Abstract
SARS-CoV-2 is a highly contagious virus that has caused serious health crisis world-wide resulting into a pandemic situation. As per the literature, the SARS-CoV-2 is known to exploit humanACE2 receptors (similar toprevious SARS-CoV-1) for gaining entry into the host cell for invasion, infection, multiplication and pathogenesis. However, considering the higher infectivity of SARS-CoV-2 along with the complex etiology and pathophysiological outcomes seen in COVID-19 patients, it seems that there may be an alternate receptor for SARS-CoV-2. I performed comparative protein sequence analysis, database based gene expression profiling, bioinformatics based molecular docking using authentic tools and techniques for unveiling the molecular basis of high infectivity of SARS-CoV-2 as compared to previous known coronaviruses. My study revealed that SARS-CoV-2 (previously known as 2019-nCoV) harbors a RGD motif in its receptor binding domain (RBD) and the motif is absent in all other previously known SARS-CoVs. The RGD motif is well known for its role in cell-attachment and cell-adhesion. My hypothesis is that the SARS-CoV-2 may be (via RGD) exploiting integrins, that have high expression in lungs and all other vital organs, for invading host cells. However, an experimental verification is required. The expression of ACE2, which is a known receptor for SARS-CoV-2, was found to be negligible in lungs. I assume that higher infectivity of SARS-CoV-2 could be due to this RGD-integrin mediated acquired cell-adhesive property. Gene expression profiling revealed that expression of integrins is significantly high in lung cells, in particular αvβ6, α5β1, αvβ8 and an ECM protein, ICAM1. The molecular docking experiment showed the RBD of spike protein binds with integrins precisely at RGD motif in a similar manner as a synthetic RGD peptide binds to integrins as found by other researchers. SARS-CoV-2 spike protein has a number of phosphorylation sites that can induce cAMP, PKC, Tyr signaling pathways. These pathways either activate calcium ion channels or get activated by calcium. In fact, integrins have calcium & metal binding sites that were predicted around and in vicinity of RGD-integrin docking site in our analysis which suggests that RGD-integrins interaction possibly occurs in calcium-dependent manner. The higher expression of integrins in lungs along with their previously known high binding affinity (~KD = 4.0 nM) for virus RGD motif could serve as a possible explanation for high infectivity of SARS-CoV-2. On the contrary, human ACE2 has lower expression in lungs and its high binding affinity (~KD = 15 nM) for spike RBD alone could not manifest significant virus-host attachment. This suggests that besides human ACE2, an additional or alternate receptor for SARS-CoV-2 is likely to exist. A highly relevant evidence never reported earlier which corroborate in favor of RGD-integrins mediated virus-host attachment is an unleashed cytokine storm which causes due to activation of TNF-α and IL-6 activation; and integrins role in their activation is also well established. Altogether, the current study has highlighted possible role of calcium and other divalent ions in RGD-integrins interaction for virus invasion into host cells and suggested that lowering divalent ion in lungs could avert virus-host cells attachment.
1. Introduction
Ever since the recent emergence of novel coronavirus (SARS-CoV-2, earlier known as 2019-nCoV) in the Wuhan city of China and its subsequent transmission in other countries has resulted into serious heatth crisis as well as breakdown of socio-economic development. Scientific community from all over the world is industriously engaged in and committed to find a potent therapeutic solution for the treatment of COVID-19. As of 10th April, 1,521,252 confirmed cases and 92,798 deaths were recorded as a cumulative data from different parts of the world (WHO Situation Report no. 81 available at who.int accessed on 12-04-2020). At the onset of the disease, the infected symptomatic patients experience hyperthermia, pharyngeal congestion, cough, and anosmia (in some cases); however, as disease progress more than fifty percent patients develop severe labored breathing, clinically referred to as dyspnoea or tachypnoea (Huang et al., 2020, Qiu et al., 2020, Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c). Advance stages are characterized by severe pulmonary inflammation, fibrosis and obstructions of the bronchioles resulting in a pneumonia-like pathophysiological condition (Huang et al., 2020, Qiu et al., 2020, Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c). In both symptomatic and asymptomatic COVID-19 patients, SARS-CoV-2 manages to cause significant damages to multiple organs before any patients could realize they are infected with SARS-CoV-2. This is because neither any neurological indications nor any signs of heart, kidney and liver failure are seen at the onset of disease in these patients (Qiu et al., 2020).
As of today, there is no specific antiviral therapy available in any form, either vaccine or drug or others, to combat COVID-19 and its infection. The neutralizing antibodies that were previously tested and found successful against SARS-CoV-1 have displayed inappreciable cross-reactivity against SARS-CoV-2. For some antibodies (such as CR3022) that could bind to SARS-CoV-2, their neutralizing efficacy has not been tested and appreciated yet (Tian et al., 2020, Yuan et al., 2020, Wrapp et al., 2020). It has been asserted that the binding sites for these monoclonal antibodies on SARS-CoV-2 are vulnerable and it is only an assumption that antibodies that could bind strongly would possibly neutralize the SARS-CoV-2 also. Moreover, cross-reacting neutralizing antibodies are also doubted for their ability to confer prolonged protection against SARS-CoV-2. So far, we have no other choice than to use previously tested drugs or to solely oblige known tactics as precautionary measures against COVID-19. Recently, some researchers suggested the use of combination of remdesivir (a broad-range antiviral drug) and chloroquine for effective control of SARS-CoV-2 infection under in vitro condition (Devaux et al., 2020, Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c). Similarly, hydroxychloroquine and azithromycin has also been referred to as a potent therapeutic weapon against the SARS-CoV-2 virus (Colson et al., 2020). Drug repurposing approach, which involved a screening of a thousand of molecules showed that HIV protease inhibitors, RNA-dependent RNA polymerase (RdRp) inhibitors and some other inhibitors and agonists such as methisazone (an inhibitor of protein synthesis), CGP42112A (an angiotensin AT2 receptor agonist) and ABT450 (an inhibitor of the non-structural protein 3-4A) could become promising treatment options for COVID-19 (Gordon et al., 2020, Li et al., 2020, Shah et al., 2020). Lu (2020) suggested some treatment options for COVID-19 that include use of nucleoside analogues, neuraminidase inhibitors, lopinavir or ritonavir, remdesivir, 3TC/TDF/EFV monotherapy or combination therapy (DNA polymerase inhibitors), anti-inflammatory or immune-suppressive drugs, just to name a few. Besides this, some traditional Chinese medicine, for instance, ShuFengJieDu and Lianhuaqingwen capsules could also be useful (Lu, 2020). Using virtual screening (Kandeel and Al-Nazawi, 2020), epigenetic dysregulation (Sawalha et al., 2020), protein–protein interaction mapping (Cava et al., 2020), integrated network pharmacological approach (Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c), and similar such approaches, a number of other drugs have also been repurposed for COVID-19 treatment. However, most of the drugs are either in developmental stages or under clinical trials (https://clinicaltrials.gov/). In the midst of this pandemic situation, it is obvious that there exists no preventive therapy for highly contagious SARS-CoV-2 and this is strikingly alarming to all of us.
A number of studies demonstrated the key role of human ACE2 in virus attachment to host cells (Wan et al., 2020, Zhao et al., 2020). The three-dimensional crystal structure of SARS-CoV-2 spike receptor binding domain complexed with its receptor, human ACE2 (Angiotensin converting enzyme 2), has been already solved (Chen et al., 2020). All other structural, functional and antigenicity related information related to SARS-CoV-2 are also available (Walls et al., 2020). Based on biophysical data, it has been demonstrated that the human ACE2 binds to SARS-CoV-2 with greater affinity than SARS-CoV-1 (Wrapp et al., 2020). However, owing to their low copy number (protein expression) (Chen et al., 2020), their high affinity (for SARS-CoV-2 than SARS-CoV-1) alone could not manifest reasonable virus-host cell attachment, at least in lung cells. When this manuscript study was under progress, Sigrist and coauthors (2020) showed that SARS-CoV-2 harbors a RGD motif and integrins (that display high affinity for RGD motifs) may be involved in facilitating virus entry into host cells. However, the study did not explain the full mechanistic state of affairs involved in RGD-integrins interaction and virus entry into the host cells. I also found RGD motif in the spike receptor binding domain of SARS-CoV-2 and studied mechanistic basis of RGD-integrin (and other ECM protein such as ICAM1) mediated virus invasion into host cells. This study is the first study to present striking evidence (substantiated by existing facts in literature) favoring the role of calcium and other divalent ions (magnesium, manganese etc.) in RGD–integrins mediated virus attachment with the host cells for and that lowering the concentration of calcium and other divalent ions in lungs could be a possible mechanism to avert SARs-CoV-2 infection and invasion. Herein, I did comparative protein sequence analysis, motif scanning, gene expression data analysis and bioinformatics based molecular docking using most trustworthy tools and techniques. The results generated in this study indicated and underscored the key role of calcium and other divalent ions in mediating virus-host cell attachment for invasion into human lung cells possibly via integrins and other cell adhesion proteins present on the host cell surface. I designed and suggested novel pulmonary EDTA chelation therapy as a technical simple (requires nebulizer/inhaler/nasal sprays that are available in all hospitals and clinics), quick (1-3hr a day, 2–3 times a week), safe (EDTA/EGTA are FDA approved for calcium chelation), affordable (estimated cost approx. 50–100 USD) and efficient (based of efficacy data from literature) treatment therapy for COVID-19 patients.
2. Materials & methods
2.1. Sequence retrieval
The coronavirus related nucleotide and protein sequences were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/), UniProtKB (https://www.uniprot.org) and SARS Coronavirus 2 data hub of the NCBI (https://www.nih.gov/coronavirus).
2.2. Sequence alignment
The protein sequence of SARS-CoV-1 and SARS-CoV-2 coronaviruses were subjected to pair-wise sequence alignment using Clustal Omega using default setting (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Madeira et al., 2019).
2.3. Motif scanning and prediction
The Pfam, Prosite and HAMAP profiles of the S protein from the coronavirus was ascertained using MyHits Motif Scan (SIB, Switzerland) (https://myhits.isb-sib.ch/cgi-bin/motif_scan). The putative N-linked glycosylation and other post-translation modification sites were predicted as Frequent Pattern in MyHits outputs (Pagni et al., 2007).
2.4. Gene expression profiling
The expression profile of ACE2 receptor protein and integrins in lungs was ascertained using Gene Expression Database at EBI (https://www.ebi.ac.uk/gxa/home) and the Human Protein Atlas (https://www.proteinatlas.org/) (Uhlén et al., 2015, Uhlen et al., 2019).
2.5. Calcium and magnesium binding sites in integrins
In order to understand the role of divalent ions in RGD-integrins mediated virus-host cells attachment. A composite calcium and magnesium binding-site in integrins were predicted using IonCom (https://zhanglab.ccmb.med.umich.edu/IonCom/), which used an integrated approach based on ab initio training and template-based transferals (Hu et al., 2016) and predicts calcium binding sites in a given protein by searching four or more oxygen atoms on protein surface arranged in a spherical manner. The input PDB file of integrins such as α5β1 (PDB ID: 3VI3) and αvβ6 (5FFG) were subjected for ion binding sites’ prediction to specifically predict calcium and magnesium binds sites.
2.6. Spike RBD docking with integrins
The PDB file of the spike receptor binding domain (PDB ID: 6LZG) and integrins such as α5β1 (PDB ID: 3VI3 and 3VI4) and αvβ6 (PDB ID: 5FFG) were retrieved from Protein Data Bank (http://www.rcsb.org/structure/). The protein–protein docking was performed using HDOCK server which rely on template-based modeling and ab initio free docking using default parameters (hdock.phys.hust.edu.cn/). The docked structures were visualized and high resolution photographs were generated in Chimera 1.10.
2.7. Spike RBD docking with EDTA
The PDB file of the spike receptor binding domain was retrieved from Protein Data Bank (as mentioned above). The PDB file of the EDTA was online obtained and converted in Mol2 file in Chimera 1.10.1 version. The docking was done using Swissdock server of SIB (http://www.swissdock.ch/docking). Protein-ligand binding modes were clustered according to their rank based on average FullFitnessin output data (Grosdidier et al., 2007).
3. Results
3.1. N-terminus of spike protein is variable
Pairwise sequence alignment of SARS-CoV-1 and SARS-CoV-2 spike protein revealed that the N-terminus (especially upto 250 aa) of SARS-CoV-2 is highly divergent than that of the SARS-CoV-1 (Fig. 1 ). The S1 Glycoprotein (from 268 to 304 aa in SARS-CoV-1 and from 282 to 317 aa in SARS-CoV-2) domain which plays an important role in recognition of host cell receptor were found divergent at 10 amino acid positions. The spike receptor binding domain is of 253 amino acid residues in SARS-CoV-1 and ranges from 317 to 569 aa; whereas, it is of 254 amino acid residues in SARS-CoV-2 and stretch from 330 to 583 aa. There were 49 mismatches and 1 insersion/deletion that render SARS-CoV-1 and SARS-CoV-2 approximately 80% sequence similarity. The S2 glycoprotein domain is a large 544 amino acid residues long region at the C-terminus of the spike protein which plays crucial role in virus fusion with host cells. This region has been predicted to be less divergent than S1 glycoprotein and RBD. There were 49 amino acids substitution which amount for approximately 9% sequence divergence between SARS-CoV-1 and SARS-CoV-2. The cysteine rich region at the C-terminus part of the SARS-CoV-2 spike protein has an additional Cys residue (actually Ala > Cys) at 1247 position (corresponds to Ala residue at 1229 position in SARS-CoV-1 spike protein in alignment). This region is also involved in virus-induced membrane fusion (Chang et al., 2000). The presence extra Cys residue results in the formation of a CX6CC motif, which is usually found in envelop proteins of some viruses, for instance Ebola virus and murine leukemia virus, and is required for virus fusion with host cell (Johnston and Radke, 2000, Lee and Saphire, 2009).
Fig. 1.
Pair-wise sequence alignment of SARS-CoV-1 and SARS-CoV-2 spike protein RBD sequence using Clustal Omega.
I also found two insertion sequences with the stretch HVSGTNGTKRFD69-80 and RFQTLLALHRSYLTPGDSSSGWTAG236-261at the N-terminus region of spike protein and these were seen as discontinuous in the amino acid sequence as predicted by the pair-wise sequence alignment. However, considering that these inserts are very short and appeared in the hypervariable region of viral spike protein, the most likely assumption for their origin is that they might have arisen naturally. Besides this, the receptor binding domain of the spike protein in SARS-CoV-2 has a stretch of sequence (TEIYQAGSTPCNGVEGF470-486) showing mismatch with SARS-CoV-1.
3.2. Increased number of phosphorylation sites in SARS-CoV-2 led to increased viral infection and pathogenesis
The number and position of putative N-linked glycosylation sites, post-translation modification sites that can induce cAMP, CK2, Tyr, PKC signaling pathways and Myristyl sites were predicted to be varying in SARS-CoV-1 and SARS-CoV-2 (Table 1 ). Although both coronaviruses have equal number of N-linked glycosylation sites but the position of sites differs (Table 1). Furthermore, phosphorylation sites of all types (except PKC phosphorylation) were predicted to be in more number in SARS-CoV-2 spike protein than in SARS-CoV-1. The phosphorylation of viral proteins is actuated by host cells’ kinases has a significant impact on viral infection, replication, multiplication and cytotoxicity (Bretaña et al., 2012, Keating and Striker, 2012, Keck et al., 2015), I assume that the increased number of phosphorylation sites in SARS-CoV-2 spike protein could be one of the major reasons for its higher infectivity as observed in case of COVID-19. This can be ascertained using in-vitro experiments. These pathways either activate calcium ion channels or get activated by calcium. This suggested that virus might be directly involved in regulating these calcium ion channels or might be dependent upon presence of Ca+2 ion or other divalent ions for attachment with host cells.
Table 1.
Prediction of putative N-linked glycosylation and other PTM sites using MyHits Motif Scan of SIB. Top scored N-linked Glycosylation sites are represented in bold.
| S.No. | Pattern | Position |
|
|---|---|---|---|
| SARS-CoV-1 | SARS-CoV-2 | ||
| 1. | ASN_Glycosylation | 29, 65, 73, 109, 118, 119, 158, 227, 269, 318, 330, 357, 589, 602, 691, 699, 783, 1056, 1080, 1116, 1140, 1155, 1176 | 17, 61, 74, 122, 149, 165, 234, 282, 331, 343, 603, 616, 657, 709, 717, 801, 1074, 1098, 1134, 1158, 1173, 1194 |
| 2. | CAMP_Phospho_Site | 343 | 356, 528 |
| 3. | CK2_Phospho_Site | 12, 20, 271, 561, 644, 716, 798, 809, 964, 1118, 1129, 1178 | 50, 108, 151, 221, 250, 284, 734, 816, 827, 982, 1136, 1147, 1160, 1196 |
| 4. | Myristyl | 86, 104, 225, 246, 298, 368, 418, 531, 587, 634, 653, 682, 739, 862, 871, 890, 953, 1113, 1153, 1228 | 72, 89, 184, 232, 311, 381, 431, 446, 476, 545, 601, 648, 667, 700, 757, 880, 889, 908, 971, 1093, 1131, 1171, 1246 |
| 5. | PKC_Phospho_Site | 36, 92, 173, 215, 289, 363, 541, 561, 670, 775, 795, 980, 1019, 1087, 1129 | 19, 76, 95, 302, 376, 415, 555, 632, 680, 813, 998, 1037, 1105, 1147 |
| 6. | Tyr_Phospho_Site | 188, 715 | 417, 733 |
3.3. SARS-CoV-2 spike receptor binding domain harbors a RGD motif which is absent in SARS-CoV-1
The spike protein sequence of SARS-CoV-1 and SARS-CoV-2 were subjected to motif scanning and prediction using MyHits Motif Scan at SIB server. A number of motifs were predicted in the spike protein sequence such as RGD (from 403 to 405 aa in receptor binding domain of SARS-CoV-2) (Table 2 ). The RGD motif (K403R substitution) was predicted in spike receptor binding domain of SARS-CoV-2 and the same was not predicted in the SARS-CoV-1 spike protein and other previous known coronaviruses. This motif was originally found in fibronectin, which is an extracellular matrix protein and this motif plays a major role in cell adhesion and attachment. A number of proteins are known to interact with these RGD motifs. One such group of proteins are integrins that have strong affinity for RGD motif (~KD = 4.0 nM) and they employ this motif to efficiently mediate cell adhesion. Eight out of approximately twenty known integrins recognize and exploit RGD motifs for cell adhesion (Ruoslahti, 1996, Teoh et al., 2016). Presence of motif RGD in SARS-CoV-2 spike receptor binding domain is a strikingly alarming as this motif is expected to facilitate even stronger attachment between virus and human target cells, and thus, rendering human cells more susceptible and vulnerable to infection. Motif scan also predicted fusion glycoprotein F0 (from 910 to 935 aa in S2 glycoprotein of SARS-CoV-1) and FMRFamide (from 437 to 447 aa in receptor binding domain of SARS-CoV-1). The absence of fusion glycoprotein F0 in SARS-CoV-2may also have potential deleterious effect because this motif acts as a potent antigen that are targeted by neutralizing antibodies induced by virus infection as a part of humoral immune response and adaptive immunity (Prabakaran et al., 2006) and its absence in SARS-CoV-2 spike protein explains why monoclonal antibodies raised against SARS-CoV-1 could not show cross-reactivity with SARS-CoV-2 spike protein. This also accounts for the reason why an appropriate and adequate immune response could not mount in COVID-19 patients, especially in aged and immunocompromised COVID-19 patients, which eventually led to patients’ death. The FMRFamide (Phe-Met-Arg-Phe-amide) like peptides (also known as FLPs) (predicted to be absent in SARS-CoV-2), are one particular group of Arg-Phe-amides (RFamides), are known to play a central role in parasite neuromuscular biology (Peymen et al., 2014); however, their role in pathogenesis of SARS-CoV-2 cannot be ascertained yet and same for other predicted motifs (Table 2).
Table 2.
Artificial neural network based predicted motifs in the protein sequence of SARS-CoV-1 and SARS-CoV-2.
| S.No. | Motifs | Position |
||
|---|---|---|---|---|
| HCoV-1 | HCoV-2 | Motif’s Role/Function | ||
| 1 | S1 Glycoprotein | 268–304 | 519–592 | Viral attachment |
| 2 | Potato Inhibitor 1 | 309–320 | Not predicted | NA |
| 3 | Spike Receptor Binding domain | 317–569 | 330–583 | Attachment with host cells |
| 4 | RGD | Not Predicted | 403–405 | Possibly attachment with host cells |
| 5 | FMRP | 437–447 | Not predicted | NA |
| 6 | S2 Glycoprotein | 641–1247; 694–1237 | 684–1265; 712–1255 | Fusion of viral and cell membrane as well as fusion of infected cells with adjoining cells. |
| 7 | Borrelia Repeat | 662–679 | Not Predicted | NA |
| 8 | DUF16 | 892–971 | Not Predicted | Protein of unknown function |
| 9 | Fusion Glycoprotein F0 | 910–935 | Not Predicted | Induces fusion of viral and cellular membranes leading to delivery of the nucleocapsid into the cytoplasm |
| 10 | BIG1 | Not predicted | 1–3 | NA |
| 11 | Cysteine-rich Domain | 1217–1236 | 1235–1254 | Membrane fusion |
3.4. ACE2 expression is high in digestive route while integrins expression is high in lung cells
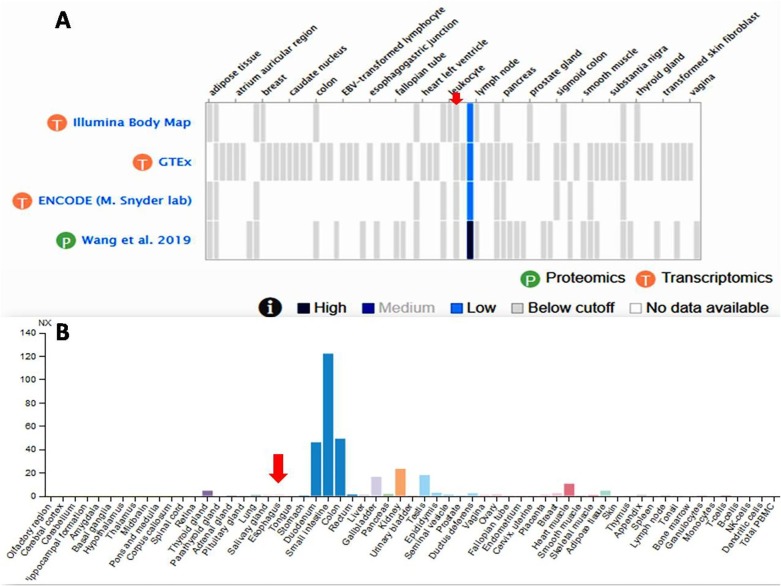
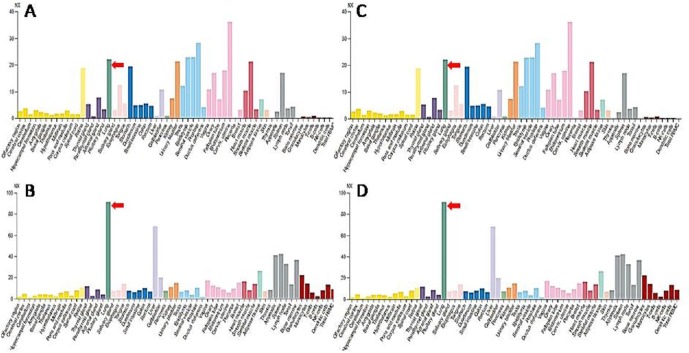
Presence of RGD motif in the spike receptor binding domain of the SARS-CoV-2 and the fact that integrins display a strong affinity for proteins harboring RGD motifs (Liu, 2009) suggested and indicated that virus possibly enters into host cells via integrins receptor (Ruoslahti, 1988, Van Agthoven et al., 2014). In order to assess this, I analyzed the gene expression data for all eight integrins at Human Protein Atlas (https://www.proteinatlas.org/) (Uhlén et al., 2015, Uhlen et al., 2019) and found that expression of three integrins (αvβ6, α5β1, α8β1) is significantly high in lung cells and other vital organs of human body that have been found to be affected by SARS-CoV-2 infection (Fig. 2 A–C). These three integrins were used for molecular docking experiments. I also found that expression of an ECM protein ICAM1 is significantly higher (even higher than integrins) in lungs (Fig. 2D) suggesting that SARS-CoV-2 spike protein might be using these integrins and/or ECM proteins such as ICAM1 (or others) for invading host cells. On the contrary, the ACE2 RNA is detected in very low quantity (almost unidentifiable) in lungs and detectable expression peaks were detected in other organs such as duodenum, small intestine, colon, gallbladder, kidney, testis, and heart muscles (Fig. 3 ). In congruence with this, Xu et al., 2020a, Xu et al., 2020b found high expression of ACE2 in oral mucosa and Zhang and co-authors (2020) demonstrated digestive route as a potential mechanism of ACE2 mediated virus infection based on the single-cell transcriptomic analysis. On the contrary, Chen and coauthors (2020) observed relatively low levels of ACE2 mRNA expression in lungs which further supports my claims.
Fig. 2.
Gene expression profile of RGD binding integrins αvβ6 (A), α5β1 (B), α8β1 (C) and human ICAM1 (D) in lungs as obtained and plotted at the Human Protein Atlas. (https://www.proteinatlas.org/). The bar shows expression level of respective protein in different human tissues and organs. The expression peaks that correspond to lungs have been shown using red arrow to ease visualization and verification of data. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Gene expression profile of ACE2 in lungs as obtained and plotted at Gene Expression Database at EBI (https://www.ebi.ac.uk/gxa/home) (A) and the Human Protein Atlas (https://www.proteinatlas.org/) (B). The expression peak that corresponds to lungs have been shown using red arrow to ease visualization and verification of data. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. SARS-CoV-2 spike RBD interacts with integrins and ICAM1 precisely at RGD motif
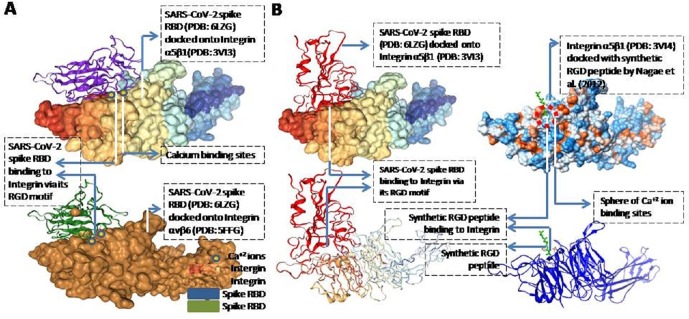
The RGD motif is the minimal indispensable requirement for integrins to bind with any viral protein which virus can use for attachment with host cells (Hussein et al., 2015). Nagae and coauthors (2012) demonstrated that synthetic RGD tripeptide binds to the integrin α5β1 (PDB ID: 3V14). In order to obtain molecular evidences for integrins mediated SARS-CoV-2 entry into lung cells, I docked the viral spike RBD domain (PDB ID: 6LZG) onto the structure of integrin α5β1 (PDB ID: 3V13 without synthetic RGD tripeptide) (Nagae et al., 2012) and integrin αvβ6 (PDB ID: 5FFG) (Dong et al., 2017) and found that spike RBD binds with integrin precisely via the RGD motif as expected (Fig. 4 A). The predicted docking energy score was −302.33 (ΔG) and the ligand RSMD score was 71.80 Å for spike RBD-integrin αvβ6 docking; and for the spike RBD-integrin α5β1, the scores were respectively −315.45 and 37.4 Å. I focused on only αvβ6, α5β1, and α8β1 as they have been found to be expressing abundantly in lungs tissues/cells. All the other integrins are also known to bind to RGD peptides with different degree of affinity and I did in-silico work on those that show higher affinity for RGD. I checked the binding of other integrins with RGD and I found that the stability of integrin-RGD is low (data not shown). I found that the spike RGD interacts with loop turns AAG199-201, GSYFWQ225-230 and RQASSIYDDA260-270 of the α5 chain of the integrin α5β1 (Nagae et al., 2012) (Fig. 4B). The latter (RQASSIYDDA260-270) has also been predicted as one of the calcium binding sites as per the UniProt KD database (ITGA5; UniProt ID: P06756) and our IonCon analysis. Presence of Ca+2 ion binding sites around and in vicinity of RGD-integrins docking site suggest that RGD-integrin interaction is indeed calcium-dependent (Fig. 4B). ICAM1 is also known to bind integrins and this mechanism is exploited by some viruses (such as Rhinoviruses) to gain entry into respiratory system for pathogenesis (Bella et al., 1998).
Fig. 4.
Molecular docking of viral spike RBD domain onto the structure of α5β1 (PDB ID: 3VI3) and αvβ6 (PDB ID: 5FFG) showing interaction via the RGD motif present in the spike RBD domain (A). The molecular docking of viral spike RBD domain onto the structure of α5β1 (PDB ID: 3VI3) showed that the spike RBD is interacting with the α5β1 through its RGD motif in the same way (B, left panel) as synthetic RGD peptide binds and interact to α5β1 (PDB ID: 3VI4) (B, right panel) as already known in literature (Nagae et al., 2012). The docking was done using HDOCK server (hdock.phys.hust.edu.cn/). Spike protein is shown in red and synthetic RGD peptide is shown in green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Molecular docking suggests EDTA chelation therapy as a novel treatment for COVID-19
The RGD motifs are well known in the field of cell biology, cell therapy and tissue engineering because of their remarkable cell-adhesive property. Extracellular matrix proteins, such as fibronectin and laminin, possess RGD motif and are frequently coated onto the surface of biomaterials (petri-dishes, T-flasks) for facilitating human cells adhesion onto the surface of the biomaterials. Bivalent ions such as Ca+2& Mg+2play indispensable role in promoting such cell adhesion and EDTA is used as a chelating agent for disaggregating Ca+2 mediated cell bondage.
Two features in SARS-CoV-2 spike protein suggest virus attachment with host cells via integrins in a calcium-dependent manner: 1) presence of RGD motif in receptor binding domain of SARS-CoV-2 and 2) presence of a number of phosphorylation sites for key pathways that regulates calcium ion channels, and secondly, Ca+2 signaling. Similarly, integrins possess calcium and magnesium binding sites (as predicted by IonCon) for interaction with RGD motif containing proteins, and the same in current context are spike protein of SARS-CoV-2. Besides this, integrins are also known to regulate cell adhesion by altering intracellular calcium dynamics and have involvement in regulation of ion channels and calcium signaling to exert pathophysiological impact in other diseases (Sjaastad and Nelson, 1997, Arcangeli and Becchetti, 2010). Altogether, it appears that SARS-CoV-2 enters into human cells via interaction of their spike receptor RGD motif with integrins in a Ca+2 dependent manner.
Altogether, the current study highlighted the possible role of calcium and other divalent ions in RGD-integrins binding for virus invasion and suggested that lowering divalent ion in lungs could avert virus-host cells binding. This can be accomplished by EDTA or EGTA, both are well known chelating agents I strongly believe that phenotypic remodeling and regulation of Ca+2 ions (Berridge, 2012) by pulmonary EDTA chelation can discourage the SARS-CoV-2 virus attachment with human lung cells which will led to attainment of a state of protection against this virus. Finally, EDTA chelation therapy targeting lungs (as in discussion part) holds immense potential in successful management and treatment of COVID-19 patients.
3.7. RGD-Integrins binding cause activation of downstream integrin-mediated cell signaling (GO:0007229)
Further, it seems that RGD-integrins binding would relay or activate downstream integrins mediated cell signaling pathway (GO:0007229) which comprise of 106 human genes related to different biological processes and cellular pathways (http://amigo.geneontology.org/; Data not shown). The experimental evidences collected from UniProt KB databases show that integrins play key role in viral attachment and TGF-β and IL-1 mediated signaling. Other signaling pathways that are known to be regulated by integrins upon interaction with different viruses are FGF1 signaling, FGF2 signaling, NRG1-ERBB signaling, CX3CL1 signaling and CD40-CD40LG signaling (Table 3 ).
Table 3.
RGD binding Integrins, their receptors and their role in virus attachment and cell signaling as sourced from UniProt KB database.
| S.N. | Integrin | UniProt ID | Receptor | Virus Attachment | Function |
|---|---|---|---|---|---|
| 1 | αvβ3 ITGAV-ITGB3 CD61 |
P06756 P05106 |
CD40LG FGF1/FGF2 CX3CL1 PLA2G2A NRG1 IGF2 IL-1B FBN1 CD40-LG IGF2 Ca+2 Binding Sites 260–268; 314–322; 379–387; 443–451 |
Herpes virus 8 HHV-8 Coxsackievirus A9 Hantaan virus Cytomegalovirus/HHV-5 Human metapneumovirus Human parechovirus 1 West nile virus HIV-1 |
FGF1 signaling FGF2 signaling NRG1-ERBB signaling CX3CL1 signaling 1L-1B signaling CD40-CD40L signaling IGF2 signaling |
| 2 | αvβ5 ITGAV-ITGB5 |
P06756 P18084 |
Fironectin | Adenovirus type c Coxsackievirus A9 CoxsackievirusB1 |
TGF-β1 singaling |
| 3 | αvβ6 ITGAV-ITGB6 |
P06756 P18564 |
Fibronectin Cytotactin TGF-β1 FBN1 |
Coxsackievirus A9 Coxsackievirus B1 Herpes simplex virus-1 HHV-1 |
TGF-β1 activation Clathrin-mediated endocytosis |
| 4 | αvβ8 ITGAV-ITGB8 |
P06756 P26012 |
Fibronectin TGF-β1 |
Data not found | TGF-β1 activation |
| 5 | α5β1 ITGA5-ITGB1 CD29 |
P08648 P05556 | Fibrinogen Fibronectin PLA2G2A CD40LG IL1B FBN1 Ca+2 Binding Sites 280–288; 334–342; 401–409; 465–473 |
Cytomegalovirus/HHV-5 Epstein-Barr virus/HHV-4 Human parvovirus B19 Mammalian reovirus HIV-1 Human metapneumovirus |
CD40-CD40L signaling IL1B signaling |
| 6 | α8β1 ITGA8-ITGB1 |
P53708 P05556 |
TNC FN1 SPP1 TGFB1/TGFB3 VTN NPNT Ca+2 Binding Sites 329–337; 395–404; 459–467 |
Data not found | TGF-β1 signaling |
4. Discussion
In any disease or pathological condition where integrin expression would be high, I can expect patient to be highly vulnerable to COVID-19. Integrin expression has been found high in case of cancer (Bianconi et al., 2016) and diabetes (Roth et al., 1993, Miller et al., 2014) and I assume the susceptibility for SARS-CoV-2 infection and COVID-19 would be high in patients suffering from any diseases in which integrin expression is high. The immune-diagnosis and profiling of COVID-19 patients showed elevated levels of pro-inflammatory cytokines including IL-6 and IL-1β in serum (Huang et al., 2020, Tan et al., 2020, Qin et al., 2020, Xu et al., 2020a, Xu et al., 2020b). They also exhibited an increased serum levels of other cytokine and chemokine molecules such as IL-2, IL-8, IL-17, G-CSF, GM-CSF, IP10, MCP1, MIP1α (also known as CCL3) and TNF (Huang et al., 2020, Shi et al., 2020, Qin et al., 2020, Xu et al., 2020a, Xu et al., 2020b). Also, C-reactive protein and D-dimer are found to be abnormally high (Cao, 2020). High levels of pro-inflammatory cytokines may lead to tissue shedding and damage in lungs, heart, liver and kidney, i.e., multiple organ failure (Cao, 2020). The most evident pathophysiological outcome of SARS-CoV-2 infection is activation of immune cells and massive infiltration of neutrophils and macrophages that eventually led to “cytokine storm” resulting in severe lung inflammations (such as pulmonary edema and alveolar damages) and can be characterized by pneumonia-like symptoms and respiratory failure (Mehta et al., 2020). Herein, I underline another strong and convincing evidence in favor of virus-host attachment through RGD-integrins, i.e., the cytokine storm, caused due to activation of pro-inflammatory cytokines such as TNF-α and IL-6 activation; and integrins role in their activation is also well established (Blobe et al., 2000, Annes et al., 2002, Lu et al., 2002, Sheppard, 2008, Hung et al., 2018). Integrin α1β1 regulates Ca+2ion transient responses to IL-1 and TGF-β (Parekh et al., 2014). Integrins also regulates calcium mobilization in associated pathways such as Go-mediated cell signaling (Erb et al., 2001). Besides this, envelop proteins of virus, including previous coronaviruses, are Ca+2 channeling viroporins that exploit host cell signaling and are involved in virus entry, replication, immune evasion leading to activation of autophagy and inflammasome responses such as that of NLRP3 (Nieto-Torres et al., 2015, Hyser and Estes, 2015). In context of SARS-CoV2 and integrins attachment, what exact role does the dynamics (in terms of mobilization, concentration, frequency, timing) of intracellular and extracellular calcium play is still not clear; however, understanding this dynamics would greatly assist us in appropriately and adequately controlling the host cells’ response to SARS-CoV-2so as to counteract virus infection and invasion. However, it is apparently clear that high Ca+2 ion concentration in lungs is detrimental in case of SARS-CoV-2 infection.
A number of studies demonstrated the key role of human ACE2 in virus attachment to host cells (Wan et al., 2020, Zhao et al., 2020). Previous coronaviruses have also been found to enter into host cells using clathrin and caveolae-independent pathway (Inoue et al., 2007, Wang et al., 2008). Integrin αvβ6 have also been implicated in mediating clathrin-dependent endocytosis (Table 3). Based on the biophysical data, it has been demonstrated that the human ACE2 binds to SARS-CoV-2 with greater affinity than SARS-CoV-1 (Wrapp et al., 2020). However, owing to their low copy number (gene expression), their high affinity (for SARS-CoV-2 than SARS-CoV-1) alone could not manifest reasonable virus-host cell attachment, at leastin lung cells. On the other hand, the affinity of integrins for RGD motif of the spike RBD is considerably high. The binding affinity of ACE2 for whole spike RBD is KD = 15.2 nM (KD = dissociation constant) (Wrapp et al., 2020) and on the other hand the binding affinity of integrins for RGD is close to KD = 4.0 nM (Bernhagen et al., 2017, Bernhagen et al., 2019, Ma et al., 2017). Since, the binding affinity has inverse relationship with KD, I can rule out that virus spike protein would prefer binding to integrins rather than ACE2, at least in lung cells. However, ACE2 expression has been found to be considerably high in the digestive tract from duodenum to rectum as well as in gall bladder and kidney. Therefore, ACE2 could be responsible for virus infection in these organs. This study presents striking evidence favoring the role of calcium and other divalent ions (magnesium, manganese etc.) in RGD–integrins mediated virus attachment with the host cells in lung cells for infection and invasion.
Chelating agents are long known for treating metal toxicity in humans in the context to several disease conditions such as coronary heart disease, neurotoxicity and arthritis etc. (Goyer et al., 1995, Bamonti et al., 2011, Ferrero, 2016, Fulgenzi and Ferrero, 2019). The EDTA is a well-known chelating agent and is a FDA approved drug for treatment of hypercalcemia conditions. In EDTA chelation therapy disodium EDTA (or similar sodium-EDTA derivative) is used as a chelating agent and is administered intravenously for the chelating divalent ions and the resulting chelates then come out in urine after passing through glomerulus filtration (Bamonti et al., 2011, Ferrero, 2016, Fulgenzi and Ferrero, 2019).
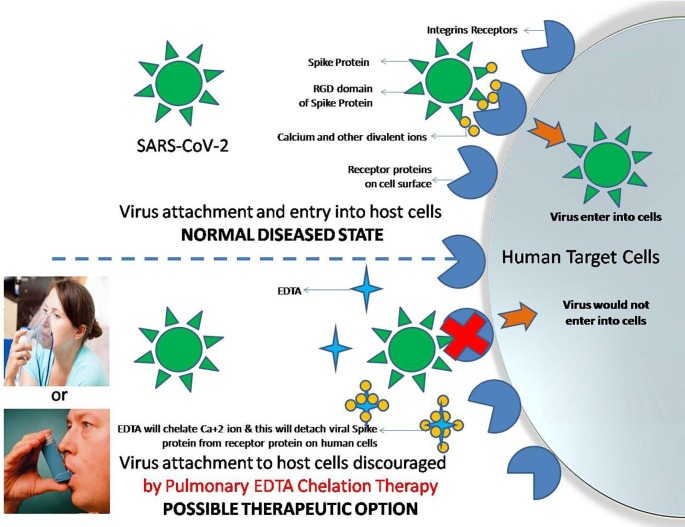
I have proposed EDTA chelation therapy as a potent anti-viral therapy against SARS-CoV-2. Pulmonary EDTA chelation therapy has already been executed clinically for asthmatic patients and is also known to increase the efficacy of nebulized bronchodilators (Asmus et al., 2001). Sodium salts of EDTA can be used for chelating Ca+2 ions as calcium-EDTA has higher stability than sodium-EDTA (Blaurock-Busch and Busch, 2014) and the resulting Ca-EDTA chelate would be soluble that can easily excrete out of body through glomerular filtration (Fig. 5 ). I have designed EDTA chelation therapy as a novel therapeutic strategy for safe, fast, cost-effective and efficient treatment of COVID-19 based on genomic understanding of SARS-CoV-2 virus spike protein. In EDTA chelation therapy, disodium EDTA will be administered in the body (preferably through nebulizer/inhaler/nasal spray or intravenously) so as to reach to its target organ, i.e., lungs as shown in Fig. 5. In normal disease cells, virus could make entry into host cells via binding of RGD motif (in their spike receptor binding domain) to the integrins receptor proteins present on the surface of human lung cells. Ca+2 ions (or other bivalent ions) would augment the binding of viral spike protein with host cells and subsequently facilitate viral invasion into cells for infection, replication and pathogenesis. On the contrary, using EDTA chelation therapy, Ca+2 or other bivalent ions can be chelated and can be made unavailable to viral spike protein for attaching to host cells’ integrins. In parallel, strict control over diet (or medication) for ensuring proper availability of Ca+2 in the body may also be required for successful phenotypic remodeling for EDTA chelation therapy. In this way, virus could not enter cells and a state of protection against SARS-CoV-2 and COVID-19 can be achieved (Fig. 5).
Fig. 5.
Pulmonary EDTA chelation therapy which can be clinically executed through a nebulizer or inhaler to allow sodium-EDTA to pass into the lungs. The sodium-EDTA will chelates Ca+2 ions and other divalent ions making them unavailable for RGD-integrin mediated virus attachment to the host cells. A novel strategy for safe, technically simple, quick, cost-effective and efficient treatment of COVID 19 patients.
Besides ceasing the RGD-integrin mediated virus attachment to host cells, treatment with EDTA or EGTA is expected to cease SARS-CoV-2 viral infection in a multifaceted way. Firstly, Iexpect that chelating agents would disintegrate the viral membrane as Ca+2 forms major component of retrovirus membrane and are known to stabilize envelop proteins and others (Wunderlich and Sydow, 1982). Second, I also expect chelation of the metal cofactors within the enzyme active site of the SARS-CoV-2, as seen in HIV-1 IN enzyme (Hicks and Gulick, 2009). Third, EDTA is expected to lower metal-ion dependent reactive oxygen species (ROS) production and would also lipid peroxidation (Roussel et al., 2009). EDTA has also been shown to induce protective antioxidant and anti-inflammatory response in case of liver fibrosis (González-Cuevas et al., 2011). Finally, as COVID-19 patients exhibit excessive cytokine secretion (called as cytokine storm) (Mehta et al., 2020), use of EDTA is expected to render an anti-inflammatory and immuno-suppressive effect which would be highly beneficial. As such there are plenty of studies that confirm EDTA safe use in chelation therapy with no potent adversity and fatality (Roussel et al., 2009). The EDTA concentrations greater than 0.6 mM may led to reduced energy metabolism in lung cells (Asmus et al., 2001). Besides this, some complications such as hypocalcemia, proteinurea and renal insufficiency are known to associate with EDTA chelation therapy and I suggest that a strict monitoring protocol, comprising routine cytokine profiling, blood examination, urine test and CT scanning etc., must be practiced while treating patients with pulmonary EDTA chelation therapy.
5. Conclusion
In combination with the EDTA/EGTAbased pulmonary chelation therapy, modulation of integrins expression using integrins inhibitors or anti-integrin antibodies could also serve as a mechanism to treat COVID-19 patients (Henderson et al., 2013, Hatley et al., 2018, Sigrist et al., 2020). The expression of such integrins can be down regulated by modulation of intracellular redox ions such as superoxide anion (O2•−) and H2O2 (Puchsaka et al., 2016). Besides this, some bioactive compounds/molecules such as Ouabain are also known to affect integrins expression in lung cells (Ninsontia and Chanvorachote, 2014). Application of synthetic RGD tripeptide or its derivatives (linear, cyclic etc.) can also specifically block integrins and could also serve as a possible mechanism to discourage virus-host attachment (Bernhagen et al., 2017, Bernhagen et al., 2019, Ma et al., 2017). Besides this, PGNase treatment is also expected to be equally effective as the mass spectrometric studies revealed that all major glycan classes of human lung glycome can be released effectively by PNGase F (Jia et al., 2020). Since, spike protein of SARS-CoV-2 has a number of sites for cAMP, CK2, PKC and Tyr phosphorylation, I believe synergistic clinical application of other potential drugs such as kinase inhibitors can also be beneficial in COVID-19 treatment. Therapies specifically targeting TNF-α and IL-6 mediated signaling to downregulate cytokines secretion seems to be a promising approach to deal with the situation of cytokine storm. In this regard, tocilizumab (anti-IL6 monoclonal antibody) alone or in combination with infliximab and Emodin are also worth testing for COVID-19 treatment (Cao, 2020). Besides this, I also believe that blocking the calcium ion channels (or others) and downregulation of integrin-mediated cell signaling (GO:0007229) pathways such as FGF1 signaling, FGF2 signaling, NRG1-ERBB signaling, CX3CL1 signaling and CD40-CD40LG signaling are also available as a potent immunotherapeutic solutions against COVID-19. Finally, in the era of multi-omics technologies, medicines/therapies based on comparative coronovirus genome analysis and understanding of individual pathophysiology could pave way towards successful development of effective therapeutic solution for combating COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
I acknowledge Mohanlal Sukhadia University, Udaipur for the infrastructure facility.
Funding statement
The work has been conducted in absence of any funding.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imbio.2020.152021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Annes J.P., Rifkin D.B., Munger J.S. The integrin alphaVbeta6 binds and activates latent TGFbeta3. FEBS Lett. 2002;511:65–68. doi: 10.1016/s0014-5793(01)03280-x. PMID:11821050. [DOI] [PubMed] [Google Scholar]
- Arcangeli A., Becchetti A. Integrin structure and functional relation with ion channels. Adv. Exp. Med. Biol. 2010;674:1–7. doi: 10.1007/978-1-4419-6066-5_1. [DOI] [PubMed] [Google Scholar]
- Asmus M.J., Barros M.D., Liang J., Chesrown S.E., Hendeles L. Pulmonary function response to EDTA, an additive in nebulized bronchodilators. J. Allergy Clin. Immunol. 2001;107:68–72. doi: 10.1067/mai.2001.111591. [DOI] [PubMed] [Google Scholar]
- Bamonti F., Fulgenzi A., Novembrino C., Ferrero M.E. Metal chelation therapy in rheumathoid arthritis: a case report. Successful management of rheumathoid arthritis by metal chelation therapy. Biometals. 2011;24:1093–1098. doi: 10.1007/s10534-011-9467-9. [DOI] [PubMed] [Google Scholar]
- Bella J., Kolatkar P.R., Marlor C.W., Greve J.M., Rossmann M.G. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. U S A. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhagen D., Jungbluth V., Quilis N.G., Dostalek J., White P.B., Jalink K., Timmerman P. Bicyclic RGD peptides with exquisite selectivity for the integrin αvβ3 receptor using a “Random Design” approach. ACS Comb. Sci. 2019;21:198–206. doi: 10.1021/acscombsci.8b00144. [DOI] [PubMed] [Google Scholar]
- Bernhagen D., De Laporte L., Timmerman P. High-affinity RGD-knottin peptide as a new tool for rapid evaluation of the binding strength of unlabeled RGD-peptides to αvβ3, αvβ5, and α5β1 integrin receptors. Anal. Chem. 2017;89:5991–5997. doi: 10.1021/acs.analchem.7b00554. [DOI] [PubMed] [Google Scholar]
- Berridge M.J. Calcium signallingremodelling and disease. Biochem. Soc. Trans. 2012;40:297–309. doi: 10.1042/BST20110766. [DOI] [PubMed] [Google Scholar]
- Bianconi D., Unseld M., Prager G.W. Integrins in the spotlight of cancer. Int. J. Mol. Sci. 2016;17(12):2037. doi: 10.3390/ijms17122037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretaña N.A., Lu C.T., Chiang C.Y., Su M.G., Huang K.Y., Lee T.Y., Weng S.L. Identifying protein phosphorylation sites with kinase substrate specificity on human viruses. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0040694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaurock-Busch E., Busch Y.M. Comparison of chelating agents DMPS, DMSA and EDTA for the diagnosis and treatment of chronic metal exposure. Br. J. Med. Med. Res. 2014;4:1821–1835. [Google Scholar]
- Blobe G.C., Schiemann W.P., Lodish H.F. Role of transforming growth factor-β in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava C., Bertoli C., Castiglioni I. In-silico discovery of candidate drugs against Covid-19. Viruses. 2020;12:404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.W., Sheng Y., Gombold J.L. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology. 2000;269:212–224. doi: 10.1006/viro.2000.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., Zhuang J.Z. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-2019. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Zhao B., Iacob R., et al. Force interacts with macromolecular structure in activation of TGF-β. Nature. 2017;542:55–59. doi: 10.1038/nature21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb L., Liu J., Ockerhausen J., Kong Q., Garrad R.C., Griffin K., Neal C., Krugh B., Santiago-Pérez L.I., González F.A., Gresham H.D., Turner J.T., Weisman G.A. An RGD sequence in the P2y2 receptor interacts with αVβ3 integrins and is required for Go-mediated signal transduction. J. Cell Biol. 2001;53:491–502. doi: 10.1083/jcb.153.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero M.E. Rationale for the successful management of EDTA chelation therapy in human burden by toxic metals. BioMed Res. Int. 2016;2016:8274504. doi: 10.1155/2016/8274504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulgenzi A., Ferrero M.E. EDTA chelation therapy for the treatment of neurotoxicity. Int. J. Mol. Sci. 2019;20:1019. doi: 10.3390/ijms20051019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Cuevas J., Navarro-Partida J., Marquez-Aguirre A.L., Bueno-Topete M.R., Beas-Zarate C., Armendáriz-Borunda J. Ethylenediaminetetraacetic acid induces antioxidant and anti-inflammatory activities in experimental liver fibrosis. Redox Rep. 2011;16:62–70. doi: 10.1179/174329211X13002357050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer R.A., Cherian M.G., Jones M.M., Reigart J.R. Role of chelating agents for prevention, intervention and treatment of exposures to toxic metals. Environ. Health Perspect. 1995;103:1048–1052. doi: 10.1289/ehp.951031048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. EADock: Docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins. 2007;67:1010–1025. doi: 10.1002/prot.21367. [DOI] [PubMed] [Google Scholar]
- Hatley R.J.D., Macdonald S.J.F., Slack R.J., Le J., Ludbrook S.B., Lukey P.T. Anαv-RGD integrin inhibitor toolbox: drug discovery insight, challenges and opportunities. Angew. Chem. Int. Ed. Engl. 2018;57:3298–3321. doi: 10.1002/anie.201707948. [DOI] [PubMed] [Google Scholar]
- Hicks C., Gulick R.M. Raltegravir: the first HIV type 1 integrase inhibitor. Clin. Infect. Dis. 2009;48:931–939. doi: 10.1086/597290. [DOI] [PubMed] [Google Scholar]
- Henderson N.C., Arnold T.D., Katamura Y., Giacomini M.M., Rodriguez J.D., McCarty J.H., Pellicoro A., Raschperger E., Betsholtz C., Ruminski P.G., Griggs D.W., Prinsen M.J., Maher J.J., Iredale J.P., Lacy-Hulbert A., Adams R.H., Sheppard D. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Dong Q., Yang J., Zhang Y. Recognizing metal and acid radical ion binding sites by integrating ab initio modeling with template-based transferals. Bioinformatics. 2016;32:3260–3269. doi: 10.1093/bioinformatics/btw396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.F., Wilson C.L., Chow Y.H., Schnapp L.M. Role of integrin alpha8 in murine model of lung fibrosis. PLOS ONE. 2018;13 doi: 10.1371/journal.pone.0197937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H.A.M., Walker L.R., Abdel-Raouf U.M., Desouky S.A., Montasser A.K.M., Akula S.M. Beyond RGD: virus interactions with integrins. Arch. Virol. 2015;160:2669–2681. doi: 10.1007/s00705-015-2579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyser J.M., Estes M.K. Pathophysiological consequences of calcium-conducting viroporins. Annu. Rev. Virol. 2015;2:473–496. doi: 10.1146/annurev-virology-100114-054846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston E.R., Radke K. The SU and TM envelope protein subunits of bovine leukemia virus are linked by disulfide bonds, both in cells and in virions. J. Virol. 2000;74:2930–2935. doi: 10.1128/jvi.74.6.2930-2935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N., Byrd-Leotis L., Matsumoto Y., Gao C., Wein A.N., Lobby J.L., Kohlmeier J.E., Steinhauer D.A., Cummings R.D. The human lung glycome reveals novel glycan ligands for influenza a virus. Sci Rep. 2020;10:5320. doi: 10.1038/s41598-020-62074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;117627 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating J.A., Striker R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev. Med. Virol. 2012;22:166–181. doi: 10.1002/rmv.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck F., Ataey P., Amaya M., Bailey C., Narayanan A. Phosphorylation of single stranded RNA virus proteins and potential for novel therapeutic strategies. Viruses. 2015;7:5257–5273. doi: 10.3390/v7102872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Saphire E.O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhou Y., Zhang M., Wang H., Zhao Q., Liu J. Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020 doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Radiolabeled cyclic RGD peptides as integrin αvβ3-targeted radiotracers: maximizing binding affinity via bivalency. Bioconjug. Chem. 2009;20:2199–2213. doi: 10.1021/bc900167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Lu M., Munger J.S., Steadele M., Busald C., Tellier M., Schnapp L.M. Integrin alpha8beta1 mediates adhesion to LAP-TGFbeta1. J. Cell Sci. 2002;115:4641–4648. doi: 10.1242/jcs.00145. [DOI] [PubMed] [Google Scholar]
- Ma Y., Ai G., Zhang C., Zhao M., Dong X., Han Z., Wang Z., Zhang M., Liu Y., Gao W., Li S., Gu Y. Novel linear peptides with high affinity to αvβ3 integrin for precise tumor identification. Theranostics. 2017;7:1511–1523. doi: 10.7150/thno.18401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:P1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.G., Pozzi A., Zent R., Schwarzbauer J.E. Effects of high glucose on integrin activity and fibronectin matrix assembly by mesangial cells. Mol. Biol. Cell. 2014;25:2315–2317. doi: 10.1091/mbc.E14-03-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M., Re S., Mihara E., Nogi T., Sugita Y., Takagi J. Crystal structure of α5β1 integrin ectodomain: atomic details of the fibronectin receptor. J. Cell Biol. 2012;197:131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninsontia C., Chanvorachote P. Ouabain mediates integrin switch in human lung cancer cells. Anticancer Res. 2014;34:5495–5502. [PubMed] [Google Scholar]
- Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C.V., Hau J., Martin O., Kuznetsov D., Falquet L. improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh R., Lorenzo M.K., Shin S.Y., Pozzi A., Clark A.L. Integrin α1β1 differentially regulates cytokine-mediated responses in chondrocytes. Osteoarthr. Cartil. 2014;22:499–508. doi: 10.1016/j.joca.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peymen K., Watteyne J., Frooninckx L., Schoofs L., Beets I. The FMRFamide-like peptide family in nematodes. Front. Endocrinol. 2014;5:1–21. doi: 10.3389/fendo.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhary V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchsaka P., Chaotham C., Chanvorachote P. α-Lipoic acid sensitizes lung cancer cells to chemotherapeutic agents and anoikis via integrin β1/β3 downregulation. Int. J. Oncol. 2016;49:1445–1456. doi: 10.3892/ijo.2016.3624. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.-S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet. 2020 doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T., Podestá F., Stepp M.A., Boeri D., Lorenzi M. Integrin overexpression induced by high glucose and by human diabetes: potential pathway to cell dysfunction in diabetic microangiopathy. Proc. Natl. Acad. Sci. USA. 1993;90(20):9640–9644. doi: 10.1073/pnas.90.20.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A.M., Hininger-Favier I., Waters R.S., Osman M., Fernholz K., Anderson R.A. EDTA Chelation therapy, without added vitamin C, decreases oxidative DNA damage and lipid peroxidation. Altern. Med. Rev. 2009;14:56–62. [PubMed] [Google Scholar]
- Ruoslahti E. Fibronectin and its receptors. Ann. Rev. Biochem. 1988;57:375–413. doi: 10.1146/annurev.bi.57.070188.002111. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. ClinImmunol. 2020;108010 doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. The role of integrins in pulmonary fibrosis. Eur. Respir. Rev. 2008;17:157–162. doi: 10.1183/09059180.00010909. [DOI] [Google Scholar]
- Shi, Y. et al., 2020.Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Preprint at medRxiv. https://doi.org/10.1101/2020.03.12.20034736. [DOI] [PMC free article] [PubMed]
- Sigrist C.J.A., Bridge A., Mercier P.L. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177 doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaastad M.D., Nelson W.J. Integrin-mediated calcium signaling and regulation of cell adhesion by intracellular calcium. Bioessays. 1997;19:47–55. doi: 10.1002/bies.950190109. [DOI] [PubMed] [Google Scholar]
- Teoh C.M., Tan S.S.L., Tran T. Integrins as therapeutic targets for respiratory diseases. Curr. Mol. Med. 2016;15:714–734. doi: 10.2174/1566524015666150921105339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Uhlen M., Karlsson M.J., Zhong W., Tebani A., Pou C., Mikes J., Lakshmikanth T., Forsström B., Edfors F., Odeberg J., Mardinoglu A., Zhang C., von Feilitzen K., Mulder J., Sjöstedt E., Hober A., Oksvold P., Zwahlen M., Ponten F., Lindskog C., Sivertsson Å., Fagerberg L., Brodin P. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366:eaax9198. doi: 10.1126/science.aax9198. [DOI] [PubMed] [Google Scholar]
- Van Agthoven J.F., Xiong J.P., Alonso J.L., Rui X., Adair B.D., Goodman S.L., Arnaout M.A. Structural basis for pure antagonism of integrin αVβ3 by a high-affinity form of fibronectin. Nat. Struct. Mol. Biol. 2014;21:383–388. doi: 10.1038/nsmb.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-X., Ma J.-R., Wang S.-Q., Zeng Y.-Q., Zhou C.-Y., Ru Y.-H., Zhang L., Lu Z.-G., Wu M.-H., Li H. Utilizing integrating network pharmacological approaches to investigate the potential mechanism of Ma Xing Shi Gan Decoction in treating COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020;24(6):3360–3384. doi: 10.26355/eurrev_202003_20704. [DOI] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., Graham, B. S., McLellan, J. S., 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020, abb2507. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed]
- Wunderlich V., Sydow G. Disintegration of retroviruses by chelating agents. Arch. Virol. 1982;73:171–183. doi: 10.1007/BF01314725. [DOI] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Hongxia D., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12 doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.-S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kang, Z., Gong, H., Xu, D., Wang, J., Li, Z., Cui, X., Xiao, J., Meng, T., Zhou, W., Liu, J., Xu, H., 2020. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020, 927806; doi: https://doi.org/10.1101/2020.01.30.927806.
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.