Abstract
Cleft alveolar is often accompanied by non-syndromic cleft lip with/without palate (NSCL/P), which could seriously affect the growth and development of the maxilla. In this study, we assessed the associations between 47 susceptible SNPs from previous GWASs of NSCL/P and cleft alveolar in Western Han Chinese population. We recruited 228 trios of NSCL/P with cleft alveolar (156 males and 72 females). The 47 SNPs were genotyped by SNPscan method; Hardy-Weinberg equilibrium test, TDT and parent-of-origin effects were analyzed by PLINK; linkage disequilibrium analysis was conducted by Haploview software. TDT analysis revealed FOXE1 rs894673 (P = 0.0071, ORtransmission = 0.35, 95% CI: 0.16-0.78) and rs3758249 (P = 0.0071, ORtransmission = 0.35, 95% CI: 0.16-0.78) were associated with NSCL/P accompanied cleft alveolar bone. Parent-of-origin effect analysis revealed a paternal special under-transmission of allele A at rs894673 (P = 0.039), allele T at rs3759249 (P = 0.039), and allele T at rs4460498 (P = 0.039) of FOXE1. Allele A at rs987525 showed a significant paternal over-transmission (P = 0.0077). Pairwise LD analysis showed strong LD among rs894673, rs3759249 and rs4460498 (r2 > 0.95, D’ = 1). To conclude, our findings indicated that FOXE1 is the susceptible gene for cleft alveolar accompanied by NSCL/P.
Keywords: Cleft alveolar, non-syndromic cleft lip with/without palate, susceptible gene, association study
Introduction
Non-syndromic cleft lip with or without cleft palate (NSCL/P), as common congenital birth defects, comprise a range of morphological abnormalities of oral and maxillofacial regions. Based on previous epidemiologic studies, Chinese have higher incidences of NSCL/P compared to other ethnicities [1-3], and the prevalence of NSCL/P was more than 1.6% in China [4,5]. In particular, the incidence rate of NSCL/P in western regions was significantly higher than that of other regions of China [5].
Cleft alveolus is a severe bony defect malformation that is present in 75% of NSCL/P [6]. The cleft alveolar refers to a space in the alveolar bone of maxilla with a discontinuity of the dental arch. This bony defect of the alveolus can prevent normal eruption of the cleft-adjacent permanent teeth, disrupt the stability of the maxillary segments, and maintain an oronasal fistula [6-8]. Each of these problems requires a special rehabilitation to avoid malocclusion, maxillary constriction or face asymmetry. Despite the mature sequential therapy involving alveolar bone grafting, the patients had maxillary growth retardation and craniofacial abnormalities thereafter [9].
Achieving a successful and well-functioning reconstruction is the ideal goal of cleft alveolar repair. Alveolar bone grafting is generally considered a gold standard for alveolar cleft treatment. However, the failure rate of this procedure was up to 15%, and normally accompanied with a serious of complications including pain, bleeding, infection, fracture, scar, or chronic pain [8-10]. Bearing in mind all the above-mentioned implications accompanying cleft alveolus repair, the pathogenesis of this deformity is worthwhile to explore.
Genome-wide association study (GWAS) has obvious advantages in the etiology of genetic diseases, and is widely used to unravel the genetic basis of complex diseases [11-13]. Previous GWASs had revealed many susceptibility genes/loci for NSCL/P, which had provided more clues for detecting the etiology of clefts [14-19]. Until now, only 20% of the heritability could be explained. NSCL/P is still an important birth defect affecting quality of the birth in China. Etiological research is crucial to the prevention and control of this disease.
The premaxilla and upper lip come from the maxillary processes and medial nasal process. Cleft alveolus, as a common phenotype of bone malformation, is often accompanied by NSCL/P. Previous GWASs referring to NSCL/P identified susceptible genes of cleft lip. Unfortunately, there are no published data regarding the genetic background of cleft alveolus. In this study, we conducted an experiment with caseparent trios design to validate the association between 47 single nucleotide polymorphisms (SNPs) and cleft alveolus. The purpose of this study was to identify the susceptible genes related to cleft alveolar.
Materials and methods
Ethics statement
All the participants signed the informed consent before being recruited in this study. For patients younger than 16 years old, the informed consents were written by their guardians. The study protocols were reviewed and approved by the Hospital Ethics Committee (HEC) of West China Hospital of Stomatology (No. 2010080), Sichuan University.
Samples description
We recruited 228 case-parent trios of NSCL/P with cleft alveolar, including 156 males and 72 females of the probands, who hospitalized between 2008 and 2013 in the Cleft Lip and Palate Surgery Department of West China Stomatology College, Sichuan University. The probands with non-syndromic cleft lip with or without cleft palate accompanied with cleft alveolus and their parents were enrolled in this study. All the patients were checked by at least 2 professional maxillofacial doctors of the West China Hospital of Stomatology. The probands with other congenital deformities or mental retardation were excluded. All participants were restricted as Western Han Chinese according to self-identification.
SNPs selection and genotyping
Based on the previous GWAS findings about NSCL/P, we selected 47 SNPs with the most significant P values in published studies. Primary information of these SNPs is shown in Table 1.
Table 1.
Primary information on 47 SNPs
| Chr | SNP | Position (Hg19) | Gene | Function |
|---|---|---|---|---|
| 1 | rs4920522 | 18940380 | PAX7 | intergenic |
| 1 | rs766325 | 18956458 | PAX7 | intergenic |
| 1 | rs6695765 | 18979320 | PAX7 | intron |
| 1 | rs742071 | 18979874 | PAX7 | intron |
| 1 | rs560426 | 94553438 | ABCA4 | intron |
| 1 | rs481931 | 94570016 | ABCA4 | intron |
| 1 | rs4147811 | 94575056 | ABCA4 | intron |
| 1 | rs6677101 | 108699730 | SLC25A24 | intron |
| 1 | rs2235371 | 209964080 | IRF6 | extron |
| 1 | rs642961 | 209989270 | IRF6 | intergenic |
| 1 | rs126280 | 210019824 | DIEXF | intron |
| 1 | rs2064163 | 210048819 | DIEXF | intergenic |
| 1 | rs12063989 | 210049893 | DIEXF | intergenic |
| 1 | rs4844913 | 210068117 | SYT14 | intergenic |
| 1 | rs9429830 | 210110537 | SYT14 | intergenic |
| 1 | rs11119388 | 210174417 | SYT14 | intron |
| 1 | rs227178 | 210216946 | SYT14 | intron |
| 1 | rs2485893 | 210348155 | SYT14 | intergenic |
| 2 | rs7590268 | 43540125 | THADA | intron |
| 3 | rs7632427 | 89534377 | EPHA3 | intergenic |
| 4 | rs12506428 | 93830884 | GRID2 | intron |
| 8 | rs6558002 | 27389542 | EPHX2 | intron |
| 8 | rs12543318 | 88868340 | DCAF4L2 | intergenic |
| 8 | rs987525 | 129946154 | LOC728724 | intergenic |
| 9 | rs894673 | 100612270 | FOXE1 | intergenic |
| 9 | rs3758249 | 100614140 | FOXE1 | intergenic |
| 9 | rs4460498 | 100620412 | FOXE1 | intergenic |
| 10 | rs7078160 | 118827560 | VAX1 | intron |
| 10 | rs4752028 | 118834991 | VAX1 | intron |
| 13 | rs9574565 | 80668874 | SPYR2 | intergenic |
| 13 | rs8001641 | 80692811 | SPYR2 | intergenic |
| 14 | rs17563 | 54417522 | BMP4 | exon |
| 15 | rs1258763 | 33050423 | FMN1 | intergenic |
| 15 | rs7179658 | 63312695 | TPM1 | intergenic |
| 16 | rs8049367 | 3980445 | CREBBP-ADCY9 | intergenic |
| 17 | rs9788972 | 8919630 | NTN1 | intergenic |
| 17 | rs4791774 | 8932119 | NTN1 | intron |
| 17 | rs9915089 | 8952894 | NTN1 | intron |
| 17 | rs8069536 | 8956285 | NTN1 | intron |
| 17 | rs8081823 | 8965551 | NTN1 | intron |
| 17 | rs17760296 | 54615617 | NOG | intergenic |
| 20 | rs6072081 | 39261054 | MAFB | intergenic |
| 20 | rs6065259 | 39261979 | MAFB | intergenic |
| 20 | rs17820943 | 39268516 | MAFB | intergenic |
| 20 | rs13041247 | 39269074 | MAFB | intergenic |
| 20 | rs11698025 | 39274083 | MAFB | intergenic |
| 20 | rs6102085 | 39281629 | MAFB | intergenic |
Note: Chr, chromosome; SNP, single-nucleotide polymorphism.
Venous blood samples were collected from all participants after recruitment in the study. Genomic DNA was extracted using the protein precipitation method [20]. All the experiments of genotyping were done by the Genesky Biopharm Technology Company (http://www.geneskies.com/) with SNPscan technology.
Statistical analysis
The Hardy-Weinberg Equilibrium (HWE) analysis and the minor allele of frequency (MAF) determination were performed to check the deviation for each SNP among the unaffected parents. The HWE analysis was calculated through chi-square test. The MAF and allelic transmission disequilibrium test (TDT) and parent-of-origin effects analyses were calculated by PLINK software (http://zzz.bwh.harvard.edu/plink/data.shtml) [21]. The TDT analysis is a family-based association analysis method, which accessed whether the probability of two different alleles passed from heterozygous parents to their affected offspring varies by 50%. Pairwise linkage disequilibrium (LD) was calculated as D’ and r2 for 10 SNPs of 3 target genes (including NTN1, FOXE1 and VAX1) to identify LD blocks by the Haploview online software (http://www.broad.mit.edu/mpg/haploview/index.php). Linkage analysis is based on the principle of meiosis chromosomal exchange and recombination. To determine the correlation between genetic markers and the disease, the separation of genetic markers in a family was observed to see whether these markers were tightly linked with each other.
Results
Except for rs481931 and rs4147811, the genotypic distribution of remaining SNPs did not deviate from the Hardy-Weinberg equilibrium (P > 0.01) (Table 2).
Table 2.
Minor Allele Frequency and Hardy-Weinberg Equilibrium test for 47 SNPs
| SNP | A1 | A2 | MAF | HWE | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| GENO | O (HET) | E (HET) | P | ||||
| rs4920522 | T | C | 23.61% | 14/76/137 | 0.33 | 0.35 | 0.45 |
| rs766325 | A | G | 21.13% | 8/75/144 | 0.33 | 0.32 | 0.84 |
| rs6695765 | C | T | 37.17% | 30/95/100 | 0.42 | 0.45 | 0.38 |
| rs742071 | T | G | 5.20% | 0/16/211 | 0.070 | 0.068 | 1 |
| rs560426 | C | T | 32.85% | 17/111/99 | 0.49 | 0.43 | 0.068 |
| rs481931 | T | G | 32.55% | 11/115/99 | 0.51 | 0.42 | 0.0025 |
| rs4147811 | T | C | 33.19% | 11/117/99 | 0.52 | 0.42 | 0.0016 |
| rs6677101 | T | G | 46.20% | 51/111/64 | 0.49 | 0.50 | 0.89 |
| rs2235371 | T | C | 33.41% | 31/97/99 | 0.43 | 0.46 | 0.38 |
| rs642961 | A | G | 24.23% | 13/82/132 | 0.36 | 0.36 | 1 |
| rs126280 | A | G | 23.81% | 11/79/134 | 0.35 | 0.35 | 1 |
| rs2064163 | T | G | 39.73% | 37/107/82 | 0.47 | 0.48 | 0.89 |
| rs12063989 | C | T | 36.38% | 35/100/91 | 0.44 | 0.47 | 0.40 |
| rs4844913 | G | A | 47.22% | 46/106/74 | 0.47 | 0.49 | 0.50 |
| rs9429830 | T | C | 49.76% | 63/87/58 | 0.42 | 0.50 | 0.019 |
| rs11119388 | G | A | 40.71% | 44/99/84 | 0.44 | 0.48 | 0.13 |
| rs227178 | C | T | 42.98% | 39/98/89 | 0.43 | 0.48 | 0.21 |
| rs2485893 | G | A | 47.35% | 45/103/79 | 0.45 | 0.49 | 0.28 |
| rs7590268 | G | T | 3.44% | 0/15/212 | 0.07 | 0.064 | 1 |
| rs7632427 | C | T | 16.48% | 4/70/153 | 0.31 | 0.28 | 0.25 |
| rs12506428 | C | T | 49.45% | 60/106/61 | 0.47 | 0.50 | 0.35 |
| rs6558002 | C | T | 16.19% | 5/58/164 | 0.26 | 0.25 | 1 |
| rs12543318 | A | C | 34.40% | 28/105/94 | 0.46 | 0.46 | 1 |
| rs987525 | A | C | 8.52% | 2/28/197 | 0.12 | 0.13 | 0.30 |
| rs894673 | A | T | 12.72% | 3/49/174 | 0.22 | 0.21 | 1 |
| rs3758249 | T | C | 13.08% | 3/50/174 | 0.22 | 0.22 | 1 |
| rs4460498 | T | C | 12.83% | 3/49/174 | 0.22 | 0.21 | 1 |
| rs7078160 | A | G | 49.11% | 57/115/55 | 0.51 | 0.50 | 0.89 |
| rs4752028 | C | T | 38.94% | 38/112/77 | 0.49 | 0.49 | 0.89 |
| rs9574565 | T | C | 12.72% | 6/44/177 | 0.19 | 0.22 | 0.12 |
| rs8001641 | A | G | 14.67% | 2/67/156 | 0.30 | 0.27 | 0.082 |
| rs17563 | G | A | 31.25% | 23/94/109 | 0.42 | 0.43 | 0.76 |
| rs1258763 | T | C | 8.33% | 2/38/186 | 0.17 | 0.17 | 1 |
| rs7179658 | C | T | 15.82% | 5/60/162 | 0.26 | 0.26 | 1 |
| rs8049367 | T | C | 34.29% | 35/96/96 | 0.42 | 0.46 | 0.20 |
| rs9788972 | A | G | 22.57% | 14/68/145 | 0.30 | 0.33 | 0.16 |
| rs4791774 | G | A | 22.62% | 11/73/143 | 0.32 | 0.33 | 0.69 |
| rs9915089 | T | C | 19.47% | 8/67/152 | 0.30 | 0.30 | 0.83 |
| rs8069536 | T | G | 3.65% | 0/13/214 | 0.057 | 0.06 | 1 |
| rs8081823 | A | G | 39.89% | 31/112/83 | 0.50 | 0.47 | 0.57 |
| rs17760296 | G | T | 1.55% | 0/8/219 | 0.035 | 0.035 | 1 |
| rs6072081 | G | A | 39.05% | 31/116/80 | 0.51 | 0.48 | 0.33 |
| rs6065259 | A | G | 36.76% | 29/107/86 | 0.48 | 0.47 | 0.67 |
| rs17820943 | T | C | 39.38% | 31/116/80 | 0.51 | 0.48 | 0.33 |
| rs13041247 | C | T | 39.38% | 31/116/80 | 0.51 | 0.48 | 0.33 |
| rs11698025 | A | G | 32.15% | 21/99/107 | 0.44 | 0.43 | 0.88 |
| rs6102085 | A | G | 42.86% | 42/115/69 | 0.51 | 0.49 | 0.69 |
Note: SNP, single-nucleotide polymorphism; A1, minor allele; A2, major allele; MAF, minor allele frequency; HWE, Hardy-Weinberg Equilibrium test; GENO, genotype; O (HET), observed heterozygosity; E (HET), expect heterozygosity.
Allelic TDT analysis was usually assessed the transmission of minor alleles from heterozygous informative parents to affected child within case-parents trios. The results showed allele A at rs894673 of FOXE1 (P = 0.0071, ORtransmission = 0.35, 95% CI: 0.16-0.78), and allele T at rs3758249 of FOXE1 (P = 0.0071, ORtransmission = 0.35, 95% CI: 0.16-0.78) were under-transmitted. Allele A at rs7078160 and allele C at rs4752028 of VAX1 showed a tendency of over-transmission (P = 0.039, ORtransmission = 1.62, 95% CI: 1.02-2.58; P = 0.024, ORtransmission = 1.73, 95% CI: 1.07-2.81; respectively) (Table 3).
Table 3.
Allelic transmission disequilibrium test results for 47 SNPs
| SNP | A1 | T:U | CHISQ | P | OR (95% CI) | Parental | COM | |||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| A:U | CHISQ | P | CHISQ | P | ||||||
| rs4920522 | T | 26:23 | 0.18 | 0.67 | 1.13 (0.65-1.98) | 69:60 | 0.46 | 0.50 | 0.64 | 0.42 |
| rs766325 | A | 24:23 | 0.02 | 0.88 | 1.04 (0.59-1.85) | 67:55 | 0.92 | 0.34 | 0.83 | 0.36 |
| rs6695765 | C | 34:29 | 0.40 | 0.53 | 1.17 (0.71-1.92) | 94:62 | 5.02 | 0.025 | 5.13 | 0.024 |
| rs742071 | T | 10:5 | 1.67 | 0.20 | 2.00 (0.68-5.85) | 31:16 | 4.41 | 0.036 | 6.06 | 0.014 |
| rs560426 | C | 42:28 | 2.80 | 0.094 | 1.50 (0.93-2.42) | 74:63 | 0.74 | 0.39 | 2.68 | 0.10 |
| rs481931 | T | 31:39 | 0.91 | 0.34 | 0.79 (0.50-1.27) | 80:60 | 2.27 | 0.13 | 0.59 | 0.44 |
| rs4147811 | T | 35:41 | 0.47 | 0.49 | 0.85 (0.54-1.34) | 83:58 | 3.57 | 0.059 | 1.44 | 0.23 |
| rs6677101 | T | 42:30 | 2.00 | 0.16 | 1.40 (0.88-2.24) | 74:81 | 0.24 | 0.63 | 0.090 | 0.77 |
| rs2235371 | T | 39:38 | 0.013 | 0.91 | 1.03 (0.66-1.60) | 70:81 | 0.62 | 0.43 | 0.37 | 0.54 |
| rs642961 | A | 23:26 | 0.18 | 0.67 | 0.88 (0.50-1.55) | 66:62 | 0.10 | 0.75 | 0.0049 | 0.94 |
| rs126280 | A | 20:28 | 1.33 | 0.25 | 0.71 (0.40-1.27) | 70:55 | 1.53 | 0.22 | 0.25 | 0.62 |
| rs2064163 | T | 32:37 | 0.36 | 0.55 | 0.86 (0.54-1.39) | 82:80 | 0.019 | 0.89 | 0.032 | 0.86 |
| rs12063989 | C | 31:38 | 0.71 | 0.40 | 0.82 (0.51-1.31) | 73:80 | 0.25 | 0.62 | 0.73 | 0.39 |
| rs4844913 | G | 40:33 | 0.67 | 0.41 | 1.21 (0.76-1.92) | 100:70 | 4.05 | 0.044 | 4.64 | 0.031 |
| rs9429830 | T | 30:25 | 0.45 | 0.50 | 1.20 (0.71-2.04) | 67:78 | 0.61 | 0.44 | 0.14 | 0.71 |
| rs11119388 | G | 38:40 | 0.051 | 0.82 | 0.95 (0.61-1.48) | 80:86 | 0.16 | 0.69 | 0.22 | 0.64 |
| rs227178 | C | 36:32 | 0.24 | 0.63 | 1.13 (0.70-1.81) | 105:67 | 6.28 | 0.012 | 5.92 | 0.015 |
| rs2485893 | G | 39:32 | 0.69 | 0.41 | 1.22 (0.76-1.95) | 108:62 | 9.36 | 0.0022 | 9.46 | 0.0021 |
| rs7590268 | G | 1:8 | 5.44 | 0.020 | 0.13 (0.016-1.00) | 14:12 | 0.15 | 0.70 | 0.71 | 0.40 |
| rs7632427 | C | 14:18 | 0.50 | 0.48 | 0.78 (0.39-1.56) | 46:58 | 1.16 | 0.28 | 1.64 | 0.20 |
| rs12506428 | C | 32:41 | 1.11 | 0.29 | 0.78 (0.49-1.24) | 78:83 | 0.11 | 0.74 | 0.66 | 0.42 |
| rs6558002 | C | 10:20 | 3.33 | 0.068 | 0.50 (0.23-1.07) | 50:42 | 0.60 | 0.44 | 0.029 | 0.86 |
| rs12543318 | A | 30:37 | 0.73 | 0.39 | 0.81 (0.50-1.31) | 70:77 | 0.24 | 0.62 | 0.73 | 0.39 |
| rs987525 | A | 15:7 | 2.91 | 0.088 | 2.14 (0.87-5.26) | 41:28 | 2.25 | 0.13 | 4.55 | 0.033 |
| rs894673 | A | 8:23 | 7.26 | 0.0071 | 0.35 (0.16-0.78) | 38:37 | 0.012 | 0.91 | 1.75 | 0.19 |
| rs3758249 | T | 8:23 | 7.26 | 0.0071 | 0.35 (0.16-0.78) | 39:38 | 0.012 | 0.91 | 1.72 | 0.19 |
| rs4460498 | T | 8:22 | 6.53 | 0.011 | 0.36 (0.16-0.82) | 37:37 | 0 | 1.00 | 1.78 | 0.18 |
| rs7078160 | A | 47:29 | 4.26 | 0.039 | 1.62 (1.02-2.58) | 69:77 | 0.35 | 0.56 | 0.38 | 0.54 |
| rs4752028 | C | 45:26 | 5.09 | 0.024 | 1.73 (1.07-2.81) | 67:82 | 1.19 | 0.28 | 0.062 | 0.80 |
| rs9574565 | T | 16:14 | 0.13 | 0.72 | 1.14 (0.56-2.34) | 47:42 | 0.25 | 0.62 | 0.37 | 0.54 |
| rs8001641 | A | 18:14 | 0.50 | 0.48 | 1.29 (0.64-2.59) | 43:53 | 0.93 | 0.34 | 0.26 | 0.61 |
| rs17563 | G | 28:29 | 0.018 | 0.89 | 0.97 (0.57-1.62) | 73:74 | 0.0056 | 0.94 | 0.017 | 0.90 |
| rs1258763 | T | 8:8 | 0 | 1.00 | 1.00 (0.38-2.67) | 29:38 | 1.08 | 0.30 | 0.89 | 0.35 |
| rs7179658 | C | 23:17 | 0.90 | 0.34 | 1.35 (0.72-2.53) | 57:54 | 0.074 | 0.79 | 0.50 | 0.48 |
| rs8049367 | T | 33:24 | 1.42 | 0.23 | 1.38 (0.81-2.33) | 69:89 | 1.96 | 0.16 | 0.46 | 0.50 |
| rs9788972 | A | 19:30 | 2.47 | 0.12 | 0.63 (0.36-1.13) | 72:60 | 0.91 | 0.34 | 0.0048 | 0.94 |
| rs4791774 | G | 21:26 | 0.53 | 0.47 | 0.81 (0.45-1.44) | 69:55 | 1.34 | 0.25 | 0.42 | 0.52 |
| rs9915089 | T | 16:30 | 4.26 | 0.039 | 0.53 (0.29-0.98) | 68:56 | 0.94 | 0.33 | 0.020 | 0.89 |
| rs8069536 | T | 4:7 | 0.82 | 0.37 | 0.57 (0.17-1.95) | 20:13 | 1.49 | 0.22 | 0.36 | 0.55 |
| rs8081823 | A | 36:37 | 0.014 | 0.91 | 0.97 (0.62-1.54) | 96:84 | 0.61 | 0.44 | 0.39 | 0.53 |
| rs17760296 | G | 2:2 | 0 | 1.00 | 1.00 (0.14-7.10) | 6:8 | 0.29 | 0.59 | 0.22 | 0.64 |
| rs6072081 | G | 35:46 | 1.49 | 0.22 | 0.76 (0.49-1.18) | 75:73 | 0.021 | 0.88 | 0.30 | 0.58 |
| rs6065259 | A | 32:42 | 1.35 | 0.25 | 0.76 (0.48-1.21) | 73:73 | 0 | 1.00 | 0.38 | 0.54 |
| rs17820943 | T | 36:46 | 1.22 | 0.27 | 0.78 (0.51-1.21) | 74:71 | 0.048 | 0.83 | 0.18 | 0.67 |
| rs13041247 | C | 36:46 | 1.22 | 0.27 | 0.78 (0.51-1.21) | 74:71 | 0.048 | 0.83 | 0.18 | 0.67 |
| rs11698025 | A | 35:41 | 0.47 | 0.49 | 0.85 (0.54-1.34) | 71:60 | 0.74 | 0.39 | 0.10 | 0.75 |
| rs6102085 | A | 36:38 | 0.054 | 0.82 | 0.95 (0.60-1.49) | 79:82 | 0.043 | 0.84 | 0.088 | 0.77 |
Note: SNP, single-nucleotide polymorphism; A1, minor allele; T:U, transmitted:untransmitted; OR, odds ratio; A:U, discordance counts; COM, combined analysis results; Bold characters indicate the items with P-value less than 0.05.
In parent-of-origin effects analysis, allele A at rs894673, allele T at rs3758249 and allele T at rs4460498 of FOXE1 showed a weak paternal special under-transmission bias (P = 0.039; P = 0.039; P = 0.039; respectively). Allele A at rs987525 (downstream of LOC728724) showed a relatively strong paternal special over-transmission bias (P = 0.0077) (Table 4). However, no significant difference was found within combined analysis between father and mother.
Table 4.
Parent-of-Origin effects analysis for 47 SNPs
| SNP | A1:A2 | Paternal | Maternal | Z | P | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| T:U | CHISQ | P | T:U | CHISQ | P | ||||
| rs4920522 | T:C | 15:11 | 0.62 | 0.43 | 11:12 | 0.043 | 0.83 | 0.69 | 0.49 |
| rs766325 | A:G | 12.5:12.5 | 0 | 1 | 11.5:10.5 | 0.045 | 0.83 | -0.16 | 0.88 |
| rs6695765 | C:T | 15.5:12.5 | 0.32 | 0.57 | 18.5:16.5 | 0.11 | 0.74 | 0.20 | 0.84 |
| rs742071 | T:G | 2:3 | 0.20 | 0.65 | 8:2 | 3.60 | 0.058 | -1.48 | 0.14 |
| rs560426 | C:T | 17:14 | 0.29 | 0.59 | 25:14 | 3.10 | 0.078 | -0.78 | 0.43 |
| rs481931 | T:G | 17:26 | 1.88 | 0.17 | 14:13 | 0.037 | 0.85 | -1.01 | 0.31 |
| rs4147811 | T:C | 19.5:24.5 | 0.57 | 0.45 | 15.5:16.5 | 0.031 | 0.86 | -0.36 | 0.72 |
| rs6677101 | T:G | 20:18 | 0.11 | 0.75 | 22:12 | 2.94 | 0.086 | -1.03 | 0.30 |
| rs2235371 | T:C | 19:20 | 0.026 | 0.87 | 20:18 | 0.11 | 0.75 | -0.34 | 0.73 |
| rs642961 | A:G | 9.5:13.5 | 0.70 | 0.40 | 13.5:12.5 | 0.038 | 0.84 | -0.74 | 0.46 |
| rs126280 | A:G | 8:12 | 0.80 | 0.37 | 12:16 | 0.57 | 0.45 | -0.20 | 0.84 |
| rs2064163 | T:G | 15:21 | 1 | 0.32 | 17:16 | 0.030 | 0.86 | -0.82 | 0.41 |
| rs12063989 | C:T | 15.5:20.5 | 0.69 | 0.40 | 15.5:17.5 | 0.12 | 0.73 | -0.33 | 0.74 |
| rs4844913 | G:A | 21.5:14.5 | 1.36 | 0.24 | 18.5:18.5 | 0 | 1 | 0.83 | 0.40 |
| rs9429830 | T:C | 13.5:13.5 | 0 | 1 | 16.5:11.5 | 0.89 | 0.34 | -0.66 | 0.51 |
| rs11119388 | G:A | 18.5:18.5 | 0 | 1 | 19.5:21.5 | 0.098 | 0.75 | 0.22 | 0.83 |
| rs227178 | C:T | 19.5:14.5 | 0.74 | 0.39 | 16.5:17.5 | 0.029 | 0.86 | 0.73 | 0.47 |
| rs2485893 | G:A | 22:13 | 2.31 | 0.13 | 17:19 | 0.11 | 0.74 | 1.32 | 0.19 |
| rs7590268 | G:T | 1:4 | 1.80 | 0.18 | 0:4 | 4 | 0.046 | NA | NA |
| rs7632427 | C:T | 7:12 | 1.32 | 0.25 | 7:6 | 0.077 | 0.78 | -0.95 | 0.34 |
| rs12506428 | C:T | 14.5:21.5 | 1.36 | 0.24 | 17.5:19.5 | 0.11 | 0.74 | -0.60 | 0.55 |
| rs6558002 | C:T | 5:12 | 2.88 | 0.09 | 5:8 | 0.69 | 0.41 | -0.52 | 0.60 |
| rs12543318 | A:C | 17:19 | 0.11 | 0.74 | 13:18 | 0.81 | 0.37 | 0.43 | 0.66 |
| rs987525 | A:C | 8.5:0.5 | 7.11 | 0.0077 | 6.5:6.5 | 0 | 1 | 1.82 | 0.069 |
| rs894673 | A:T | 3.5:11.5 | 4.27 | 0.039 | 4.5:11.5 | 3.06 | 0.080 | -0.30 | 0.76 |
| rs3758249 | T:C | 3.5:11.5 | 4.27 | 0.039 | 4.5:11.5 | 3.06 | 0.080 | -0.30 | 0.76 |
| rs4460498 | T:C | 3.5:11.5 | 4.27 | 0.039 | 4.5:10.5 | 2.4 | 0.12 | -0.41 | 0.68 |
| rs7078160 | A:G | 24.5:13.5 | 3.18 | 0.074 | 22.5:15.5 | 1.29 | 0.26 | 0.47 | 0.64 |
| rs4752028 | C:T | 18.5:13.5 | 0.78 | 0.38 | 26.5:12.5 | 5.03 | 0.025 | -0.88 | 0.38 |
| rs9574565 | T:C | 7:10 | 0.53 | 0.47 | 9:4 | 1.92 | 0.17 | -1.50 | 0.13 |
| rs8001641 | A:G | 11.5:5.5 | 2.12 | 0.15 | 6.5:8.5 | 0.27 | 0.61 | 1.37 | 0.17 |
| rs17563 | G:A | 10.5:12.5 | 0.17 | 0.68 | 17.5:16.5 | 0.029 | 0.86 | -0.43 | 0.67 |
| rs1258763 | T:C | 3:4 | 0.14 | 0.71 | 5:4 | 0.11 | 0.74 | -0.50 | 0.62 |
| rs7179658 | C:T | 13.5:9.5 | 0.70 | 0.40 | 9.5:7.5 | 0.24 | 0.63 | 0.18 | 0.86 |
| rs8049367 | T:C | 17.5:11.5 | 1.24 | 0.27 | 15.5:12.5 | 0.32 | 0.57 | 0.38 | 0.70 |
| rs9788972 | A:G | 9:13 | 0.73 | 0.39 | 10:17 | 1.82 | 0.18 | 0.28 | 0.78 |
| rs4791774 | G:A | 10:13 | 0.39 | 0.53 | 11:13 | 0.17 | 0.68 | -0.16 | 0.87 |
| rs9915089 | T:C | 7:16 | 3.52 | 0.061 | 9:14 | 1.09 | 0.30 | -0.62 | 0.54 |
| rs8069536 | T:G | 0:3 | 3 | 0.083 | 4:4 | 0 | 1 | NA | NA |
| rs8081823 | A:G | 19.5:23.5 | 0.37 | 0.54 | 16.5:13.5 | 0.30 | 0.58 | -0.81 | 0.42 |
| rs17760296 | G:T | 1:1 | 0 | 1 | 1:1 | 0 | 1 | 0 | 1 |
| rs6072081 | G:A | 21.5:25.5 | 0.34 | 0.56 | 13.5:20.5 | 1.44 | 0.23 | 0.54 | 0.59 |
| rs6065259 | A:G | 17:25 | 1.52 | 0.22 | 15:17 | 0.13 | 0.72 | -0.55 | 0.58 |
| rs17820943 | T:C | 20:24 | 0.36 | 0.55 | 16:22 | 0.95 | 0.33 | 0.30 | 0.76 |
| rs13041247 | C:T | 20:24 | 0.36 | 0.55 | 16:22 | 0.95 | 0.33 | 0.30 | 0.76 |
| rs11698025 | A:G | 16.5:22.5 | 0.92 | 0.34 | 18.5:18.5 | 0 | 1 | -0.67 | 0.50 |
| rs6102085 | A:G | 20.5:18.5 | 0.10 | 0.75 | 15.5:19.5 | 0.46 | 0.50 | 0.71 | 0.48 |
Note: SNP, single-nucleotide polymorphism; A1, minor allele; A2, major allele; T:U, transmitted:undertransmitted; Z, vector of the large sample Z statistic; NA, not available; Bold characters indicate the items with P-value less than 0.05.
We calculated the pairwise LD of SNPs on 3 target genes (including NTN1, FOXE1 and VAX1) based the association results. Pairwise LD analysis indicated that the SNPs of FOXE1 (rs894673, rs3758249, and rs4460498) were significantly linked with each other (r2 > 0.95, D’ = 1) (Figure 1).
Figure 1.
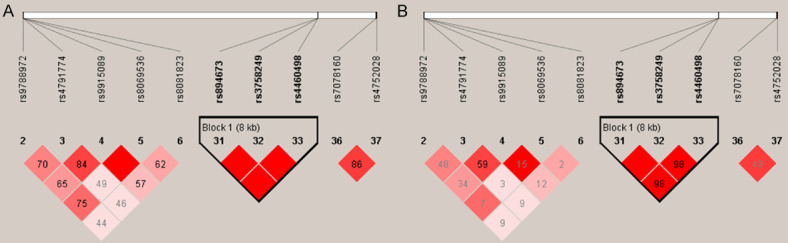
Pairwise LD analysis of the target genes (NTN1, FOXE1 and VAX1) for cleft alveolus (A, D’; B, r2).
Discussion
Phenotype is an observable or measurable trait of a disease. Some typical phenotype (e.g. lower lip pits for Van der Woude syndrome; bilateral zygomatic and malar hypoplasia for Treacher Collins syndrome) is important and usually will lead to a clinical diagnosis, which could provide information to infer the candidate genes with phenotypic similarities and consult the phenotype-genotype heterogeneous network [22,23]. Previous genetic studies of NSCL/P paid more attention to NSCLO, NSCPO and NSCLP, and even combined analysis with mixed samples. As a specific phenotype, cleft alveolus has not been studied by itself.
The maxillary processes fuse with the medial nasal process to form the upper lip, alveolus, and primary palate during the 5th and 6th weeks of embryonic development [1,24]. The process of forming alveolar bone has extensive similarity to the process in the upper lip, which arises from the same embryonic origin. Thus, we infer that cleft alveolus might share the same gene regulatory network with cleft lip.
FOXE1, located on 9q22, was initially suggested as a candidate gene through genome scanning and linkage analysis in some multiplex CL/P families [25]. Subsequently, significant SNPs that linked 9q22 region were replicated in CL/P families of Colombia, USA, and the Philippines. Rs3758249 and rs4460498 showed highly significant signals, which were located inside a 70 kb high linkage disequilibrium block containing FOXE1. Then, expression of foxe1 was detected in caudal epithelium of medial nasal and maxillary processes at E11.5. Especially, foxe1 is obviously expressed in the stage of fusion between the medial nasal and maxillary processes [26]. Animal experiments had demonstrated that FOXE1 in knockout mice will lead to developmental malformation, including thyroid dysgenesis and cleft palate [27]. Then, FOXE1 overexpression experiments in a mouse model also displayed a phenotype of cleft palate, and FOXE1 was highly expressed in the medial edge epithelium (MEE) [28]. This evidence suggested that FOXE1 might regulate the pathogenesis of cleft lip and palate.
Notably, the impact of FOXE1 on the NSCL/P should be validated in different ethnic groups in order to exclude the limitation of single populations’ or regions’ mutation. In a follow-up association study of fifty SNPs at 9q22, rs894673 and rs3758249 near FOXE1 were genotyped in 291 multiplex cleft families. However, no significant association of FOXE1 was found in the test populations [29]. Subsequently, the association of target SNPs (rs894673, rs3758249 and rs4460498) near FOXE1 with NSCL/P were validated among distinct populations [30-33]. These results supported that FOXE1 gene was a positive risk factor for orofacial cleft. In our study, rs894673 and rs3758249 showed a significant association with cleft alveolus, and rs4460498 showed a moderate association with cleft alveolus (Table 3). Meanwhile, marked parent of origin effects were seen with rs894673, rs3758249, and rs4460498 alleles. Under-transmission was shown preferentially from comparing fathers to mothers (Table 4). Pair-wise LD analysis also showed a strong LD between rs894673, rs3758249 and rs4460498 (Figure 1), indicating they were tightly linked with each other. All these findings suggested the pathogenesis of cleft alveolus involved FOXE1 gene expression and regulation.
Rs894673 and rs3758249 are located in the 5’-upstream region of FOXE1, speculating they might affect FOXE1 transcription by altering transcription factor binding sites. The entire promoter region of FOXE1 was screened with 35 cleft palate patients and 160 unaffected people to identify the suspicious variants A novel non-coding variant in the 5’-untranslated region of FOXE1 was found. Based on later cell experiments, the variant could prevent the binding of MYF-5 to FOXE1 promoter and affect the FOXE1 expression [34]. Rs4460498, located in the 3’-downstream region of FOXE1, was first reported to have an association with NSCL/P among Caucasian and Asian populations with P value 6.51E-12 [26]. Three potentially functional SNPs of FOXE1 (two in the 5’-upstream and one in 3’-UTR) were replicated among central Chinese population in a following study. They found that the target SNP in 3’-UTR contributed to altering binding ability with target miRNA in in vitro studies [35].
Numerous studies had reported a significant association between rs987525 and NSCL/P in more than one population origin [14,15,18]. In this study, we found rs987525 had a mild association with parents and cases in combined TDT analysis (Table 3). But the association did not present in the cases. Meanwhile, an allele of rs987525 showed a significant over-transmission from fathers compared to mothers (Table 4). Rs987525 located in 8q24, which had been demonstrated this region containing a remote Myc-regulated enhancer. Deletion of this region would lead to alternation of facial morphology, and even to CL/P phenotype [36]. Myc expression also can be regulated by such cis-enhancer element interacting with the Myc promoter by transcription factor Tcf-4 binding [37].
Rs481931 and rs4147811 deviated from the Hardy-Weinberg equilibrium (P < 0.01) in our samples, indicating it may need larger sample size to validate its significance. Previously, we had validated the association between these two SNPs and NSCL/P among 440 orofacial cleft trios. Unfortunately, rs481931 and rs4147811 were not compatible with the Hardy-Weinberg equilibrium as well [38]. Maybe there was a higher genetic load from parents to the probands in the case-parents design of the research.
In summary, we replicated 47 SNPs contributing to NSCL/P to investigate their roles in cleft alveolus in a western Han Chinese population. Based on the current study, we confirmed FOXE1 as a susceptible gene for the cleft alveolus, which provides a new research direction for the development of alveolar bone.
Acknowledgements
We are particularly thankful to patients and their families who participated in this study. This work was supported by the National Science Funds of China (No. 81600849) and Sichuan Province Science and Technology support program (2020YJ0211).
Disclosure of conflict of interest
None.
References
- 1.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 2.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800. doi: 10.12688/f1000research.9503.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai L, Zhu J, Mao M, Li Y, Deng Y, Wang Y, Liang J, Tang L, Wang H, Kilfoy BA, Zheng T, Zhang Y. Time trends in oral clefts in Chinese newborns: data from the Chinese National Birth Defects Monitoring Network. Birth Defects Res Part A. 2010;88:41–47. doi: 10.1002/bdra.20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan D, Wu S, Liu L, Xia Q, Tian G, Wang W, Ye S, Wang L, Rao J, Yang X, Yu Z, Xin L, Li S, Duan Z, Zhang T, Wu S, Guo X, Liu Z. Prevalence of non-syndromic orofacial clefts: based on 15,094,978 Chinese perinatal infants. Oncotarget. 2018;9:13981–13990. doi: 10.18632/oncotarget.24238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho-Lee GY, García-Díez EM, Nunes RA, Martí-Pagès C, Sieira-Gil R, Rivera-Baró A. Review of secondary alveolar cleft repair. Ann Maxillofac Surg. 2013;3:46–50. doi: 10.4103/2231-0746.110083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiago PE, Schuster LA, Levy-Bercowski D. Management of the alveolar cleft. Clin Plastic Surg. 2014;41:219–232. doi: 10.1016/j.cps.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Fudalej P, Janiszewska-Olszowska J, Wedrychowska-Szulc B, Katsaros C. Early alveolar bone grafting has a negative effect on maxillary dental arch dimensions of pre-school children with complete unilateral cleft lip and palate. Orthod Craniofac Res. 2011;14:51–57. doi: 10.1111/j.1601-6343.2011.01507.x. [DOI] [PubMed] [Google Scholar]
- 9.Gładysz D, Hozyasz KK. Stem cell regenerative therapy in alveolar cleft reconstruction. Arch Oral Biol. 2015;60:1517–1532. doi: 10.1016/j.archoralbio.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Stasiak M, Wojtaszek-Słomińska A, Racka-Pilszak B. Current methods for secondary alveolar bone grafting assessment in cleft lip and palate patients-A systematic review. J Craniomaxillofac Surg. 2019;47:578–585. doi: 10.1016/j.jcms.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Shete S. Effects of population structure on genetic association studies. BMC Genet. 2005;6:S109. doi: 10.1186/1471-2156-6-S1-S109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo X, Rotter JI. Genome-wide association studies. JAMA. 2019;322:1705–1706. doi: 10.1001/jama.2019.16479. [DOI] [PubMed] [Google Scholar]
- 13.Mills MC, Rahal C. The GWAS diversity monitor tracks diversity by disease in real time. Nat Genet. 2020;52:242–243. doi: 10.1038/s41588-020-0580-y. [DOI] [PubMed] [Google Scholar]
- 14.Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, de Assis NA, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Pötzsch S, Steegers-Theunissen RP, Pötzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nöthen MM, Mangold E. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 15.Grant SA, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, Frackelton EC, Otieno FG, Chiavacci RM, Nah HD, Kirschner RE, Hakonarson H. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4 . Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, Du Y, Wang M, Lan F, Hu Z, Wang G, Jiang M, Ma J, Zhang X, Ma H, Ma J, Zhang W, Huang Q, Zhou Z, Ma L, Li Y, Jiang H, Xie L, Jiang Y, Shi B, Cheng J, Shen H, Wang L, Yang Y. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414. doi: 10.1038/ncomms7414. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, Meng L, Wang W, Song Y, Cheng Y, Zhou F, Chen G, Zheng X, Wang X, Liang B, Zhu Z, Fu X, Sheng Y, Hao J, Liu Z, Yan H, Mangold E, Ruczinski I, Liu J, Marazita ML, Ludwig KU, Beaty TH, Zhang X, Sun L, Bian Z. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L, Jia Z, Shi Y, Du Q, Shi J, Wang Z, Mou Y, Wang Q, Zhang B, Wang Q, Ma S, Lin H, Duan S, Yin B, Lin Y, Wang Y, Jiang D, Hao F, Zhang L, Wang H, Jiang S, Xu H, Yang C, Li C, Li J, Shi B, Yang Z. Genetic factors define CPO and CLO subtypes of nonsyndromicorofacial cleft. PLoS Genet. 2019;15:e1008357. doi: 10.1371/journal.pgen.1008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q, He S, Zeng N, Ma J, Zhang B, Shi B, Jia Z. BMP7 gene involved in nonsyndromic orofacial clefts in Western Han Chinese. Med Oral Patol Oral Cir Bucal. 2015;20:e298–304. doi: 10.4317/medoral.20335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MR, Bender D, Maller J, Sklar P, de Bakker PW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang SH, Wu C, Li X, Chen X, Jiang W, Gong BS, Li J, Yan YQ. From phenotype to gene: detecting disease-specifi c gene functional modules via a text-based human disease phenotype network construction. FEBS Lett. 2010;584:3635–3643. doi: 10.1016/j.febslet.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 23.Yao X, Hao H, Li Y, Li S. Modularity-based credible prediction of disease genes and detection of disease subtypes on the phenotype-gene heterogeneous network. BMC Syst Biol. 2011;5:79. doi: 10.1186/1752-0509-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifeldin SA. Is alveolar cleft reconstruction still controversial? Saudi Dent J. 2016;28:3–11. doi: 10.1016/j.sdentj.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Adela Mansilla M, Leigh Field L, Liu Y, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski DF, Bailey-Wilson JE, Albacha-Hejazi H, Beaty TH, McIntosh I, Hetmanski JB, Tuncbilek G, Edwards M, Harkin L, Scott R, Roddick LG. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, Johnson MK, Brauer D, Krahn K, Daack-Hirsch S, L’Heureux J, Valencia-Ramirez C, Rivera D, Maria López A, Moreno MA, Hing A, Lammer EJ, Jones M, Christensen K, Lie RT, Jugessur A, Wilcox AJ, Chines P, Pugh E, Doheny K, Arcos-Burgos M, Marazita ML, Murray JC, Lidral AC. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum Mol Genet. 2009;18:4879–4896. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Felice M, Ovitt C, Biffali E, Rodriguez-Mallon A, Arra C, Anastassiadis K, Macchia PE, Mattei MG, Mariano A, Schöler H, Macchia V, Di Lauro R. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat Genet. 1998;19:395–398. doi: 10.1038/1289. [DOI] [PubMed] [Google Scholar]
- 28.Meng T, Shi JY, Wu M, Wang Y, Li L, Liu Y, Zheng Q, Huang L, Shi B. Overexpression of mouse TTF-2 gene causes cleft palate. J Cell Mol Med. 2012;16:2362–2368. doi: 10.1111/j.1582-4934.2012.01546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letra A, Menezes R, Govil M, Fonseca RF, McHenry T, Granjeiro JM, Castilla EE, Orioli IM, Marazita ML, Vieira AR. Follow-up association studies of chromosome region 9q and nonsyndromic cleft lip/palate. Am J Med Genet Part A. 2010;152A:1701–1710. doi: 10.1002/ajmg.a.33482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig KU, Böhmer AC, Rubini M, Mossey PA, Herms S, Nowak S, Reutter H, Alblas MA, Lippke B, Barth S, Paredes-Zenteno M, Muñoz-Jimenez SG, Ortiz-Lopez R, Kreusch T, Hemprich A, Martini M, Braumann B, Jäger A, Pötzsch B, Molloy A, Peterlin B, Hoffmann P, Nöthen MM, Rojas-Martinez A, Knapp M, Steegers-Theunissen RP, Mangold E. Strong association of variants around FOXE1 and orofacial clefting. J Dent Res. 2004;93:376–381. doi: 10.1177/0022034514523987. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, Lu Y, Ai L, Jiao B, Yu J, Zhang B, Liu Q. Association between FOXE1 and non-syndromic orofacial clefts in a northeastern Chinese population. Br J Oral Maxillofac Surg. 2015;53:705–710. doi: 10.1016/j.bjoms.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Lammer EJ, Mohammed N, Iovannisci DM, Ma C, Lidral AC, Shaw GM. Genetic variation of FOXE1 and risk for orofacial clefts in a California population. Am J Med Genet Part A. 2016;170A:2770–2776. doi: 10.1002/ajmg.a.37871. [DOI] [PubMed] [Google Scholar]
- 33.Duan SJ, Zhang BH, Shi B, Shi YJ, He S, Jiang SY, Zeng N, Jia ZL. Thyroid cancer causal gene FOXE1 involved in orofacial clefts in Western Han Chinese. Int J Clin Exp Med. 2017;10:5398–5405. [Google Scholar]
- 34.Venza M, Visalli M, Venza I, Torino C, Tripodo B, Melita R, Teti D. Altered binding of MYF-5 to FOXE1 promoter in non-syndromic and CHARGE-associated cleft palate. J Oral Pathol Med. 2009;38:18–23. doi: 10.1111/j.1600-0714.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 35.Yin X, Zhang H, Zhu Z, Wang H, Du Y, Li S, Zhang Z, Fan W, Pan Y. FOXE1 polymorphisms and non-syndromic orofacial cleft susceptibility in a Chinese Han population. Oral Diseases. 2016;22:274–279. doi: 10.1111/odi.12435. [DOI] [PubMed] [Google Scholar]
- 36.Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 2014;46:753–758. doi: 10.1038/ng.2971. [DOI] [PubMed] [Google Scholar]
- 37.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan SJ, Shi B, Shi JY, Feng F, Jia ZL. Further evidence of GWAS signals in non-syndromic orofacial clefts from Western Han Chinese. J Interdiscipl Med Dent Sci. 2016;4:1. [Google Scholar]


