Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Viral load
Abstract
Background
It is necessary to know the viral kinetics and conduct epidemiological investigations of confirmers to prevent the spread of the new infectious disease COVID-19 to the community. To date, no study has been published on viral kinetics during the preclinical and clinical periods of SARS-CoV-2.
Methods
A confirmed case was defined as a patient with positive results by real-time reverse transcription polymerase chain reaction (RT-PCR) assay for SARS-CoV-2. Both specimen types were collected over the whole clinical course in all patients. Asymptomatic patients who had been screened for COVID-19 due to a strong epidemiological link were also enrolled. The study population included 54 hospitalized patients with confirmed COVID-19.
Results
COVID-19 shows a very high viral load on the day of symptom development, which then decreases overall. Rapid viral proliferation was observed 0–5 days before symptoms developed. Cycle threshold (Ct) value was the lowest in the clinical course from 5 days before symptoms to 10 days after symptoms occurred (Ct < 30). The rRT-PCR results were negative approximately 3 weeks after the onset of symptoms. However, there was a continuous pattern that was negative and positive for up to 6 weeks and more.
Conclusion
Considering the characteristic that COVID-19 has a high viral load before symptoms appear, it is necessary to consider to expand the scope of epidemiological investigations. As there is a very low possibility of transmission after 10 days of symptom occurrence, it may be considered to release isolation after 10 days of symptom occurrence in limited resource situations. This study allows for the planning of epidemiological investigations, patient's ward supply, and follow-up of patients through sequential changes in viral loads over the entire clinical course. In addition, it is possible to estimate the clinical time at which the patient is present.
Introduction
Coronavirus disease-2019 (COVID-19), which started in Wuhan in December 2019, has spread worldwide. As of 20 May 2020, the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (genus Betacoronavirus, family Coronaviridae) has been reported in 227 countries with more than 4,789,205 confirmed cases and 318,789 deaths (World Health Organization, 2020a). Contact screening and extensive testing are proposed methods of blocking the transmission of infectious diseases during pandemics.
It is necessary to know the viral kinetics and conduct epidemiological investigations of confirmers to prevent the spread of COVID-19. To date, there have been no published data on viral loads before and after the onset of symptoms. The purpose of this study was to infer viral kinetics, including preclinical, clinical, and post-clinical periods, to assist in epidemiological investigations, prevent transmission, and predict the patient's progress.
Methods
Case definition
A confirmed case was defined as a patient with positive results by rRT-PCR assay for SARS-CoV-2 in upper respiratory specimens (nasopharyngeal and oropharyngeal swabs), with or without a lower respiratory specimen (sputum). Both specimen types were collected over the whole clinical course in all patients. Patients who had no symptoms, but had been screened for COVID-19 due to a strong epidemiological link, such as family members, were also included upon laboratory confirmation.
RNA extraction
rRT-PCR was used to detect SARS-CoV-2. RNA was extracted from clinical samples with the QIAamp Viral RNA Mini kit (QIAGEN, Hilden, Germany), QIAsymphony RNA Kit (QIAGEN, Hilden, Germany), and ExiPrep 16 Dx (Bioneer, Korea); manufacturer’s instructions were followed. All specimens were handled in accordance with the laboratory biosafety guidelines of the Korea CDC for SARS-CoV-2.
rRT-PCR
The optimal concentration of primers and probes was synthesized by using published sequences determined with the RNA transcripts of SARS-CoV-2. The primer and probe sequences for RNA-dependent RNA polymerase gene detection were as follows: 5′-GTGARATGGTCATGTGTGGCGG-3′ (Forward), 5′-CAR ATGTTAAASACACTATTAGCATA-3′ (Reverse) and 5′-CAGGTGGAACCTCATCAGGAGATGC-3′ (probe in 5-FAM/3′-BHQ format). Primer and probe sequences for the E gene detection were: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ (Forward), 5′-ATATTGCAGCAGTACGCACACA-3′ (Reverse), and 5′-ACACTAGCCATCCTTACTGCGCTTCG-3′ (probe in 5-FAM/3′-BHQ format). A 20 μL reaction contained 5 μL of template RNA, 11 μL of RT-PCR Premix, and 4 μL of each primer/probe mix. Thermal cycling was performed at 50 °C for 30 min for reverse transcription, followed by the inactivation of the reverse transcriptase at 95 °C for 10 min. PCR amplification was performed with 40 cycles of 95 °C for 15 s and 60 °C for 1 min in the CFX-96 real-time PCR detection system (Bio-Rad). Ct values were reported.
Ethics statement
The Institutional Review Board (IRB) at Dankook University Hospital reviewed and approved the study protocol (IRB registration No. 2020-03-029−001); the requirement for written consent was waived.
Results
The study population included 54 hospitalized patients with confirmed COVID-19 (Table 1 ). The median age of the 54 patients was 45 years (interquartile range, 40–52 years; range, 2–81 years) and 33 (61.1%) were women. Of the 54 patients, 17 (31.5%) had one or more comorbid medical conditions; 41.2% of patients were hypertensive. Eleven cases (20.4%) were asymptomatic when they were confirmed to have COVID-19.
Table 1.
Clinical characteristics of 54 persons with diagnosed coronavirus diseasea.
| Characteristics | No. (%) |
|---|---|
| Age, y (mean + standard deviation) | 47.0 + 13.0 |
| Sex | |
| M | 21 (38.9) |
| F | 33 (61.1) |
| Preexisting conditions | 17 (31.5) |
| Hypertension | 7 (13.0) |
| Diabetes | 6 (11.1) |
| Chronic obstructive pulmonary disease | 1 (1.9) |
| Congestive heart failure | 0 |
| Chronic kidney disease | 0 |
| Chronic liver disease | 2 (3.7) |
| Malignancy | 0 |
| Body mass index, kg/m2b | 23.6 + 3.2 |
| Symptoms at admission | |
| Fever (>37.5 °C) | 17 (31.5) |
| Chills | 9 (16.7) |
| Cough | 21 (38.9) |
| Sputum | 14 (25.9) |
| Dyspnea | 5 (9.3) |
| Rhinorrhea | 9 (16.7) |
| Sore throat | 9 (16.7) |
| Nausea | 1 (1.9) |
| Diarrhea | 4 (7.4) |
| Abdominal pain | 1 (1.9) |
| Myalgia | 14 (25.9) |
| Headache | 5 (9.3) |
| Anosmia or taste abnormality | 4 (7.4) |
| MEWS(Modified Early Warning Score)† | 1.5 + 0.91 |
| Laboratory findings | |
| Blood leukocyte count, reference range 4.0–11.0 × 109/L | |
| <4.0 × 109/L | 9 (16.7) |
| >4.0 × 109/L | 45 (83.3) |
| Lymphocyte count, reference range 1.0–3.4 × 109/Lb | 1.463 + 0.634 × 109/L |
| Lymphopenia, <1.0 × 109/L | 12 (22.2) |
| Platelet count, reference range 182–369 × 109/L | |
| <150 × 109/L | 7 (13.0) |
| >150 × 109/L | 47 (87.0) |
| Hemoglobin, reference range 11.2–15.7 g/dLb | 13.8 + 1.23 g/dL |
| C-reactive protein level >10 mg/L | 12 (22.2) |
| Procalcitonin level >0.5 ng/mL | 1 (1.9) |
| Lactate dehydrogenase >250 IU/L | 24 (44.4) |
| Creatinine >133 μmol/L | 1 (1.9) |
| Alanine aminotransferase >40 IU/L | 7 (13.0) |
| Infiltration in chest x-ray | 26 (48.1) |
| Infiltration in computed tomography | 28 (51.9) |
| Supplementary oxygen | 6 (11.1) |
Characteristics and clinical findings at the time of isolation or hospital admission.
Mean + standard deviation.
During hospitalization, pneumonia was present in 28 patients (51.85%). Six patients (11.1%) required oxygen supplement therapy: four with nasal cannula, one with facial mask, and one with high flow nasal cannula oxygen therapy. No patients required mechanical ventilator or ECMO therapy. Of 28 patients treated with pneumonia, 1 patient received systemic corticosteroid for the management of acute respiratory distress syndrome.
By May 29, 48 patients were off isolation or discharged. The median day of off-isolation/discharge was 25.0 days after symptom onset (range, 12–49 days).
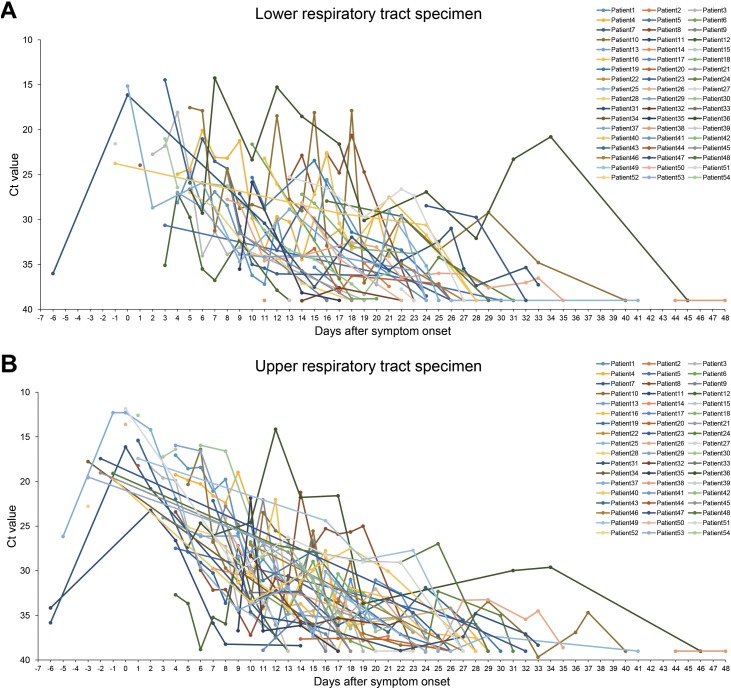
We evaluated viral kinetics by serial rRT-PCR of respiratory specimens from 54 patients (Figure 1 ). All patients were sampled regularly by nasopharyngeal swabs, lower respiratory specimens, and within the sputum (Figure 1). RT-PCR results from 6 days before symptoms to 48 days after symptoms were confirmed.
Figure 1.
Changes in the SARS-CoV-2 Ct value of rRT-PCR in respiratory specimens. (A) Changes in the Ct value of SARS-CoV-2 RNA (RNA-dependent RNA polymerase gene, RdRP) in lower respiratory specimens in 54 patients with COVID-19. (B) Changes in the Ct value of SARS-CoV-2 RNA (RdRP) in nasopharyngeal specimens.
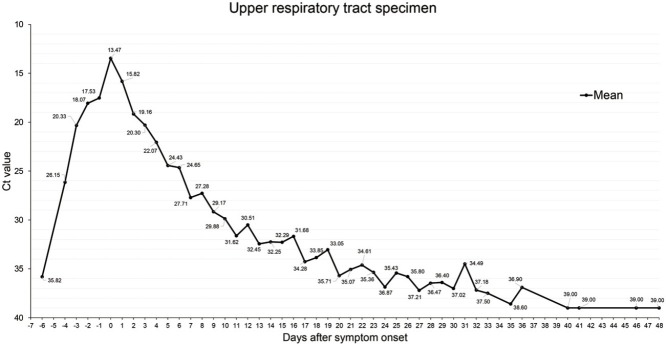
We classified the patient's test results from the date of symptom occurrence and calculated the average Ct value (Figure 2). The Ct value on the day of symptom development was the lowest at 13.47 (11.85∼16.13). The Ct value was lowest in the clinical course from 5 days before symptoms to 10 days after the occurrence of symptoms (Ct < 30).
Figure 2.
Changes in the SARS-CoV-2 Ct value of rRT-PCR in respiratory specimens. The calculated value as the mean of the Ct value of SARS-CoV-2 RNA(RdRP) in the nasopharyngeal specimens. The Ct value shows the lowest value on the day of symptoms and negative 3 weeks from the date of symptoms. (Negative > Ct value 35).
Out of the twelve people who tested positive for SARS-CoV-2 before symptoms appeared, six people were family members of confirmatory patients and the other six were in close contact with an instructor who was identified as COVID-19-positive in the dance fitness class. The other was a Korean immigrant from the United States and was confirmed by laboratory screening for visits. In one case, a test conducted six days before the symptom occurred showed negative findings (Ct value was 35.82).
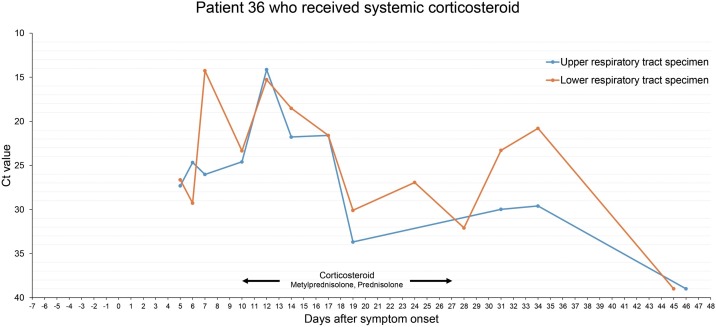
In the 36th patient who received systemic corticosteroid, Ct value decreased 10 days after methylprednisolone administration. In the rRT-PCR test performed with the lower respiratory tract specimen, the Ct value decreased from 32.09 to 20.8 (Figure 3 ).
Figure 3.
Changes in the SARS-CoV-2 Ct value of rRT-PCR in respiratory specimens of patient 36. A 71-year-old man with a history of diabetes was hospitalized on the 5th day of cough and fever. On the 10th day of symptom onset, dyspnea worsened and tachypnea persisted even after high flow nasal cannula oxygen therapy. On the 10th day of symptom development, methylprednisolone was administered at 0.5 mg/kg for 3 days and then stopped. Oxygen demand was reduced, but it was worsened on the 4th day after discontinuation, so methylprednisolone was taken at 1 mg/kg for 3 days. (The concomitant drugs used during the period of use of methylprednisolone were nafamostat, piperacillin/tazobactam, levofloxacin, and lansoprazole.) After clinical improvement, the prednisolone dose was reduced and finally stopped after 10 days. In the rRT-PCR test after the discontinuation of corticosteroid administration, the Ct value was continuously decreased and negative was confirmed on the 19th day after the final discontinuation.
Discussion
COVID-19 shows a very high viral load on the day of symptom development, which then decreases overall. Rapid viral proliferation was observed from 0 to 6 days before the onset of symptoms, suggesting that the risk of transmission is extremely high due to a significant amount of viral load in the early stages of symptoms and preclinical period. A recent study found that a Ct value of 30 or less showed positive findings in viral culture, although viral culture positive Ct does not mean an infectious dose (Arons et al., 2020, Young et al., 2020). In our finding, the Ct value is as low as 30 or less from 5 days before symptom onset to 10 days after symptom onset.
The epidemiological investigation currently being conducted by the WHO (World Health Organization) guideline is set as of the second day before patient symptoms are confirmed (World Health Organization, 2020b). However, our results show it is possible to see a rapid increase in the virus 5 days before symptom occurrence and a very high viral load that can occur 5 days before to 10 days after symptoms occur. Within this Ct range, the virus contains a sufficient amount of virus transmission ability and is sufficient to confirm the contact by more than 3 days before symptoms occur. COVID-19 has a high viral load before symptoms; epidemiological investigations from two days before the symptoms of diagnosed patients according to the WHO guidelines are thought to be difficult to prevent the spread of hidden infections as much as possible. Considering the characteristic that COVID-19 has a high viral load before symptoms appear, it is necessary to consider the expansion of the scope of epidemiological investigations.
In addition, 10 days after symptom onset, the Ct value exceeds 30. As there is a very low possibility of transmission after 10 days of symptom occurrence, it may be considered to release isolation after 10 days of symptom occurrence in limited resource situations.
Based on our findings, even a patient who is currently asymptomatic can deduce the present clinical condition of the patient with the patient's rRT-PCR results, and the typical COVID-19 patient's hospitalization can be determined based on the patient’s rRT-PCR results. Recently, a patient was confirmed after returning from the United States and was admitted to the hospital. The patient had no symptoms at that time, but 2 months earlier the patient had severe cold symptoms. At the time of hospitalization, the Ct value of rRT-PCR test performed outside the hospital was as high as 33.64, and the Ct value of all tests performed during hospitalization for two consecutive days was negative. This patient was thought to be highly likely to be “PCR detection after isolation” published by the Korean Central Disaster and Safety Countermeasures Headquarters.
Ultimately, rRT-PCR turned negative after ∼3 weeks; however, a negative and positive pattern can occur repeatedly for approximately 3 more weeks. Many COVID-19 patients were retested after discharge, confirmed repositivity, and requarantined in Korea (World Health Organization, 2020a, Jiang et al., 2020). Until May 19, the Korean public health center conducted rRT-PCR tests again two weeks after COVID-19 patients were discharged to confirm repositivity. Based on our study, there was a continuous pattern, which showed negative and positive results for 6 weeks and longer, with the persistence of Ct values of 33 or more in discharged patients with COVID-19 for 6 weeks and longer. Three PCR detection after isolation patients were excluded from this study. Up to 11 weeks after the onset of symptoms, rRT-PCR positive findings were observed.
In addition, it was shown that a high viral load can be maintained for a long time in patients who were administered with corticosteroids in this study. A recent study reported that the use of corticosteroids may delay virus removal and develop bacterial infections caused by immunosuppression (Yang et al., 2020). Therefore, we need to treat this result with caution and selectively apply systemic corticosteroid.
This study is limited by a small sample size (n = 54). In addition, a more comprehensive graph can be created if the results of rRT-PCR from the contact before the onset of symptoms can be added. Further research is needed to determine how much of the viral load is infectious.
In conclusion, changes in viral load allow us to estimate when patients will develop symptoms and determine the scope of epidemiological investigations. This study of viral kinetics provides a basis for estimating the inpatient discharge time and the patient's isolation release time, which can make the supply and demand of the hospital room predictable.
Disclosure
J-Y Rhee reports grants from 2020 Development Fund of the Dankook University College of Medicine. All other authors have no potential conflicts to disclose.
CRediT authorship contribution statement
Sukbin Jang: Data curation, Formal analysis, Investigation, Writing - original draft, Visualization. Ji-Young Rhee: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing. Yu Mi Wi: Data curation, Investigation. Bo Kyeung Jung: Data curation, Writing - original draft.
Acknowledgments
This work was supported by the Dankook University College of Medicine. We appreciate the efforts of all the staff and their families at the isolation unit at Dankook University Hospital. We would like to thank all the members of the Cheonansi Government, Seochogu Government, and Korea Centers for Disease Control and Prevention for their dedication.
References
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Li Y., Han M., Wang Z., Zhang Y., Du X. Recurrent PCR positivity after hospital discharge of people with coronavirus disease 2019 (COVID-19) J Infect. 2020 doi: 10.1016/j.jinf.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Novel coronavirus (2019-nCoV): situation report—121.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200520-covid-19-sitrep-121.pdf Accessed May 21, 2020. [Google Scholar]
- World Health Organization . 2020. Contact tracing in the context of COVID-19: interim guidance.https://apps.who.int/iris/rest/bitstreams/1277571/retrieve Accessed May 21, 2020. [Google Scholar]
- Yang Z., Liu J., Zhou Y., Zhao X., Zhao Q., Liu J. The effect of corticosteroid treatment on patients with coronavirus infection: a systematic review and meta-analysis. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Ng L.F., Anderson D.E., Chia W.N., Chia P.Y. 2020. Immunological and Viral Correlates of COVID-19 Disease Severity: A Prospective Cohort Study of the First 100 Patients in Singapore. [Google Scholar]