMSF plots from the individual replica simulations showed consistent results, suggesting a single simulation is sufficient to reproduce the dynamics of the system (Fig. S2).
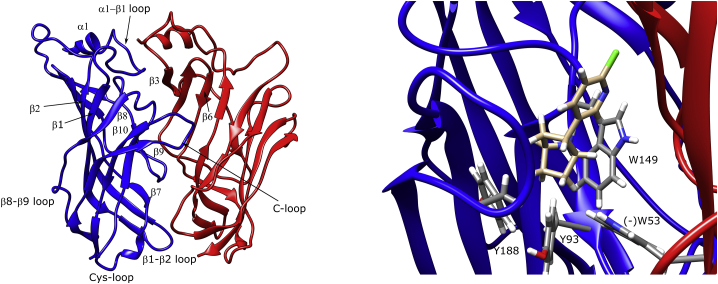
Figure 2.
Names and locations of the important α7 ECD domains shown on the α7 ECD homology model (left) and the epibatidine starting binding configuration at the orthosteric site with aromatic cage residues labeled (right). The blue color stands for the positive face subunit, and the red color stands for the complementary (negative) face subunit. The α-denotation applies to the helical secondary structure, and the β-denotation applies to the b-sheet secondary structure. To see this figure in color, go online.
AChBP C-loops move asymmetrically in the unbound simulations
Although the mobility profiles of the AChBP domains were similar, with the exception of the C-loop, this could be an artifact of averaging the mobilities of multiple subunits from multiple simulations. Per-residue RMSD-values were investigated to assess the tertiary symmetry of the subunits both over the individual simulations and between the epibatidine-bound and unbound states of the protein. Trajectories of all the individual chains were aligned for each simulation, and per-residue RMSD-values were calculated in reference to the same point. The per-residue RMSD-values obtained this way allow us to compare whether the subunits went through asymmetric motions at the tertiary level during the simulations whereby large per-residue RMSD-values are indicative of disconcerted motions.
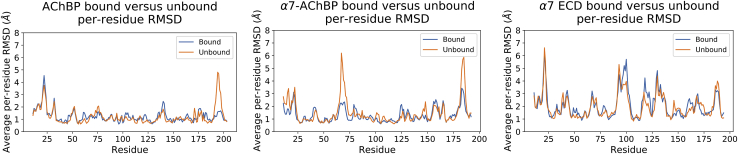
Among the replicate simulations belonging to the same protein ligation state, there were only minor differences in average per-residue RMSD-values (Fig. S3, top row). The only region that showed a per-residue RMSD difference between the epibatidine-bound and unbound simulations was the C-loop, suggesting disconcerted motion of this region among the subunits of the unbound simulations (Fig. 3, left). The region between residues 19 and 24 had a high average per-residue RMSD in both states, although the values were slightly higher in the epibatidine-bound simulations. Similar disconcerted motions at this region were previously reported for lobeline-bound AChBP simulations (37).
Figure 3.
The average per-residue RMSD-values calculated for epibatidine-bound and unbound AChBP (left), α7-AChBP (middle), and α7 ECD (right). The blue line stands for the epibatidine-bound simulations, and the orange line stands for the unbound simulations. To see this figure in color, go online.
C-loops are consistently closed in epibatidine-bound simulations but may be open or closed in unbound simulations
To quantify variations in C-loop positions in the bound and unbound simulations, we calculated C-loop openings (see Materials and Methods) for the chains of the individual simulations with epibatidine-bound and unbound AChBP (Table S2, top row). The results showed that the C-loops were overall more closed with opening distances around 9–11 Å in the epibatidine-bound simulations. The difference between the most open and most closed C-loops was ∼1.5 Å. In the unbound simulations, the opening distances were typically over 11 Å, with large variations among the different subunits. Comparison of these values with the epibatidine-bound values showed that one, two, and three C-loops were closed in the three unbound replica simulations despite the absence of a ligand molecule.
AChBP DCCMs show no noticeable difference between the epibatidine-bound and unbound states
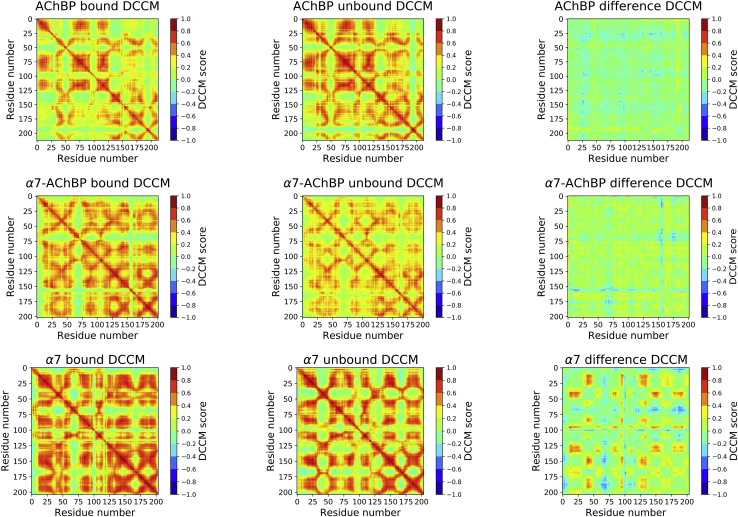
We calculated DCCMs to investigate coupled motions throughout the protein. DCCM calculations involve a calculation of the covariance matrix using the RMSD-values to determine pairs of residues that move together in the course of an MD simulation (38). DCC-values of +1 correspond to completely correlated motions between a pair of residues, and values of −1 correspond to anticorrelated motions.
To assess the effects of agonist binding on the correlation profile, we used the replicate simulations as the input for DCCM calculations with and without a bound epibatidine. Correlations were observed between multiple regions, including the β1–β2 loop, C-loop, and the Cys-loop in both the epibatidine-bound and unbound DCCMs (Fig. 4, top row). The difference map of the two states, on the other hand, showed little difference between the strength of coupling between these two states.
Figure 4.
Dynamic cross-correlation maps (DCCMs) calculated for the AChBP (top row), α7-AChBP (middle row), and α7 ECD (bottom row) replica simulations in the epibatidine-bound state (left), epibatidine unbound state (middle), and the difference between the epibatidine-bound and unbound states (right). To see this figure in color, go online.
α7-AChBP epibatidine-bound and unbound simulations have similar properties
The α7-AChBP RMSD-values were within 0.8 Å of the epibatidine-bound replica calculations with an average RMSD of 2.4 Å and was within 0.4 Å with an average RMSD of 2.2 Å for the unbound calculations (Fig. 1, B and E). Similar to the AChBP calculations, there was little variation in the 2D RMSD plots of the epibatidine-bound calculations, although one simulation had a relatively larger RMSD difference between the earlier and later frames (Fig. S1 C). The unbound simulations showed the same profile as the epibatidine-bound simulations (Document S1. Figs. S1–S5 and Tables S1–S3, Document S2. Article plus Supporting Material).
The JSD-values were slightly lower for the unbound simulations, although both epibatidine-bound and unbound simulations showed marked differences among each other and were larger than the values calculated for AChBP (Table S1, middle row).
Interestingly, there was virtually no difference between the replica-averaged RMSF-values of the epibatidine-bound and unbound α7-AChBP MD simulations even at the C-loop region, which became more mobile upon the removal of the bound epibatidine molecules in AChBP simulations (Fig. 1 H). Comparison of the RMSF-values from the individual simulations showed differences at two regions. For the epibatidine-bound calculations, there was a slight difference between the residues 126–132 (Cys-loop). For the unbound calculations, the difference was at the C-loop (Fig. S2, middle row).
α7-AChBP C-loops are more open compared with that of the AChBP
The C-loop openings also reflected the trend observed for the RMSF calculations. Different than the AChBP simulations, the C-loop openings of the epibatidine-bound simulations were typically between 9 and 13 Å. Similar results were observed for the unbound calculations (Table S2, middle row). When the 11-Å opening distance from the AChBP simulations was taken as the reference for a closed loop, all simulations were found to have a combination of apparently open and closed C-loops, although lack of a well-defined reference as in AChBP prevented us from making precise comments regarding whether these C-loops should be considered open or closed.
Increased asymmetry is observed at two α7-AChBP regions based on per-residue RMSD calculations
Despite the lack of a large difference between the RMSF-values of the epibatidine-bound and unbound simulations, the average per-residue RMSD-values showed large variations in two regions, specifically between the residues 62–73 and 182–187 (Fig. 3, middle). The 62–73 region at the top part of the protein corresponds to the main immunogenic region (MIR) of the α1 nAChR subtype (39,40). Because of its position that was similar to the MIR of α1 nAChR, this region was called “the MIR-like domain” for the rest of the manuscript. The 182–187 region corresponds to the tip of the C-loop. Based on these results, the C-loops of α7-AChBP go through disconcerted motions in the absence of epibatidine, although the average mobility remains similar in both states.
α7-AChBP DCCMs show increased coupling of two region pairs in the unbound simulations
Similar to the AChBP case, there were very few differences between the plotted DCCM-values of the epibatidine-bound and unbound simulations (Fig. 4, middle row). The unbound simulations showed correlations at the same region as the epibatidine-bound simulations, but the strength of correlation was weaker. As a result, the difference DCCM had a larger baseline value overall. However, two regions of increased correlation in the unbound simulations were observed.
The first region indicated a correlation between the movement of the residues 53–81 (MIR-like domain) and 142–150. The 142–150 region covers the β7–β8 loop. This loop hosts the critical tryptophan residue (α7-AChBP W143; human α7 nAChR W149), whose mutation significantly diminishes ligand binding to α7 (41). The second region indicated a correlation between the residues 62 and 74 (MIR-like domain) and 173–179. The 173–179 region corresponds to the portion of the β9 domain that forms the lower part of the C-loop. Overall, epibatidine binding to α7-AChBP was found to have no effect on the replica-averaged RMSF-values of the protein but resulted in more disconcerted motions at two domains.
α7 ECD RMSD-values show variations among the replica trajectories
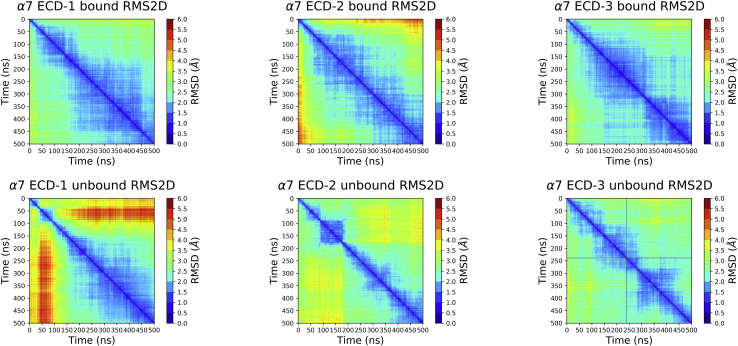
The α7 ECD had the most complex RMSD and RMSF profiles as can be expected because of its highly dynamic structure, potential artifacts associated with homology modeling, and the truncated model lacking an intracellular domain and TMD regions. The replica-averaged RMSD-value calculated for the epibatidine-bound simulations was 3.9 Å, and the RMSD-value calculated for the unbound simulations was 3.6 Å. The difference between the highest and lowest RMSD-values among the replica simulations was 1.3 Å for the epibatidine-bound calculations, and 0.8 Å for the unbound simulations. The larger variation observed for the epibatidine-bound calculations was caused by the larger RMSD-value of a single simulation compared with the other two replica simulations (Fig. 1 C, orange line).
The α7 ECD 2D-RMSD-values demonstrated the difference between the epibatidine-bound and unbound simulations in terms of the sampled conformational space. The epibatidine-bound simulations overall had large regions of similar RMSD-values suggestive of few discernable different intermediate states (Fig. 5, top row). The unbound simulations showed much narrower regions around the diagonal axis, and the difference between later and earlier frames of the simulations were more noticeable (Fig. 5, bottom row). Both simulations 1 and 2 had regions before 200 ns that largely differed from the later frames, but the differences became more subtle after this point.
Figure 5.
2D RMSD plots of the epibatidine-bound (top row) and epibatidine unbound (bottom row) α7 ECD replica simulations. To see this figure in color, go online.
The differences between the individual simulations were also reflected in the calculated JSD-values (Table S1, bottom row). The α7 ECD simulations had the highest divergence values among the three proteins tested, indicative of largest variations among the replica simulations.
α7 ECD RMSF-values are similar for the epibatidine-bound and unbound states
Despite the differences in the RMSD-values, the RMSF plots of the epibatidine-bound and unbound simulations showed marked similarities (Fig. 1 I). The baseline of the unbound RMSF-values were higher than the epibatidine-bound simulations. To better delineate the effect of different baselines, we normalized the unbound RMSF-values to epibatidine-bound RMSF-values by dividing the unbound RMSF-values by the ratio of the average RMSFs between the two states (Fig. S4). The normalized results were highly similar except for two regions. One region was the vestibular loop between 94 and 99, which was associated with allosteric ligand binding in previous studies (20,42, 43, 44). The other region was the three-amino-acid segment of the tip of the β8–β9 loop (residues 167–170).
Contrary to the AChBP and α7-AChBP simulations, the RMSF plots of the replica simulations showed variations in the α7 ECD simulations (Fig. S2, bottom row). The most remarkable examples were regions such as the α1-β1 loop, β1–β2 loop, vestibular loop, β8–β9 loop, and Cys-loop to different degrees in the epibatidine-bound and unbound simulations.
α7 C-loops are a mixture of closed and open states
The C-loop opening distances of the epibatidine-bound simulations were akin to that of the α7-AChBP simulations whereby the distances were typically within 9–13 Å. On the other hand, the unbound simulations had C-loop opening distances close to epibatidine-bound values observed for AChBP, indicating that multiple C-loops may be closed even in the unbound state of the α7 ECD model.
α7 ECD DCCMs show a clear coupling difference between the epibatidine-bound and unbound states
The majority of the observed residue-residue correlations were larger in the epibatidine-bound simulations (Fig. 4, bottom row). The β1–β2 loop residues 42–50 had correlated motions with three regions. The first one was the 84–92 region (β3–β4 loop), the second one was the 145–158 region including the β7–β8 loop and the critical residue W149, and the third one was the 182–196 region (C-loop). The vestibular residues 97–100 had correlations with the C-loop and the 145–158 region. Finally, the Cys-loop residues 128–137 had correlations with 84–92, the C-loop (182–196), and the 145–158 region. In the unbound simulations, the MIR-like domain residues 65–74 had larger correlated motions with the C-loop (182–196) and the 145–158 region.
Overall, α7 ECD simulations showed variability among the replica simulations, but there was little consistent difference between the epibatidine-bound and unbound simulations when the average behavior of the replica simulations was considered. On the other hand, epibatidine had a clear effect on coupling the critical domains of the protein, unlike the AChBP and α7-AChBP simulations.
A standardized residue numbering based on homologous regions was created for the comparison of AChBP, α7-AChBP, and α7 ECD trajectories
Next, we focused on the mobile regions of the three proteins to determine the effect of increasing the α7 nAChR amino acid content on the RMSF-values of the AChBP, α7-AChBP, and α7 ECD by comparing the RMSF-values of the homologous regions. Because AChBP, α7-AChBP, and α7 ECD have different numbers of amino acids and several gap regions, the sequences of the three proteins were aligned based on the conserved residues to create an artificial “standard residue numbering” (SRN) (Fig. S5) to simplify the comparison of the functionally similar regions using AChBP as the reference because it has the longest amino acid sequence. The SRN residues 195–196 correspond to the C-loop disulfide cysteines of all three proteins. Note that this residue numbering does not correspond to the numbering scheme of any single protein, and the relevant regions of each protein is denoted as mentioned later on.
α7 ECD has different mobilities than AChBP and α7-AChBP at multiple regions
Although the α7-AChBP amino acid sequence has 64% homology with the α7 ECD sequence and only 38% homology with the AChBP sequence, the replica-averaged RMSF profile of α7-AChBP was closer to the AChBP than the α7 ECD in both epibatidine-bound and unbound simulations (Fig. 6, blue and orange lines). The first 20 N-terminus residues were excluded from the analyses because the N-terminus of the AChBP is longer than α7-AChBP and α7 ECD by 10 amino acids. AChBP had larger RMSF-values at the C-loop region (SRN 197–200, AChBP 195–200) of the unbound simulations. On the other hand, α7-AChBP had larger RMSF-values between the residues 65–70 (MIR-like domain, SRN 74–79) and 155–161 (portion of the β8–β9 loop across the C-loop, SRN 168–174) in both simulations.
Figure 6.
Baselined, replica-averaged RMSF-values calculated for AChBP, α7-AChBP, and α7 ECD in the epibatidine-bound (left) and epibatidine unbound (right) forms. The first and last 10 residues were omitted because of the large gap regions between AChBP and the other two proteins. To see this figure in color, go online.
α7 ECD simulations had larger RMSF-values compared with AChBP and α7-AChBP mostly at the residues in contact with the TMD in the native α7 structure and at the residues forming the subunit interfaces. The α7 residues 42–50 (β1–β2 loop, SRN 47–55), 95–103 (vestibular loop, SRN 100–108), and 132–136 (Cys-loop, SRN 137–141), and, to a lesser extent, 170–175 (β8–β9 loop, SRN 177–182) had larger replica-averaged RMSF-values than AChBP and α7-AChBP in both epibatidine-bound and unbound calculations. The RMSF differences of α7 ECD with respect to the other two proteins were larger than the differences between α7-AChBP and AChBP.
The regions with large mobility differences have low sequence identity between α7 ECD and its homologs
To understand the relation between α7 amino acid composition and structural mobility, we looked for a correlation between sequence similarity and RMSF variation (Table S3). Eight out of the nine residues in the α7 42–50 range were identical, and one residue was similar despite the large difference in the RMSF-values. Only a single residue out of six was identical between α7 170–175, and two residues were similar. None of the residues in the α7 95–103 and α7 132–136 ranges were identical between the two proteins, and there was a single-residue gap. Finally, the MIR-like domain (α7-AChBP 65–70, α7 66–71), which showed a large RMSF-value in both α7-AChBP and α7 ECD simulations, but not the AChBP simulations, had the same sequence in α7-AChBP and α7 ECD but had only two identical residues out of seven with respect to the AChBP sequence in this region.
Discussion
A single MD simulation is insufficient to sample the conformational space α7 ECD
Identification of the residues involved in α7 channel gating has been the subject of many experimental and computational studies. In this work, we compared the dynamics of epibatidine-bound and unbound states of the α7 ECD with that of its homologs AChBP and α7-AChBP to identify structural and dynamical changes unique to α7 nAChR. The latter two proteins bind α7 ligands but are not ion channels, and chimeric proteins combining the AChBP (ECD) and 5-HT3A receptor (TMD) cannot be activated by agonists unless significant loop mutations are made (21).
We sought to identify structural and dynamic changes unique to α7 between the epibatidine-bound and unbound states through replica simulations and to identify and account for functionally relevant motions of the α7 subunits. The replica MD simulations ran for the epibatidine-bound and unbound AChBP showed consistent RMSD-values at the end of each 500-ns simulation, although variations were observed at the earlier stages. The α7-AChBP simulations showed some variations in RMSD-values within a range of 0.4–0.8 Å. For the α7 ECD simulations, the RMSD-values of the replica simulations differed by up to 1 Å. In addition to the larger average RMSD-value, the α7 ECD replica simulations showed variations in average per-residue RMSD-values and RMSF-values among the different simulations.
In summary, our results suggest that a single MD simulation with AChBP and α7-AChBP is sufficient to sample protein dynamics over 500 ns, whereas a single MD simulation with α7 ECD is not enough based on the RMSD and RMSF profile differences observed among the replica simulations. An inconsistency of results from independent MD simulations has been reported in a recent study focusing on the recently crystallized α4β2 nAChR (ECD-TMD), showing that these inconsistencies are not confined to homology models (45).
The replica-averaged RMSF-values show minor differences between the epibatidine-bound and unbound states
RMSF-values are frequently used to compare the dynamics of different states of proteins. The RMSF-values of the AChBP simulations showed a marked difference between the epibatidine-bound and unbound forms only at the C-loop. To our surprise, α7-AChBP showed no difference between the epibatidine-bound and unbound simulations, not even at the C-loop, which was more mobile in the unbound simulations with AChBP and, to some extent, α7 ECD. C-loop opening distances were also consistent with these observations whereby the AChBP epibatidine-bound simulations had consistently smaller opening distances than the unbound simulations, and the latter showed a mixture of open and closed C-loops. For the α7-AChBP simulations, the opening distances varied among the simulations, and there was no clear pattern change based on epibatidine binding.
For the α7 ECD, the replica-averaged RMSF-values of important domains such as the β1–β2 loops, Cys-loops, and β8–β9 loops were similar in epibatidine-bound and unbound simulations, suggesting that mobility changes in these loops are not exclusively induced by agonist binding. The high mobility of these loops independent of ligand binding is consistent with the low electron density fits observed for the β1–β2 loop, Cys-loop, and β8–β9 loop in both the agonist-bound crystal structures of α7 nAChR homologs α4β2 (46), α7-AChBP (28), and apo AChBP and α7-AChBP structures (28,47). Further, multiple computational studies with α7 models have previously shown large RMSF-values for these regions in ECD-only (15,48,49) and ECD-TMD (18,19,50,51) simulations in agonist-bound, antagonist-bound, and unbound states of the protein.
C-loop closure may be a slow event in α7-AChBP and α7 ECD
A comparison of the epibatidine-bound (PDB: 3SQ6) and epibatidine unbound (PDB: 3SQ9) crystal structures of α7-AChBP shows that the C-loop opening distance of a single interface is the same between the two states, but the C-loops of all the remaining interfaces are more open in the unbound structure. On the other hand, our α7-AChBP and α7 ECD simulations showed no drastic difference between the epibatidine-bound and the epibatidine unbound simulations in terms of C-loop opening distances. What may be the reason behind this discrepancy?
We suggest two possible reasons for this phenomenon. One possibility is dissociation of epibatidine molecules from the orthosteric sites of the α7-AChBP and α7 ECD during the MD simulations, which would result in increased C-loop opening distances. Although all epibatidine molecules remained bound over 500 ns in both α7-AChBP and α7 ECD simulations, some epibatidine molecules shifted away from their starting conformation, which may be one of the earlier steps of the dissociation equilibrium. These shifts are associated with an increase in the C-loop opening distances, partly explaining the increases compared with the AChBP simulations.
The other answer may be related to the slower C-loop dynamics of these two proteins compared with AChBP. The unbound models in our simulations were created by removing the epibatidine molecules of the bound models. If the C-loop dynamics of these proteins is slow enough because of the rigidity of the C-loop, the 500-ns timescale used in these simulations may not be sufficient to allow a full opening of all the C-loops, contrary to what was observed with the AChBP simulations. The C-loops of α7-AChBP and α7 ECD are identical in sequence, but half of the residues at the tip region of the C-loop (α7 182–193) are different between AChBP and α7 ECD. Therefore, the slower dynamics for the opening and closure of the α7 C-loop may be related to its amino acid sequence.
Disconcerted subunit motions are more observable in unbound simulations
Subunit asymmetry at the quaternary and tertiary levels was previously observed in MD studies with the α7 ECD (15,16,49) and α7 ECD-TMD models (18,51). Asymmetric arrangement of subunits and large differences in the calculated subunit RMSD-values were more frequent in simulations with antagonist-bound and unbound models. However, these studies were based on single MD simulations, and they had no controls to deconvolute the effects of stochastic fluctuations or the effect of ligand binding to the α7 ECD. We accounted for these motions by looking at the average behavior of multiple simulations and by comparing parameters such as average per-residue RMSD-values.
Disconcerted motions were observed at the C-loops of the unbound AChBP and α7-AChBP simulations in comparison with the epibatidine-bound simulations. The α7-AChBP also showed disconcerted motions at the MIR-like domain in the unbound simulations. For the α7 ECD, both states of the protein showed large average per-residue RMSD-values in multiple regions. Some differences were observed between the two states, but the differences were small considering the large scale of the calculated per-residue RMSD-values.
What is the reason for the apparent lack of more frequent disconcerted residue motions in the unbound α7 simulations with respect to the epibatidine-bound simulations? It was previously shown that unbound α7 ECD simulations can yield subunit conformations consistent with the presence of a bound ligand (49). On the other hand, loss of ligand-protein interactions during the simulations due to ligand dissociation may yield subunit conformations consistent with unbound simulations. Further, our per-residue RMSD-values are calculated as the average of multiple subunits, which may not reflect drastic structural changes if only a small number of subunits go through such changes. Combining all these factors, similarities between the two states may have masked the differences at the subunit level.
AChBP and α7-AChBP have similar dynamics
The comparisons between the AChBP, α7-AChBP, and α7 ECD simulations shed light on the effect of α7 amino acid sequence on protein stability and mobility. The marked similarity between the AChBP and α7-AChBP RMSF-values despite that the high α7 residue content of α7-AChBP indicates that the majority of the α7 residues make no significant contribution toward the dynamic nature of α7 nAChR.
The only region that showed a mobility difference between the AChBP and α7-AChBP MD simulations was the MIR-like domain, whose amino acid sequence in α7-AChBP was different than the AChBP sequence and identical to that of the α7 receptor. The high mobility of the MIR was also observed in calculations with the muscle-type nAChR structure (52). Although there was no replica-averaged RMSF difference at the MIR-like domain between the epibatidine-bound and unbound α7-AChBP simulations, the average per-residue RMSD-values were larger in the unbound simulations compared with the epibatidine-bound simulations. Although there is no function attributed to this domain yet, co-crystallization studies with α7-AChBP have shown ligand binding at this region, and the molecules that bound to the MIR-like domain of α7-AChBP acted as noncompetitive antagonists of the α7 receptor (42). Therefore, higher mobility of the MIR-like domain in α7-AChBP and α7 ECD compared with AChBP may point to a role the MIR plays in allosteric regulation of α7 nAChR.
The dynamic nature of the α7 ECD is caused by a small number of intrinsically mobile regions
The comparisons of the α7 ECD with its homologs showed that α7 has a larger mobility than its two homologs, which do not function as ion channels. The majority of these high-mobility domains were confined to the parts of the α7 ECD that would be in contact with the TMD in the native structure, which is consistent with a mechanism whereby these mobile regions transmit the structural changes at the ECD to the TMD. The exceptional region outside the TMD was the vestibular loop, whose exact effect on receptor activation has not been established yet.
Motions of the Cys-loop, vestibular loop, and β1–β2 loop regions were also directly correlated with the motions of the C-loop based on our DCCM calculations. Although correlated motions of these regions were also observed for the AChBP and α7-AChBP simulations, the strength of coupling and the length of the coupled regions were highest in the α7 ECD simulations. On the other hand, these regions went through no significant structural change in the epibatidine-bound α7 ECD simulations based on our RMSF and per-residue RMSD calculations. These results suggest that ligand binding to α7 nAChR may not trigger large-scale motions that result in channel opening but rather stabilize one of the activatable states of the receptor that occur stochastically because of the intrinsic mobility of these domains. This idea is supported by other computational studies that revealed that modes associated with channel opening can be observed for unbound models as well (53).
Movement of the β1–β2 loop may be triggered by the Cys-loop or the β8–β9 loop
The amino acid sequences of these high-mobility domains were typically unique to α7. The exception to this was the β1–β2 loop that had a similar sequence in α7 ECD and α7-AChBP yet showed a larger RMSF-value in α7 ECD simulations. The β1–β2 loop sits next to the Cys-loop and the β8–β9 loop, which were also high mobility. Further, our DCCM calculations demonstrated the correlation between these three regions in both epibatidine-bound and unbound states. Because the β1–β2 amino acid sequence has no mobility solely caused by its amino acid sequence as observed in the α7-AChBP simulations, it is reasonable to argue that the direction of the structural changes that result in β1–β2 motion are triggered by the Cys-loop or the β8–β9 loop. Previous principal component analysis calculations demonstrated the coupled motions of the Cys-loop and β1–β2 loop with an unbound model (53).
Our model of opening is consistent with previous experimental and computational studies
Many experimental and computational studies aiming at identifying the channel-opening mechanism of nAChR have been done so far, providing important insight into this process (54). A quaternary twist model was suggested based on normal mode analysis (NMA) calculations with α7 ECD-TMD homology models (17,55). The common finding of these studies was that the ECD and the TMD rotate in opposite directions, which results in a “twist” of the protein. Hinge points for the twisting motion at the ECD were identified as the β1–β2 and β8–β9 loops along with the Cys-loop. The β1–β2 and the Cys-loops were more rigid than our calculations indicate, but they went through correlated motions, consistent with our findings. The β1–β2 loops and Cys-loops were found to interact with the M2-M3 linker, which in turn results in channel opening (56). Although direct comparisons with these studies and our findings are hard because of the lack of a TMD in our study, the correlations we identified between the C-loop and the β1–β2 loops and Cys-loops are consistent with such a channel-opening mechanism.
Limitations of the study
Our 500-ns simulations are among the longest simulations run with α7 models to date based on a survey on modeling literature of α7 nAChR (14). However, nAChR channel activation and other functional changes can take place at the millisecond scale, if not second scale, in reality (56). Therefore, the calculations run in this study may not be sufficient to explore the full conformational space of the α7 receptor. On the other hand, longer simulations with ECD-only homology models may affect the stability and quaternary structure of the system because of the lack of a stabilizing TMD and membrane environment in the calculations. With this consideration, instead of running longer simulations, we ran three replica simulations to sample a total of 1.5 μs for each of the studied systems and looked at the average behavior of α7 and its homologs based on these calculations.
In addition to issues related to the timescale, five equivalent binding sites of the α7 nAChR make analyses challenging. For the sake of symmetry and consistency, fivefold epibatidine occupancy was assumed in our calculations. On the other hand, the total number of ligands bound, and their relative binding positions yield a total of 31 binding permutations that may behave differently. Seeing the effect of the number of bound ligands and permutations would benefit model-building procedures immensely and should be considered in future studies aiming at analyzing α7 nAChR mode of action.
Conclusions
Overall, our results show important effects of agonist binding on the receptor dynamics and structure and highlight some limitations of the MD simulations with α7 ECD models. α7 ECD subunits went through different motions at a larger extent in comparison with the AChBP and α7-AChBP simulations. This asymmetry indicates that conclusions from analyses on individual α7 subunits are not generalizable, and an aggregate approach involving all the subunits should be taken for a reliable analysis of the protein dynamics and structure. Correlated motions unique to α7 were identified at regions including the MIR-like domain, the vestibular loop, C-loop, and the ECD loops in contact with the TMD. Of these domains, the exact function of the MIR-like domain or the vestibular loop has not been established yet, and experimental data on these regions are limited. Our findings suggest involvement of these domains in α7 activation, which should be explored in future experimental and computational studies. In addition, future studies to identify finer details of the α7 dynamics could benefit from better sampling, Markov models, and statistical analyses. Finally, the inclusion of the TMD and perhaps intracellular domain in such analyses would allow us to get a better picture of the α7 receptor mechanism of activation. We believe that consideration of these observations in future α7 modeling studies will help produce improved α7 nAChR models.
Author Contributions
A.G. designed and performed the research, analyzed the data, and wrote the manuscript. J.M. wrote the manuscript. N.A.H. designed the research and wrote the manuscript.
Acknowledgments
The authors wish to thank Dr. R. L. Papke for helpful discussions.
Portions of this work were supported under NIH grant GM57481, NIH R01 HL144131, and NIH NIGMS R01 GM080403.
Editor: Marta Filizola.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.09.006.
Supporting Material
References
- 1.Séguéla P., Wadiche J., Patrick J.W. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J. Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couturier S., Bertrand D., Ballivet M. A neuronal nicotinic acetylcholine receptor subunit (α 7) is developmentally regulated and forms a homo-oligomeric channel blocked by α-BTX. Neuron. 1990;5:847–856. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- 3.Palma E., Bertrand S., Bertrand D. Neuronal nicotinic α 7 receptor expressed in Xenopus oocytes presents five putative binding sites for methyllycaconitine. J. Physiol. 1996;491:151–161. doi: 10.1113/jphysiol.1996.sp021203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen N., Corradi J., Bouzat C. Stoichiometry for activation of neuronal α7 nicotinic receptors. Proc. Natl. Acad. Sci. USA. 2013;110:20819–20824. doi: 10.1073/pnas.1315775110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams D.K., Stokes C., Papke R.L. The effective opening of nicotinic acetylcholine receptors with single agonist binding sites. J. Gen. Physiol. 2011;137:369–384. doi: 10.1085/jgp.201010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beinat C., Banister S.D., Kassiou M. The therapeutic potential of α7 nicotinic acetylcholine receptor (α7 nAChR) agonists for the treatment of the cognitive deficits associated with schizophrenia. CNS Drugs. 2015;29:529–542. doi: 10.1007/s40263-015-0260-0. [DOI] [PubMed] [Google Scholar]
- 7.Martin L.F., Freedman R. Schizophrenia and the α 7 nicotinic acetylcholine receptor. In: Abi-Dargham A., Guillin O., editors. International Review of Neurobiology. Academic Press; 2007. pp. 225–246. [DOI] [PubMed] [Google Scholar]
- 8.Wallace T.L., Ballard T.M., Wettstein J.G. Drug targets for cognitive enhancement in neuropsychiatric disorders. Pharmacol. Biochem. Behav. 2011;99:130–145. doi: 10.1016/j.pbb.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Mineur Y.S., Mose T.N., Picciotto M.R. Hippocampal α7 nicotinic ACh receptors contribute to modulation of depression-like behaviour in C57BL/6J mice. Br. J. Pharmacol. 2018;175:1903–1914. doi: 10.1111/bph.13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao D., Xu X., Wang J. Pharmacologic activation of cholinergic alpha7 nicotinic receptors mitigates depressive-like behavior in a mouse model of chronic stress. J. Neuroinflammation. 2017;14:234. doi: 10.1186/s12974-017-1007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borovikova L.V., Ivanova S., Tracey K.J. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. 2000;405:458–462. doi: 10.1038/35013070. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Yu M., Tracey K.J. Nicotinic acetylcholine receptor α7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 13.Horenstein N.A., Papke R.L. Anti-inflammatory silent agonists. ACS Med. Chem. Lett. 2017;8:989–991. doi: 10.1021/acsmedchemlett.7b00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulsevin A., Papke R.L., Horenstein N. In silico modeling of the α7 nicotinic acetylcholine receptor: new pharmacological challenges associated with multiple modes of signaling. Mini Rev. Med. Chem. 2020;20:841–864. doi: 10.2174/1389557520666200130105256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henchman R.H., Wang H.-L., McCammon J.A. Asymmetric structural motions of the homomeric alpha7 nicotinic receptor ligand binding domain revealed by molecular dynamics simulation. Biophys. J. 2003;85:3007–3018. doi: 10.1016/S0006-3495(03)74720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henchman R.H., Wang H.L., McCammon J.A. Ligand-induced conformational change in the α7 nicotinic receptor ligand binding domain. Biophys. J. 2005;88:2564–2576. doi: 10.1529/biophysj.104.053934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X., Lu B., McCammon J.A. Channel opening motion of α7 nicotinic acetylcholine receptor as suggested by normal mode analysis. J. Mol. Biol. 2006;355:310–324. doi: 10.1016/j.jmb.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 18.Law R.J., Henchman R.H., McCammon J.A. A gating mechanism proposed from a simulation of a human α7 nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiodo L., Malliavin T.E., Ciccotti G. A structural model of the human α7 nicotinic receptor in an open conformation. PLoS One. 2015;10:e0133011. doi: 10.1371/journal.pone.0133011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulsevin A., Papke R.L., Horenstein N.A. Allosteric agonism of α7 nicotinic acetylcholine receptors: receptor modulation outside the orthosteric site. Mol. Pharmacol. 2019;95:606–614. doi: 10.1124/mol.119.115758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouzat C., Gumilar F., Sine S.M. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- 22.Frisch M.J., Trucks G.W., Fox D.J. Gaussian Inc.; Wallingford, CT: 2016. Gaussian 09, Revision A.02. [Google Scholar]
- 23.Wang J., Wang W., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Case D., Betz R., Kollman P. University of California; San Francisco, CA: 2016. Amber 16. [Google Scholar]
- 25.Wang J., Wolf R.M., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson M.P., Friesner R.A., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson M.P., Pincus D.L., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 28.Li S.-X., Huang S., Chen L. Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011;14:1253–1259. doi: 10.1038/nn.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Hansen S.B., Sulzenbacher G., Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier J.A., Martinez C., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 33.Madeira F., Park Y.M., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindorff-Larsen K., Ferkinghoff-Borg J. Similarity measures for protein ensembles. PLoS One. 2009;4:e4203. doi: 10.1371/journal.pone.0004203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiberti M., Papaleo E., Lindorff-Larsen K. ENCORE: software for quantitative ensemble comparison. PLoS Comput. Biol. 2015;11:e1004415. doi: 10.1371/journal.pcbi.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambrughi M., Tiberti M., Papaleo E. Analyzing biomolecular ensembles. In: Bonomi M., Camilloni C., editors. Biomolecular Simulations: Methods and Protocols. Springer; 2019. pp. 415–451. [DOI] [PubMed] [Google Scholar]
- 37.Mohammad Hosseini Naveh Z., Malliavin T.E., Ciccotti G. Conformational changes in acetylcholine binding protein investigated by temperature accelerated molecular dynamics. PLoS One. 2014;9:e88555. doi: 10.1371/journal.pone.0088555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hünenberger P.H., Mark A.E., van Gunsteren W.F. Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations. J. Mol. Biol. 1995;252:492–503. doi: 10.1006/jmbi.1995.0514. [DOI] [PubMed] [Google Scholar]
- 39.Barkas T., Gabriel J.M., Ballivet M. Monoclonal antibodies to the main immunogenic region of the nicotinic acetylcholine receptor bind to residues 61-76 of the α subunit. J. Biol. Chem. 1988;263:5916–5920. [PubMed] [Google Scholar]
- 40.Tzartos S.J., Kokla A., Conti-Tronconi B.M. Localization of the main immunogenic region of human muscle acetylcholine receptor to residues 67-76 of the α subunit. Proc. Natl. Acad. Sci. USA. 1988;85:2899–2903. doi: 10.1073/pnas.85.9.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galzi J.L., Bertrand D., Changeux J.P. Functional significance of aromatic amino acids from three peptide loops of the α 7 neuronal nicotinic receptor site investigated by site-directed mutagenesis. FEBS Lett. 1991;294:198–202. doi: 10.1016/0014-5793(91)80668-s. [DOI] [PubMed] [Google Scholar]
- 42.Spurny R., Debaveye S., Ulens C. Molecular blueprint of allosteric binding sites in a homologue of the agonist-binding domain of the α7 nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2015;112:E2543–E2552. doi: 10.1073/pnas.1418289112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delbart F., Brams M., Ulens C. An allosteric binding site of the α7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein. J. Biol. Chem. 2018;293:2534–2545. doi: 10.1074/jbc.M117.815316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papke R.L., Garai S., Thakur G.A. Differing activity profiles of the stereoisomers of 2,3,5,6TMP-TQS, a putative silent allosteric modulator of α7 nAChR. Mol. Pharmacol. 2020;98:292–302. doi: 10.1124/mol.120.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu R., Tae H.S., Kaas Q. Molecular dynamics simulations of dihydro-β-erythroidine bound to the human α4β2 nicotinic acetylcholine receptor. Br. J. Pharmacol. 2019;176:2750–2763. doi: 10.1111/bph.14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales-Perez C.L., Noviello C.M., Hibbs R.E. X-ray structure of the human α4β2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulens C., Akdemir A., de Esch I.J.P. Use of acetylcholine binding protein in the search for novel alpha7 nicotinic receptor ligands. In silico docking, pharmacological screening, and X-ray analysis. J. Med. Chem. 2009;52:2372–2383. doi: 10.1021/jm801400g. [DOI] [PubMed] [Google Scholar]
- 48.Yu R., Craik D.J., Kaas Q. Blockade of neuronal α7-nAChR by α-conotoxin ImI explained by computational scanning and energy calculations. PLoS Comput. Biol. 2011;7:e1002011. doi: 10.1371/journal.pcbi.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi M., Tjong H., Zhou H.-X. Spontaneous conformational change and toxin binding in alpha7 acetylcholine receptor: insight into channel activation and inhibition. Proc. Natl. Acad. Sci. USA. 2008;105:8280–8285. doi: 10.1073/pnas.0710530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiodo L., Malliavin T.E., Cottone G. A possible desensitized state conformation of the human α7 nicotinic receptor: a molecular dynamics study. Biophys. Chem. 2017;229:99–109. doi: 10.1016/j.bpc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Chiodo L., Malliavin T.E., Cottone G. Closed-locked and apo-resting state structures of the human α7 nicotinic receptor: a computational study. J. Chem. Inf. Model. 2018;58:2278–2293. doi: 10.1021/acs.jcim.8b00412. [DOI] [PubMed] [Google Scholar]
- 52.Belfield W.J., Cole D.J., Chau P.L. Constrained geometric simulation of the nicotinic acetylcholine receptor. J. Mol. Graph. Model. 2014;52:1–10. doi: 10.1016/j.jmgm.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Cheng X., Ivanov I., McCammon J.A. Nanosecond-timescale conformational dynamics of the human α7 nicotinic acetylcholine receptor. Biophys. J. 2007;93:2622–2634. doi: 10.1529/biophysj.107.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gulsevin A. Nicotinic receptor pharmacology in silico: insights and challenges. Neuropharmacology. 2020;177:108257. doi: 10.1016/j.neuropharm.2020.108257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taly A., Delarue M., Changeux J.P. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys. J. 2005;88:3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Changeux J.-P. The nicotinic acetylcholine receptor: a typical ‘allosteric machine.’. Philos. Trans. R. Soc. Lond B Biol. Sci. 2018;373:20170174. doi: 10.1098/rstb.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.