Abstract
Background
Serological severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody assays differ in the target antigen specificity, e.g. of antibodies directed against the viral spike or the nucleocapsid protein, and in the spectrum of detected immunoglobulins. The aim of the study was to evaluate the performance of two different routinely used immunoassays in hospitalized and outpatient COVID-19 cases.
Methods
The test characteristics of commercially available spike1 protein-based serological assays (Euroimmun, EI-assays), determining IgA or IgG and nucleocapsid-based assays (Virotech, VT-assays) determining IgA, IgM or IgG were compared in 139 controls and 116 hospitalized and outpatient COVID-19 cases.
Results
Hospitalized COVID-19 patients (n = 51; 115 samples) showed significantly higher concentrations of antibodies against SARS-CoV-2 and differed from outpatient cases (n = 65) by higher age, higher disease severity scores and earlier follow up blood sampling. Sensitivity of the two IgG assays was comparable in hospitalized patients tested ≥ 14 days (EI-assay: 88%, CI95% 67.6–99.9; VT-assay: 96%, CI95% 77.7–99.8). In outpatient COVID-19 cases sensitivity was significantly lower in the VT-assay (86.2%, CI95% 74.8–93.1) compared with the EI-assay (98.5%, CI95% 90.6–99.9). Assays for IgA and IgM demonstrated a lack of specificity or sensitivity.
Conclusions
Our results indicate that SARS-CoV-2 serological assays may need to be optimized to produce reliable results in outpatient COVID-19 cases who are low or even asymptomatic. Assays for IgA and IgM have limited diagnostic performance and do not prove an additional value for population-based screening approaches.
Keywords: SARS-CoV-2, Serological assay, COVID-19
Abbreviations: CD, celiac disease; COVID-19, Corona Virus Disease 2019; NPV, negative predictive value; PPV, positive predictive value; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
1. Introduction
In December 2019 pneumonia cases were increasingly caused by a novel coronavirus, later termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in the city of Wuhan, Hubei province, in the People’s Republic of China [1]. The World Health Organization (WHO) officially named the related respiratory tract infections Corona Virus Disease 2019 (COVID-19) [2]. In the following months, COVID-19 spread worldwide and became a true pandemic with currently more than 34.8 million confirmed infections and more than 1 million deaths [3]. According to WHO recommendations, identification of infectious cases should rely on molecular methods [4]. These tests detect the presence of the virus, aiming to identify whether an individual is currently infected and potentially contagious.
Large-scale test strategies differ from country to country. Moreover, there still is a considerable number of false-negative results in the detection of viral nucleic acids due to pre-analytical differences in sample collection, transportation, RNA extraction methods, enzyme inhibitors and PCR chemistries applied. Detection is particularly difficult in clinically mild courses or asymptomatic patients [5]. Population based serologic testing might help to understand and countervail spreading mechanism of the pandemic.
Due to the high demand of reliable confirmatory tests, numerous serological assays were developed within a short period of time. Available tests differ in the epitope specificity of detected antibodies directed either against the spike protein, the receptor binding domain or the nucleocapsid of SARS-CoV-2 [6]. Thus, test formats were designed to discriminate between IgA, IgM and IgG, or IgA and IgG, total immunoglobulins or IgG only. However, clinical performance strongly depends on the design and the characteristics of the test applied [7], [8]. Of note, assays of various manufacturers are not standardized nor harmonized so far. Hence, it has been shown that cutoff optimization may reduce inter-assay differences [9] and improve sensitivity [7]. Moreover, most studies on the performance of serological tests have so far been conducted in hospitalized patients with severe disease progression [8], [10], [11], whereas the assay characteristics regarding asymptomatic carriers or patients with milder symptoms would be crucial for determining a more realistic prevalence within a population. To improve the knowledge about the applicability in the general population, we compared serological tests in hospitalized as well as outpatient COVID-19 cases with milder or no symptoms. We compared tests that have been validated in recent publications to discriminate between IgA or IgG against the spike protein (Euroimmun, EI-assay) [12], [13], [14], [15] or to distinguish between IgA, IgM or IgG directed against the nucleocapsid protein (Virotech, VT-assay) [16].
2. Methods
2.1. Ethics statement
The ethics committee of the Saxonian medical chamber approved the study (registry number EK-allg-37/10–1). All procedures utilized in this study were in agreement with the 1964 Declaration of Helsinki and its later amendments and written informed consent was obtained from all study participants.
2.2. COVID-19 patients
A diagnosis of COVID-19 was settled according to the WHO guidelines [4]. Serum samples from two COVID-19 disease groups with different clinical severity and progression were applied as follows:
-
(i)
The first group consists of hospitalized COVID-19 patients admitted between March 6 and May 2, 2020 to the Department of Infectious Diseases/Tropical Medicine, Nephrology and Rheumatology at Hospital St. Georg in Leipzig, Germany. Clinical severity was classified according to WHO: Moderate (1): uncomplicated upper airway symptoms without requirement of supplemental oxygen, none respiratory symptoms (vomiting/diarrhea/fever); Severe (2): receiving supplemental oxygen, Sp02 ≤ 90%, PaFiO2 ratio <300 mmHg, respiratory rate >30/min; Critical (3): receiving ventilatory support (nasal highflow canula, noninvasive ventilation, invasive ventilation), multiple organ failure. During hospitalization, we obtained 105 individual sera of 51 cases at different time intervals.
-
(ii)
The second COVID-19 group comprises individuals with asymptomatic and moderate symptoms who appointed our outpatient department (n = 65, one sample per patient).
Taken together, 170 specimens from 116 confirmed COVID-19 cases were analyzed. Duration from onset of symptoms was calculated by reviewing the electronical medical record. Furthermore, we assessed symptoms including shortness of breath, cough, fever, loss of taste or smell, headache and sore throat as published by the Centers for Disease Control and Prevention (CDC) [17].
2.3. Control cohort
Controls included 57 specimens of employees of the central fire brigade in Leipzig, Germany (blood withdrawal between March 28 and April 4, 2020) who were subjected to strict hygiene measures to prevent SARS-CoV-2 spreading and without any known contact to SARS-CoV-2 positive individuals. A second control group included and eight specimens from patients with other infectious respiratory diseases (influenza A/H1N1, one with human coronavirus HKU1, two with human coronavirus NL63, one with Mycoplasma pneumoniae, one with respiratory syncytial virus [RSV], and two with adenoviruses, all confirmed using a commercial respiratory multiplex-polymerase chain reaction (PCR) system). Both control groups were negatively tested for the presence of SARS-CoV-2 by real-time reverse transcriptase PCR (RT-PCR) seven to ten days before blood sampling. Furthermore, 50 serum samples of children and adolescents ≥5 years of age with acute or chronic gastrointestinal complaints, excluding patients under immunosuppressive therapy and those with known autoimmune diseases, and 17 serum samples of celiac disease (CD) patients with low (n = 5, 1–5 × the upper limit of normal [xULN)), moderate (n = 6, 6–20 xULN) and high levels (n = 6, 21–252 xULN) of IgA against tissue transglutaminases (IgA-TTG, Euroimmun, Lübeck, Germany) were included [18]. These samples were collected before the emergence of SARS-CoV-2 between 2011 and 2012 and were kept frozen at −80 °C.
2.4. SARS-CoV-2 RT-PCR
To detect SARS-CoV-2 virus particles, either nasopharyngeal swabs (Copan Liquid Amies eSwabs, Brescia, Italy) or pharyngeal lavage specimens were analyzed by RT-PCR. Specimens were subjected to cellular lysis and RNA extraction on a MagNA Pure 24 System (Roche, Mannheim, Germany) or QiaSymphony (Qiagen, Hilden, Germany). Real-time RT-PCR was conducted using LightCycler Multiplex RNA Virus Master Mix on a Lightcycler 480 RT system (both Roche, Mannheim, Germany) or a ViiA7 system (Applied Biosystems, Foster City, USA).
For SARS-CoV-2 analysis, the Sarbecovirus specific LightMix Modular SARS-CoV (COVID-19) E gene assay was used (TIB Molbiol, Berlin, Germany). EAV control (TIB Molbiol, Berlin, Germany) was used as extraction and internal PCR control. Samples from control patients with acute respiratory disease were additionally analyzed molecularly on the Biofire Filmarray system (bioMérieux, Marcy-l’Étoile, France) with the respiratory panel or the pneumonia plus panel. All (RT)-PCR reactions were performed according to manufacturer’s protocol.
2.5. Serological assays
We compared two different commercially available, CE-IVD labelled enzyme-linked immunosorbent assays (ELISA) for detection of antibodies against SARS-CoV-2 antigens. The EI-assays detect IgA or IgG antibodies that are directed against the S1 domain of SARS-Cov-2 spike protein (Euroimmun, Lübeck, Germany), whereas the VT-assays determine IgA, IgM or IgG directed against the nucleocapsid protein (Virotech, Rüsselsheim, Germany). Measurements were performed on an automated ELISA processor (DSX, Dynex Technologies, UK).
In case of discrepancies between these two assays in specimen of COVID-19 patients, showing no seroconversion during the course of disease, or lack of follow-up samples, an electrochemiluminescent SARS-CoV-2 immunoassay (herein referred as ‘Elecsys’, Roche, Mannheim, Germany) was applied. The assay detects IgG and IgM antibodies specifically directed against the nucleocapsid protein. The test results were reported as cutoff indices (COI). For this study, the indicated manufacturer’s cutoffs for borderline results were reported as positives (EI-assay: COI ≥ 0.8; VT-assay: COI ≥ 9; Elecsys: COI ≥ 1). All assays were performed according to manufacturers’ specifications.
2.6. Statistics
Diagnostic sensitivity was calculated based on molecular testing as gold standard to identify COVID-19 positive individuals. Specimens of hospitalized COVID-19 patients were grouped according to following time intervals for blood sampling after symptoms onset: 0–3 days, 4–7 days, 8–10 days, 11–13 days and ≥14 days.
Numerical variables were summarized as median and compared by non-parametric Mann-Whitney-U test. Categorical variables were given as frequencies or percentages with 95% Wilson-confidence intervals (CI95%). The McNemar's Test was used to compare diagnostic properties for two tests used on a single population and Fleiss’ kappa was chosen as a measure of agreement.
Positive predictive values (PPV) and negative predictive values (NPV) were estimated in dependency to prevalence using Bayes formula. For estimation of PPV and NPV, the entire control group as well as all COVID-19 cases sorted by blood sampling ≥14 days after symptoms onset were included. Yates correction was applied for tests with either 100% sensitivity and/or specificity to calculate PPV and NPV with CI95%. SPSS version 21 (IBM, Armonk, NY, USA) and GraphPad PRISM version 5 (GraphPad Software, San Diego, CA, USA) were used for statistical calculations and generation of figures.
3. Results
Demographics of COVID-19 cases are depicted in Table 1 . Hospitalized patients were of higher age and the disease grading was severe (23/51) or even critical (12/51), compared to moderate (60/65) or asymptomatic (5/65) in the outpatient COVID-19 cohort. Median blood sampling following onset of symptoms was 9 days (0–60) in hospitalized patients, and 36 days (24–67) in outpatient cases (Table 1).
Table 1.
Demographic characteristics of COVID-19 cases and the control cohort.
| COVID-19 hospitalized | COVID-19 outpatient | p value | fire brigade | children and adolescents (2011–2012) | respiratory infection | coeliac disease | |
|---|---|---|---|---|---|---|---|
| N | 51 | 65 | 64 | 50 | 8 | 17 | |
| Males (%) | 64.7 | 67.7 | n.s. | 89.1 | 36.0 | 87.5 | 35.3 |
| Median age | 64 (25–90) | 46 (14–64) | 0.012 | 38 (19–65) | 13 (5–18) | 57 (17–88) | 13 (7–55) |
| Blood sampling | 9 (0–60) | 36 (24–67) | <0.001 | ||||
| CT values | 26.7 (11.8–34.1) | 21.6 (15.4–32.4) | n.s. | ||||
| Severity of disease | |||||||
| 0 – asymptomatic | 2 | 5 | n.s. | ||||
| 1 – moderate | 14 | 60 | <0.001 | ||||
| 2 – severe | 23 | – | <0.001 | ||||
| 3 – critical | 12 | – | <0.001 |
Data is presented as the median with minimum and maximum. Clinical severity was classified as asymptomatic (0); moderate (1): uncomplicated upper airway symptoms without requirement of supplemental oxygen, none respiratory symptoms (vomiting/diarrhea/fever); severe (2): receiving supplemental oxygen, Sp02 ≤90%, PaFiO2 ratio <300 mmHg, respiratory rate >30/min; critical (3): receiving ventilatory support (nasal high-flow, noninvasive ventilation, invasive ventilation), multiple organ failure. Mann-Whitney-U test was used to analyze differences. n.s. = not significant.
3.1. Performance of antibody assays in hospitalized and outpatient individuals
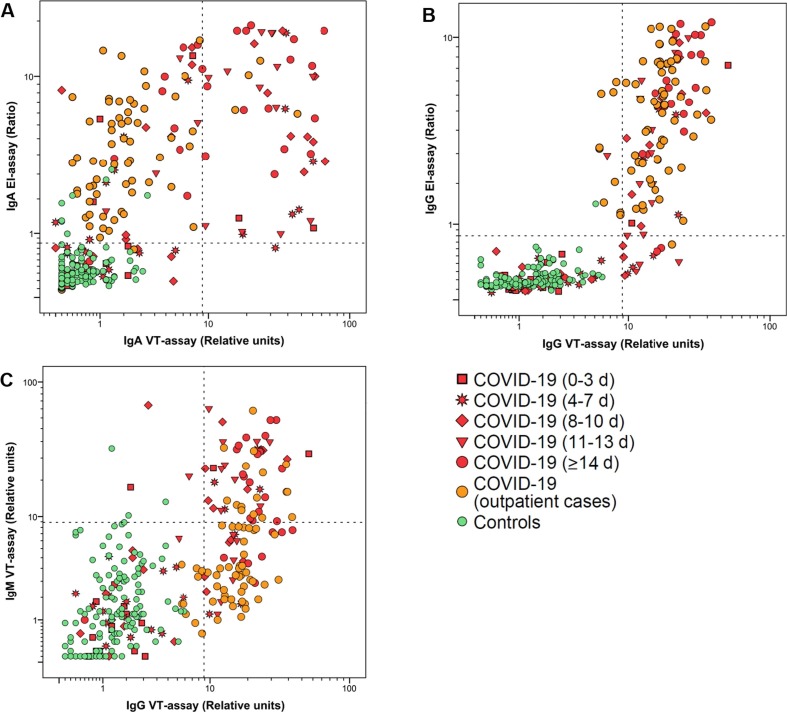
Distribution of IgA, IgG and IgM antibody results are depicted in Fig. 1 and calculated sensitivity and specificity values are shown in Table 2 . Comparison of specificity between IgA for the EI-assay (94.3%; 88.6–97.3%) and the VT-assay (100%; 96.7–100%) revealed significant differences, whereas performance of the IgG EI-assay and the VT-assay was comparable (99.3%; 95.5–99.9% and 100%; 96.7–100%). Of note, the IgM specific VT-assay resulted in 4 false-positive tested samples (97.1%; 92.3–99.1%). False-positive samples for IgA in the EI-assay were especially high in CD patients with low IgA-TTG concentrations (4/17).
Fig. 1.
Distribution of antibody results. Scatterplots visualizing the relation between IgA (A) and IgG (B) antibodies detected in EI- and Nucleocapsid-assays for SARS-CoV-2. Results of IgM antibodies directed against nucleocapsid were plotted against IgG antibodies (C). Dashed lines indicate cutoff values.
Table 2.
Performance of serological assays in dependence of time after onset of symptoms.
| IgA |
IgG |
IgM | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1-assay |
N-assay |
S1-assay |
N-assay |
N-assay |
|||||||||||
| n | pos. | % (CI95%) | pos. | % (CI95%) | p | κ | pos. | % (CI95%) | pos. | % (CI95%) | p | κ | pos. | % (CI95%) | |
| Sensitivity0-3 d | 16 | 5 | 31.2 (12.1–58.5) |
2 | 12.5 (2.2–39.6) |
n.s. | 0.470 | 2 | 12.5 (2.2–39.6) |
2 | 12.5 (2.2–39.6) |
n.s. | 0.772 | 3 | 18.8 (4.9–46.3) |
| Sensitivity4-7 d | 23 | 12 | 52.2 (31.1–72.6) |
7 | 30.4 (14.1–53.0) |
n.s. | 4 | 17.4 (5.7–39.5) |
7 | 30.4 (14.1–53.0) |
n.s. | 4 | 17.4 (5.7–39.5) |
||
| Sensitivity8-10 d | 24 | 16 | 66.7 (44.7–83.7) |
9 | 37.5 (19.6–59.2) |
0.016 | 11 | 45.8 (26.2–66.8) |
14 | 58.3 (36.9–77.2) |
n.s. | 10 | 41.7 (22.8–63.1) |
||
| Sensitivity11-13 d | 17 | 17 | 100 (0.77–100) |
13 | 76.5 (49.8–92.2) |
n.s. | 13 | 76.5 (49.8–92.2) |
15 | 88.2 (62.3–97.8) |
n.s. | 10 | 58.8 (33.4–80.1) |
||
| Sensitivity≥14 d | 25 | 24 | 96.0 (77.7–99.8) |
16 | 64.0 (42.6–81.2) |
0.008 | 22 | 88.0 (67.6–96.8) |
24 | 96.0 (77.7–99.8) |
n.s. | 17 | 68.0 (46.4–84.3) |
||
| Sensitivityoutpat. | 65 | 63 | 96.9 (88.4–99.5) |
4 | 6.2 (1.9–15.5) |
<0.001 | 0.004 | 64 | 98.5 (90.6–99.9) |
56 | 86.2 (74.8–93.1) |
0.021 | nd | 16 | 24.6 (15.1–37.1) |
| Specificity | 139 | 8 | 94.3 (88.6–97.3) |
0 | 100 (96.7–100) |
<0.001 | nd | 1 | 99.3 (95.5–99.9) |
0 | 100 (96.7–100) |
n.s. | nd | 4 | 97.1 (92.3–99.1) |
Seropositivity for IgA, IgG and IgM in 139 expected negative specimens and 170 specimens from 51 hospitalized and 65 outpatients with PCR-positive COVID-19 relative to days from onset of symptoms. Values for sensitivity and specificity are given as percentages with 95% Wilson-confidence intervals. McNemar's Test was used to compare diagnostic properties for two tests used on a single population and Fleiss’ kappa was chosen as a measure of agreement. pos. = number of positive tested samples; n.d. = not determinable, n.s. = not significant.
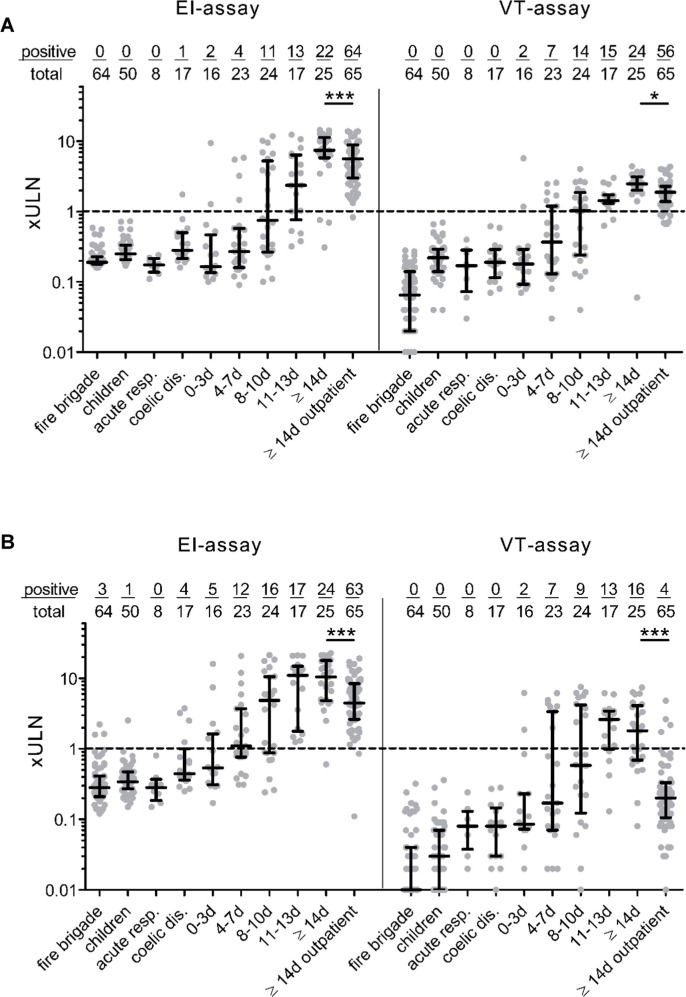
Sensitivities of the assays strongly depend on the time-point of blood-sampling after onset of symptoms (Table 2). To address the question if test performance also depends on the study population itself, we compared the different assays in severely-diseased hospitalized patients and low-symptomatic non-hospitalized COVID-19 individuals. In hospitalized patients, sensitivity of the IgA EI- and the VT-assay continuously increased from 31.2% (CI95% 12.1–58.5) and 12.5% (CI95% 2.2–39.6) at <3 days to 96.0% (CI95% 77.7–99.8) and 64.0% (CI95% 42.6–81.2) at ≥14 days after onset of symptoms. Differences at ≥14 days after onset of symptoms were statistically significant. Furthermore, a difference between the performances of both assays was observed in the outpatient group. Whereas the sensitivity of the IgA EI-assay was comparable between outpatient and hospitalized COVID-19 cases at ≥14 days (96.9% and 96%), the VT-assay returned a considerably reduced sensitivity in the outpatient cohort (6.2%; CI95% 1.9–15.5). Regarding analysis of the two different IgG assays, we observed a continuous increase in sensitivity characteristics in both EI-assay and VT-assay ranging from 12.5% (CI95% 2.2–39.6; both assays) at <3 days to 88.0% (CI95% 67.6–96.8) and 96.0% (CI95% 77.7–99.8) at ≥14 days after onset of symptoms. Again, we observed significant differences in the sensitivity when outpatients were tested with 98.5% (CI95% 90.6–99.9) for the IgG EI-assay and 86.2% (CI95% 74.8–91.1) for the VT-assay. Sensitivity of the IgM VT-assay ranged from 18.8% (CI95% 4.9–46.3) at <3 days to 68% (CI95% 46.4–84.3) at ≥14 days after onset of symptoms in hospitalized patients. In the outpatient COVID-19 cases, sensitivity of the IgM assay was limited to 24.6% (CI95% 15.1–37.1). Data are shown in Supplementary Fig. 1. Comparison of median values between hospitalized patients ≥14 days and outpatient cases revealed significant lower IgA and IgG levels values for outpatient COVID-19 cases in both the EI- and VT-assay (Fig. 2 ).
Fig. 2.
Clinical performance of IgA and IgG SARS-CoV-2 EI- and Nucleocapsid immunoassays Seropositivity for IgA (A) or IgG (B) in 139 control samples and 170 specimens from 51 hospitalized and 65 outpatient COVID-19 cases in relation to onset of symptoms. The control cohort consists of 4 subgroups, i.e. a local fire brigade (n = 57), patients with other acute respiratory diseases (n = 8), children and adolescents with acute and chronic gastrointestinal complaints (n = 50) and celiac disease patients with increased IgA concentrations (n = 17). Borderline results were reported as positives. For better visualization, COI-results were divided by the cutoff values for borderline results. Values are given as upper limit of normal (xULN). Dotted line represents cutoff value for positively tested samples. Data are given as the median and interquartile range. Mann-Whitney-U test was used to analyze differences between hospitalized COVID-19 patients with blood sampling ≥14 days after onset of symptoms and outpatient cohort, *p < 0.05, ***p < 0.001.
Concordances between IgA assays for hospitalized and outpatient individuals were 72.4% and 9%, respectively, and differed significantly between the EI-assay and the VT-assay (p < 0.001, κ = 0.470 and p < 0.001, κ = 0.004). For the IgG assays, the proportions of agreement were generally higher for both inpatients (88.6%, p = 0.006, κ = 0.772) and outpatients (84.6%, p = 0.021, κ not determinable).
3.2. Receiver operating curve (ROC) analysis and optimization of cutoff values
The distribution of IgA, IgM and IgG values show a high abundance of false-negative results when samples were analyzed with the VT-assay (Figs. 1 and 2), indicating that recommended cutoff values might be inadequate. We performed a ROC analysis to verify cutoff values of EI- and VT-assays (Supplementary Figure 2). The analysis of given samples revealed that the cutoff values for IgA and IgG VT-assays may be optimized to a COI of 3.5 and 6.4 without loss of specificity >99%, respectively.
3.3. Predictive values for IgG SARS-CoV-2 assays
IgG assays may represent a useful tool for population-based testing, however, the interpretation of the test results strongly depends on the prevalence of anti-SARS-CoV-2 positive individuals in the population. We observed a PPV for the IgG EI- and VT-assay of 98.9 and 98.8%, with a NPV of 97.2% and 93.3%, respectively, for a prevalence of 39% in our COVID-19 study cohort. Supplementary Table 1 depicts PPV and NPV in dependency of selected prevalence values to be assumed in large-scale screening scenarios.
3.4. Discrepant COVID-19 cases for IgG assays
Comparing results of the IgG assays (Fig. 1B), 12 specimens of 11 COVID-19 cases showed positivity for the VT-assay but were negative in the EI-assay if the optimized COI for the IgG VT-assay was applied. Six of 10 hospitalised patients showed seroconversion at a later time point for IgG detected in the EI-assay. Five patients had no follow-up serum sample to assess seroconversion, but seropositivity was confirmed in three cases (including one outpatient COVID-19 patient) using the electrochemiluminescent assay detecting cumulative antibodies including IgG (Supplementary Table 2). Two hospitalized patients (one case with an additional follow-up serum) with positivity in the VT-assay could not confirmed by the electrochemiluminescence assay. One was a 73-year-old male patient who was appointed to the hospital two days after positive PCR test with pronounced headache and nausea. Besides anosmia and Ageusia, the patient did not described any further symptoms. The second represented a 73-year-old male patient with diabetes type II which was primary hospitalized due to an abdominal phlegmon. Furthermore, an obstructive dyspnea (D-dimer 10315 µg/l) was observed. SARS-CoV-2 PCR screening revealed a Ct value of 35.2. In both cases, samples for PCR screening and antibody testing were obtained on the day of admission.
Three outpatient cases remained negative with the VT-assay but showed positivity for the EI-assay. The electrochemiluminescent assay confirmed the results of the EI-assay (Supplementary Table 1).
4. Discussion
Recently, a multitude of CE-IVD labelled immunoassays for antibody detection to SARS-CoV-2 were released. Proper assay sensitivity and specificity is highly relevant for epidemiologic analyses to estimate infection rates and to monitor the progression of the epidemic [19]. Here, we compared the serological testing performance of two commercially available anti-SARS-CoV-2 assays with different target antigens in severely-diseased hospitalized and low-symptomatic non-hospitalized COVID-19 patients.
Using the recommended thresholds, we identified significant discrepancies in sensitivity between the EI- and VT-assays, especially when outpatient COVID-19 cases were tested. In general, our data confirm that sensitivity of the assays increases with the time of blood sampling and produces reliable results when samples from hospitalized COVID-19 patients are tested ≥14 days after symptom onset [12], [13], [14]. Importantly, we detected relevant differences when outpatients were tested for both IgA and IgG, respectively. Compared to hospitalized COVID-19 patients, outpatient cases were younger, had lower disease severity and blood sampling was performed at later time points. We also observed nonsignificantly lower Ct values in outpatients compared to hospitalized patients. The reason for this observation could be the timing of PCR testing. Based on national test strategies, outpatient cases are identified very early after symptom onset or even before. In contrast, hospitalized patients get tested during admission into the hospital. Of note, in most patients the viral load in the upper respiratory tract already declines after day five from onset of symptoms [20].
Using the recommended thresholds, the sensitivity of the VT-assay against IgA and IgG was markedly lower (6.2% and 86.2%) for the outpatient group as compared to the hospitalized patients tested ≥14 days (64% and 96%). In contrast, sensitivity of the EI-assay against IgA and IgG was comparable between hospitalized patients (96% and 92%) and outpatient cases (96.9%% and 98.5%). Similar results were detected by Theel et al. who used an outpatient cohort to evaluate performance characteristics of two S1-assays and two N-assays to detect IgG antibodies against SARS-CoV-2 [15]. Of note, the lowest sensitivity (56.6%) was observed in the outpatient cohort using a nucleocapsid assay, whereas the other three assays produced comparable results (91.3–95.7%) [15].
Overall concordance between both tests were low for IgA but still acceptable for IgG. IgM was tested only in the VT-assay, resulting in high rates of false-negative results in hospitalized patients tested ≥14 days (sensitivity: 68%) and outpatient cases (sensitivity: 24.6%). Our study indicates that performance of the VT-assay strongly depends on the analyzed patient cohort, yielding a lower IgG or even inacceptable low IgA performance when outpatient cases with lower disease severity and later blood sampling were included. Recent findings indicate that hospitalized patients are characterized by an increased immune response compared to outpatients [21], [22]. Accordingly, in critical and non-critical hospitalized COVID-19 patients, no differences were found when seven different IgG assays were compared [11]. As serologic response differs between severely diseased hospitalized patients and low or even asymptomatic outpatients [21], [22], [23], our results indicate that outpatients with lower disease severity may prove a challenge for available assays. Moreover, our results confirm the findings of Schnurra et al., who have shown that sensitivity of the VT-assay is lower than other nucleocapsid-based assays when patients with mild symptoms were tested [16]. Accordingly, we conclude that observed discrepancies are based on the specific assay design.
Moreover, it has been shown that levels of IgA and IgM begin to decline within two months after symptom onset, whereas IgG against spike-protein and nucleocapsid stay relatively constant [6], [24], [25]. As observed differences in the outpatient cases might depend on declined antibody levels, we compared levels of IgA or IgG between the first and the last quintile regarding time point of blood sampling in the outpatients (Supplementary Fig. 3). Our results confirm that there is a nonsignificant decline of IgA. However, in both assays levels of IgG are constant in the upper and the lower quintile. Based on these results, we conclude that reduced sensitivity of the IgG VT-assay is rather caused by the specific assay then by declined antibody levels in the outpatients.
Concerning the high abundance of false negative results of the VT-assay, we performed a ROC analysis to optimize clinical performance. Applying the calculated optimal ROC cutoffs, performance of the IgG VT-assay distinctly increased and was comparable with the IgG EI-assay. We assumed that a control group with higher assay background was used for validation resulting in higher cutoffs to reach specificities of >99%. However, the cut offs were defined by the manufacturer and published in the manuals of the ICD-CE approved tests. The reason for this profound discrepancy remains ambiguous. Our results indicate that available serological tests may need to be optimized to produce reliable results in real populations with a high degree of individuals who were low or even asymptomatic and who have fully recovered from SARS-CoV-2 infection.
Regarding test specificity, we compared performance of serological tests in different control cohorts. Specificity was highly comparable between the EI-assay against IgG and the VT-assays to detect IgA, IgG and IgM, indicating a low number of false-positive test results. Otherwise, specificity was lower in the EI-assay for IgA (94.3%), confirming specifications of the manufacturer (given specificity: 92.5%). Notably, the small CD control group (n = 17) as a representative of frequent autoimmune diseases in Germany with a (sero)prevalence of 1.6% [26] displayed 4 false positive results for IgA and one for IgG in the EI-assay. We hypothesize that other auto-antibodies may cross-react in the EI-assay.
Given the early decline of anti-SARS-CoV-2 IgA and IgM [6], [24], [25], it is questionable if serological testing of IgA or IgM is useful for population based testing approaches. Since an antibody response takes several days [24], [27], serologic analyses are not indicated in the diagnosis and screening for active early infection. Thus, there is no clinical benefit to measure IgA or IgM.
As interpretation of serological testing largely depends on the prevalence in the population, we estimated and compared the NPV and PPV of the IgG EI- and VT-assay. Applying the modified cutoff for the IgG VT-assay, we estimated comparable PPV of 87.5% and 87.6% and NPV of 99.8% for both assays for a realistic prevalence of 5% [28]. Our results reveal that clinical performance of both assays highly depends on the tested population. Whereas there are large discrepancies if outpatients are tested, who have a high pre-test probability; differences are almost neglectable when optimized cutoffs are applied for population based screening approaches with low pre-test probability. However, reliable serological tests are urgently required in the event of a secondary or even seasonal recurrence of the virus. Assuming that increased antibody levels correlate with the recovery and protection of COVID-19 [29], public testing could help governments to determine when to ease lockdown measures or to allocate healthcare workers. Therefore, a specificity or PPV > 99% for serological tests seems necessary. However, it is unclear if antibodies against the SARS-CoV-2 nucleocapsid or the spike protein have protective characteristics over a long period of time [30].
A limitation of our study is that symptom onset was subjectively reported and that this information was retrieved by manual medical record review. The study was limited by the sample size and the number of serum samples was not equally distributed over the observational period, consequently influencing the accuracy. Moreover, only two manufacturers were compared. Hence, obtained results cannot be generalized to other SARS-CoV-2 serologic assays. It also needs to be considered that cutoff optimization may improves detection of outpatient COVID-19 cases at the expense of other applicability including identification of convalescent plasma donors.
In conclusion, our study indicates that the compared SARS-CoV-2 serological assays differ in clinical performance, especially when low or even asymptomatic outpatients are tested. Furthermore, our results revealed clear limitations for additional anti-SARS-CoV-2 IgA or IgM measurements for screening approaches.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to express their gratitude to the clinicians and nurses that handled and cared for COVID-19 patients included in this study. Furthermore, we recognize the willingness of all included individuals to participate in our investigation. We would like to particularly appreciate the supporting activity of Jeannine Dulz, Katrin Grabietz, Jeannette Hofmann, Michelle Karius, Sandra Reinhardt, Kerstin Rolle, Kathrin Schneider, Anne Tako, Katharina Wald, and Kristina Winter for serological and confirmatory testing at the Department of Laboratory Medicine at Hospital St. Georg Leipzig. We also thank David Petroff for statistical advice.
Contributions
J.W., R.B., S.B. and S.P. wrote the main manuscript text. S.B. and J.W. designed the research. J.W., R.B., and S.B. conducted the research. C.L., S.K., B.A., J.E., S.S., L.B., O.N. provided study material and collected medical records. J.W. and R.B. analyzed data and performed statistical analysis. B.I., T.K., and C.L. contributed with the valuable advice and by editing the manuscript. All authors read and approved the final manuscript. All authors reviewed the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.10.035.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO, Coronavirus disease Coronavirus disease, Situation Report. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201005-weekly-epi-update-8.pdf (accessed October 08, 2020).
- 4.WHO. Laboratory testing strategy recommendations for COVID-19. https://www.who.int/publications/i/item/laboratory-testing-strategy-recommendations-for-covid-19-interim-guidance (accessed October 08, 2020).
- 5.Y. Yang, M. Yang, C. Shen, F. Wang, J. Yuan, J. Li, et al., Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections, medRxiv [Epub ahead of print], unrefereed preprint as https://doi.org/10.1101/2020.02.11.20021493.
- 6.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. Favresse, C. Eucher, M. Elsen, T. Marie, J. Dogné, J. Douxfils, Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies, Clin. Chem. https://doi.org/10.1093/clinchem/hvaa131 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 8.M.S. Tang, K.G. Hock, N.M. Logsdon, J.E. Hayes, A.M. Gronowski, N.W. Anderson, C.W. Farnsworth. Clinical performance of two SARS-CoV-2 serologic assays, Clin. Chem. https://doi.org/10.1093/clinchem/hvaa120 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 9.Plebani M., Padoan A., Negrini D., Carpinteri B., Sciacovelli L. Diagnostic performances and thresholds: The key to harmonization in serological SARS-CoV-2 assays? Clin. Chim. Acta. 2020;509:1–7. doi: 10.1016/j.cca.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.P.D. Burbelo, F.X. Riedo, C. Morishima, S. Rawlings, D. Smith, S. Das, et al., Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients, J. Infect. Dis. https://doi.org/10.1093/infdis/jiaa273 [Epub ahead of print].
- 11.J. van Elslande, B. Decru, S. Jonckheere, E. van Wijngaerden, E. Houben, P. Vandecandelaere, et al., Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infecti. https://doi.org/10.1016/j.cmi.2020.07.038 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 12.Hörber S., Soldo J., Relker L., Jürgens S., Guther J., Peter S., et al. Evaluation of three fully-automated SARS-CoV-2 antibody assays. Clin. Chem. Lab. Med. (CCLM) 2020 doi: 10.1515/cclm-2020-0975. [DOI] [PubMed] [Google Scholar]
- 13.Jääskeläinen A.J., Kuivanen S., Kekäläinen E., Ahava M.J., Loginov R., Kallio-Kokko H., et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129:104512. doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.N. Kohmer, S. Westhaus, C. Rühl, S. Ciesek, H.F. Rabenau, Clinical performance of different SARS‐CoV‐2 IgG antibody tests, J. Med. Virol. https://doi.org/10.1002/jmv.26145 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 15.Theel E.S., Harring J., Hilgart H., Granger D., McAdam A.J. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58:565. doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnurra C., Reiners N., Biemann R., Kaiser T., Trawinski H., Jassoy C. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J. Clin. Virol. 2020;129:104544. doi: 10.1016/j.jcv.2020.104544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CDC, Symptoms of Coronavirus. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed October 08, 2020).
- 18.Wolf J., Petroff D., Richter T., Auth M.K.H., Uhlig H.H., Laass M.W., et al. Validation of antibody-based strategies for diagnosis of pediatric celiac disease without biopsy. Gastroenterology. 2017;153 doi: 10.1053/j.gastro.2017.04.023. 410-419.e17. [DOI] [PubMed] [Google Scholar]
- 19.S. Stringhini, A. Wisniak, G. Piumatti, A.S. Azman, S.A. Lauer, H. Baysson, et al., Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study, Lancet https://doi.org/10.1016/S0140-6736(20)31304-0 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 20.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 21.M. Becker, M. Strengert, D. Junker, T. Kerrinnes, P.D. Kaiser, B. Traenkle, et al., Going beyond clinical routine in SARS-CoV-2 antibody testing - a multiplex corona virus antibody test for the evaluation of cross-reactivity to endemic coronavirus antigens (2020).
- 22.M. Dogan, L. Kozhaya, L. Placek, C. Gunter, M. Yigit, R. Hardy, et al., Novel SARS-CoV-2 specific antibody and neutralization assays reveal wide range of humoral immune response during COVID-19, medRxiv htpps://doi.org/10.1101/2020.07.07.20148106 [Epub ahead of print].
- 23.Wellinghausen N., Plonné D., Voss M., Ivanova R., Frodl R., Deininger S. SARS-CoV-2-IgG response is different in COVID-19 outpatients and asymptomatic contact persons. J. Clin. Virol. 2020;130:104542. doi: 10.1016/j.jcv.2020.104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. Ma, W. Zeng, H. He, D. Zhao, D. Jiang, P. Zhou, et al., Serum IgA, IgM, and IgG responses in COVID-19, Cell Mol. Immunol. https://doi.org/10.1038/s41423-020-0474-z [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 25.K. Röltgen, O.F. Wirz, B.A. Stevens, A.E. Powell, C.A. Hogan, J. Najeeb, et al., SARS-CoV-2 antibody responses correlate with resolution of RNAemia but are short-lived in patients with mild illness, medRxiv https://doi.org/10.1101/2020.08.15.20175794 [Epub ahead of print].
- 26.Händel N., Mothes T., Petroff D., Baber R., Jurkutat A., Flemming G., Kiess W., Hiemisch A., Körner A., Schlumberger W., Thiery J., Wolf J. Will the real coeliac disease please stand up? Coeliac disease prevalence in the German LIFE Child Study. J. Pediatr. Gastroenterol. Nutr. 2018;67:494–500. doi: 10.1097/MPG.0000000000002052. [DOI] [PubMed] [Google Scholar]
- 27.J. Zhao, Q. Yuan, H. Wang, W. Liu, X. Liao, Y. Su, et al., Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019, Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa344 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 28.U.S. Food and Drug Administration, EUA Authorized Serology Test Performance, https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance (accessed October 08, 2020).
- 29.Casadevall A., Pirofski L. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130 doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Q. Long, X. Tang, Q. Shi, Q. Li, H. Deng, J. Yuan, et al., Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections, Nat. Med. https://doi.org/10.1038/s41591-020-0965-6 [Epub ahead of print]. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.