Abstract
Aberrant epigenetic regulation is critically involved in the pathogenesis of nasopharyngeal carcinoma (NPC), where abnormal histone methylation can be found in polycomb repressive complex-2 (PRC2) related cancer gene loci. This study investigated some novel combinational strategies against NPC in vitro using PRC2-targeting agents as a backbone. PRC2 subunit proteins were overexpressed in over 70% of NPC tumors and enhancer of zeste homolog-2 (EZH2) expression correlated with more advanced T-stage. Basal expression of EZH2 and embryonic ectoderm development (EED) was higher in Epstein-Bar virus (EBV)+ NPC cells than EBV- cells. Treatment with an EED inhibitor (EED226) led to reduced levels of H3K27me3 with minimal inhibitory effect on NPC cell growth. The combination of an EZH2 inhibitor (EPZ-6438) and trichostatin-A (TSA) yielded the highest synergy score (12.64) in NPC cells in vitro than combinations using EED226 and agents like chemotherapy and azacitadine. Global gene expression analysis showed that EED226 predominantly affects the expression of major histocompatibility complex (MHC) class I genes and cell cycle-related genes in NPC cells. Furthermore, treatment with EED226 resulted in increased MHC-I proteins in vitro. Based on the prediction of an artificial neural network, a synergistic inhibitory effect on growth was found by combining EED226 with cyclin dependent kinase (CDK) 4/6 inhibitor (LEE011) in NPC cells. In summary, this study found that PRC2-targeting agents could exert synergistic effect on growth inhibition when combined with TSA or LEE011 in NPC cells. Since MHC-I genes alterations are found in a third of NPC tumors, the effect of EED226 on MHC-I genes expression on response to immunotherapy in NPC warrants further investigations.
Keywords: EZH2 inhibitor, EED inhibitor, MHC-I, CDK4/6 inhibitor, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is endemic in Asia and North Africa especially Southern China. According to the International Agency for Research on Cancer in 2018, there were an estimated 129,079 new cases of NPC accounting for 0.7% of all cancers [1]. Over 50% of NPC patients present with stage III to IV disease [2] where concurrent chemo-radiotherapy (CRT) is a standard of care [3]. However, over 25% of patients developed distant failure following CRT and thus newer treatment is needed [4].
The Polycomb Repressive Complex-2 (PRC2) is an essential epigenetic effector which consists of three subunits: enhancer of zeste homolog-2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 (SUZ12). These PRC2-related proteins mediate the transcriptional silencing of tumor suppressor genes (TSGs) by inducing modifications in histone H3 (H3K27me3). EZH2 is the catalytic subunit that requires the presence of SUZ12 and EED in maintaining the integrity and activity of the PRC2 complex [5,6]. Perturbations of PRC2 activity have been found in many cancers and pharmacological inhibitors of PRC2 are currently under evaluation [7].
Two classes of agents that have shown promise in early phase clinical trials of solid cancers. These include a small molecule targeting the cofactor S-adenosylmethionine (SAM) of EZH2, and an allosteric inhibitor of EED that directly binds to the H3K27me3 binding pocket. Tazemetostat (EPZ-6438) is a first-in-class SAM-competitive EZH2 inhibitor that has shown promising clinical activity in patients with refractory B-cell non-Hodgkin lymphoma (NHL) with favorable toxicity profile [8]. EED226 is a first-in-class EED inhibitor and its analogue, MAK683 is being evaluated in phase 1 basket trial I lymphoma, NPC and other solid tumors (Clinical trial.gov: NCT02601950) [9].
Aberrant hypermethylation and enrichment of de novo methylation at PRC2-related gene loci are commonly found in NPC [10]. EZH2 overexpression has been found in 46-80% of NPC tumor and has been associated with advanced disease stage [11-13]. However, the prognostic significance of other PRC2-related proteins and H3K27me3 remain to be defined. In NPC models, EZH2 overexpression has been shown to suppress the expression of genes that regulate important cellular functions such as Bcl-2, c-Myc, CDK4 and CDK6 [11]. Downregulation of EZH2 via microRNA (miRNA) has been shown to inhibit cell growth and cell-cycle progression [14], while EZH2 knockdown could inhibit cellular invasiveness in NPC models [12,15].
This study hypothesized that components of the PRC2 complex, namely, EZH2 and EED are potential therapeutic targets in NPC, given their prevalent expression in NPC tumors and suppressive effect on the transcription of important genes that regulate growth, cell cycle progression and invasiveness. Furthermore, given the crosstalk between DNA methylation and PRC2 pathway in transcriptional silencing, a combinatorial approach to targeting PRC2 activity together with DNA methyl-transferase inhibitors or histone deacetylase (HDAC) inhibitors may potentially be synergistic in suppressing NPC cell growth. The effect of targeting PRC2 activity on the growth inhibitory effect of platinum-based chemotherapy also warrants investigation given the important role of chemotherapy in the treatment of NPC.
Materials and methods
Immunohistochemistry
Archival, paraffin-embedded primary NPC tumors were retrieved retrospectively, and survival data were updated. This study was approved by the New Territory-East Cluster-Chinese University of Hong Kong Ethics committee. Tumor samples were cut into a thickness of 4 µm from archived paraffin blocks by using a commercial kit (ultraView Universal DAB Detection Kit, Roche). The antibodies used for immunohistochemical (IHC) analysis were as follow: EZH2 (D2C9) (#5246) and H3K27me3 (C36B11) (#9733) were from Cell Signaling Technology (CST); anti-EED antibody (HPA061140) was from Sigma-Aldrich; anti-SUZ12 antibody [SUZ220A] (ab126577) was from Abcam. EZH2, EED, SUZ12 and H3K27me3 expression in the tumor samples were evaluated in three fields (200 × magnification) or by counting 100-200 cells. A pathologist (JT) performed the scoring and the staining pattern was graded as 0 when there was completed absence of cell nuclear staining. The positive tumor samples were graded as 1+, 2+ and 3+ according to the degree of cell nuclear staining. Tonsil tissue was used as a positive control. Negative controls were obtained by omission of the primary antibody in NPC tissue with unstained slides.
Cell lines and culture methods
The NPC cell lines used in this study included three Epstein Barr virus (EBV)-positive cell lines (C666-1, NPC43 and C17C), an EBV-negative cell line (HK1) and an immortalized normal nasopharyngeal epithelial cell line (NP69). C666-1 and HK1 cells were cultured in RPMI-1640 medium (Hyclone, Thermo Fisher Scientific-TFS, Logan, UT) with 10% of fetal bovine serum (FBS, Gibco, TFS, Logan, UT). NPC43 and C17C cells were cultured in RPMI-1640 medium with 10% of fetal bovine serum and 4 μM Rho-associated kinase inhibitor Y-27632. NP69 was cultured in keratinocyte serum-free medium supplemented with Keratinocyte-SFM (Gibco, TFS, Logan, UT), 50 μg/ml bovine pituitary extract, and 5 ng/ml EGF human recombinant epidermal growth (Gibco, TFS, Logan, UT). All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Drug preparation
The EED inhibitor (EED226) and cyclin-dependent kinase 4 and 6 (CDK4/6) inhibitor (LEE011) were purchased from Cayman Chemical (Michigan, USA). EPZ-6438 (Tazemetostat) was purchased from Selleckchem (Houston, TX, USA). Trichostatin (TSA) and azacitidine (AZA) were from Sigma-Aldrich (Darmstad, Germany). Reconstituted cisplatin and gemcitabine were obtained from leftover reconstituted drugs in vials from a hospital pharmacy. Aliquots were thawed and diluted to concentration required in the corresponding growth media before adding to cell cultures.
Assay of cytotoxicity
Cell cytotoxicity was assessed by a colorimetric assay using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) at 72 hours. C666-1, NPC43, C17C, HK1 and NP69 were cultured in 48-well plates (1,000-5,000 cells per well) in the respective culture medium. Drugs were added to the wells after cell plating and incubated at 37°C with 5% CO2 before detection. Each experiment on drug treatment was repeated in triplicates. Cell growth inhibition was expressed as the percentage of the absorbance of control cultures measured at 570 nm with a microplate reader (PerkinElmer 1420 Multilabel Counter VICTOR3, Waltham, Massachusetts, USA) and the 50% of the maximum growth inhibition (IC50) was calculated using GraphPad (PRISM 7.0, San Diego, CA).
Synergistic effect assay
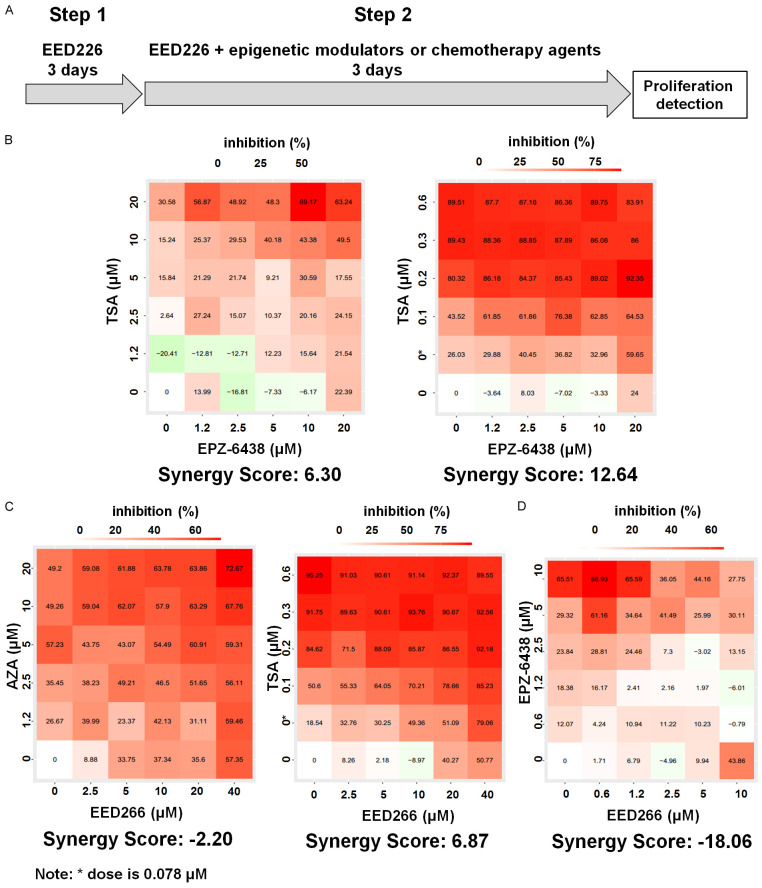
Cells were cultured in 96-well plates (500-1,000 cells per well) in culture medium, and each drug was added sequentially (Figure 4A) after the cells were plated and then incubated at 37°C with 5% CO2 before detection. Experiments were repeated in triplicates. Cell proliferation was assessed by colorimetric assay using WST-1 reagent at the 6th day [16]. Cell growth inhibition was measured at 450 nm and 690 nm with a microplate reader. The Synergistic Score (SS) of each drug combination was calculated by the SynergyFinder web (https://synergyfinder.fimm.fi) with the Bliss and Loewe model [17,18]. Negative values of SS indicate that there is an antagonistic effect between the two drugs on cell growth, while positive SS values indicate synergism.
Figure 4.
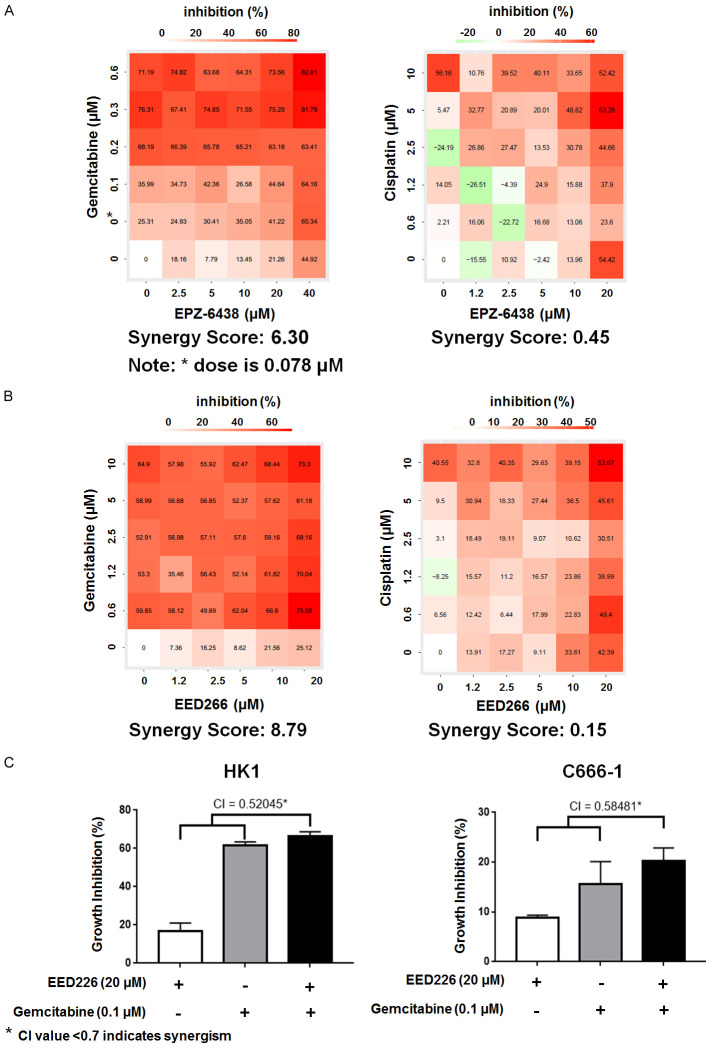
Synergistic effect of PRC2 inhibitors with chemotherapy agents. A. Combinatory effect of EPZ-6438 & gemcitabine and EPZ-6438 & cisplatin on proliferation of C666-1 cells in 6-day assay. B. Combinatory effect of EED226 & gemcitabine and EED226 & cisplatin on proliferation of C666-1 cells in 6-day assay. C. CI analysis of EED226 and gemcitabine in HK1 and C666-1 cell lines at 6 day.
WST-1 based colorimetric assay was used to study the effect of EED226 on cell proliferation in combination with gemcitabine (at day 6 of exposure), or with LEE011 (at 72 hours of drug exposure). The Combination index (CI) value was calculated by CompuSyn software (ComboSyn, Inc., Paramus, NJ 07652, USA) based on the median-effect principle of the mass-action law [19,20] and combination index theorem described by Chou et al. [20]. CI less than 0.7 indicates that the drug combination is synergistic.
Western blot analysis
Cells were lysed with RIPA buffer and the antibodies used were: EZH2 (D2C9), H3K27me3 (C36B11), GAPDH (D16H11), PCNA (D3H8P), Cdk4 (D9G3E), Cdk6 (DCS83), p21 Waf1/Cip1 (12D1), Phospho-Rb (s807/811), Rb (4H1), anti-mouse IgG, HRP-linked Antibody (#7076) and anti-rabbit IgG, HRP-linked Antibody (#7074) were all purchased from CST. Phospho-CDK4 (Thr172) antibody (PA5-64482) was from TFS. Phospho-CDK6 (Y24) (ab131469) and MHC-II (ab55152) were from Abcam. MHC-I (W6/32) was from Dako. Anti-EED (09-774) was from Millipore. Anti-β-Actin antibody (A2228) was from Sigma-Aldrich.
RNA sequencing analysis and bioinformatics analysis
The effect of EED226 (at concentration of 5 μM) on gene expression was evaluated at 3 time points (3, 7 and 14 days) with DMSO as a control. The experiments performed at each time-point were repeated in triplicates. Gene Ontology (GO) enrichment analysis, KEGG pathway enrichment analysis and artificial neural network (ANN) were used for bioinformatics analysis.
Statistical analysis
SPSS version 15.0 and GraphPad PRISM 7.0 software were used for statistical analysis. For the IHC analysis, Spearman rank test was used to evaluate any correlation between PRC2-related proteins. The clinical primary endpoints were overall survival (OS) and progression-free survival (PFS). Secondary end points were correlation with disease stage [21], distant metastasis-free survival (DMFS) and locoregional recurrence-free survival (LRFS). OS was calculated from the date of diagnosis to the date of death from any cause. PFS was calculated from the date of diagnosis to the date of first occurrence of locoregional and/or distant relapse or death from any cause. DMFS was defined as the time from diagnosis to primary local recurrence and distant metastasis. LRFS was defined as time from diagnosis to loco-regional failure. A diagnosis of disease progression was confirmed histologically, endoscopically and/or radiologically. The Cox proportional regression model was used to investigate the relationship between survival endpoints and key prognostic covariates. The mean, standard deviation (SD) and P-values were calculated by the x2 test or Fisher’s Exact Test (n < 5), with a two-sided P-value of < 0.05 as considered to be statistically significant [21,22].
Results
The prognostic significance of EED, SUZ12, EZH2 and H3K27me3 expression
A total of 93 paraffin-embedded NPC samples from 93 consecutive patients were retrieved and survival data were updated and frozen in December 2019. The characteristics of the patients who gave consent to these samples are summarized in Table S1. There were 20 locoregional and 17 distant recurrences; 27 patients died of NPC and 14 patients died of treatment-related complication or other causes (Table S2). There was no statistical association between EZH2, EED and SUZ12 expression with any of the survival endpoints (Table S3). Positive EZH2 expression correlated with more advanced T stage (x2 test, P = 0.021), whereas none of the markers correlated with distant-metastatic stage, overall stage and local failure (Table S4).
Correlation between expression of PRC2-related proteins and H3K27me3
Of the 93 NPC samples analyzed, overexpression of EZH2 was found in 72.0%, EED in 88.2%, SUZ12 in 74.2% and H3K27me3 in 74.2% of the samples (Figure 1A and 1B; Table S5). EZH2, EED and SUZ12 are positively correlated with each other, while EZH2 (r = 0.409, P < 0.000) and EED (r = 0.225, P < 0.05) correlated with H3K27me3 in NPC tumors (Table S6).
Figure 1.
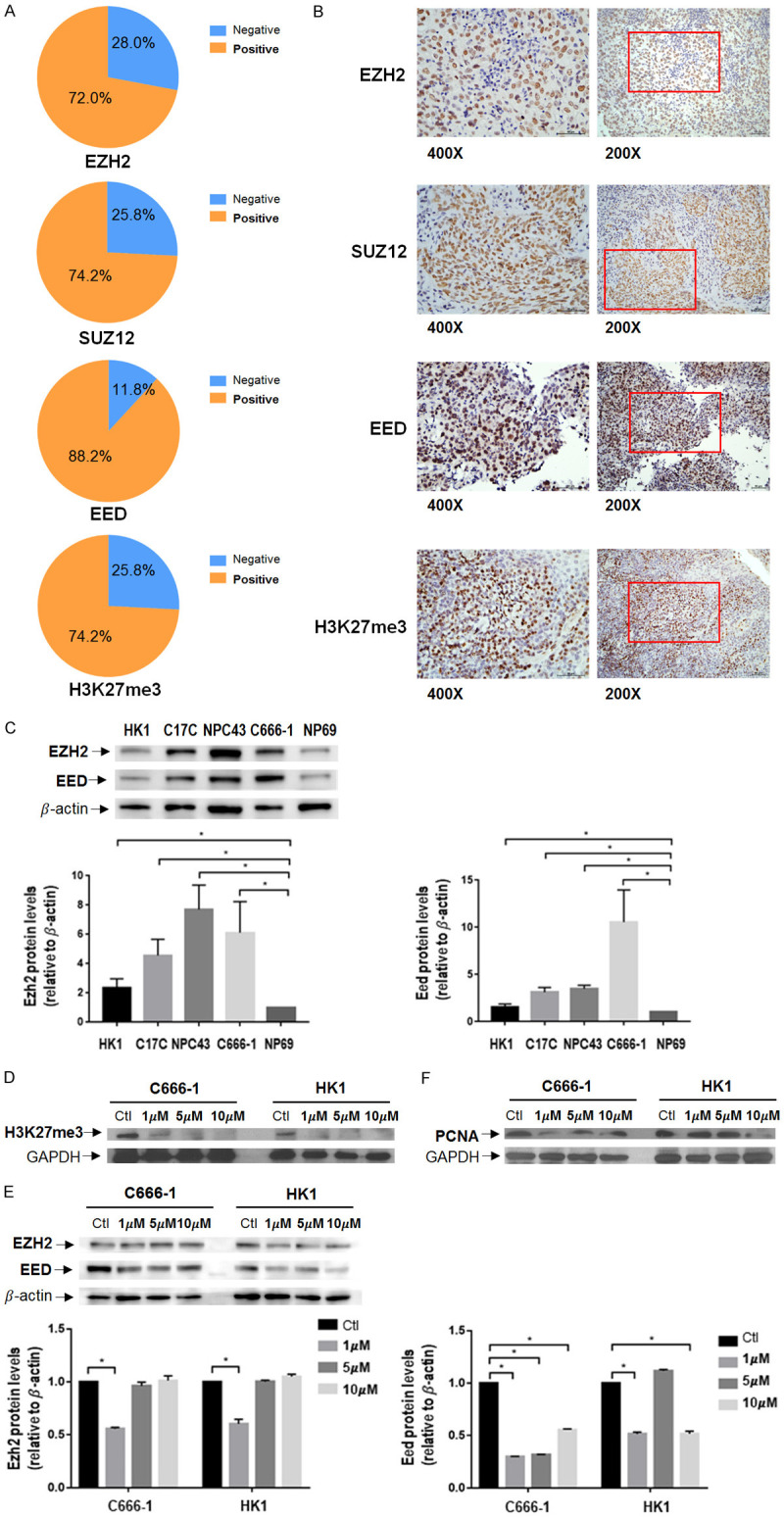
Basal levels of PRC2 related proteins in NPC and effect of EED226 in NPC cells. (A) Percentage of positive EZH2, SUZ12, EED and H3K27me3 in NPC tumors. (B) Representative immunohistochemical staining images from NPC tissue. (C) Basal PRC2 subunits protein expression of cancer cell lines HK1, C17C, NPC43, C666-1, and an immortalized epithelial nasopharyngeal epithelial cell line NP69 as normal control. EZH2 and EED were expressed in all cell lines. Cells were treated with EED226 in 3 concentrations at 72 hours. H3K27me3 (D), EZH2, EED (E) and PCNA (F) were reduced by EED226 treatment. * represent P < 0.05.
Expression of PRC2-related proteins at basal condition and after treatment with PRC2-targeting agents in NPC cell lines
The basal expression of PRC2-related proteins was determined in HK1, C17C, NPC43 and C666-1 cell lines, with an immortalized epithelial nasopharyngeal epithelial cell line NP69 serving as a control. EZH2 and EED are expressed in all 4 NPC cell lines, while the expression levels of EZH2 and EED in EBV-positive NPC cell lines (C17C, NPC43 and C666-1) appeared to be higher than in the EBV-negative NPC cell lines (HK1) and NP69 cells (Figure 1C). Following treatment with EED226 at 1 μM, 5 μM and 10 μM for up to 72 hours, H3K27me3 expression was significantly reduced in both C666-1 and HK1 cell lines in a dose-response relationship (Figure 1D). EED226 could significantly reduce EED expression, but the effect on EZH2 was relatively modest (Figure 1E). Using proliferating cell nuclear antigen (PCNA) as an indicator, EED226 at low concentrations could inhibit cellular proliferation in C666-1 cells, while higher concentrations of EED226 were needed to inhibit proliferation in HK1 cells (Figure 1F).
Effect of PRC2-targeting agents (EPZ-6438, EED226) and other epigenetic modulators as single agents on NPC cell growth
The EBV-positive C666-1 cell line and the EBV-negative HK1 cell lines were chosen for evaluating the effect of EPZ-6438, EED226, TSA and AZA as single agents on cell growth. TSA has the most potent growth inhibitory effect among all the four agents, with IC50 values at 72 hours observed in the low micro-molar range, while treatment with EED226 alone has the least growth-inhibitory effect (Figure 2). The growth-inhibitory effect of PRC2-targeting agents seemed to be lower than AZA and TSA. HK1 cells were most sensitive to the growth-inhibitory effect of EPZ-6438, AZA and TSA, while C666-1 was comparatively more sensitive to EED226. Since in these experiments, C666-1 cells were found to be more sensitive to the effect of EED226 and moderately sensitive to EPZ-6438 when compared with other cell lines, C666-1 cells were chosen for subsequent experiments on drug combinations.
Figure 2.
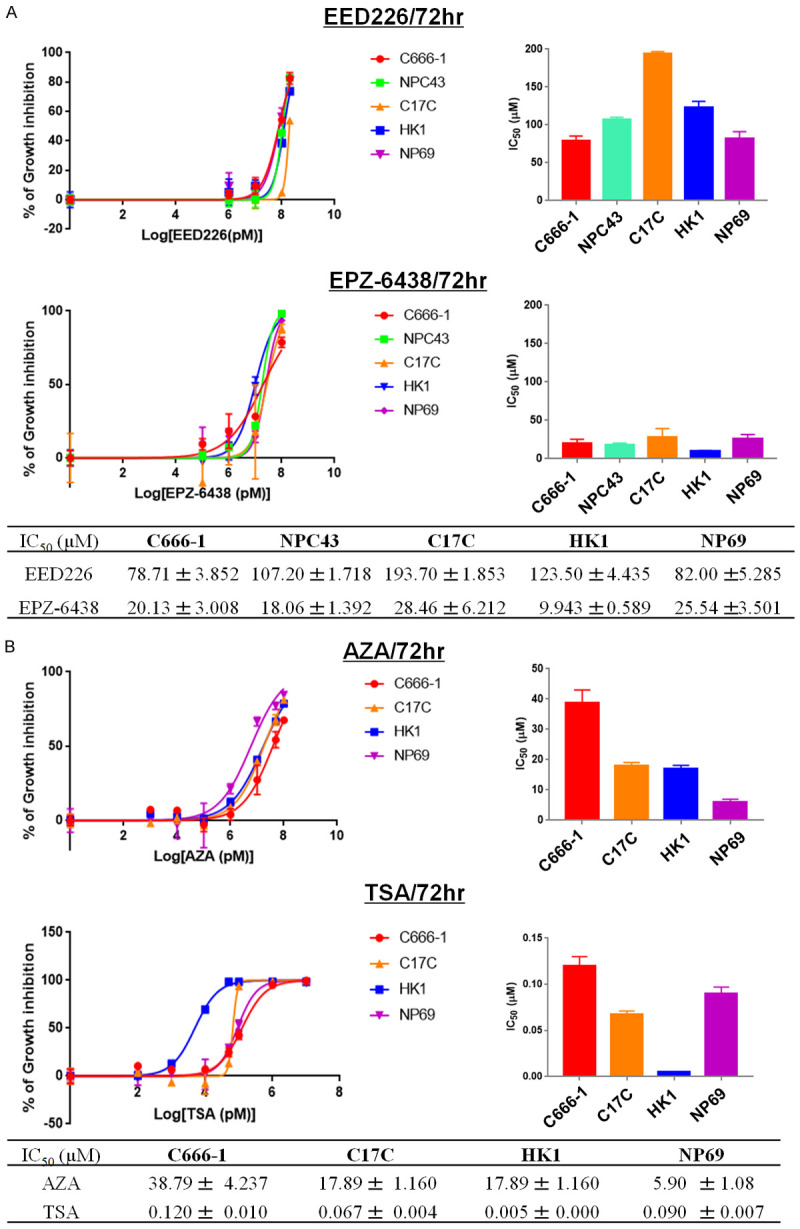
Cytotoxicity of PRC2-related inhibitors and other epigenetic modulators at 72 hours. A. Representative dose-response curves showing the cytotoxicity effect of EED226 and EPZ-6438 in C666-1, NPC43, C17C, HK1, and NP69 by MTT assay. All samples were carried out in triplicate. B. Representative dose-response curves showing the cytotoxicity effect of AZA and TSA in C666-1, C17C, HK1, and NP69 cell lines.
Effect of combining PRC2-targeting agents with other epigenetic modulators
The effect of PRC2 inhibitors in combination with other epigenetic modulators were evaluated at, and near their IC50 concentrations in C666-1 cells for up to 6 days (Figure 3A) [16]. The effect of EED226 and AZA were antagonistic on cell growth, while EPZ-6438 and TSA resulted in the highest synergy score (SS = 12.64) among all the other combinations (Figure 3B-D). Concurrent treatment of EED226 with AZA or EPZ-6438 resulted in antagonistic effects on cell growth, suggesting that EED226 should not be combined with other epigenetic modulators.
Figure 3.
Synergistic effect of PRC2 inhibitors with other epigenetic modulators. A. Schematic representation of drug sequence. B. Combinatory effect of EPZ-6438 & AZA and EPZ-6438 & TSA on proliferation of C666-1 cells in 6-day assay. C. Combinatory effect of EED226 & AZA and EED226 & TSA on proliferation of C666-1 cells in 6-day assay. D. Combinatory effect of EPZ-6438 & EED226 and on proliferation of C666-1 cells in 6-day assay.
Effect of combining PRC2-targeting agents with chemotherapy
Cisplatin and gemcitabine were chosen in this experiment because they are the standard first-line treatment for recurrent/metastatic NPC. The SS was higher when EED226 was combined with cisplatin or gemcitabine, compared with when EED226 was combined with AZA or EPZ-6438. The SS of EED226 and gemcitabine was 8.79 (Figure 4B) in C666-1 cells, and the CI value of this combination were < 0.7 - indicating a synergistic effect on cell growth inhibition (Figure 4C). This synergistic effect observed in C666-1 cells could also be observed in a confirmatory experiment using HK1 cells.
Effect of EED inhibitor on global gene expression profile
Previous studies have shown that treatment with low-dose epigenetic-modulating agents can result in the reprogramming of RNA transcription of tumor cells [23]. Qi et al. demonstrated that EED226 could inhibit H3K27 methylation of PRC2 target genes and inducing regression of human lymphoma xenograft tumors [16]. Thus, the current study hypothesized that EED226 also could change the global gene expression profile of NPC cells. C666-1 cells were treated with EED226 at 5 μM concentration at 3, 7 and 14 days. RNA seq analysis was used to investigate any time-dependent effect on gene expression (Figure 5A).
Figure 5.
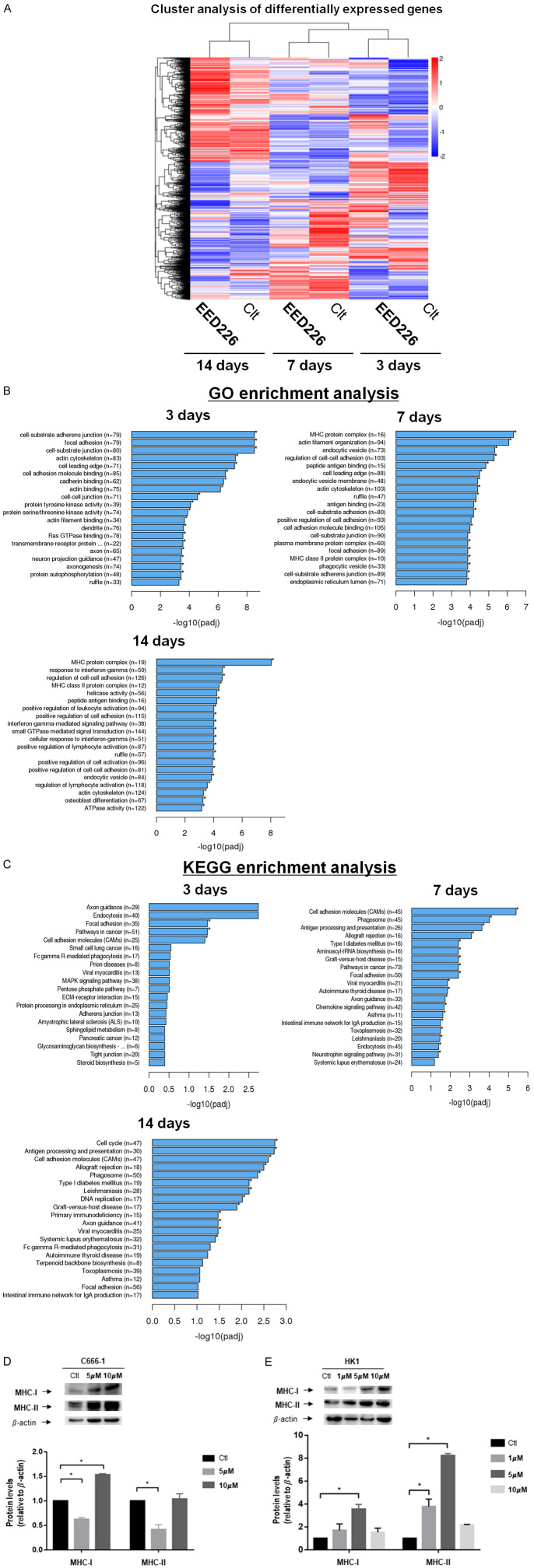
EED226 gene expression profile. A. Cluster analysis of differential genes. For samples’ gene expression, the significant criteria of differential expression genes are: |log2 fold| > 1 and padj < 0.005, and up-regulated genes show in red, down-regulated genes show in blue. B. Top GO enrichment genes by 5 μM EED226 at day 3, 7, 14. C. Top KEGG pathway enrichment by 5 μM EED226. D. MHC-I and MHC-II expression levels up-regulated after the treatment of EED226 in C666-1 cells at 7 day. E. MHC-I and MHC-II expression levels up-regulated after the treatment of EED226 in HK1 cells at 3 day. * represent P < 0.05.
GO and KEGG pathway enrichment analysis showed that EED226 could significantly enhance the expression of MHC class 1-related genes (P < 0.00001) and MHC class 2-related genes (P < 0.001) in NPC cells following prolonged exposure at 7 and 14 days (Figures 5B, S1A and S1B). Notable examples of changes in gene expression included NLRC5 (log2 fold = 0.77761), HLA-A (log2 fold = 1.2155), HLA-B (log2 fold = 1.3036), HLA-C (log2 fold = 1.4081), B2M (log2 fold = 0.74116), TAP1 (log2 fold = 0.59951) and TAP2 (log2 fold = 0.72345). Exposure to EED226 at 5 μM concentration also resulted in significant changes in the expression of cell cycle-related genes (P < 0.00001) at 14 days (Figures 5C and S1C).
Since RNA profile showed that EED226 enhanced the expression of MHC related genes at 7 and 14 days of exposure, the levels of MHC-I and MHC-II related proteins were analyzed after treatment with EED226 in C666-1 cells at day 7 first. The data showed that 10 M of EED226 could up-regulate the expression of MHC-1 proteins in C666-1 cells at day 7 day of exposure (Figure 5D), but 5 μM of EED226 resulted in down-regulation of the expression of MHC-I proteins. The expression of MHC-II proteins decreased after cells were treated with 5 μM of EED226, but no statistically significant changes were detected at 10 μM of EED226 when comparing with the control. RNA sequencing data indicated that MHC-I and MHC-II genes were up-regulated after treatment with EED226, but the MHC-I and MHC-II related proteins expression did not increase in vitro. This may be because changes in protein expression may take longer time to become detectable following EED226-induced changes in MHC-I gene expression. To confirm this finding, the experiment was repeated in another NPC cell line-HK-1. Western blot data showed that EED226 exposure increased the expression of MHC-I and MHC-II protein in HK1 cells as early as at day 3 of treatment-earlier than it took in C666-1 cells (Figure 5E).
Effect on combining EED226 with LEE011 on cell growth and cell cycle
Deep learning is currently used in Bioinformatics and Computational Medicine including analysis of RNA Seq gene expression data [24]. ANN are computing systems which have been used for classification and prediction task, such as in the unsupervised data-mining of molecular data [25]. Having demonstrated the effect of EED226 on cell cycle-related genes in C666-1 cells, this study tested the hypothesis that combining EED226 with inhibitors of the cell cycle could be more effective. ANN was used to predict the sensitivity of C666-1 cells towards 97 drugs with or without the presence of EED226 and found a potential complementary effect when combining EED226 with CDK4/6 inhibitors (Figure 6A). LEE011 (Ribociclib) is a selective CDK4/6 inhibitor that induces G1 arrest by blocking the formation of cyclin D1-CDK4/6 complex and inhibiting retinoblastoma (Rb) phosphorylation. The authors have previously shown that LEE011 has preclinical activity in NPC cell lines [26]. This prediction by ANN was confirmed when a synergistic effect on growth inhibition was observed when EED226 was combined with LEE011 in C666-1 cells (Figure 6B).
Figure 6.
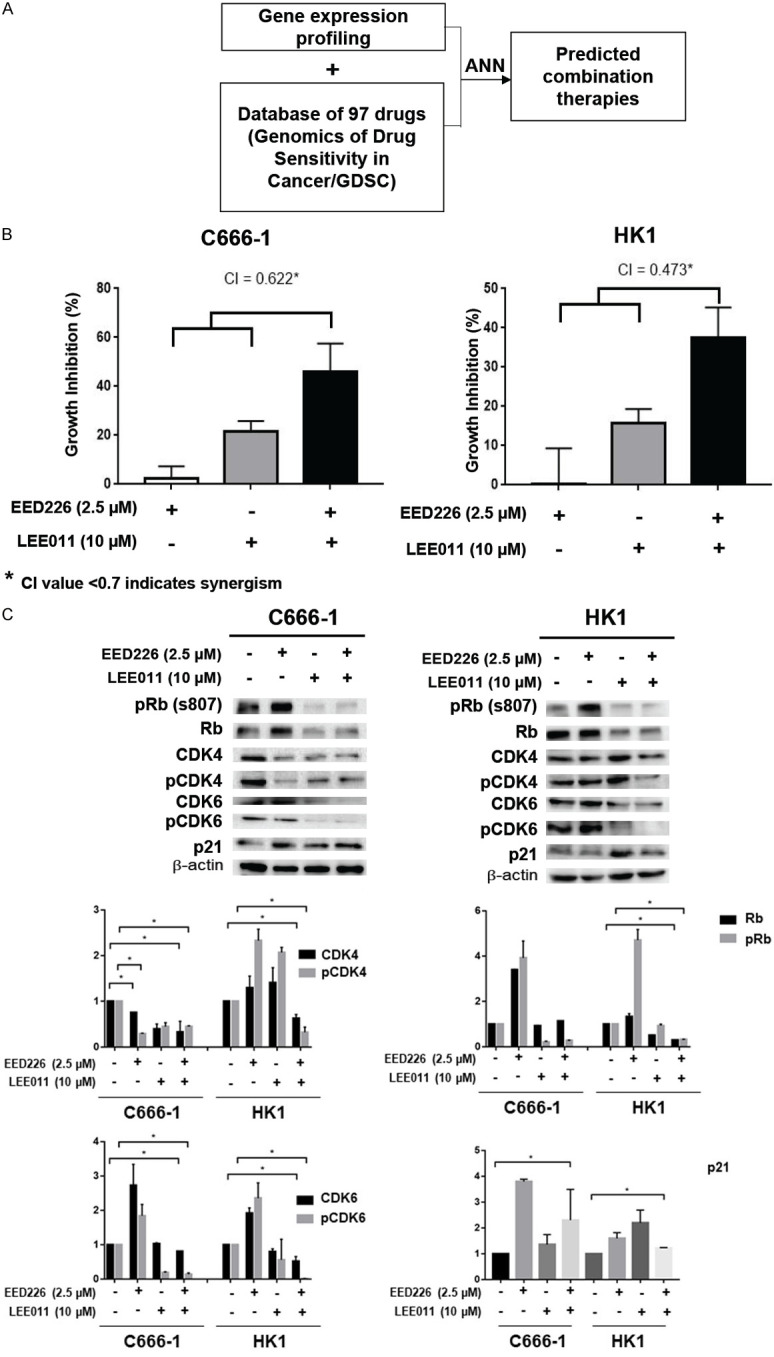
Synergistic effect of EED226 with LEE011. A. Schematic representation of ANN assay method. B. CI analysis of EED226 and LEE011 in HK1 and C666-1 at 72 hours. C. Western blot for cell cycle related proteins on HK1 and C666-1 cell lines treated with combination therapies at 24 hours. * represent P < 0.05.
Rb is hypophosphorylated by the CDK4/cyclin D complex, which controls whether cells will transit through the G1/S checkpoints. The CDK inhibitor p21 also binds to and inhibits the activity of cyclin-CDK4/6 complexes, therefore promoting cell cycle arrest [27]. To investigate the underlying mechanism of the synergistic effect of combining EED226 and LEE011 observed in this study, an immunoblot assay showed that combination therapy of EED226 and LEE011 down-regulate the expression of CDK4, pCDK4, CDK6 and pCDK6 in C666-1 cells. This observation could be replicated in HK1 cells. Furthermore, co-treatment with EED226 and LEE011 resulted in the upregulation of p21 expression in C666-1 cells. EED226 and LEE011 combination have better suppression effect of the expression of Rb and pRb in HK1 cells (Figure 6C).
Discussion
The findings of this study support the therapeutic targeting of PRC2-related proteins in NPC, when used in combination with other anti-cancer agents such as HDAC inhibitor or CDK4/6 inhibitor. This study found that PRC2-related proteins are commonly expressed in NPC cell lines and tumors but were not associated with survival in NPC patients-although EZH2 was associated with advanced T-stage. When used as single agents, EED and EZH2 inhibitors have modest effect on inhibiting NPC cell growth, even though EED inhibition could significantly reduce the level of H3K27me3 in vitro at relatively low concentrations. Subsequent exploratory gene expression analysis revealed that EED inhibition could significantly affect the expression of genes and proteins which regulating the antigen-presentation pathways and the cell-cycle. Furthermore, the combined inhibition of CDK4/6 and EED activity was synergistic on inhibiting cell growth in NPC in vitro. Likewise, the combination of EZH2 inhibitor and other epigenetic modulators could result in additive to synergistic inhibitory effect on NPC cell growth.
Overexpression of EZH2 has previously been shown to be an independent prognostic factor in patients with NPC, but this was not observed in the current study [12]. It may caused by the different patient characteristics in different studies. In the current study, 47 patients received chemotherapy for NPC and 6 patients developed distant metastasis (Table S2). Considering PRC2-related proteins are involved in chemotherapy resistance [28,29], prior chemotherapy could have influenced the expression levels of PRC2 subunits. Furthermore, EZH2 expression has not been shown to have prognostic significance in some cancers like prostate cancer [30], because PRC2-related proteins may have a pleotropic effect in different human cancers.
The observation that EED226 could significantly reduce the expression of H3K27me3 supports the hypothesis that PRC2-targeting may potentially ‘re-program’ gene expression in NPC. In this study, the most effective combination appeared to be the co-targeting of EZH2 and HDAC in NPC, probably because EZH2 could form a co-repressor complex with HDAC1/HDAC2 induced cell growth inhibition and invasiveness in NPC models [10,13]. This is supported by studies in other cancers, showing that the combination of EZH2 and HDAC inhibitors (or triple combination of AZA, EZH2 inhibitor and TSA) have additive to synergistic effect on inhibiting cell growth in lung cancer and leukemia [31-33]. In this study, although EED226 showed synergistic effect when combined with gemcitabine, the effective dose of EED226 used (20 μM) in CI assay will be too high to be used in vivo or clinical setting.
Whole exome sequencing (WES) of NPC has shown that up to a third of NPC harbor MHC class 1-related gene alterations which contribute to defective antigen-presentation in NPC [36]. Given the promising activity of immune-checkpoint inhibitors (ICP) in NPC patients [22], the effect of EED226 on increasing the expression of MHC-1 related genes may have important clinical implications on modulating response to ICP in NPC. This finding is supported by a previous study showing that PRC2-targeting agents can up-regulate immune response related pathways in Karpas422 cells [16], as well as enhancing the expression levels of B2M and CXCL10 in some tumors [34,35]. Treatment with EZH2 inhibitors have been shown to increase the level of effector T-cell tumor infiltration in ovarian tumor-bearing mice [37]. Many head and neck squamous cancers are defective in MHC class 1-mediated antigen presentation [7], suggesting that PRC2-targeting agents may also potentially influence response to immunotherapy.
However, the effect of PRC2 inhibitors in modulating response to ICP in NPC may be influenced by the EBV. Previous studies have shown that the inhibition of multiple MHC-I genes was strongly correlated with the increase in EBV gene expression especially EBNA1 [38]. The link between MHC-I and EBV may possibly decrease the effect of PRC2 inhibitors in the re-expression of cell surface antigens.
The synergistic effect observed in the co-targeting of CDK4/6 and EED activity has provided another promising strategy that warrants further investigation, especially when these agents have shown tolerable toxicity profiles in clinical trials. This study has demonstrated a proof-of-concept for this combinatorial strategy, by showing that an EED inhibitor could suppress CDK4/6 and pCDK4/6. This observation is supported by a precious observation that EZH2 downregulation could suppress the expression of CDK4/6 [14].
In conclusion, EED inhibition can significantly down-regulate the level of H3K27me3 at relatively low concentrations for prolonged period of exposure, and therefore affect global gene expression in NPC. This study supports several combinatorial approaches of combining PRC2-targeting agents in NPC which warrants further investigation in animal models of NPC. The most promising combinations included EED and CDK4/6 inhibition, as well as EZH2 and HDAC inhibition. The mechanism of EED inhibition on influencing response to ICI in tumors that are known to be defective in MHC class-I antigen presentation should also be investigated.
Acknowledgements
This study was supported and funded by the Direct grant for research, The Chinese University of Hong Kong (#2018.012) and National Natural Science Foundation of China (NSFC #81772869), and Hong Kong Research Grants Council (GFR grant #14161317). We want to thank Prof. SW Tsao from HKU for providing us the cell lines NP69, NPC43 and C17C, and Dr. Choy Chi Tung help us to finish the ANN analysis in this project.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: current practice and future perspective. J. Clin. Oncol. 2015;33:3356–3364. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 3.Colevas AD, Yom SS, Pfister DG, Spencer S, Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ, Cmelak AJ, Eisele DW, Fenton M, Foote RL, Gilbert J, Gillison ML, Haddad RI, Hicks WL, Hitchcock YJ, Jimeno A, Leizman D, Maghami E, Mell LK, Mittal BB, Pinto HA, Ridge JA, Rocco J, Rodriguez CP, Shah JP, Weber RS, Witek M, Worden F, Zhen W, Burns JL, Darlow SD. NCCN guidelines insights: head and neck cancers, version 1.2018. J Natl Compr Canc Netw. 2018;16:479–490. doi: 10.6004/jnccn.2018.0026. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD, Yang KY, Jin F, Shi M, Chen YP, Hu WH, Cheng ZB, Wang SY, Tian Y, Wang XC, Sun Y, Li JG, Li WF, Li YH, Tang LL, Mao YP, Zhou GQ, Sun R, Liu X, Guo R, Long GX, Liang SQ, Li L, Huang J, Long JH, Zang J, Liu QD, Zou L, Su QF, Zheng BM, Xiao Y, Guo Y, Han F, Mo HY, Lv JW, Du XJ, Xu C, Liu N, Li YQ, Chua MLK, Xie FY, Sun Y, Ma J. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. N Engl J Med. 2019;381:1124–1135. doi: 10.1056/NEJMoa1905287. [DOI] [PubMed] [Google Scholar]
- 5.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33:903–935. doi: 10.1101/gad.325050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, Coindre JM, Blakemore SJ, Clawson A, Suttle B, McDonald AA, Woodruff M, Ribich S, Hedrick E, Keilhack H, Thomson B, Owa T, Copeland RA, Ho PTC, Ribrag V. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Zhang J, Yu Z, Zhang H, Wang Y, Lingel A, Qi W, Gu J, Zhao K, Shultz MD, Wang L, Fu X, Sun Y, Zhang Q, Jiang X, Zhang J, Zhang C, Li L, Zeng J, Feng L, Zhang C, Liu Y, Zhang M, Zhang L, Zhao M, Gao Z, Liu X, Fang D, Guo H, Mi Y, Gabriel T, Dillon MP, Atadja P, Oyang C. Discovery of first-in-class, potent, and orally bioavailable embryonic ectoderm development (EED) inhibitor with robust anticancer efficacy. J Med Chem. 2017;60:2215–2226. doi: 10.1021/acs.jmedchem.6b01576. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, Tsao SW, Loh TK, Sun H, Chan AT, Zeng YX, Chan WY, Chan FK, Tao Q. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics. 2015;7:155–173. doi: 10.2217/epi.14.79. [DOI] [PubMed] [Google Scholar]
- 11.Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, Yue S, O’Sullivan B, Liu FF. Enhancer of zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010;1:e85. doi: 10.1038/cddis.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang CF, Huang HY, Chen CH, Chien CY, Hsu YC, Li CF, Fang FM. Enhancer of zeste homolog 2 overexpression in nasopharyngeal carcinoma: an independent poor prognosticator that enhances cell growth. Int J Radiat Oncol Biol Phys. 2012;82:597–604. doi: 10.1016/j.ijrobp.2010.11.062. [DOI] [PubMed] [Google Scholar]
- 13.Shu XS, Li L, Ji M, Cheng Y, Ying J, Fan Y, Zhong L, Liu X, Tsao SW, Chan AT, Tao Q. FEZF2, a novel 3p14 tumor suppressor gene, represses oncogene EZH2 and MDM2 expression and is frequently methylated in nasopharyngeal carcinoma. Carcinogenesis. 2013;34:1984–1993. doi: 10.1093/carcin/bgt165. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Wang X, Niu X, Wang X, Jiang R, Xu T, Liu Y, Liang L, Ou X, Xing X, Li W, Hu C. EZH2 suppresses the nucleotide excision repair in nasopharyngeal carcinoma by silencing XPA gene. Mol Carcinog. 2017;56:447–463. doi: 10.1002/mc.22507. [DOI] [PubMed] [Google Scholar]
- 15.Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, Liu TH, Bian XW, Guan XY, Lin MC, Zeng MS, Zeng YX, Kung HF, Xie D. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 16.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, Zhang M, Zhang J, Yu Z, Li L, Teng L, Chuai S, Zhang C, Zhao M, Chan H, Chen Z, Fang D, Fei Q, Feng L, Feng L, Gao Y, Ge H, Ge X, Li G, Lingel A, Lin Y, Liu Y, Luo F, Shi M, Wang L, Wang Z, Yu Y, Zeng J, Zeng C, Zhang L, Zhang Q, Zhou S, Oyang C, Atadja P, Li E. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol. 2017;13:381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 17.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;3:585–615. [Google Scholar]
- 19.Chou TC. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J Theor Biol. 1976;59:253–276. doi: 10.1016/0022-5193(76)90169-7. [DOI] [PubMed] [Google Scholar]
- 20.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 21.Ma BB, Poon TC, To KF, Zee B, Mo FK, Chan CM, Ho S, Teo PM, Johnson PJ, Chan AT. Prognostic significance of tumor angiogenesis, Ki 67, p53 oncoprotein, epidermal growth factor receptor and HER2 receptor protein expression in undifferentiated nasopharyngeal carcinoma--a prospective study. Head Neck. 2003;25:864–872. doi: 10.1002/hed.10307. [DOI] [PubMed] [Google Scholar]
- 22.Ma BBY, Lim WT, Goh BC, Hui EP, Lo KW, Pettinger A, Foster NR, Riess JW, Agulnik M, Chang AYC, Chopra A, Kish JA, Chung CH, Adkins DR, Cullen KJ, Gitlitz BJ, Lim DW, To KF, Chan KCA, Lo YMD, King AD, Erlichman C, Yin J, Costello BA, Chan ATC. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: an international, multicenter study of the mayo clinic phase 2 consortium (NCI-9742) J. Clin. Oncol. 2018;36:1412–1418. doi: 10.1200/JCO.2017.77.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding JR, Condon A, Shah SP. Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat Commun. 2018;9:13. doi: 10.1038/s41467-018-04368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choy CT, Wong CH, Chan SL. Embedding of genes using cancer gene expression data: biological relevance and potential application on biomarker discovery. Front Genet. 2018;9:682. doi: 10.3389/fgene.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CH, Ma BBY, Hui CWC, Lo KW, Hui EP, Chan ATC. Preclinical evaluation of ribociclib and its synergistic effect in combination with alpelisib in non-keratinizing nasopharyngeal carcinoma. Sci Rep. 2018;8:8010. doi: 10.1038/s41598-018-26201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulappa T, Reddy RS, Suman S, Nyakeriga AM, Damodaran C. Molecular interplay between cdk4 and p21 dictates G0/G1 cell cycle arrest in prostate cancer cells. Cancer Lett. 2013;337:177–183. doi: 10.1016/j.canlet.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aries IM, Bodaar K, Karim SA, Chonghaile TN, Hinze L, Burns MA, Pfirrmann M, Degar J, Landrigan JT, Balbach S, Peirs S, Menten B, Isenhart R, Stevenson KE, Neuberg DS, Devidas M, Loh ML, Hunger SP, Teachey DT, Rabin KR, Winter SS, Dunsmore KP, Wood BL, Silverman LB, Sallan SE, Van Vlierberghe P, Orkin SH, Knoechel B, Letai AG, Gutierrez A. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J Exp Med. 2018;215:3094–3114. doi: 10.1084/jem.20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Duff M, Gouju J, Jonchere B, Guillon J, Toutain B, Boissard A, Henry C, Guette C, Lelievre E, Coqueret O. Regulation of senescence escape by the cdk4-EZH2-AP2M1 pathway in response to chemotherapy. Cell Death Dis. 2018;9:199. doi: 10.1038/s41419-017-0209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, Atadja P, Bhalla K. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–3104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 32.Momparler RL, Cote S, Momparler LF, Idaghdour Y. Epigenetic therapy of acute myeloid leukemia using 5-aza-2’-deoxycytidine (decitabine) in combination with inhibitors of histone methylation and deacetylation. Clin Epigenetics. 2014;6:19. doi: 10.1186/1868-7083-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takashina T, Kinoshita I, Kikuchi J, Shimizu Y, Sakakibara-Konishi J, Oizumi S, Nishimura M, Dosaka-Akita H. Combined inhibition of EZH2 and histone deacetylases as a potential epigenetic therapy for non-small-cell lung cancer cells. Cancer Sci. 2016;107:955–962. doi: 10.1111/cas.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong H, Liu S, Zhang X, Chen S, Kang L, Chen Y, Ma S, Fu X, Liu Y, Zhang H, Zou B. An allosteric PRC2 inhibitor targeting EED suppresses tumor progression by modulating the immune response. Cancer Res. 2019;79:5587–5596. doi: 10.1158/0008-5472.CAN-19-0428. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Mudianto T, Ma X, Riley R, Uppaluri R. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti-PD-1 resistance in head and neck cancer. Clin Cancer Res. 2020;26:290–300. doi: 10.1158/1078-0432.CCR-19-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li YY, Chung GT, Lui VW, To KF, Ma BB, Chow C, Woo JK, Yip KY, Seo J, Hui EP, Mak MK, Rusan M, Chau NG, Or YY, Law MH, Law PP, Liu ZW, Ngan HL, Hau PM, Verhoeft KR, Poon PH, Yoo SK, Shin JY, Lee SD, Lun SW, Jia L, Chan AW, Chan JY, Lai PB, Fung CY, Hung ST, Wang L, Chang AM, Chiosea SI, Hedberg ML, Tsao SW, van Hasselt AC, Chan AT, Grandis JR, Hammerman PS, Lo KW. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat Commun. 2017;8:14121. doi: 10.1038/ncomms14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, Sun Y, Zhao E, Vatan L, Szeliga W, Kotarski J, Tarkowski R, Dou Y, Cho K, Hensley-Alford S, Munkarah A, Liu R, Zou W. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, Cheng YJ, Westra WH, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.