Abstract
MicroRNA-451 (miR-451) is lowly expressed in stomach cancer cells and improves their metastatic ability by down-regulating extracellular signal-regulated kinase 2 (ERK2). Many studies have found that caveolin-1 (CAV1) plays an important role in cancer progression. Additionally, miR-451 has been reported to regulate the expression of CAV1 in chronic obstructive pulmonary disease. Therefore, this study aims to determine if miR-451 regulates the biological functions of stomach cancer cells by regulating CAV1 expression. Through a bioinformatics analysis, we found that miR-451a regulates CAV1 expression, and miR-451a expression is relatively low in stomach cancer cells. Next, we confirmed that miR-451a negatively regulates CAV1 expression using a dual-luciferase reporter assay. Then MTT, 5-ethynyl-2’-deoxyuridine (EdU), propidium iodide (PI), an Annexin V-FITC/PI apoptosis kit, and transwell assays were used to measure the changes in cell proliferation, the cell cycle, apoptosis, cell migration, and invasiveness in stomach cancer cells overexpressing miR-451a or both miR-451a and CAV1. It was found that increasing the miR-451a expression in stomach cancer cells inhibits cell growth, migration, and invasiveness, and promotes apoptosis. After restoring the CAV1 expression, these biological processes resumed. In summary, in stomach cancer cells, the overexpression of miR-451a can restrain cell growth and promote apoptosis, so it is a potential treatment for stomach cancer.
Keywords: MicroRNA-451a, Caveolin-1, stomach cancer
Introduction
Stomach cancer is one of the most common malignant tumors and has a high mortality rate [1]. Patients with early stage stomach cancer do not present with specific symptoms, so stomach cancer is often not diagnosed until the later stages [1]. Clinical data show that the 5-year survival rate of patients with advanced stomach cancer is only 30% to 40%, compared to 70% to 90% for those with early stage stomach cancer [1]. Therefore, to optimize the treatment strategy, predict the efficacy of treatment, and prolong patient survival time, it is necessary to identify new markers for stomach cancer.
The microRNA-451 (miR-451) family is located on chromosome 17 and is highly evolutionarily conserved. This family consists of miR-451a and miR-451b, each containing 22 nucleotides in their mature sequence [2]. MiR-451 plays a vital role in many biological processes [2]. Additionally, miR-451 has recently been identified as a valuable biomarker for cancer detection, prognosis, and treatment [2]. Multiple studies have shown that miR-451 is widely dysregulated in breast cancer, lung cancer, stomach cancer, colorectal cancer, glioma, renal cell carcinoma, pancreatic cancer, and leukemia [2-5]. These findings prove that miR-451 plays a key role in tumorigenesis.
Caveolin-1 (CAV1) is a protein located in the cell membrane that is abundantly expressed in fat cells, endothelial cells, pneumocytes, fibroblasts, and muscle cells [6,7]. Signal transduction involving CAV1 regulates many biological processes, such as the cell cycle, cell senescence, proliferation, invasion, cell death, membrane composition, lipid homeostasis, and metabolism [6,8-12]. The resistance of solid tumors to chemotherapy and radiation therapy remains the main obstacle to anticancer therapy [6]. CAV1 is highly expressed in many tumors, and high levels of CAV1 are associated with tumor progression, invasion, and metastasis, making it the focus of improved solid tumor treatment strategies [6,13-16]. Previous studies have found that CAV1 is overexpressed in liver cancer, colon cancer, breast cancer, kidney cancer, lung cancer, and, depending on the type and stage of the tumor, it can play a role in promoting or inhibiting tumor progression [17-19]. A high expression of CAV1 has been reported to drive tumorigenesis by inhibiting apoptosis and promoting adherent growth, drug resistance, and metastasis [17,19,20].
MiR-451 inhibits tumor progression by targeting MIF, c-myc, MDR-1, and the RAS-related protein 14 (RAB14) [2,21]. It has been found that in chronic obstructive pulmonary disease, hsa-miR-451 may regulate CAV1 expression [22]; however, in stomach cancer, there are few studies implicating miR-451 in the regulation of CAV1. Therefore, this study aims to explore the regulatory relationship between miR-451 and CAV1 in stomach cancer.
Materials and methods
Bioinformatics analysis
The UpSetR package was used to analyze the microRNA (miRNA) data targeting CAV1 downloaded from the microT-CDS, StarBase, miRDB, TargetScan, and mirDIP databases in R version 3.5.1. RNA-seq data from stomach cancer patients with individual clinical information were downloaded from The Cancer Genome Atlas (TCGA) database. An in-depth analysis of miR-451a expression in unpaired or paired gastric carcinoma samples was performed using GraphPad Prism 8.2.1 (GraphPad Software, Inc., La Jolla, California, United States).
Cell lines
The stomach cancer cell lines HGC-27 and MKN-45 were purchased from XIAMEN Anti-HeLa Biological Technology Trade Co. Ltd. (Xiamen, Fujian, China) and cultured in Dulbecco’s modified eagle medium (DMEM) (GIBCO, catalog no. 11965-092, Shanghai, China) containing 10% fetal bovine serum (FBS) (GIBCO, catalog no. 10091-148, Shanghai, China) and 1% penicillin-streptomycin (GIBCO, catalog no. 15070-063, Shanghai, China).
Real-time quantitative PCR (RT-qPCR)
RNA was extracted from the cells using Trizol (Ambion, catalog number: 15596-026, Shanghai, China) and reverse transcribed into cDNA using Hiscript Reverse Transcriptase (VAZYME, catalog number: R101-01/02, Nanjing, Jiangsu, China). cDNA was then used for the RT-qPCR with SYBR Green Master Mix (VAZYME, catalog number: Q111-02, Nanjing, Jiangsu, China) and an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) to determine the relative RNA expression levels. The primers for the RT-qPCR were designed using DNAMAN 10.0 (https://dnaman.software.informer.com/) and are shown in Table 1. cDNA was pre-denatured at 94°C for 30 minutes, followed by 40 cycles of denaturation at 94°C for 15 seconds, annealing at 60°C for 20 seconds, and extension at 72°C for 20 seconds. Finally, the target gene expression was quantified using the 2-ΔΔCt method.
Table 1.
The primers for RT-qPCR
| Name | Sequence (5’-3’) | GenBank accession |
|---|---|---|
| Forward primer | CCAAGGAGATCGACCTGGTCAA | NM_001172895.1 |
| Reverse primer | GCCGTCAAAACTGTGTGTCCCT |
Western blot
The proteins were extracted from the cells using an ice-cold RIPA buffer (Beyotime Biotechnology, catalog no. P0013C, Shanghai, China) and quantified with a BCA protein assay kit (Beyotime Biotechnology, catalog no. P0012S, Shanghai, China). Subsequently, western blotting was performed as previously described [23]. The primary antibodies used were anti-CAV1 (Proteintech, catalog no. 66067-1-Ig, 1:3,000, Wuhan, Hubei, China) and anti-GAPDH (Proteintech, catalog no. 60004-1-lg, 1:20,000, Wuhan, China). The secondary antibody conjugated with ECL was anti-mouse (Cell Signaling Technology, catalog no. 9309, 1:2,000, Boston, Massachusetts, USA).
Dual-luciferase reporter assay
Wild type or mutant versions of the CAV1 3’Untranslated Regions (3’UTR) were cloned into a pYr-MirTarget vector (Hunan Changsha Yingrun Biotechnology Co., Ltd., Changsha, Hunan, China) and named CAV1 3’UTR-wt and CAV1 3’UTR-mut, respectively. The mutation in the CAV1 3’UTR-mut sequence is shown in red letters in Figure 1D. The corresponding primers were designed using DNAMAN 10.0 and are shown in Table 2. The HGC-27 and the MKN cells were incubated overnight in 24-well plates at a density of 5×104 cells per well. CAV1 3’UTR-wt or CAV1 3’UTR-mut vector was co-transfected into the HGC-27 and the MKN cells with the negative control miR-451a mimic (miRNA NC) or the miR-451a mimic (miR-451a) using Lipofectamine 2000 (Invitrogen, catalog no. 11668-019, Shanghai, China) according to the manufacturer’s instructions. Dual-Luciferase Reporter Assay (Promega, catalog no. E1910, Beijing, China) was performed 48 hours after the transfection following the manufacturer’s instructions. The differences in the transfection efficiency were normalized by dividing the firefly luciferase activity by the values of the Renilla luciferase.
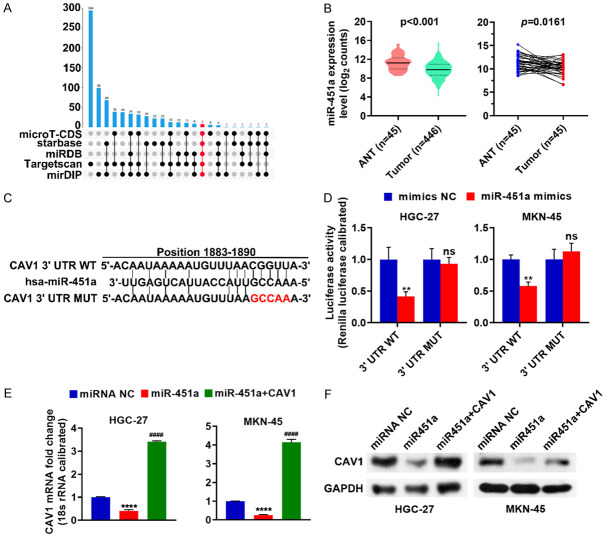
Figure 1.
MiR-451a is lowly expressed in stomach cancer cells and negatively regulates CAV1. (A) MiRNAs targeting CAV1 in the microT-CDS, StarBase, miRDB, TargetScan, and mirDIP databases were analyzed using the UpSetR package in R version 3.5.1. (B) The differences in the miR-451a mRNA levels in the stomach cancer tissues and adjacent normal tissues were analyzed either unpaired or paired using GraphPad Prism 8.2.1. (C) Schematic of the CAV1 3’UTR point mutation used in this study. (D) The regulatory relationship between miR-451a and CAV1 was analyzed by a dual-luciferase reporter assay. (E, F) After miR-451a was transfected either alone or with the CAV1 expression plasmid into HGC-27 or MKN-45 cells, the CAV1 expression was determined using RT-qPCR (E) and western blot (F). ns, not significant (P > 0.05); **P < 0.01; ****P < 0.0001, miR-451a vs miRNA NC; ####P < 0.0001, miR-451a + CAV1 vs miR-451a. ANT: adjacent normal tissues; miR-451a+CAV1: miR-451a was co-transfected with the CAV1 expression plasmid; CAV1: caveolin-1; miRNA NC: the negative control of miR-451a.
Table 2.
The primers for CAV1 3’UTR-wt and CAV1 3’UTR-mut
| Name | Sequence (5’-3’) | GenBank accession |
|---|---|---|
| 3’UTR-wt-F | CTCGCTAGCCTCGAGATGACATTTCAAGGATAGAAG | NM_001172895.1 |
| 3’UTR-wt-R | CATGCCTGCAGGTCGACTTTTTTTTTTTGACTGTTTAACCG | |
| 3’UTR-mut-F | TCATCTATTGCCAACAGTCTGAATTTTTAAAACCC | |
| 3’UTR-mut-R | AGACTGTTGGCAATAGATGAAATAGCTCAGAAGAG |
Construction of the plasmids
CAV1 was cloned into pCDH-EF1α-MCS-T2A-puro vector purchased from Anti-HeLa BioTech (Xiamen, Fujian, China) to prepare the CAV1 expression plasmid (Gene ID: 857). The corresponding primer sequences were designed using DNAMAN 10.0 (Table 3).
Table 3.
The primers for CAV1 expression plasmid
| Name | Sequence (5’-3’) | GenBank accession |
|---|---|---|
| CAV1-F | CTAGAGCTAGCGAATTCATGGCAGACGAGCTGAGCGAG | NM_001172895.1 |
| CAV1-R | TGTCGTCATCGTCTTTGTAGTCTATTTCTTTCTGCAAGTTGATG |
MTT assay
HGC-27 and MKN-45 cells were cultured overnight in 96-well plates at a density of 5×103 cells per well. Mir-451a and miRNA NC were transfected into the HGC-27 and MKN-45 cells either alone or with plasmids expressing CAV1 using Lipofectamine 2000. After 48 hours, the MTT assay was performed as previously described [24]. The results were determined by measuring the absorbance at OD450 on a microplate reader (Thermo, Shanghai, China).
EdU assay
The HGC-27 and MKN-45 cells transfected with miR-451a alone or co-transfected with miR-451a and the CAV1 expression plasmid were evenly spread across the 96-well plates at a density of 5×103 cells per well. After 24 hours, EdU (RiboBio, Guangzhou, Guangdong, China) was used to measure the cell proliferation and DNA synthesis according to the manufacturer’s instructions and as previously described [25]. Images were obtained using a fluorescence microscope (MOTIC, Hongkong, China).
Cell cycle assay
The HGC-27 and MKN-45 cells were evenly plated and incubated in 6-well plates overnight. After transfection with miR-451a alone or co-transfection with miR-451a and the CAV1 expression plasmid, the cells were collected and fixed in 70% ethanol at 4°C overnight. After two washes with phosphate buffer saline (PBS), the fixed cells were incubated in PBS with 0.2% Triton X-100 and 10 μg/mL RNase at 37°C for 30 minutes. Then, the cells were stained with propidium iodide (PI, 50 μg/mL) (Nanjing Kaiji Biotechnology Development Co., Ltd., catalog no. KGA512, Nanjing, Jiangsu, China) at 4°C for 30 minutes in the dark and analyzed using a flow cytometer (ACEA Biosciences, Inc., San Diego, CA, USA).
Apoptosis assay
The HGC-27 and MKN-45 cells were cultured overnight in 6-well plates at a density of 5×105 cells per well. After transfection with miR-451a alone or co-transfection with miR-451a and the CAV1 expression plasmid for 48 hours, the cells were stained using an Annexin V-FITC/PI apoptosis detection kit (Nanjing Kaiji Biotechnology Development Co., Ltd., catalog no. KGA108, Nanjing, Jiangsu, China) in the dark for 10 minutes at room temperature. A subsequent analysis was performed using a flow cytometer (ACEA Biosciences, Inc., San Diego, CA, USA).
Transwell assay
After the HGC-27 and MKN-45 cells were cultured overnight, mir-451a and miRNA NC were transfected into these cells either alone or with the CAV1 expression plasmid using Lipofectamine 2000. After 48 hours, the cells were used for a transwell assay. The migration was measured using Matrigel-free transwell plates with 8 μm porous membranes, and the invasion was measured using transwell plates with Matrigel. The cells in the serum-free medium were seeded into the upper chamber of the transwell plate at a density of 4×103 cells per well and 500 μl of medium supplemented with 10% FBS was added to the lower chamber. After 24 hours, the cellular migration or invasion was evaluated using 0.5% crystal violet staining. Images were captured using a lighted microscope (MOTIC, Hongkong, China) and the invasive/motile cells were counted.
Statistical analysis
SPSS 22.0 (IBM SPSS, Armonk, NY, USA) was used for all the statistical analyses. Comparisons between two groups were performed using Student’s t-tests (unpaired). Comparisons between multiple groups were performed using analyses of variance (ANOVA). P < 0.05 indicates a statistically significant difference, and the graphs were created using GraphPad Prism 8.2.1. The data are representative of at least three independent experiments and are shown as the mean ± standard deviation (SD).
Results
MiR-451a is lowly expressed in stomach cancer cells and negatively regulates CAV1
By analyzing potential miRNAs targeting CAV1, we identified 20 miRNAs in the microT-CDS, StarBase, miRDB, TargetScan, and mirDIP databases (Figure 1A), among which mir-451a was found to relatively underexpressed in stomach cancer tissues (Figure 1B). A sequence alignment revealed a miR-451a recognition sequence in the 3’UTR of CAV1 (Figure 1C). To determine whether this was an active regulatory sequence, CAV1 3’UTR-wt or CAV1 3’UTR-mut were co-transfected with miR-451a into the HGC-27 and MKN-45 cells. A dual-luciferase reporter assay revealed that miR-451a reduced the expression of WT CAV1 but not of the CAV1 mutant (Figure 1D), both confirming that miR-451a can regulate the CAV1 expression and identify the regulatory binding site. MiR-451a and miRNA NC were transfected into the HGC-27 and MKN-45 cells either alone or with the CAV1 expression plasmid, and the CAV1 expression was evaluated using RT-qPCR and western blot. This analysis demonstrated that increased miR-451a expression reduced both the mRNA and protein levels of CAV1 (Figure 1E, 1F). These results illustrate that miR-451a acts as a negative regulator of CAV1 expression in stomach cancer cells.
MiR-451a suppresses stomach cancer cell proliferation
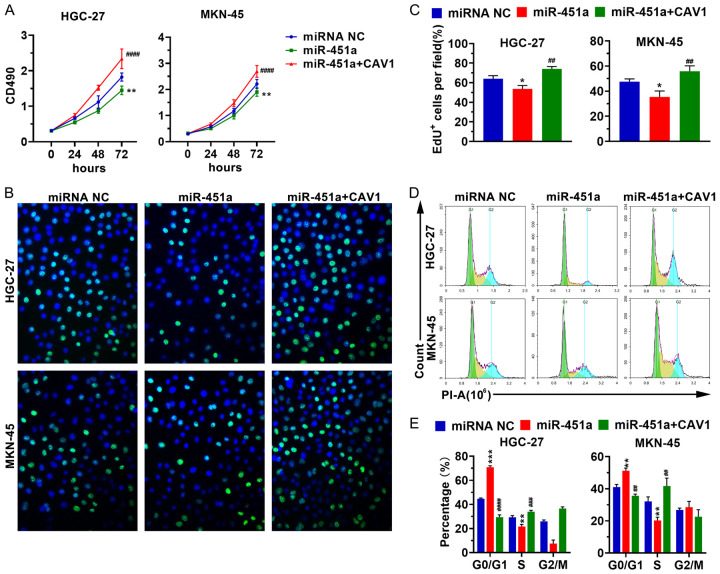
To investigate how the miR-451a regulation of CAV1 regulates the biological processes in stomach cancer cells, mir-451a and miRNA NC were transfected into HGC-27 and MKN-45 cells either alone or with the CAV1 expression plasmid. The MTT, EdU, and cell cycle analyses suggest that increasing miR-451a expression inhibits cellular proliferation (Figure 2A-C) and arrests the cell cycle in the G0/G1 phase (Figure 2D, 2E). These data imply that the overexpression of miR-451a leads to a decrease in CAV1expression, leading to the impaired proliferation of stomach cancer cells.
Figure 2.
MiR-451a suppresses stomach cancer cell proliferation. After miR-451a was transfected alone or with the CAV1 expression plasmid into the HGC-27 or MKN-45 cells, the cellular proliferation was analyzed using MTT (A) and EdU assays (B, C), and the cell cycle phase was determined by PI staining (D, E). *P < 0.05, miR-451a vs miRNA NC; **P < 0.01, miR-451a vs miRNA NC; ##P < 0.01, miR-451a + CAV1 vs miR-451a; ***P < 0.001, miR-451a vs miRNA NC; ###P < 0.001, miR-451a + CAV1 vs miR-451a; ####P < 0.0001, miR-451a + CAV1 vs miR-451a. miR-451a+CAV1: miR-451a was co-transfected with the CAV1 expression plasmid; CAV1: caveolin-1; miRNA NC: the negative control of miR-451a.
MiR-451a promotes stomach cancer cell apoptosis
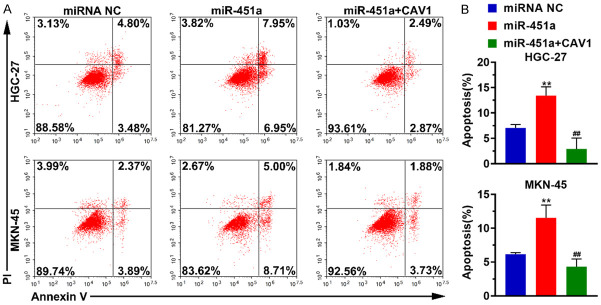
Next, we examined the effect of miR-451a on apoptosis in stomach cancer cells. We found that an increased expression of mir-451a leads to more apoptosis. Interestingly, the CAV1 supplementation in the HGC-27 and MKN-45 cells blocked the increase in apoptosis (Figure 3). This demonstrates that increased miR-451a expression suppressed the CAV1 expression to enhance apoptosis in stomach cancer cells.
Figure 3.
MiR-451a promotes apoptosis in stomach cancer cells. After the HGC-27 and MKN-45 cells were transfected with miR-451a alone or with miR-451a and the CAV1 expression plasmid, the apoptosis was detected using the Annexin V-FITC/PI apoptosis detection kit. A. Representative images of the apoptosis analysis. B. Histogram representing the percentage of apoptotic cells under various conditions. **P < 0.01, miR-451a vs miRNA NC; ##P < 0.01, miR-451a + CAV1 vs miR-451a. miR-451a+CAV1: miR-451a was co-transfected with the CAV1 expression plasmid; CAV1: caveolin-1; miRNA NC: the negative control of miR-451a.
Mir-451a interferes with the migration and invasion of stomach cancer cells
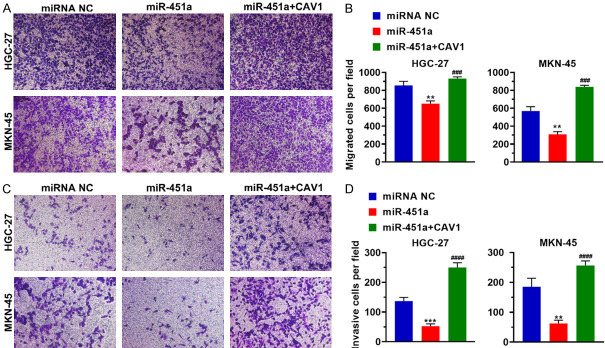
After HGC-27 and MKN-45 were transfected with miR-451a alone or along with the CAV1 expression plasmid, the migration and invasiveness of the cells were tested. The results showed that increased miR-451a expression reduced the number of migrating cells compared to the control, but the number of migrating cells increased following the CAV1 supplementation (Figure 4). These findings demonstrate that the upregulation of mir-451a inhibits the CAV1 expression of CAV1, leading to impaired migration and invasiveness in stomach cancer cells.
Figure 4.
Mir-451a interferes with the migration and invasion of stomach cancer cells. MiR-451a was transfected alone or with the CAV1 expression plasmid into HGC-27 or MKN-45 cells. A. The effect of miR-451a on cellular migration was examined using a transwell assay. B. Histogram representing the number of migrated cells per field of view under various treatment conditions. C. Representative images of the invasion analysis. D. Histogram representing the number of invasive cells per field of view under various conditions. **P < 0.01, miR-451a vs miRNA NC; ***P < 0.001, miR-451a vs miRNA NC; ###P < 0.001, miR-451a + CAV1 vs miR-451a; ####P < 0.0001, miR-451a + CAV1 vs miR-451a. miR-451a+CAV1: miR-451a was co-transfected with the CAV1 expression plasmid; CAV1: caveolin-1; miRNA NC: the negative control of miR-451a.
Discussion
The latest global cancer data shows that stomach cancer is currently the fifth most common cancer in the world, and its mortality rate ranks third among cancer deaths [26]. The 5-year survival rate of stomach cancer is less than 30% [27]. MiRNAs are essential for the maintaining the homeostasis of the body [28]. MiRNAs have also been identified as tumor markers with important roles in the early diagnosis of tumors, targeted therapy, and prognosis evaluation [28]. MiR-451 expression levels have been found to be relatively reduced in stomach cancer cells [2,29-33]. MiR-451 downregulation is often directly related to lymph node metastasis, the patient’s clinical stage, and overall survival [2]. However, the ectopic expression of miR-451 in stomach cancer cells has no significant effect on cell growth, but it does reduce cellular migration and invasion [2,32,34]. Consistent with this, this study further verified that miR-451a is expressed in stomach cancer tissues and can inhibit the migration and invasiveness of stomach cancer cells by negatively regulating the expression of CAV1. In contrast, the overexpression of miR-451a in stomach cancer cells was found to downregulate cell proliferation. In addition, miR-451 has been reported to effectively regulate a variety of metastatic phenotypes, including cell growth, migration, invasion, and the epithelial-mesenchymal transition (EMT) in stomach cancer by down-regulating the target gene MAPK pathway member extracellular signal-regulated kinase 2 (ERK2) [2]. This gene has also been shown to enhance cancer metastasis [2,33]. In this study, we found that miR-451a can also regulate these biological processes by down-regulating CAV1. This demonstrates that miR-451 can regulate the development of stomach cancer by regulating the expression of various genes.
CAV1 has been shown to be overexpressed or mutated in many human solid tumors and is known to play an important role in the carcinogenic process [6,13,15]. Surprisingly, CAV1 is both an anti-apoptotic protein and a pro-apoptotic protein, both a tumor promoter and a tumor suppressor [6]. In this study, CAV1 was confirmed to promote the proliferation, migration, and invasiveness of stomach cancer cells. Additionally, CAV1 stimulates metastasis and is considered to be a prognostic indicator [6]. Analyses at the transcriptional level revealed that many oncogenes down-regulate CAV1 expression [6], and miR-451 is able to inhibit the proliferation, migration, and invasiveness of various tumor cells [2]. Further, we confirmed that miR-451a down-regulated CAV1 expression. Additional work has demonstrated that the gene encoding CAV1 is located at the putative tumor suppression site [6]. However, for several tumors, CAV1 up-regulation will promote the survival and growth of cancer cells, which is conducive to tumor progression [6,35]. Similarly, we also found that up-regulating CAV1 can inhibit apoptosis in stomach cancer cells, promote stomach cancer cell growth, and promote the progression of stomach cancer.
In summary, miR-451a expression is low in stomach cancer tissues, and the overexpression of miR-451a in stomach cancer cells can reduce the expression of CAV1 to inhibit cell growth and promote apoptosis. Restoring CAV1 expression can cause the cells to resume their growth. Therefore, miR-451a can inhibit the proliferation of stomach cancer cells by reducing the expression of CAV1; however, the detailed molecular mechanism underlying this requires further study.
Acknowledgements
The authors hank the Research on Application of Startup Fund for Scientific Research, Fujian Medical University (2017XQ1220 and 2018QH1231), the National Clinical Key Specialty Construction Program of China, the National Science Foundation Project of Fujian Science and Technology Department (2017J01264 and 2018Y0015), and the Foundation for Fujian Provincial Health Technology Project (2019-ZQN-16, 2019-CXB-9 and 2019006).
Disclosure of conflict of interest
None.
References
- 1.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khordadmehr M, Jigari-Asl F, Ezzati H, Shahbazi R, Sadreddini S, Safaei S, Baradaran B. A comprehensive review on miR-451: a promising cancer biomarker with therapeutic potential. J Cell Physiol. 2019;234:21716–21731. doi: 10.1002/jcp.28888. [DOI] [PubMed] [Google Scholar]
- 3.Khordadmehr M, Shahbazi R, Ezzati H, Jigari-Asl F, Sadreddini S, Baradaran B. Key microRNAs in the biology of breast cancer; emerging evidence in the last decade. J Cell Physiol. 2019;234:8316–8326. doi: 10.1002/jcp.27716. [DOI] [PubMed] [Google Scholar]
- 4.Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 5.Baradaran B, Shahbazi R, Khordadmehr M. Dysregulation of key microRNAs in pancreatic cancer development. Biomed Pharmacother. 2019;109:1008–1015. doi: 10.1016/j.biopha.2018.10.177. [DOI] [PubMed] [Google Scholar]
- 6.Ketteler J, Klein D. Caveolin-1, cancer and therapy resistance. Int J Cancer. 2018;143:2092–2104. doi: 10.1002/ijc.31369. [DOI] [PubMed] [Google Scholar]
- 7.Xu H, Zhang L, Chen W, Xu J, Zhang R, Liu R, Zhou L, Hu W, Ju R, Lee C, Lu W, Kumar A, Li X, Tang Z. Inhibitory effect of caveolin-1 in vascular endothelial cells, pericytes and smooth muscle cells. Oncotarget. 2017;8:76165–76173. doi: 10.18632/oncotarget.19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Qu X, Li H, Li C, Liu J, Zheng H, Liu Y. Src/caveolin-1-regulated EGFR activation antagonizes TRAIL-induced apoptosis in gastric cancer cells. Oncol Rep. 2014;32:318–324. doi: 10.3892/or.2014.3183. [DOI] [PubMed] [Google Scholar]
- 9.Meyer C, Liu Y, Kaul A, Peipe I, Dooley S. Caveolin-1 abrogates TGF-β mediated hepatocyte apoptosis. Cell Death Dis. 2013;4:e466. doi: 10.1038/cddis.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudrand R, Gupta N, Garza AE, Vaidya A, Leopold JA, Hopkins PN, Jeunemaitre X, Ferri C, Romero JR, Williams J, Loscalzo J, Adler GK, Williams GH, Pojoga LH. Caveolin 1 modulates aldosterone-mediated pathways of glucose and lipid homeostasis. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart PC, Ratti BA, Mao M, Ansenberger-Fricano K, Shajahan-Haq AN, Tyner AL, Minshall RD, Bonini MG. Caveolin-1 regulates cancer cell metabolism via scavenging Nrf2 and suppressing MnSOD-driven glycolysis. Oncotarget. 2016;7:308–322. doi: 10.18632/oncotarget.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Rojo MA, Gongora M, Fitzsimmons RL, Martel N, Martin SD, Nixon SJ, Brooks AJ, Ikonomopoulou MP, Martin S, Lo HP, Myers SA, Restall C, Ferguson C, Pilch PF, McGee SL, Anderson RL, Waters MJ, Hancock JF, Grimmond SM, Muscat GE, Parton RG. Caveolin-1 is necessary for hepatic oxidative lipid metabolism: evidence for crosstalk between caveolin-1 and bile acid signaling. Cell Rep. 2013;4:238–247. doi: 10.1016/j.celrep.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Guan L, Li S, Jiang Y, Xiong N, Li L, Wu C, Zeng H, Liu Y. Mechanosensitive caveolin-1 activation-induced PI3K/Akt/mTOR signaling pathway promotes breast cancer motility, invadopodia formation and metastasis in vivo. Oncotarget. 2016;7:16227–16247. doi: 10.18632/oncotarget.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassar ZD, Hill MM, Parton RG, Parat MO. Caveola-forming proteins caveolin-1 and PTRF in prostate cancer. Nat Rev Urol. 2013;10:529–536. doi: 10.1038/nrurol.2013.168. [DOI] [PubMed] [Google Scholar]
- 15.Ayala G, Morello M, Frolov A, You S, Li R, Rosati F, Bartolucci G, Danza G, Adam RM, Thompson TC, Lisanti MP, Freeman MR, Vizio DD. Loss of caveolin-1 in prostate cancer stroma correlates with reduced relapse-free survival and is functionally relevant to tumour progression. J Pathol. 2013;231:77–87. doi: 10.1002/path.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer. 2015;15:225–237. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- 17.Nwosu ZC, Ebert MP, Dooley S, Meyer C. Caveolin-1 in the regulation of cell metabolism: a cancer perspective. Mol Cancer. 2016;15:71. doi: 10.1186/s12943-016-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta R, Toufaily C, Annabi B. Caveolin and cavin family members: dual roles in cancer. Biochimie. 2014;107:188–202. doi: 10.1016/j.biochi.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Wang N, Liu P, Peng F, Tang H, Chen Q, Xu R, Dai Y, Lin Y, Xie X, Peng C, Situ H. Caveolin-1, a stress-related oncotarget, in drug resistance. Oncotarget. 2015;6:37135–37150. doi: 10.18632/oncotarget.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faggi F, Chiarelli N, Colombi M, Mitola S, Ronca R, Madaro L, Bouche M, Poliani PL, Vezzoli M, Longhena F, Monti E, Salani B, Maggi D, Keller C, Fanzani A. Cavin-1 and Caveolin-1 are both required to support cell proliferation, migration and anchorage-independent cell growth in rhabdomyosarcoma. Lab Invest. 2015;95:585–602. doi: 10.1038/labinvest.2015.45. [DOI] [PubMed] [Google Scholar]
- 21.Pan X, Wang R, Wang ZX. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol Cancer Ther. 2013;12:1153–1162. doi: 10.1158/1535-7163.MCT-12-0802. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Cai W, Chen Y, Dong L. The CAV1 gene 3’untranslated region single nucleotide polymorphisms are associated with the risk of pulmonary hypertension in Chinese Han chronic obstructive pulmonary patients. Genet Test Mol Biomarkers. 2019;23:634–643. doi: 10.1089/gtmb.2019.0053. [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Lee W, Shen J. Site-2 protease responds to oxidative stress and regulates oxidative injury in mammalian cells. Sci Rep. 2014;4:6268. doi: 10.1038/srep06268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei J, Li W, Yang Y, Lu Q, Zhang N, Bai G, Zhong D, Su K, Liu B, Li X, Wang Y, Wang X. TC-1 overexpression promotes cell proliferation in human non-small cell lung cancer that can be inhibited by PD173074. PLoS One. 2014;9:e100075. doi: 10.1371/journal.pone.0100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han W, Shi L, Ren L, Zhou L, Li T, Qiao Y, Wang H. A nanomedicine approach enables co-delivery of cyclosporin A and gefitinib to potentiate the therapeutic efficacy in drug-resistant lung cancer. Signal Transduct Target Ther. 2018;3:16. doi: 10.1038/s41392-018-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goetze OT, Al-Batran SE, Chevallay M, Mönig SP. Multimodal treatment in locally advanced gastric cancer. Updates Surg. 2018;70:173–179. doi: 10.1007/s13304-018-0539-z. [DOI] [PubMed] [Google Scholar]
- 27.Mahawongkajit P, Tomtitchong P. A survey of early and advanced gastric cancer treatment by surgeons in Thailand. Oncol Rev. 2018;12:369. doi: 10.4081/oncol.2018.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Bu Z, Zhao F, Xiao D. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. 2018;109:65–73. doi: 10.1111/cas.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren C, Chen H, Han C, Fu D, Wang D, Shen M. High expression of miR-16 and miR-451 predicating better prognosis in patients with gastric cancer. J Cancer Res Clin Oncol. 2016;142:2489–2496. doi: 10.1007/s00432-016-2243-z. [DOI] [PubMed] [Google Scholar]
- 31.Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM, Araya JC, Roa JC. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr) 2016;39:23–33. doi: 10.1007/s13402-015-0247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Z, Zhao J, Rong Z, Geng W, Wang Z. MiR-451, a potential prognostic biomarker and tumor suppressor for gastric cancer. Int J Clin Exp Pathol. 2015;8:9154–9160. [PMC free article] [PubMed] [Google Scholar]
- 33.You W, Xu L, Zhang X, Zou H, Shi D, Zhang H, Li J, Chen W, Li R. High-throughput screening identifies miR-451 as a pleiotropic modulator that suppresses gastric cancer metastasis. SLAS Technol. 2017;22:136–143. doi: 10.1177/2211068216675858. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Gong JM, Zhou LL, Sheng JH. MiR-451 as a new tumor marker for gastric cancer. Oncotarget. 2017;8:56542–56545. doi: 10.18632/oncotarget.17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz-Valdivia N, Bravo D, Huerta H, Henriquez S, Gabler F, Vega M, Romero C, Calderon C, Owen GI, Leyton L, Quest AF. Enhanced caveolin-1 expression increases migration, anchorage-independent growth and invasion of endometrial adenocarcinoma cells. BMC Cancer. 2015;15:463. doi: 10.1186/s12885-015-1477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]