Abstract
CCAAT/enhancer binding proteins (CEBPs, including CEBPA, CEBPB, CEBPD, CEBPE, CEBPG, and CEBPZ) play critical roles in a variety of physiological and pathological processes. However, the molecular characteristics and biological significance of CEBPs in esophageal squamous cell carcinoma (ESCC) have rarely been reported. Here, we show that most of the CEBPs are upregulated and accompanied with copy number amplifications in ESCC. Of note, high CEBPG expression is regulated by the ESCC specific transcription factor TP63 and serves as a prognostic factor for poor survival in ESCC patients. Functionally, CEBPG significantly promotes the proliferation and migration of ESCC cells both in vitro and in vivo. Mechanistically, CEBPG activates the PI3K-AKT signaling pathway through directly binding to distal enhancers and/or promoters of genes involved in this pathway, including genes of CCND1, MYC, CDK2, etc. These findings provide new insights into CEBPs dysregulation in ESCC and elucidate a crucial role for CEBPG in the progression of ESCC, highlighting its potential therapeutic value for ESCC treatment.
Keywords: CEBPG, ESCC, TP63, PI3K-AKT signaling
Introduction
Esophageal cancer, histologically classified as either esophageal squamous cell carcinoma (ESCC) or esophageal adenocarcinoma (EAC), is the sixth leading cause of cancer mortality and the eighth highest in cancer incidence worldwide [1]. EAC is common in Western countries, while ESCC is much more frequently diagnosed in East Asia, particularly in China [2]. Due to a lack of understanding of the molecular bases underlying ESCC and limited effective targeted regimens, the clinical outlook for ESCC patients remains dismal, with a 5-year survival rate of approximately 17% [1]. Therefore, the alarming rise in the incidence of ESCC imposes an urgent need to further determine its molecular pathogenesis and establish successful treatment strategies.
The CCAAT/enhancer binding proteins (CEBPs, including CEBPA, CEBPB, CEBPD, CEBPE, CEBPG, and CEBPZ) are a family of leucine-zipper transcription factors that participate in physiological processes such as energy metabolism, cell proliferation, and differentiation [3-5]. Recently, CEBPs have gained considerable attention owing to their potential as molecular markers for cancer prognosis [6-11]. For example, CEBPA has been extensively studied and generally confirmed as a tumor suppressor in breast cancer, lung cancer, acute myelogenous leukemia, etc [9,11]. Both CEBPB and CEBPD play important roles in mediating cell viability of multiple cancer cell lines [12]. In hematopoietic diseases, mutations of CEBPE have been found in patients with neutrophil-specific granule deficiency [10]; CEBPG deregulation results in differentiation arrest in acute myeloid leukemia [13,14]; CEBPZ is imperative for the maintenance of leukemic state [15]. Despite biological functions of CEBPs have been revealed in several cancer types, their expression patterns and molecular changes in ESCC remain unresolved.
Here, we firstly applied The Cancer Genome Atlas (TCGA) ESCC samples to elucidate the expression patterns and molecular characteristics of CEBPs in ESCC. We found that overexpression of CEBPG presents an unfavorable prognostic indicator for ESCC patients. Importantly, the ESCC specific transcription factor, TP63, activates the expression of CEBPG by directly binding to its distal enhancer region. Moreover, CEBPG promotes ESCC cell proliferation and migration by activating PI3K-AKT signaling pathway, thus accelerating the progression of ESCC. These findings suggest that CEBPG could act as a biomarker and potential therapeutic target for ESCC patients.
Materials and methods
Cell lines and culture
ESCC cell lines were provided by the Medical College of Shantou University. The KYSE150, KYSE140, KYSE70, TE5, and COLO68N cell lines were cultured in RPMI-1640 medium and the KYSE450 cell line was in DMEM medium. All of the cell lines were supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL), and were maintained at 37°C with 5% CO2.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from cells using RNAiso Plus (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. 1 μg RNA was used for high-fidelity cDNA synthesis by the PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan). qRT-PCR was performed on an iQ5 real-time detection system (Bio-Rad Laboratories, Hercules, CA, USA) with the SYBR Kit (TaKaRa, Tokyo, Japan). Relative expression levels of genes among different samples were calculated using the ΔΔCt method, with GAPDH being used as the normalization control. Primer sequences are listed in Table 1.
Table 1.
Primers used for qRT-PCR analyses
| Name | Sequence (5’-->3’) |
|---|---|
| CEBPG Forward | ACTCCAGGGGTGAACGGAAT |
| CEBPG Reverse | CATGGGCGAACTCTTTTTGCT |
| TP63 Forward | TTCGGACAGTACAAAGAACGG |
| TP63 Reverse | GCATTTCATAAGTCTCACGGC |
| IRS1 Forward | ACAAACGCTTCTTCGTACTGC |
| IRS1 Reverse | AGTCAGCCCGCTTGTTGATG |
| MYC Forward | GGCTCCTGGCAAAAGGTCA |
| MYC Reverse | CTGCGTAGTTGTGCTGATGT |
| CCND1 Forward | GCTGCGAAGTGGAAACCATC |
| CCND1 Reverse | CCTCCTTCTGCACACATTTGAA |
| CDK2 Forward | CCAGGAGTTACTTCTATGCCTGA |
| CDK2 Reverse | TTCATCCAGGGGAGGTACAAC |
| LAMC1 Forward | GGACTCCGCCCGAGGAATA |
| LAMC1 Reverse | ACTTGAGACGCACATAGGTGA |
| FGF2 Forward | AGAAGAGCGACCCTCACATCA |
| FGF2 Reverse | CGGTTAGCACACACTCCTTTG |
| GAPDH Forward | CGACCACTTTGTCAAGCTCA |
| GAPDH Reverse | TTACTCCTTGGAGGCCATGT |
Abbreviations: CCND1, cyclin D1.
Western blotting
Total protein was extracted from cells by the RIPA buffer placed on ice. Protein concentrations of the total lysates were measured using the Bradford method. Western blotting was performed by using the SDS-PAGE and conventional wet gel transfer systems. PVDF membranes (Millipore, New Bed-ford, MA, USA) were incubated with primary antibodies overnight at 4°C, followed by incubation of secondary antibodies for 1 h at room temperature on the next day. At last, the chemiluminescence kit (Thermo Fisher Scientific) was used to detect immunoreactive protein bands. Information about antibodies used is as follows: CEBPG (Proteintech, No. 12997-1-AP; 1:500), TP63 (Proteintech, No. 12143-1-AP; 1:1000), IRS1 (Proteintech, No. 17509-1-AP; 1:1000), CDK2 (CST, No. 2546; 1:1000), AKT (CST, No. 4691; 1:1000), P-AKT (CST, No. 5315; 1:1000), MYC (TRAN, No. M21217; 1:1000), CCND1 (CST, No. 2922; 1:1000), GAPDH (Proteintech, No. 10494-1-AP; 1:1000), Tubulin (Abiocode, No. R0742-3; 1:1000).
siRNAs and transfection
Two different siRNAs were used for CEBPG knockdown by using the Lipofectamine RNAiMAX (Thermo Fisher Scientific). The target sequences of CEBPG siRNAs are: CEBPG-siRNA1, 5’-GACCCATTGGAGGCTATTT-3’; CEBPG-siRNA2, 5’-CTGACCAAGGAATTAAGTGTA-3’. For retroviral infection experiments, double-stranded oligonucleotides were firstly inserted into the pLKO.1-TRC (Addgene#10878) vector. Retrovirus was then generated in 293T cells by cotransfection of the recombinant vectors with psPAX2 (Addgene#12260) and pMD2.G (Addgene#12259). Supernatants were collected at 24, 48, and 72 h post-transfection. To obtain stable cell lines, ESCC cells were infected at a multiplicity of infection and selected with the presence of 2 mg/ml puromycin. The sequences of CEBPG shRNAs are identical to those of the CEBPG siRNAs. The shRNA sequences for TP63 are: shTP63-1, 5’-GGACAGCAGCATTGATCAA-3’; shTP63-2, 5’-CCGTTTCGTCAGAACACACAT-3’.
Cell growth and migration
For in vitro cell growth assays, a total of 4000 cells transfected with either scramble or siRNAs were seeded into each well of the 96-well plates. The cell proliferation rate was determined by the CCK8 (Cell Counting Kit-8) method. For cell migration assays, transwell chambers were used to assess cell motility. Briefly, transfected cells in serum-free medium were plated into the upper chamber, while medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, migratory cells were fixed with 4% formalin, stained with 0.1% crystal violet, photographed under a microscope, and counted in five random fields per well. Experiments were repeated independently three times.
Animal xenograft experiment
Eighteen 4-week-old BALB/c-nude mice were obtained from the Experimental Animal Center of the Chinese Academy of Science (Guangzhou, China). Under the Institutional Animal Care and Use Committee of Sun Yat-Sen University, the mice were randomly divided into three groups. Then a total of 5×106 KYSE150 cells transfected with either scramble or CEBPG shRNAs were injected subcutaneously into the right flank of each mouse. When tumor xenografts were palpable, tumor volumes were measured every 4 days and continuously monitored over the next 24 days. Tumor volumes were calculated using the following formula: Volume = length×width2/2. Tumor tissues were dissected for further analyses after mice were sacrificed.
Chromatin Immunoprecipitation PCR (ChIP-PCR)
For ChIP assays, cells were crosslinked with 1% formaldehyde for 10 min at room temperature and quenched by glycine. Cells were then subjected to SDS lysis buffer and sheared in a Bioruptor Sonicator (Diagenode, USA) to achieve genomic fragments of approximately 500 base pairs. After sonication, cell debris was removed, supernatant was incubated with indicated antibodies overnight at 4°C. On the next day, magnetic beads were added to pull down the antibody-chromatin complexes. Finally, DNA from the precipitated immune-complex was extracted and purified. ChIP-PCR was used to detect the degree of DNA enrichment in each group. Primer sequences are listed in Table 2.
Table 2.
Primers used for ChIP-PCR analyses
| Name | Sequence (5’-->3’) |
|---|---|
| CEBPG enhancer Forward | TGCAGAGGTCATCTGTAATAA |
| CEBPG enhancer Reverse | AAACCACTTGAGTCACACAGGAA |
| CEBPG promoter Forward | TCAGAAAATGCTTCCAAAGGCAC |
| CEBPG promoter Reverse | GACGGTTACGAAACCGGCAGCCT |
| CCND1 promoter Forward | TTTGATCTTTGCTTAACAACAGT |
| CCND1 promoter Reverse | CTCGGCTCTCGCTTCTGCT |
| MYC promoter Forward | GCAGAGGGCGTGGGGGAAAAGAA |
| MYC promoter Reverse | TTATACTCAGCGCGATCCCTCCC |
| MYC enhancer Forward | TGACAAGGCTTTGGTGAGAGCCT |
| MYC enhancer Reverse | ATTTAGTGACATAGTTTGTACTG |
| NC Forward | GGGCCAAGGACTCTTACTGT |
| NC Reverse | AGGAATGGGTGGGAAGTCAG |
Abbreviations: CCND1, cyclin D1; NC, negative control.
RNA-Seq data analysis
The 50 bp single-end clean reads were aligned to the hg19 Ensemble (V82) transcriptome using the HISAT2 (v2.0.4) [16,17]. Under their default parameter settings, RSEM (v1.2.12) and DESeq (v3.4) were used for differential gene expression analyses [18,19]. DAVID database was used for pathway enrichment analyses of identified differentially expressed genes (DEGs, Log2 |fold change|>1, P<0.05) [20,21].
Data and materials availability
The RNA-Seq data of KYSE150 cells with the absence and presence of CEBPG generated in this study have been deposited into the GEO under accession number “GSE156257” (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156257). Other RNA-Seq data, as well as the copy number data and clinical data of ESCC were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). The ATAC-Seq data of ESCC were obtained from https://gdc.cancer.gov/about-data/publications/ATACseq-AWG. The ChIP-Seq data of H3K27ac, SOX2, and TP63 in ESCC cells and CEBPG in HepG2 cells were obtained from GEO under accession numbers “GSE106563” and “GSE96281” [22,23].
Statistical analysis
The relations between expressions of CEBPs with overall survival of ESCC patients were evaluated using the Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=background). Comparisons between two groups were carried out using the Student’s t-test or the Mann-Whitney U test. Statistical analyses were performed by the GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Statistically significant P values are indicated as *P<0.05, **P<0.01, and ***P<0.001.
Results
Identification of CEBPG as an unfavorable prognostic factor for ESCC patients
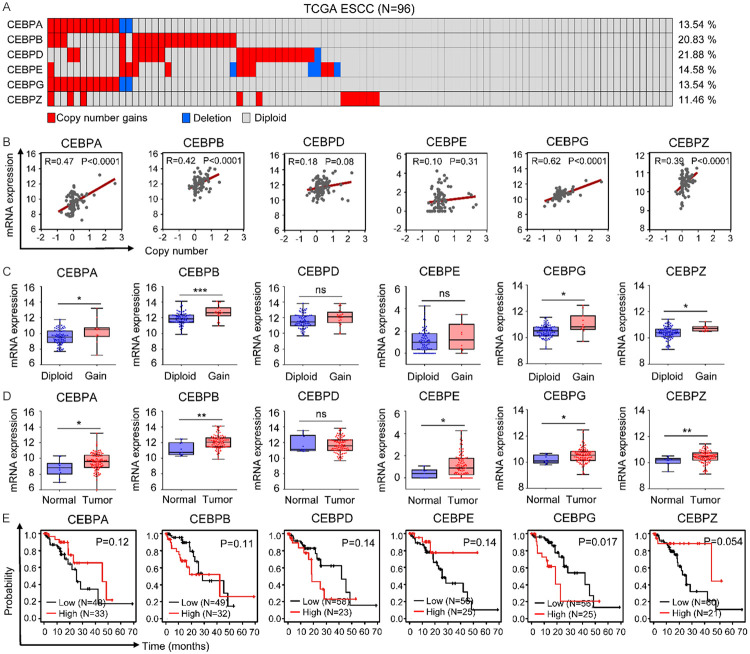
To illuminate the molecular characteristics of CEBPs in ESCC, we firstly applied cBioPortal to identify their mutational status in TCGA ESCC samples. Among all CEBPs, CEBPZ was the only molecule found to have somatic mutations, with a frequency of 1% (Figure S1). Since major sources of genetic diversity also include copy number variations (CNVs), we next analyzed CNVs of CEBPs in ESCC. Notably, various degrees of copy number gains (defined as log2 copy number (tumor/normal)>0.5) [24] of CEBPs were observed in ESCC. In addition, the frequency of CNVs was higher than 10% for each CEBP. Specifically, CNVs of CEBPA, CEBPB, CEBPD, CEBPE, CEBPG, and CEBPZ were detected in 13 (13.54%), 20 (20.83%), 21 (21.88%), 14 (14.58%), 13 (13.54%), and 11 (11.46%) of the ESCC samples, respectively (Figure 1A).
Figure 1.
Identification of CEBPG as an unfavorable prognostic factor for ESCC patients. A. The copy number variations of CEBPs in TCGA ESCC samples. B. Pearson correlations between copy number of each CEBP and their respective mRNA expression. C. The mRNA expressions of CEBPs were compared between TCGA ESCC samples, which were stratified into groups of Diploid and Gain based on the gene dosage of each CEBP. Mann-Whitney U test. *P<0.05; **P<0.01; ***P<0.001; ns, not significant. D. The mRNA expressions of CEBPs in TCGA ESCC samples and adjacent normal tissues. Mann-Whitney U test. *P<0.05; **P<0.01; ns, not significant. E. Kaplan-Meier plots showing the associations between expressions of CEBPs and the overall survival of ESCC patients.
To determine whether copy number gains affect the expressions of CEBPs in ESCC, correlation analyses were performed based on gene expression and copy number data from TCGA. Interestingly, the mRNA expression levels of CEBPA, CEBPB, CEBPG, and CEBPZ were positively correlated with their copy number gains (Figure 1B). We further categorized TCGA ESCC samples into groups of diploid and copy-number gain (Gain) based on the gene dosage of each CEBP, and found that relative to the diploid groups, the mRNA expressions of CEBPA, CEBPB, CEBPG, and CEBPZ were significantly up-regulated in their Gain groups (Figure 1C). These results suggested that copy number gains contribute to high expressions of CEBPA, CEBPB, CEBPG, and CEBPZ in ESCC.
After illuminating the genetic features of CEBPs, we then analyzed their expression patterns in ESCC. Interestingly, compared to normal tissues, the expression levels of most CEBPs, including CEBPA, CEBPB, CEBPE, CEBPG, and CEBPZ, were up-regulated in ESCC (Figure 1D). To further identify their potential prognostic values, we evaluated the overall survival of ESCC patients in relation to each CEBP expression using the Kaplan Meier plotter. Notably, the overall survival of ESCC patients with higher CEBPG expression was significantly poorer than that of the patients with lower CEBPG expression (Figure 1E), suggesting that CEBPG represents a negative prognostic factor for ESCC patients. Therefore, CEBPG was prioritized for further biological and mechanistic studies.
TP63 directly activates CEBPG by binding to its distal enhancer
Besides genetic alterations, epigenetic modifications, such as DNA methylation, also play an important role in ESCC progression [25,26]. In order to investigate whether DNA methylation affects CEBPG expression in ESCC, we analyzed the methylation status of CEBPG using TCGA ESCC DNA methylation data (HumanMethylation450). Unfortunately, the methylation level of CEBPG promoter region showed no difference between tumor and normal samples (Figure S2).
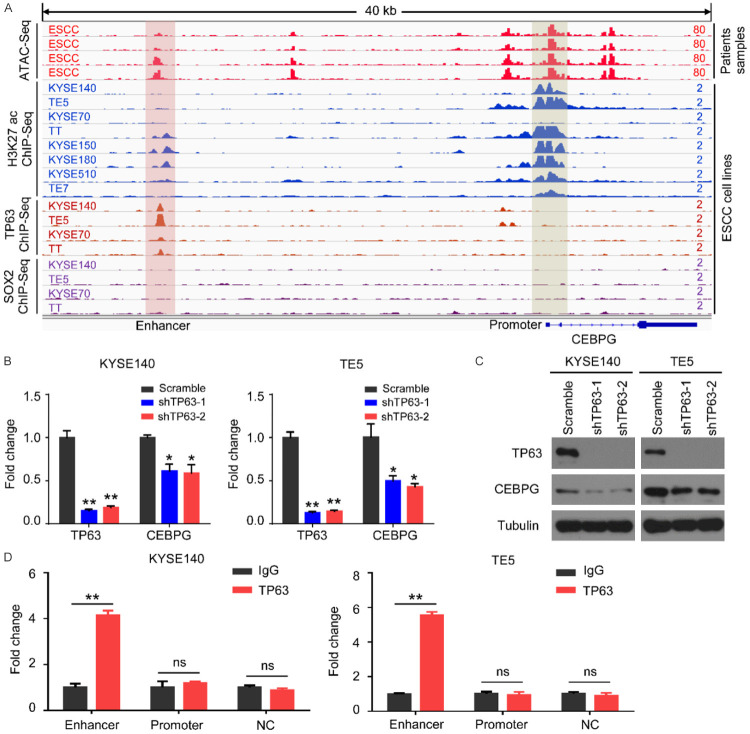
On the other hand, the expression patterns of genes are strictly modulated by the transcriptional regulation process. We therefore attempted to elucidate the molecular basis underlying CEBPG transcriptional activation in ESCC. Importantly, the ATAC-Seq data from TCGA showed that three promoter regions and two distal regions of CEBPG exhibited high chromatin accessibility in ESCC samples (top 4 red tracks, Figure 2A). Further interrogating the ChIP-Seq data of ESCC cell lines, we found that two of these regions (one at the promoter region and the other at the enhancer region) had coincident H3K27ac signals (tracks 5-12, Figure 2A), suggesting their potential transcriptional regulatory roles. Since TP63 and SOX2 are the two best characterized ESCC specific transcription factors [22,27], we reasonably suspected whether they could regulate CEBPG expression. Accordingly, we individually knocked down TP63 and SOX2 in two different ESCC cell lines (KYSE140 and TE5) and found that CEBPG mRNA expression was unaffected upon SOX2 knockdown in both cell lines (Figure S3). In contrast, both the mRNA and protein levels of CEBPG were significantly down-regulated when silencing TP63 (Figure 2B, 2C). These data demonstrated that the ESCC specific transcription factor TP63 activates CEBPG expression in ESCC.
Figure 2.
TP63 activates CEBPG by directly binding to its distal enhancer. A. The ATAC-Seq and indicated ChIP-Seq signals at CEBPG locus. The ATAC-Seq data of ESCC patients were downloaded from the TCGA database. The ChIP-Seq data of H3K27ac, TP63, and SOX2 of ESCC cell lines were obtained from GSE106563. B. The relative CEBPG mRNA expression following knockdown of TP63 in KYSE140 and TE5 cell lines. Data were shown as Mean ± SD. Unpaired t test. *, P<0.05; **, P<0.01; ns, not significant. C. Western blotting showing the expression changes of CEBPG and TP63 when silencing TP63 in KYSE140 and TE5 cell lines. D. Confirmation of the TP63 binding sites at the CEBPG locus by ChIP-qPCR. NC was used as a negative control. Unpaired t test. **, P<0.01; ns, not significant.
We next asked whether the regulation of TP63 on CEBPG was mediated by the two putative regulatory elements on CEBPG (tracks 1-12, Figure 2A). As anticipated, ChIP-Seq data showed that TP63 had readily discernible signals at the CEBPG enhancer region (tracks 13-16, Figure 2A). However, TP63 binding at the CEBPG promoter was hardly detectable (tracks 13-16, Figure 2A). In addition, weak ChIP-seq signals of SOX2 were consistently observed along the whole body of CEBPG (bottom 4 tracks, Figure 2A). We further performed TP63 ChIP-PCR in KYSE140 and TE5 cells lines for verification. Congruent with the ChIP-Seq data, TP63 was highly enriched at the CEBPG enhancer, while absent at its promoter in both cell lines (Figure 2D). Collectively, these data demonstrated that the ESCC specific transcription factor TP63 directly activates CEBPG expression through binding to its distal enhancer.
CEBPG promotes ESCC cell proliferation, migration, and tumor growth
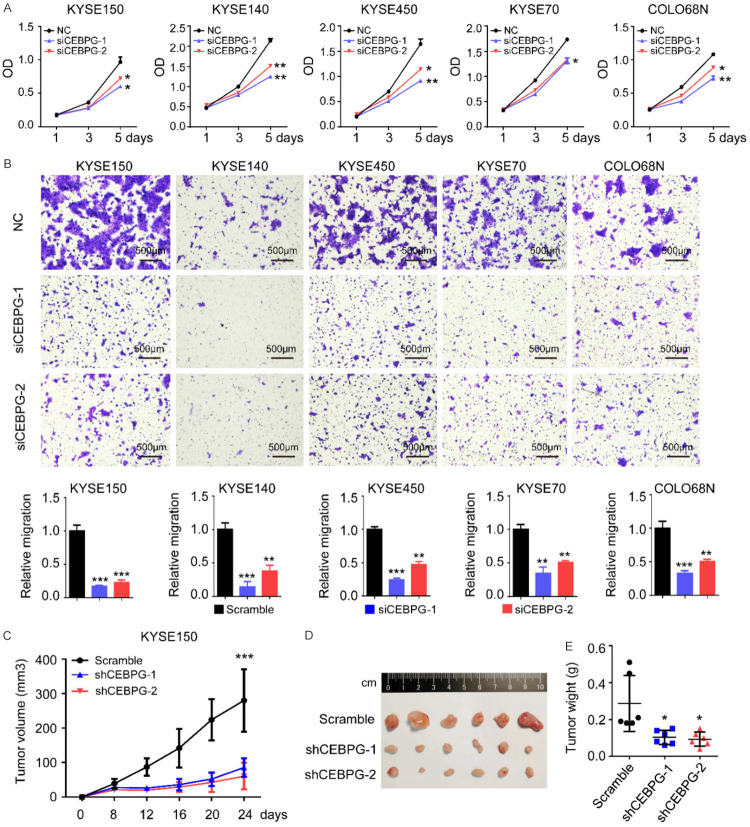
After elucidating the molecular mechanisms underlying CEBPG overexpression in ESCC, we were particularly interested in exploring its biological significance. To address this, we selected 5 ESCC cell lines (KYSE150, KYSE140, KYSE450, KYSE70, and COLO68N) with relatively high expression of CEBPG and knocked down CEBPG in each of them by using two independent siRNAs (Figure S4A). The knockdown efficiency of CEBPG was monitored by Western blotting (Figure S4B). Importantly, knockdown of CEBPG significantly inhibited the proliferation rates of all 5 ESCC cell lines (Figure 3A). In order to investigate the function of CEBPG on cell motility, transwell assays were performed. Notably, upon silencing CEBPG, cell migration abilities of all 5 ESCC cell lines were dramatically attenuated (Figure 3B). Furthermore, a subcutaneous xenograft tumor model of ESCC in nude mice was established to confirm the oncogenic role of CEBPG in vivo. Importantly, silencing of CEBPG by shRNAs conspicuously inhibited xenograft tumor growth (Figure 3C-E), corroborating the in vitro results. Altogether, these results strongly suggested that CEBPG is an oncogenic factor, contributing to the proliferation and migration of ESCC cells.
Figure 3.
CEBPG promotes ESCC cell proliferation, migration and tumor growth in vivo. (A) Proliferation curves and (B) representative images of transwell assay results of KYSE150, KYSE140, KYSE450, KYSE70, and COLO68N cell lines with the presence or absence of CEBPG knockdown. Above: the schematic diagram; Bottom: a statistical graph. Data were shown as Mean ± SD. Unpaired t test. *, P<0.05; **, P<0.01; *** P<0.001. (C) Growth curves of the xenograft tumors, (D) tumor images and (E) tumor weight measured at the end point of the in vivo experiments. N=6. Unpaired t test. *, P<0.05; ***, P<0.001.
CEBPG activates the PI3K-AKT signaling pathway in ESCC
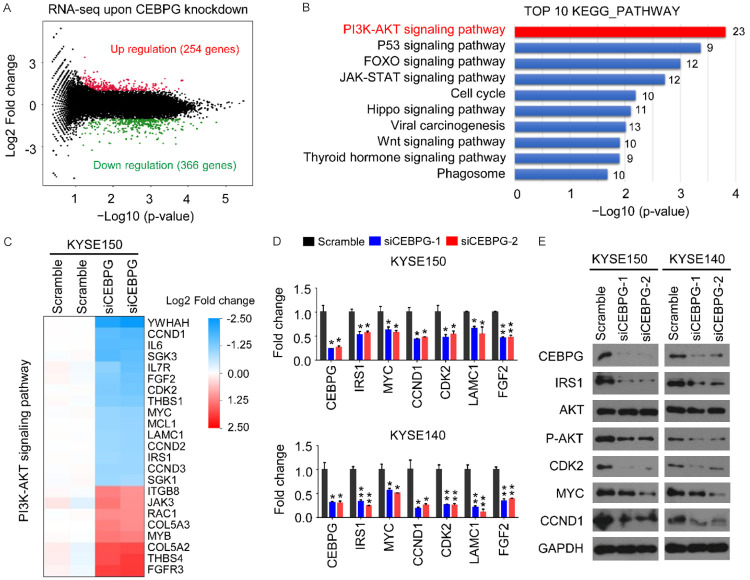
To reveal possible signaling pathways responsible for CEBPG-promoted ESCC cell proliferation and migration, RNA-seq analyses were performed on KYSE150 cell line in either the absence or presence of CEBPG. Compared to the control group, a total of 620 DEGs were identified upon CEBPG knockdown, including 366 downregulated genes and 254 upregulated genes (Log2 |fold change|>1, P<0.05, Figure 4A). We then conducted a functional enrichment analysis of all of these DEGs by using the KEGG Pathway Database. Of note, results showed that they were significantly enriched in tumor growth-related pathways such as the PI3K-AKT signaling (ranking 1st), the P53 signaling (ranking 2nd), the FOXO signaling (ranking 3rd), the JAK-STAT signaling (ranking 4th), etc (Figure 4B). These were highly congruent with our previous findings that CEBPG promoted ESCC cell proliferation as well as cell migration (Figure 3).
Figure 4.
CEBPG activates the PI3K-AKT signaling pathway in ESCC. A. The volcano plot showing that a total of 620 genes were differentially expressed (366 downregulated v.s. 254 upregulated) upon CEBPG knockdown in KYSE150 cell line. B. KEGG pathway analysis of the differentially expressed genes (DEGs, Log2 fold change>1 or <-1, P<0.05). C. Heatmap of DEGs that are associated with the PI3K-AKT signaling pathway. D. The mRNA expression changes of indicated genes following knockdown of CEBPG in KYSE150 and KYSE140 cell lines. Data were shown as Mean ± SD. Unpaired t test. *, P<0.05; **, P<0.01. E. Western blotting of indicated proteins in KYSE140 and TE5 cells following knockdown of CEBPG.
Being the highest ranking pathway of the enrichment analysis result, the PI3K-AKT signaling was selected for in-depth investigation. Notably, compared to the control group, 23 DEGs (8 upregulated and 15 downregulated) identified in the siCEBPG group were associated with the PI3K-AKT signaling pathway, including genes of CCND1, CDK2, IRS1, MYC, etc (Figure 4C). To further determine the regulation of CEBPG on the PI3K-AKT signaling pathway, a total of 6 genes (IRS1, MYC, CCND1, CDK2, LAMC1, and FGF2) from Figure 4C were randomly selected for qRT-PCR validation. In accordance with the RNA-Seq results, the mRNA levels of all of these 6 genes were downregulated in ECSS cell lines when silencing CEBPG (Figure 4D). Additionally, the protein levels of some of these genes (IRS1, MYC, CCND1, and CDK2) were consistently reduced after CEBPG knockdown (Figure 4E). Moreover, reduced levels of P-AKT, a key node in the PI3K-AKT signaling pathway, in the siCEBPG-treated cells further confirmed that targeting CEBPG indeed inhibited the activation of PI3K-AKT signaling pathway. These results strongly indicated that CEBPG activates the PI3K-AKT signaling pathway in ESCC.
CEBPG directly binds to regulatory element of genes involved in the PI3K-AKT signaling pathway
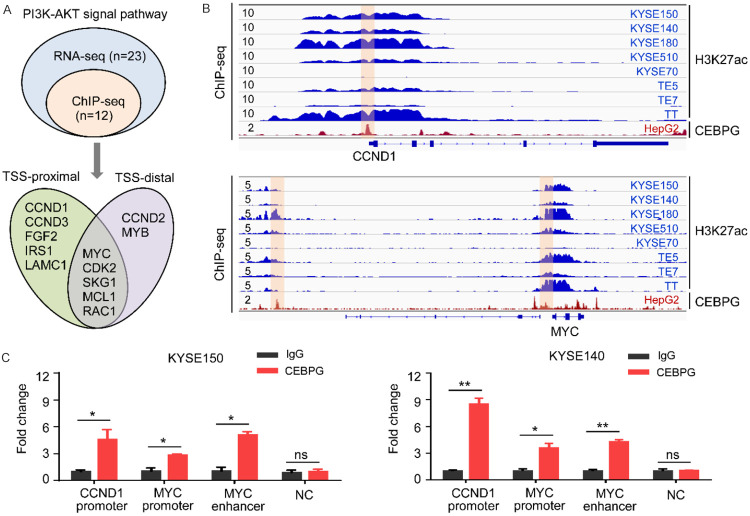
In order to clarify the molecular mechanisms of how CEBPG was affecting the PI3K-AKT signaling pathway, we analyzed publicly available ChIP-Seq data of CEBPG from the ENCODE database and performed ChIP-PCR experiments for further verification. Focusing on the PI3K-AKT signaling pathway, we found that more than 50% (12/23) of the DEGs identified in the RNA-Seq analyses were directly occupied by CEBPG (Figure 5A). Amongst them, five genes, including CCND1, CCND3, FGF2, IRS1, and LAMC1, were bound by CEBPG only at their TSS-proximal regions (Figure 5A), while two genes (CCND2 and MYB) were bound by CEBPG only at their TSS-distal regions (Figure 5A). Moreover, binding peaks of CEBPG were observed at both the TSS-proximal and TSS-distal regions of genes of MYC, CDK2, SKG1, MCL1, and RAC1 (Figure 5A). Notably, the binding profiles of CEBPG on representative target genes (CCND1 and MYC) have been shown in Figure 5B. To further confirm the ChIP-Seq results, we also conducted CEBPG ChIP-PCR experiments in KYSE150 and KYSE140 cell lines. Consistently, we found that CEBPG was highly enriched at the TSS-proximal region of CCND1, and indeed occupied at both the TSS-proximal and TSS-distal regions of MYC (Figure 5C). These results suggested that CEBPG activates PI3K-AKT signaling pathway through directly binding to the distal enhancers and/or promoters of genes involved in this pathway.
Figure 5.
CEBPG directly binds to the regulatory elements of genes involved in the PI3K-AKT signaling pathway. A. Based on the ChIP-Seq and RNA-Seq data, Venn diagrams showing the numbers of overlapped genes associated with the PI3K-AKT signaling pathway. The CEBPG ChIP-Seq data of HepG2 cell line was downloaded from GSE96281. TSS-proximal, <1 kb of TSS; TSS-distal, >1 kb of TSS; TSS, Transcription Start Site. B. Various ChIP-Seq profiles of CCND1 and MYC gene loci. The H3K27ac ChIP-Seq data of various ESCC cell lines were downloaded from GSE106563. The CEBPG ChIP-Seq data of HepG2 cells line was downloaded from GSE96281. C. The ChIP-qPCR for CEBPG at the CCND1 and MYC loci in KYSE150 and KYSE140 cell lines. NC was used as a negative control. Unpaired t test. *, P<0.05; **, P<0.01; ns, not significant.
Discussion
As a major pathological subtype of esophageal cancer, ESCC maintains a low 5-year survival rate due to its propensity to recur [1]. Therefore, the molecular mechanisms underlying ESCC progression need to be better understood, so as to accelerate the development of more effective therapeutic strategies. As a crucial class of DNA binding factors, CEBPs have been reported to play vital roles in many physiological and pathological processes [3-5,28]. However, the molecular features and biological significance of CEBPs in ESCC remain unclear. Here, we revealed the molecular alterations of CEBPs in ESCC and found that most of the CEBPs were upregulated and accompanied by DNA copy number amplifications in ESCC. We also confirmed the biological functions and molecular mechanisms of CEBPG in ESCC and identified it as a negative prognostic factor for ESCC patients.
Many studies indicated that both genetic and epigenetic variations play important roles in regulating gene expression. For instance, gene suppression could be caused by either DNA mutations or hypermethylation, while gene activation could be induced by DNA copy number gains and transcription factors [26,29,30]. In this study, we identified the recurrent copy number gains of CEBPs in ESCC. However, somatic mutations on CEBPs occurred rarely in ESCC, with the exception of CEBPZ, of which a mutation frequency of 1% was found. Moreover, significant positive correlations were identified between the mRNA expressions and copy number gains of CEBPA, CEBPB, CEBPG, and CEBPZ in ESCC, suggesting that copy number gains account for the high expression of these CEBPs. Interestingly, among all CEBPs, although CEBPD possessed the highest frequency of copy number gains in ESCC, these copy number gains did not result in its mRNA upregulation. The undefined reasons for this phenomenon await further investigation.
Recently, a variety of genes that are transcriptionally activated by tissue-specific transcription factors have been found to contribute greatly to human cancers, including ESCC [22,31]. Here, we firstly identified that the ESCC specific transcription factor TP63 could directly interact with the distal enhancer region of CEBPG and upregulate its expression. Mechanistically, we presumed that TP63 might remove the Polycomb Repressive Complex from CEBPG and then recruit additional transcriptional cofactors to finally activate the transcription of CEBPG. However, the exact molecular mechanisms need to be investigated further.
In the physiological state, CEBPG plays a role in the ATF5-regulated vomeronasal sensory neuron differentiation during early postnatal development [32]. While in the pathological state, CEBPG is a critical regulator involved in cellular stress response, DNA repair, and endoplasmic reticulum stress response [33-35]. Moreover, CEBPG is also associated with heroin addiction, chronic obstructive pulmonary disease, senescence, and inflammatory [36-38]. In the context of cancers, the functional significance of CEBPG has been extensively studied in acute myeloid leukemia [8,14,39]. For example, CEBPG is known to mediate the myeloid differentiation arrest induced by CEBPA deficiency [14]. Although several studies have also indicated that CEBPG increases the risk of lung cancer [33,40], its functional contribution to ESCC remains hitherto unknown. Here, we firstly demonstrated that CEBPG was required for ESCC cell proliferation and migration as well as xenograft growth in vivo, adding further evidence to confirm that CEBPG manifests oncogenic activity in solid tumors.
Increasing studies have reported that CEBPG is closely related to tumorigenesis via regulation of antioxidant and DNA repair genes, such as ERCC5 [41,42]. In the present study, we found that CEBPG promoted ESCC tumorigenesis through enhancing the PI3K-AKT signaling pathway. Mechanistically, acting as a transcription factor [43], CEBPG directly occupies the TSS-proximal and/or TSS-distal regions of downstream target genes associated with PI3K-AKT signaling, such as MYC, CDK2, SKG1, MCL1, etc.
In summary, we systematically analyzed the molecular characteristics of CEBPs in ESCC and identified CEBPG as a prognostic factor for poor survival of ESCC patients. As a highly expressed member of CEBPs in ESCC, CEBPG was directly activated by the ESCC specific transcription factor TP63. Functionally, CEBPG promoted ESCC cell proliferation and migration in vitro and accelerated tumor growth in vivo. Mechanistically, CEBPG activated the PI3K-AKT signaling pathway via directly binding to the distal enhancers and/or promoters of target genes. Thus, our findings provided novel insights into the CEBPs dysregulation in ESCC and elucidated a crucial role for CEBPG in promoting ESCC progression.
Acknowledgements
This study was supported by Natural Science Foundation of China (81872140, 81572484, 81420108026, 81872306 and 81621004); Guangdong Science and Technology Department (2019B020226003); Guangzhou Bureau of Science and Information Technology (201704030036).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–1187. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 4.Fong Y, Shen KH, Chen LJ, Cheng JT. Changes of CCAAT enhancer-binding proteins (CEBPs) in the lung of streptozotocin-induced diabetic rats. Horm Metab Res. 2011;43:261–267. doi: 10.1055/s-0030-1270452. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Croniger CM, Lekstrom-Himes J, Zhang P, Fenyus M, Tenen DG, Darlington GJ, Hanson RW. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J Biol Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- 6.Roe JS, Vakoc CR. C/EBPα: critical at the origin of leukemic transformation. J Exp Med. 2014;211:1–4. doi: 10.1084/jem.20132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Reebye V, Hitchen P, Fan J, Jiang H, Sætrom P, Rossi J, Habib NA, Huang KW. Mechanisms involved in the activation of C/EBPα by small activating RNA in hepatocellular carcinoma. Oncogene. 2019;38:3446–3457. doi: 10.1038/s41388-018-0665-6. [DOI] [PubMed] [Google Scholar]
- 8.Akasaka T, Balasas T, Russell LJ, Sugimoto KJ, Majid A, Walewska R, Karran EL, Brown DG, Cain K, Harder L, Gesk S, Martin-Subero JI, Atherton MG, Brüggemann M, Calasanz MJ, Davies T, Haas OA, Hagemeijer A, Kempski H, Lessard M, Lillington DM, Moore S, Nguyen-Khac F, Radford-Weiss I, Schoch C, Struski S, Talley P, Welham MJ, Worley H, Strefford JC, Harrison CJ, Siebert R, Dyer MJ. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109:3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 9.Avellino R, Havermans M, Erpelinck C, Sanders MA, Hoogenboezem R, van de Werken HJ, Rombouts E, van Lom K, van Strien PM, Gebhard C, Rehli M, Pimanda J, Beck D, Erkeland S, Kuiken T, de Looper H, Gröschel S, Touw I, Bindels E, Delwel R. An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation. Blood. 2016;127:2991–3003. doi: 10.1182/blood-2016-01-695759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shyamsunder P, Shanmugasundaram M, Mayakonda A, Dakle P, Teoh WW, Han L, Kanojia D, Lim MC, Fullwood M, An O, Yang H, Shi J, Hossain MZ, Madan V, Koeffler HP. Identification of a novel enhancer of CEBPE essential for granulocytic differentiation. Blood. 2019;133:2507–2517. doi: 10.1182/blood.2018886077. [DOI] [PubMed] [Google Scholar]
- 11.Lourenço AR, Coffer PJ. A tumor suppressor role for C/EBPα in solid tumors: more than fat and blood. Oncogene. 2017;36:5221–5230. doi: 10.1038/onc.2017.151. [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Jefferson P, Zhou Q, Angelastro JM, Greene LA. Dominant-negative ATF5 compromises cancer cell survival by targeting CEBPB and CEBPD. Mol Cancer Res. 2020;18:216–228. doi: 10.1158/1541-7786.MCR-19-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown AL, de Smith AJ, Gant VU, Yang W, Scheurer ME, Walsh KM, Chernus JM, Kallsen NA, Peyton SA, Davies GE, Ehli EA, Winick N, Heerema NA, Carroll AJ, Borowitz MJ, Wood BL, Carroll WL, Raetz EA, Feingold E, Devidas M, Barcellos LF, Hansen HM, Morimoto L, Kang AY, Smirnov I, Healy J, Laverdière C, Sinnett D, Taub JW, Birch JM, Thompson P, Spector LG, Pombo-de-Oliveira MS, DeWan AT, Mullighan CG, Hunger SP, Pui CH, Loh ML, Zwick ME, Metayer C, Ma X, Mueller BA, Sherman SL, Wiemels JL, Relling MV, Yang JJ, Lupo PJ, Rabin KR. Inherited genetic susceptibility to acute lymphoblastic leukemia in down syndrome. Blood. 2019;134:1227–1237. doi: 10.1182/blood.2018890764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberich-Jordà M, Wouters B, Balastik M, Shapiro-Koss C, Zhang H, Di Ruscio A, Radomska HS, Ebralidze AK, Amabile G, Ye M, Zhang J, Lowers I, Avellino R, Melnick A, Figueroa ME, Valk PJ, Delwel R, Tenen DG. C/EBPγ deregulation results in differentiation arrest in acute myeloid leukemia. J Clin Invest. 2012;122:4490–4504. doi: 10.1172/JCI65102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, Vakoc CR, Vassiliou GS, Kouzarides T. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, Xiao JF, Li CQ, Huang ML, Ding LW, Sun QY, Xu L, Kanojia D, Jeitany M, Deng JW, Liao LD, Soukiasian HJ, Berman BP, Hao JJ, Xu LY, Li EM, Wang MR, Bi XG, Lin DC, Koeffler HP. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619. doi: 10.1038/s41467-018-06081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin DC, Wang MR, Koeffler HP. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology. 2018;154:374–389. doi: 10.1053/j.gastro.2017.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: In the right place at the right time. Science. 2018;361:1336–1340. doi: 10.1126/science.aat6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, Fox C, Francis JM, Pedamallu CS, DeLuca DS, Brooks AN, Wang S, Que J, Rustgi AK, Wong KK, Ligon KL, Liu XS, Marto JA, Meyerson M, Bass AJ. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 2014;124:1636–1645. doi: 10.1172/JCI71545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Xia X, Mao L, Wang S. The CCAAT/enhancer-binding protein family: its roles in MDSC expansion and function. Front Immunol. 2019;10:1804. doi: 10.3389/fimmu.2019.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kojima S, Cimini D. Aneuploidy and gene expression: is there dosage compensation? Epigenomics. 2019;11:1827–1837. doi: 10.2217/epi-2019-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 31.Sonawane AR, Platig J, Fagny M, Chen CY, Paulson JN, Lopes-Ramos CM, DeMeo DL, Quackenbush J, Glass K, Kuijjer ML. Understanding tissue-specific gene regulation. Cell Rep. 2017;21:1077–1088. doi: 10.1016/j.celrep.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano H, Iida Y, Murase T, Oyama N, Umemura M, Takahashi S, Takahashi Y. Co-expression of C/EBPγ and ATF5 in mouse vomeronasal sensory neurons during early postnatal development. Cell Tissue Res. 2019;378:427–440. doi: 10.1007/s00441-019-03070-2. [DOI] [PubMed] [Google Scholar]
- 33.Blomquist T, Crawford EL, Mullins D, Yoon Y, Hernandez DA, Khuder S, Ruppel PL, Peters E, Oldfield DJ, Austermiller B, Anders JC, Willey JC. Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 2009;69:8629–8635. doi: 10.1158/0008-5472.CAN-09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, Kasoji M, Haines DC, Quiñones OA, Johnson PF. C/EBPγ is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol Cell Biol. 2015;36:693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang JW, Zhang W, Yeh HS, Park M, Yao C, Shi Y, Kuang R, Yong J. An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic Acids Res. 2018;46:5996–6008. doi: 10.1093/nar/gky340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo J, Morales DA, Chen T, Crawford EL, Zhang X, Blomquist TM, Levin AM, Massion PP, Arenberg DA, Midthun DE, Mazzone PJ, Nathan SD, Wainz RJ, Nana-Sinkam P, Willey PFS, Arend TJ, Padda K, Qiu S, Federov A, Hernandez DR, Hammersley JR, Yoon Y, Safi F, Khuder SA, Willey JC. RNAseq analysis of bronchial epithelial cells to identify COPD-associated genes and SNPs. BMC Pulm Med. 2018;18:42. doi: 10.1186/s12890-018-0603-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen SJ, Liao DL, Shen TW, Yang HC, Chen KC, Chen CH. Genetic signatures of heroin addiction. Medicine (Baltimore) 2016;95:e4473. doi: 10.1097/MD.0000000000004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huggins CJ, Malik R, Lee S, Salotti J, Thomas S, Martin N, Quiñones OA, Alvord WG, Olanich ME, Keller JR, Johnson PF. C/EBPγ suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPβ. Mol Cell Biol. 2013;33:3242–3258. doi: 10.1128/MCB.01674-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haghi N, Brody J, Mahmood N, Gheewala D, Allen SL, Sreekantaiah C, Zhang X. B-cell precursor acute lymphoblastic leukemia with isolated t(14;19)(q32;q13) abnormality involving the CEBPG gene. Leuk Lymphoma. 2015;56:2978–2981. doi: 10.3109/10428194.2015.1011639. [DOI] [PubMed] [Google Scholar]
- 40.Blomquist TM, Brown RD, Crawford EL, de la Serna I, Williams K, Yoon Y, Hernandez DA, Willey JC. CEBPG exhibits allele-specific expression in human bronchial epithelial cells. Gene Regul Syst Bio. 2013;7:125–138. doi: 10.4137/GRSB.S11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullins DN, Crawford EL, Khuder SA, Hernandez DA, Yoon Y, Willey JC. CEBPG transcription factor correlates with antioxidant and DNA repair genes in normal bronchial epithelial cells but not in individuals with bronchogenic carcinoma. BMC Cancer. 2005;5:141. doi: 10.1186/1471-2407-5-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford EL, Blomquist T, Mullins DN, Yoon Y, Hernandez DR, Al-Bagdhadi M, Ruiz J, Hammersley J, Willey JC. CEBPG regulates ERCC5/XPG expression in human bronchial epithelial cells and this regulation is modified by E2F1/YY1 interactions. Carcinogenesis. 2007;28:2552–2559. doi: 10.1093/carcin/bgm214. [DOI] [PubMed] [Google Scholar]
- 43.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.