Abstract
Biological pathway analysis provides new insights for cell clustering and functional annotation from single-cell RNA sequencing (scRNA-seq) data. Many pathway analysis algorithms have been developed to transform gene-level scRNA-seq data into functional gene sets representing pathways or biological processes. Here, we collected seven widely-used pathway activity transformation algorithms and 32 available datasets based on 16 scRNA-seq techniques. We proposed a comprehensive framework to evaluate their accuracy, stability and scalability. The assessment of scRNA-seq preprocessing showed that cell filtering had the less impact on scRNA-seq pathway analysis, while data normalization of sctransform and scran had a consistent well impact across all tools. We found that Pagoda2 yielded the best overall performance with the highest accuracy, scalability, and stability. Meanwhile, the tool PLAGE exhibited the highest stability, as well as moderate accuracy and scalability.
Keywords: scRNA-seq, Pathway analysis, Gene expression, Benchmark
1. Introduction
With the advance of single-cell RNA sequencing (scRNA-seq) technologies, a growing and large number of studies [1], [2], [3] have been reported for revealing heterogeneity of cellular populations at unprecedented resolution. scRNA-seq analysis enables researchers to uncover more refined and novel cell clusters [4], which have greatly advanced our understanding of cellular states. There were many state-of-art computational tools developed for clustering cells, identifying marker genes, and visualizing scRNA-seq data [5], [6], [7]. However, biological interpretation of the clustering results remains a big challenge [8], [9].
Pathway analysis has been widely used to depict transcriptional heterogeneity and classify disease subtypes [10], [11]. In single-cell studies, pathway activity scores (PASs) analysis has been applied to transform the gene-level data into explainable gene sets representing biological processes or pathways to uncover the potential mechanism of cell heterogeneity [12], [13], [14]. Although GSVA [15] and ssGSEA [16] were designed for bulk RNA-seq data to estimate pathway activity variation of a single sample across all samples, both tools have been extensively applied to perform functional enrichment analyses for scRNA-seq data [17], [18]. Recently, Pagoda2 [19], [20] and Vision [21] were developed for parsing single cell transcriptome data to identify potential cell-type-specific heterogeneity by incorporating prior information from biological pathways or functional gene sets. Both bulk-based and single-cell-based pathway activity transformation algorithms enable to identify functionally analogous cell types [12], stable cell types for characterizing ovarian cells [22], and a gene-set that potential drive cellular differentiation heterogeneity of white adipocytes [14]. Most recently, a web-based platform scTPA (http://sctpa.bio-data.cn/sctpa) was developed to explore transcriptional heterogeneity of cell populations integrating these widely applied PAS analysis tools [23].
A recently published benchmarking study [24] compared the performance of three transcription factor estimators (DoRothEA [25], SCENIC/AUCell [26], and metaVIPER [27]) and three pathway activity estimators (PROGENy [28], GSEA [29], [42] and AUCell [26]) on scRNA-seq data, and found transcription factor and pathway activities effectively preserve cell type-specific variability. To the best of our knowledge, there were no systematic benchmark studies to evaluate the performance of unsupervised PAS transformation algorithms which could enable us to analyze scRNA-seq data on PASs instead of individual genes expression.
To fill this knowledge gap, we systematically evaluated the accuracy, stability, and scalability of seven widely-used PAS tools with 32 real scRNA-seq datasets. We concentrated on the performance of these tools on their ability to dissect meaningful cellular heterogeneity which still retain in reduced dimensionality space and could determine cell types alone through supervised and unsupervised classification. We hope our study can provide a useful guidance for researchers to choose the appropriate method to effectively and accurately analyze their scRNA-seq data from a pathway functional annotation insight.
2. Materials & methods
2.1. Pathway activity score calculation tools
AUCell [26] (Version 1.8.0) is a recovery-based method that allows identity cells with activity score. This statistical method calculates PAS using an area under the recovery curve (AUC) score among all ranked genes in a particular cell. The AUC score estimates the proportion of highly expressed genes in each gene set.
Vision [21] (Version 2.1.0) is an annotation toolkit that uses autocorrelation statistics to identify biological variations across cells. Vision starts to identify closest K-nearest neighbors of each cell for generating a cell–cell K-nearest-neighbor (KNN) graph. PASs in Vision are calculated by averaging expressed genes for each gene set. To account for the influence of sample-level metrics (the number of UMIs/reads per cells), PASs are then corrected by their means and standard deviations. Expression data used in Vision could be scaled and normalized, but not log-transformed.
Pathway and gene set overdispersion analysis (Pagoda2) (Version 0.1.1) [19] is a computational framework that aims to detect cell heterogeneity from scRNA-seq data. This method fits an error model for each cell to depict its properties, and residual variance of each gene in the cell is re-normalized subsequently. Then, the PAS of each gene set is quantified by its first weighted principal component.
Gene set variation analysis (GSVA) (Version 1.35.6) [15] assesses the variation of gene set using the Kolmogorov-Smirnov (K-S) like random walk statistic. GSVA first estimate kernel-based cumulative density for each gene, which uses classical maximum deviation method by default. Then, PASs are calculated from gene density profile by K-S-like statistic.
Single sample gene set enrichment analysis (ssGSEA) (Version 1.35.6) [16] is an extension method of Gene Set Enrichment Analysis (GSEA) which could transform gene expression into PAS profile without phenotype labeling. ssGSEA ranks gene expression within each cell separately, then the PAS of each pair of cell and gene set is calculated by an enrichment score using K-S like random walk statistic.
Combined z-score (z-score) (Version 1.35.6) [30] is a classical strategy to aggregate expression of several genes. The gene expression is scaled by mean and standard deviation over cells. Then, the PASs for each cell are calculated by averaging scaled gene expression of all genes within each gene set.
Pathway level analysis of gene expression (PLAGE) (Version 1.35.6) [31] captures PASs from singular value decompositions (SVD) strategy. PLAGE first standardizes gene expression matrix across cells. For a submatrix which genes in a particular gene set, the first coefficient of right-singular vector in SVD of this matrix is extracted as PAS.
2.2. Datasets
We collected 32 data sets containing distinct cell compositions, which were widely used in benchmark studies on scRNA-seq (Supplemental Table S1). All data sets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) or hemberg-lab (https://hemberg-lab.github.io/scRNA.seq.datasets/). These data sets were derived from six different biological organs of pancreas, liver, lung, stem cell, peripheral blood, and brain across human and mouse based on 16 scRNA-seq experimental protocols (Supplemental Table S1). Of note, 15 data sets were generated from Unique Molecule identifier (UMI) -based protocols and the other 17 data sets were generated without UMI. These collected gene expression profiles had different matrices including counts data and normalized data (TPM, CPM, or RPM). The mean number of detected genes of each cell across all data sets ranged from 2,180 to 11,006. The resource of cell types of each dataset including mixed cell lines, sorted by fluorescence activated cell sorting (FACS), gathered from different time points, or clustering were provided by their original studies. Details for each data set are described in Supplemental Table S1.
2.3. Accuracy
To evaluate ability of these tools on meaningfully extracting transcriptional heterogeneity, we assessed that the cell-type-specific obtained from PAS should be retained in dimensional reductional space and could assign cells into cell populations through unsupervised clustering or supervised classification [20], [24], [26], [32], [33], [34], [35], [36], [37], [38], [39], [40]. Therefore, the accuracy of PAS transformation methods were assessed by three methods of dimensional reduction, clustering and cell type annotation. The pathway used in evaluation of accuracy incorporates 186 KEGG pathways generated from MSigDB (Version 7.1) (Supplemental Table S3).
2.3.1. Dimensional reduction
We applied the R package Seurat to perform dimensional reduction on PAS matrix. Two dimensions were achieved by Uniform Manifold Approximation and Projection (UMAP) with “method = ‘umap-learn’” on the first 10 principal component (PCs). We then used the silhouette function in R package cluster to execute silhouette analysis. Averaged silhouette width across all cells was used to evaluate the performance of dimensional reduction for each data set.
2.3.2. Clustering accuracy
We accessed the performance of seven tools on unsupervised clustering using Louvain clustering which is a hierarchical algorithm in igraph R package. Louvain clustering algorithm executed on first 10 PCs which was also used in dimensional reduction. In each data set, the number of predicted clusters were set to the same number of the known cell types. We used the adjustedRandIndex function in mclust package to calculate adjusted rand index (ARI).
2.3.3. Cell type annotation
We evaluated the utility of PASs using a multinormal logistic regression model and stratified cross-validation implemented in the python package scikit-learn. The inverse of regularization strength of multinormal logistic regression model was set to 1. The parameter k of cross-validation was set to 5. Before training and evaluating model, PASs of training set was scaled to acquire values between 0 and 1, and the parameters of scaler were spread to test set in the stage of validation.
2.4. Best choice of preprocessing procedure
The impact of preprocessing procedures of PAS transformation algorithms was assessed by their accuracy. This preprocessing includes two steps as follow:
-
(1)
Filtering for genes expressed in<5% of cells, as previous reported methods [41], [42].
-
(2)
Three typical methods of normalization:
-
i)
Log: standard log-normalization with subsequent scaling included in Seurat package (Version 3.1.4) [7];
-
ii)
Scran: a deconvolution strategy implemented by scran package (Version 1.10.2) [43];
-
iii)
Sctransform: variance-stabilizing transformation wrapper in Seurat package (Version 3.1.4) [44]. We also investigated whether the performance of seven tools differed between the filtering strategies using student’s t test and normalizations using two-way ANOVA test. For the detailed information of these normalization procedures, please refer to official tutorials and our Github repository (https://github.com/sulab-wmu/PASBench).
2.5. Stability
2.5.1. Correlation between two matrices
The commonly detected rows and columns among two matrices were used for computing their Spearman correlation. For each row, considering the diverse distribution of matrices, the Spearman correlation was calculated. Then, the median of correlation score across all rows was represented as the correlation score of these two matrices.
2.5.2. Correlations between runs of drop-out events
We randomly dropped a given percent of expressed genes per cell and calculated the correlations between PAS matrices generated from dropped data sets and original data sets. The correlation between PAS profiles was computed as described in Section 2.6.1. To eliminate stochastic effects, genes were randomly dropped out 10 times for a given drop-ratio.
2.5.3. Correlations between different sequencing technologies
We assessed the stability of seven tools by calculating the correlations across 13 data sets based on 13 sequencing technologies. Before calculating correlations between each pair of technologies, PASs across cells in each cell population were averaged. Then the correlation between technologies, with the row in pathways and col in cell clusters, was calculated as described previously (see Section 2.6.1). For each tool, the correlation scores across all pairs of different technologies were averaged as the final correlation score. In the end, the corrplot package in R (Version 0.84) was used to visualize the correlations across technologies for seven tools, respectively.
2.6. Scalability
Total six data sets were randomly sampled from two data sets generated by UMI-based 10x Genomics protocol and nonUMI-based Smart-Seq2 protocol. UMI-based data set was collected from Zheng et al. 68 k data set [45] and nonUMI-based data set was collected from Lei et al. Smart-Seq2 data set [22]. These sampled data sets included 10 k (k = 1,000) and 20 k genes with 5 k, 10 k and 20 k cells, which were widely applied in existing scRNA-seq analyses. In addition, two gene sets collections (Gene set #1, N = 50 pathways, and Gene set #2, N = 200 pathways) were randomly selected from chemical and genetic perturbations collection in MSigDB (Version 7.1) [46]. Each task was equally allocated four CPU core of centos. For each tool, memory usage was measured by maximum memory-used after every step. Memory consumption information was captured by the gc() function in R. When calculating running time, time consuming from data and package loading, pre- and post-processing steps was excluded. Running time was measured by the System.time() function in R.
2.7. Overall performance score
The overall performance score was aggregated three different performance scores yielded from estimating the accuracy, stability and scalability of each PAS transformation algorithm. For accuracy, scaled mean silhouette widths, scaled ARI, and scaled classification accuracy were aggregated to obtain the accuracy score. For stability, aggregated score across different runs (dropouts and technologies) was represented the stability score of each tool. For the scalability, we first scaled running time and memory usage to obtain a value between 0 and 1, respectively. Then, we averaged scaled running time and memory usage for assessing the scalability score. Finally, both averaged and median performance scores were yielded to represent the overall performance of each PAS calculator, respectively.
3. Results
3.1. Evaluation scheme
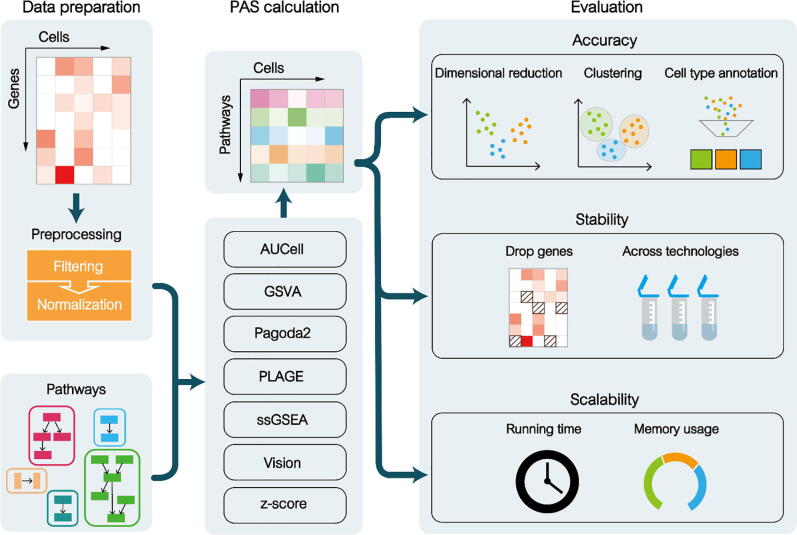
The schematic of the benchmarking framework is shown in Fig. 1. We systematically searched the literature from the PubMed database to gather all functional analysis tools that could aggregated an ensemble of related genes for scRNA-seq data in an unsupervised manner. By using a reviewing process with appropriate exclusion criteria, we examined methods with functional analysis algorithms for possible inclusion. The exclusion criteria used in the current benchmark study as follows (see Table 1 for details): (1) superseded by an updated method; (2) running too slow (cost >2 h for Test Data, see Table 1 for details); (3) not implemented in R/Python; (4) non-extensible. Therefore, a total of seven state-of-art methods that could combine gene profiles with pathways (or gene sets) for calculating pathway activity scores (PAS) were included. We performed a systematic evaluation and comparison of their performances from three different aspects: accuracy, stability, and scalability.
Fig. 1.
An evaluation framework for benchmarking pathway activity score (PAS) calculators. The seven widely applied PAS inference algorithms were assessed on 32 well-defined benchmark data sets. These algorithms combined prior knowledge (biological pathways or functional gene sets) with a statistic method to aggregate gene-level matrix into PAS-level matrix. The accuracy (take into account three downstream applications), stability of results (in the presence of dropout events and across technologies), and scalability (running time and memory usage) were used to systematically evaluate these algorithms.
Table 1.
Overview of available PAS tools included in this benchmarking.
| Name | Date | Platform | Description / Exclusion reason | Inclusion | Reference |
|---|---|---|---|---|---|
| PLAGE | 2005 | R* | Singular value decomposition | True | [31] |
| z-score | 2008 | R* | Combined z-score | True | [30] |
| ssGSEA | 2009 | R* | Kolmogorov-Smirnov-like rank statistic based on gene expression of single sample | True | [16] |
| GSVA | 2013 | R | Kolmogorov-Smirnov-like rank statistic based on kernel estimation of the cumulative density | True | [15] |
| Pagoda2 | 2017 | R | First principal component of gene sets | True | [19] |
| AUCell | 2017 | R | Area under the ranked gene expression curve | True | [26] |
| Vision | 2019 | R | Summarizing the normalized expression of genes in the gene sets | True | [21] |
| ROMA | 2016 | R/Python/Matlab | Running time is too slow (costs 2.8 h on Test Data* with 4 cores) | False | [55] |
| f-scLVM | 2017 | R | Running time is too slow (costs 4.3 h on Test Data*) | False | [56] |
| PROGENY | 2018 | R | Non-extensible (This method only inferred pathway activity scores for predefined 14 signaling pathways) | False | [28] |
| Single Cell Signature Explore | 2019 | GO | not implemented in R/Python | False | [57] |
Note: R*: original article did not have implemented it, cooperated in R package GSVA; Test Data*: 33,694 genes × 10000 cells, combining with KEGG database.
3.2. Impact of RNA-seq data preprocessing
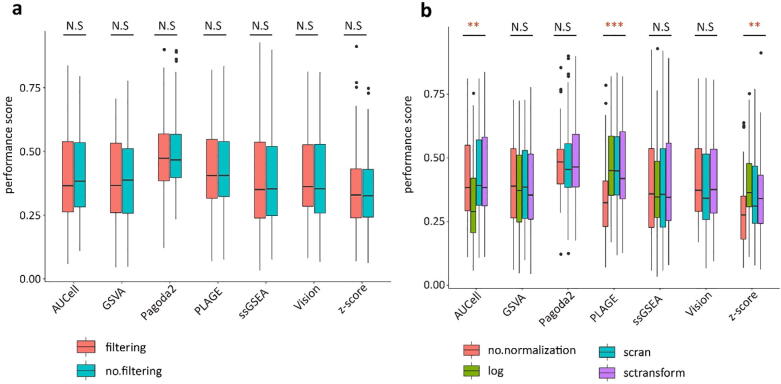
Because of the high abundance of zeros and poor signal-to-noise ratio in scRNA-seq data, it is in general assumed that preprocessing including filtering low quality genes and data normalization contribute to uncover cellular identities. Thus, we determined whether preprocessing before PAS calculation would improve the performance of PAS tools. We compared the performance of seven tools with two filtering strategies and four normalization strategies across all curated data sets. We found filtering had non-significant impact on the performance of PAS transformation algorithms (Fig. 2a, all p-values > 0.05). The performance of AUCell, PLAGE and z-score were significantly influenced by different procedures of normalization (Fig. 2b, p-values = 3.3 × 10−4, 3.1 × 10−6, and 2.8 × 10−4, separately), while the other four tools showed non-significant differences among four normalization procedures. Compared with log-transform and without normalization, the sctransform and scran normalization yielded better performances across all tools (Fig. 2b). Furthermore, Pagoda2 and Vision had a potent risk to report error when using log-transform method to normalize expression data for calculating PAS. An earlier study mentioned that sctransform outperformed other normalization tools in scRNA-seq analysis [47]. Therefore, we examined the performance of these PAS tools based on sctransform-normalized data without gene filtering in this study.
Fig. 2.
Distribution of averaged performance score across filtering strategies (a) or normalization strategies (b). Results of significance testing are characteristic in dash line for each tool. N.S no significance, *p < 0.0, ** p < 0.001, *** p < 0.0001.
3.3. Evaluation of accuracy of methods
3.3.1. Dimensional reduction
By using the UMAP method to generate two dimensions for seven tools, we used dimensional reduction (DR) to investigate the accuracy of these PAS transformation algorithms. We applied mean silhouette widths, as the metric for assessing the separation between cell types based on PASs. In all datasets, we found the Pagoda2 achieved the best performance of DR (DR = 0.82), and the PLAGE (DR = 0.6), AUCell (DR = 0.59), and Vision (DR = 0.53) performed reasonably better than the other three tools (Fig. 3). Notably, the Pagoda2 remained to be the best one in both UMI-based datasets (DR = 0.89) and nonUMI-based datasets (DR = 0.76). Pagoda2, PLAGE, and Vision gained a slightly better performance in UMI-based dataset than that in nonUMI-based datasets, while AUCell and GSVA outperformed their performances in nonUMI-based datasets compared with in UMI-based datasets (Fig. 3). The z-score method showed the worst performance in all datasets, UMI-based datasets, and nonUMI-based datasets (DR < 0.2). Furthermore, in silico evaluation (Supplemental Fig. S1, Supplemental Note 1, and Supplemental Table S2), Pagoda2 was demonstrated to be the most robust tool, which was slightly influenced by the library size. Four tools (z-score, ssGSEA, GSVA, and PLAGE) developed for bulk-seq data sets were more sensitive to the library size compared with others (Vision, AUCell, Pagoda2) designed for single-cell data sets. The number cells have non-significant influence on the performance of PAS tools (Supplemental Fig. S2).
Fig. 3.
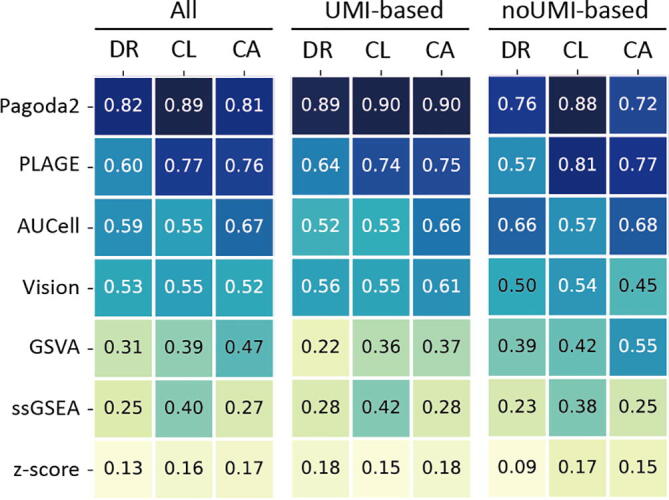
Evaluation accuracy of seven PAS tools on all data sets, UMI-based data sets and nonUMI-based data sets. Accuracy of seven PAS tools were assessed in dimensional reduction (DR), clustering (CL), and cell type annotation (CA). Heatmap of three metrics – mean silhouette width, adjusted rand index (ARI) and classification accuracy – averaged across all datasets. To compare the performance of seven tools, metrics was scaled to a value between 0 and 1 before averaged. A higher scaled score represents a better performance. PAS tools ranked by their performance of DR across all data sets are shown in the plot.
3.3.2. Clustering
We compared the performance of these PAS transformation algorithms for clustering (CL) analysis. Graph-based Louvain algorism was used for unsupervised clustering, and adjusted rand index was selected as evaluation metric. We found Pagoda2 had the best performance of CL among all datasets (CL = 0.89), UMI-based datasets (CL = 0.90), and nonUMI-based datasets (CL = 0.88). PLAGE, AUCell, and Vision yielded a relatively better performance than GSVA, ssGSEA, and z-score among all datasets (Fig. 3). For these seven tools, there was no prominent difference in performances for CL between UMI-based and nonUMI-based datasets.
3.3.3. Cell type annotation
The ability of these PAS transformation algorithms was assessed for cell type annotation (CA). A multinormal logistic regression model with cross validation strategy was employed to train these PASs matrices. Averaged classification accuracy was determined as an evaluation metric for annotating original cell types. Among the seven tools, Pagoda2 had the highest classification accuracy for CA in all datasets (CA = 0.81) and UMI-based datasets (CA = 0.90), while PLAGE performed the best for CA in nonUMI-based datasets (CA = 0.77). AUCell and Vision achieved a moderate classification accuracy for CA in all datasets, UMI-based datasets, and nonUMI-based datasets (CA > 0.5). The GSVA reached a moderate classification accuracy in nonUMI-based datasets (CA = 0.55). Both z-score and ssGSEA showed worse performances for CA in all datasets (CA < 0.3). Altogether, we found Pagoda2 exhibited the best performance from three different aspects of the assessment of accuracy. And PLAGE, AUCell, and Vision reached a moderate performance among all accuracy assessment. Our results indicated that different tools manifested a distinct performance according to UMI-based and nonUMI-based datasets.
3.4. Evaluation of stability of methods
3.4.1. Dropout genes in gene expression profiles
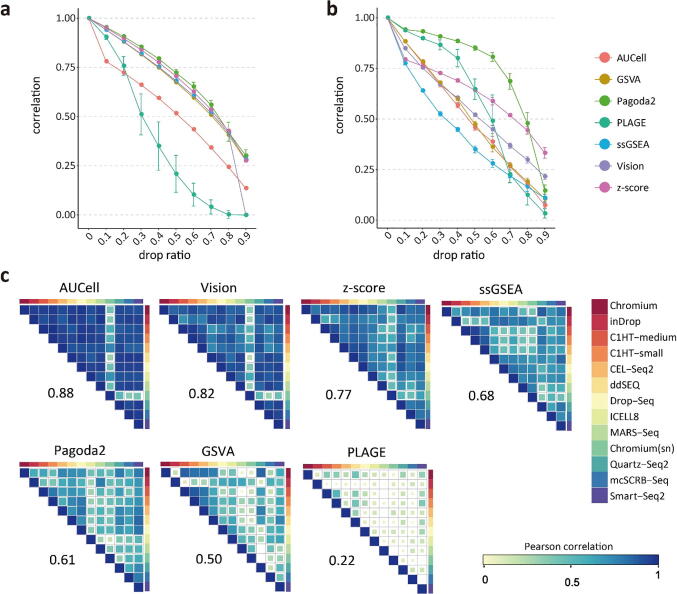
We applied the gene coverage reduction to evaluate the stability of different PAS transformation algorithms. Considering that the number of detected genes were varied across different single-cell data sets, these PAS tools were expected to cope with data sets with various library size [24]. We randomly dropped expressed genes in each cell and calculated average pairwise correlation between PASs derived from dropped data set and original data set (see “Methods”). Generally, compared with other tools, we observed that Pagoda2 had the best stability with genes dropout in both UMI-based (Fig. 4a) and nonUMI-based datasets (Fig. 4b). The standard deviation of Pagoda2 was relatively small. In UMI-based datasets, GSVA, ssGSEA, Vision, and z-score showed a more stable performance than AUCell and PLAGE. In nonUMI-based datasets, the performances of ssGSEA, AUCell, GSVA, and ssGSEA were dropped remarkably with the drop ratio increased.
Fig. 4.
Evaluation stability of PAS tools. a) Spearman correlation between the PASs inferred from original UMI-based data set and dropped data set using seven tools; b) Spearman correlation between the PASs inferred from original nonUMI-based data set and dropped data set using seven tools; c) Spearman correlation of PASs between 13 technologies for each tool.
3.4.2. Stability of different scRNA sequencing protocols
It is not only important that a PAS tool could produce similar results when given sparse matrix, but also that it is able to retrieve relatively similar PASs across different technologies, as they were transformed from same biological context [24], [48]. To do so, we downloaded the human peripheral blood mononuclear cells (PBMCs) from Human Cell Atlas Project, which were generated from 13 different scRNA-seq protocols. To further assess these tools’ stability, Spearman correlations between 13 technology protocols for each PAS tool were estimated. We found that AUCell and Vision showed the higher correlations across 13 technologies (median r = 0.88 and 0.82, respectively). The z-score, ssGSEA, Pagoda2, and GSVA had the moderate correlations across 13 technologies (median r = 0.77, 0.68, 0.61, and 0.5, respectively). The PLAGE was very sensitive to the technology protocols and data distribution (Fig. 4c). Besides, we found that with the increasing drop ratio of genes in pathways/gene sets, the performance of these tools was decreasing. (Supplemental Fig. S7). Altogether, combining the results from dropout genes with correlations across different technologies, we found Pagoda2 showed the most stability than the other tools (Fig. 6).
Fig. 6.
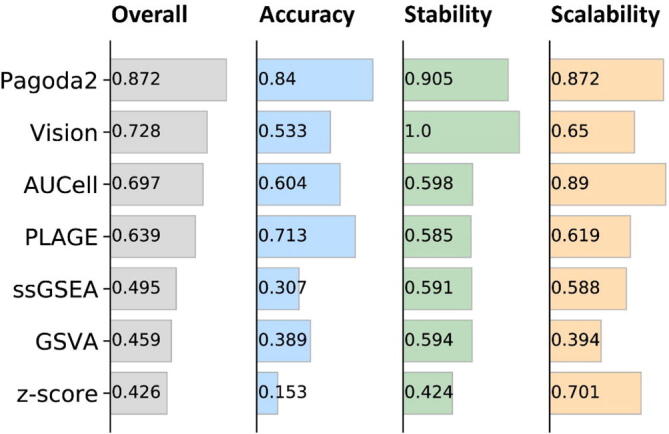
Overall summary of results evaluating PAS calculators for scRNA-seq data. Methods are ranked by overall performance score which was averaged across three categories: accuracy, stability and scalability. Accuracy was averaged across mean silhouette width, ARI and classification accuracy. Stability was averaged across correlations between PASs inferred from data sets across drop-outs and technologies. Running time and memory usage were scaled to a value [0, 1] before averaged as scalability.
3.5. Evaluation of scalability of methods
With the number of cells produced from current protocols were increased gradually, current analysis tools were expected to deal with hundreds of thousands of cells [49], [50]. To assess the scalability, we randomly generated three gene expression data sets consisting of different number of genes and cells from nonUMI-based and UMI-based gene expression profiles, separately. These two parts of data sets were designed to investigate the effect of data sparsity on assessment of scalability (see “Methods”). Cells sampled from Smart-Seq2 expressed 38% genes on average (Fig. 5a), while cells generated from 10x Genomics platform expressed 2% genes on average (Fig. 5b).
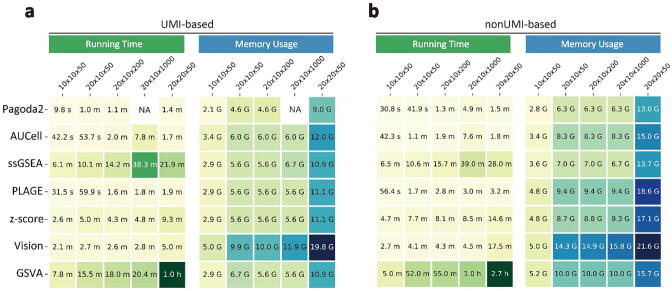
Fig. 5.
Evaluation scalability of PAS tools. Running time and memory usage were evaluated based on varying number of genes, cells, and gene sets (no. of genes × no. of cells × no. of gene sets). The data sets were randomly generated from nonUMI-based data set (a) and UMI-based data set (b). The value with in the brackets represented average percentage of genes detected per cell. NA (not applicable) indicated an error was produced during calculation.
Overall, the maximum running time of seven tools across nonUMI-based data sets (range from 1.5 min to 2.7 h) were longer than that across UMI-based data sets (range from 1.4 min to 1 h). The memory usage of seven tools across nonUMI-based data sets were larger than that across UMI-based data sets. The number of gene sets have no impact on the memory usage in either UMI-based or nonUMI-based data sets (Fig. 5a and 5b). However, we observed that these algorithms required longer time when cope with more gene sets. Running times of AUCell, Pagoda2 and ssGSEA were increased six-fold, four-fold and three-fold separately when the number of gene sets reaching to 1000, while other four tools (GSVA, Vision, PLAGE, and z-score) increased no more than one-fold.
Pagoda2 has the best performance of scalability according to running time and memory usage which cost 1.4 min and 13 Gb when the number of cells reaching to 20,000. GSVA was the most time-consuming method, which takes approximate 45-fold time longer than Pagoda2. Vision used the largest memory overhead value during calculation. The maximum memory usage of Vision was up to 21 Gb on 20 × 20 × 1000 (20,000 genes × 20,000 cells × 1000 gene sets) UMI-based data set while Pagoda2 only need 13 Gb.
Together, these chosen tools had varied performances of the scalability. Pagoda2 was estimated to be the best-performing tool based on running time and memory usage. The number of genes, cells, gene sets collections, and data sparsity would influence the running time of all tools, and the memory consumption mainly be attributed to dimensionality of gene expression profile. The running time of AUCell, Pagoda2 and ssGSEA was more sensitive to the number of gene sets compare to other four tools.
3.6. Overall performance
By aggregating three metrics from the assessment of accuracy, stability, and scalability through mean or median, we yielded an overall performance score for each tool (Fig. 6, Supplemental Figs. S3 and S4). We demonstrated that the relative accuracy of PAS tools was consistent regardless of the choice of dimensional reduction technique, clustering method, or supervised classifier (Supplemental Fig. S5). We found that Pagoda2 achieved the best overall performance score with the highest accuracy, scalability, and stability regardless of the UMI-based data sets or nonUMI-based data sets (Supplemental Figs. S3 and S6). Meanwhile, the tool PLAGE exhibited the highest stability, as well as moderate accuracy and scalability. However, the performance score of these tools varied across evaluation criteria, for example, PLAGE had a better performance than AUCell in term of accuracy while it showed weaker performance in the assessment of stability.
4. Discussion
In this study, we systematically collected seven widely-accepted pathway activity transformation algorithms including four bulk-based and three single-cell-based tools with 32 benchmark datasets. For these benchmark datasets, there were 15 UMI-based and 17 nonUMI-based datasets included. Here, we presented a comprehensive benchmark evaluation to examine the accuracy, stability, and scalability of these seven PAS tools on scRNA-seq data.
Multiple lines of evidence have demonstrated [10], [11] that pathway-based enrichment analysis could help to elucidate biological implications and molecular mechanisms. Thus, a growing number of single-cell-based studies [12], [13], [14] have applied the pathway-based activity score analysis to discover cells heterogeneities. One of advantages of using PASs is that the transformation is based on widely-accepted canonical pathways curated from literature resources such as the KEGG database, which provide a reasonable biological interpretation of cell identities and avoid the process of marker genes selections before training classifiers. In the current benchmarking study, we adopted seven PAS tools based on the KEGG pathways to evaluate their performances on scRNA-seq analysis, and found that Pagoda2 yielded the best performance. Meanwhile, we noticed that different PAS tools achieved a distinct performance according to UMI-based and nonUMI-based datasets, suggesting that researchers should consider the resource of scRNA-seq data before selecting the tools for analysis.
Many studies showed that bulk RNA-seq tools could be applied to analyze scRNA-seq data [24], [51]. For example, z-score, which is embedded in matchScore2 [32], was used for calculating the combined marker-gene-sets activity scores to train a classification model. By using the z-score method, Lee et al. [32] established a classification model to annotate cell populations with a prediction accuracy of 0.9. Recently, Holland et al. [24] evaluated the performance of three pathway analysis tools: PROGENy [28], GSEA [29], [46] and AUCell in their benchmarking study, and found bulk-seq functional analysis tools that rely on manually curated gene sets are effective in calculating pathway activities from scRNA-seq data and partially better than scRNA-seq tools. In the current study, our results showed that the summarized performance across single-cell-based tools (Pagoda2, AUCell, and Vision) outperformed the performance of bulk-based tools (ssGSEA, GSVA, and z-score) from three different aspects of accuracy, stability and scalability, while the bulk-based tool of PLAGE yielded a relatively better performance than Vision. Although Vision showed a moderate performance than Pagoda2, AUCell, and PLAGE, it is an annotation toolkit that could capture biological variation with or without priori labeling of cells. Therefore, researchers could use Vision to investigate differential pathway analysis or other stratification-based analysis.
Since the influence of low-quality counts and systematic noise, filtering and normalization are generally thought to be essential steps in scRNA-seq analysis. In the present study, we found filtering low quality genes had non-significant influence on the performance of PAS transformation tools, but AUCell, PLAGEA, and z-score were remarkably affected by normalization with distinct methods. Our results suggested that PAS methods based on a subset of functional genes might be stable for cell clustering compared with these based on gene-level expression. Furthermore, we found that in a noisy data set, Pagoda2 mitigated the scRNA-seq batch effects well and obtained the differences between biological cell identities (Supplemental Fig. S8, Supplemental Note 1).
Several studies showed that gene regulator network (GRN) could extract an ensemble of genes that represents a particular combination of transcription factors to be used for functional interpreting and annotating scRNA-seq data [24], [25]. These GRN tools enables to combine with PAS tools to offer more dataset-specific information [26], such as SCENIC. Although there were some outstanding benchmarking studies evaluated the performance of GRN tools [24], [25], [50], they only focused on the accuracy of GRN tools or included a subgroup of GRN tools and PAS tools. Further benchmarking studies are warranted to systematically compare the performance between GRN tools and PAS tools or investigate which combinations of these state-of-art tools could provide more efficient and robust transcriptional heterogeneity. To have a glance at this field, we extended our research on more well-defined pathways/gene sets, as well as the dataset-specific gene sets generated from SCENIC. As shown in Supplemental Fig. S9, there were no consistent best pathways or gene sets for every tools. Though, Pagoda2 still achieved the best performance across most pathways and gene sets. Benefiting from the establishment of Human Cell Atlas [52], Mouse Cell Atlas [53], and Tubula Muris [54], researchers could unbiasedly investigate the ability of PASs on supervised cell type annotation across technologies, organs and species, which might further facilities the automatic functional annotation of single cells from transcriptomics perspective.
In summary, we performed a systematical evaluation of seven widely-used PAS tools from three different aspects of accuracy, stability, and scalability based on 32 well-defined benchmark datasets. Compared with other tools, Pagoda2 showed the best overall performance. AUCell and PLAGE achieved a relatively better overall performance. We hope our current benchmarking study will assist computational researchers to select appropriate methods and design high-quality researches that will contribute to scientific advances.
CRediT authorship contribution statement
Yaru Zhang: Conceptualization, Methodology, Software, Writing - original draft, Writing - review & editing. Yunlong Ma: Methodology, Software, Writing - original draft, Writing - review & editing. Yukuan Huang: Investigation, Resources, Data curation. Yan Zhang: Investigation, Resources, Data curation. Qi Jiang: Investigation, Resources, Data curation. Meng Zhou: Investigation, Resources, Data curation. Jianzhong Su: Conceptualization, Methodology, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (61871294 and 61902352), and Science Foundation of Zhejiang Province (LR19C060001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 2.Method of the Year 2013. Nat Methods 2014;11:1. https://doi.org/10.1038/nmeth.2801. [DOI] [PubMed]
- 3.Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018;13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 4.Beumer J., Puschhof J., Bauzá-Martinez J., Martínez-Silgado A., Elmentaite R., James K.R. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell. 2020:1–16. doi: 10.1016/j.cell.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Lafzi A., Moutinho C., Picelli S., Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 2018;13 doi: 10.1038/s41596-018-0073-y. [DOI] [PubMed] [Google Scholar]
- 6.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lähnemann D., Köster J., Szczurek E., Mccarthy D.J., Hicks S.C. Eleven grand challenges in single-cell data science. Genome Biol. 2020 doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiselev V.Y., Andrews T.S., Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2020;217:1–15. doi: 10.1084/jem_20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M., Zhang M., Hu S., Hu S., Min M., Ni Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut. 2020:1–12. doi: 10.1136/gutjnl-2019-320368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding H., Blair A., Yang Y., Stuart J.M. Biological process activity transformation of single cell gene expression for cross-species alignment. Nat. Commun. 2019;10:1–6. doi: 10.1038/s41467-019-12924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S., Zheng Y., Li J., Yu Y., Zhang W., Song M. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 2020;180(585–600) doi: 10.1016/j.cell.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez A.K., Dankel S.N., Rastegarpanah B., Cai W., Xue R., Crovella M. Single-cell transcriptional networks in differentiating preadipocytes suggest drivers associated with tissue heterogeneity. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-16019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänzelmann S., Castelo R., Guinney J.G.S.V.A. Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbie DA, Tamayo P, Boehm JS, Kim SY, Susan E, Dunn IF, et al. Processing-a-Programming-Handbook-for-Visual-Designers-and-Artists.Pdf 2010;462:108–12. https://doi.org/10.1038/nature08460.Systematic.
- 17.Celiku O., Gilbert M.R., Lavi O. Computational modeling demonstrates that glioblastoma cells can survive spatial environmental challenges through exploratory adaptation. Nat. Commun. 2019;10:5704. doi: 10.1038/s41467-019-13726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang R, Cheng S, Luo N, Gao R, Yu K, Kang B, et al. Distinct epigenetic features of tumor- reactive CD8 + T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis 2020:1–13. [DOI] [PMC free article] [PubMed]
- 19.Yung YC, Duong TE, Gao D, Chun J, Kharchenko P V. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain 2018;36:70–80. https://doi.org/10.1038/nbt.4038.Integrative. [DOI] [PMC free article] [PubMed]
- 20.Kaper F, Fan J, Zhang K, Chun J, Peter V. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis 2016;13:241–4. https://doi.org/10.1038/nmeth.3734.Characterizing. [DOI] [PMC free article] [PubMed]
- 21.DeTomaso D., Jones M.G., Subramaniam M., Ashuach T., Ye C.J., Yosef N. Functional interpretation of single cell similarity maps. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-12235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181(442–459) doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 23.Su J, Zhang Y, Yu F, Zhang Y, Zhang J, Guo F, et al. scTPA: A web tool for single-cell transcriptome analysis of pathway activation signatures. 2020. https://doi.org/10.1101/2020.01.15.907592. [DOI] [PubMed]
- 24.Holland C.H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M.P., Mereu E. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:1–19. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aibar S, González-blas CB, Moerman T, Huynh-thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-cell regulatory network inference and clustering 2018;14:1083–6. https://doi.org/10.1038/nmeth.4463.02200317. [DOI] [PMC free article] [PubMed]
- 27.Ding H., Douglass E.F., Sonabend A.M., Mela A., Bose S., Gonzalez C. Quantitative assessment of protein activity in orphan tissues and single cells using the metaVIPER algorithm. Nat. Commun. 2018;9:1471. doi: 10.1038/s41467-018-03843-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018;9 doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 30.Lee E., Chuang H.Y., Kim J.W., Ideker T., Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomfohr J., Lu J., Kepler T.B. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinf. 2005;6:1–11. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mereu E., Lafzi A., Moutinho C., Ziegenhain C., McCarthy D.J., Álvarez-Varela A. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020;38:747–755. doi: 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
- 33.Cole M.B., Risso D., Wagner A., DeTomaso D., Ngai J., Purdom E. Performance assessment and selection of normalization procedures for single-cell RNA-Seq performance assessment and selection of normalization procedures for single-cell RNA-Seq. Cell. Syst. 2019;8(315–328) doi: 10.1016/j.cels.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S., Zhu J., Ma Y., Zhou X. Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 2019;20:1–21. doi: 10.1186/s13059-019-1898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou W., Ji Z., Ji H., Hicks S.C. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 2020;21:218. doi: 10.1186/s13059-020-02132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germain P.L., Sonrel A., Robinson M.D. pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single cell RNA-seq preprocessing tools. Genome Biol. 2020;21:227. doi: 10.1186/s13059-020-02136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L., Dong X., Freytag S., Lê Cao K.A., Su S., JalalAbadi A. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods. 2019;16:479–487. doi: 10.1038/s41592-019-0425-8. [DOI] [PubMed] [Google Scholar]
- 38.Li C., Liu B., Kang B., Liu Z., Liu Y., Chen C. SciBet as a portable and fast single cell type identifier. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-15523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao Z., Moreno P., Huang N., Papatheodorou I., Brazma A., Teichmann S.A. Putative cell type discovery from single-cell gene expression data. Nat. Methods. 2020;17:621–628. doi: 10.1038/s41592-020-0825-9. [DOI] [PubMed] [Google Scholar]
- 40.Tsuyuzaki K., Sato H., Sato K., Nikaido I. Benchmarking principal component analysis for large-scale single-cell RNA-sequencing. Genome Biol. 2020;21:1–17. doi: 10.1186/s13059-019-1900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Praktiknjo S.D., Obermayer B., Zhu Q., Fang L., Liu H., Quinn H. Tracing tumorigenesis in a solid tumor model at single-cell resolution. Nat. Commun. 2020;11. doi: 10.1038/s41467-020-14777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lun ATL, Mccarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor [version 2 ; referees : 3 approved , 2 approved with reservations]. F1000Research 2016. [DOI] [PMC free article] [PubMed]
- 44.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:1–15. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8 doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Germain P.-L., Sonrel A., Robinson M.D. pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single-cell RNA-seq preprocessing tools. BioRxiv. 2020;2020(02) doi: 10.1101/2020.02.02.930578. 02.930578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mereu E, Lafzi A, Moutinho C, Ziegenhain C, Maccarthy DJ, Alvarez A, et al. Benchmarking Single-Cell RNA Sequencing Protocols for Cell Atlas Projects n.d. [DOI] [PubMed]
- 49.Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 50.Pratapa A., Jalihal A.P., Law J.N., Bharadwaj A., Murali T.M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods. 2020;17:147–154. doi: 10.1038/s41592-019-0690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stegle O., Teichmann S.A., Marioni J.C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 52.Paper W. The human cell atlas [October 2018] 2017.
- 53.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S. Mapping the mouse cell atlas by microwell-Seq. Cell. 2018;172(1091–1107) doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martignetti L., Calzone L., Bonnet E., Barillot E., Zinovyev A. ROMA: Representation and quantification of module activity from target expression data. Front. Genet. 2016;7:1–12. doi: 10.3389/fgene.2016.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buettner F., Pratanwanich N., McCarthy D.J., Marioni J.C., Stegle O. f-scLVM: Scalable and versatile factor analysis for single-cell RNA-seq. Genome Biol. 2017;18:1–13. doi: 10.1186/s13059-017-1334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pont F., Tosolini M., Fournié J.J. Single-cell signature explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.