Abstract
Glioblastoma (GBM) is the most common and malignant brain tumor in adults. Genomic and epigenomic alterations of multiple cancer-driving genes are frequent in GBM. To identify molecular alterations associated with epigenetic aberrations, we performed whole exome sequencing-based analysis of DNA copy number variations in 55 adult patients with IDH-wild-type GBM. Beside mutations in common GBM driver genes such as TERTp (76%), TP53 (22%) and PTEN (20%), 67% of patients were affected by amplifications of genes associated with RTK/Rb/p53 cell signaling, including EGFR (45%), CDK4 (13%), and MDM2/4 (both 7%). The minimal deleted region at chromosome 10 was detected at the DNA demethylase TET1 (93%), mainly due to a loss-of-heterozygosity of complete chromosome 10 (53%) or by a mono-allelic microdeletion at 10q21.3 (7%). In addition, bi-allelic TET1 deletions, detected in 18 patients (33%), frequently co-occurred with EGFR amplification and were associated with low levels of TET1 mRNA expression, pointing at loss of TET1 activity. Bi-allelic TET1 loss was not associated with global concentrations of 5-hydroxymethylcytosine, indicating a site-specific effect of TET1 for DNA (de)methylation. Focal amplification of EGFR positively correlated with overall mutational burden, tumor size, and poor long-term survival. Bi-allelic TET1 loss was not an independent prognostic factor, but significantly associated with poor survival in patients with concomitant EGFR amplification. Rates of genomic TET1 deletion were significantly lower in a cohort of IDH1-mutated patients. Despite the relevance of TET1 for DNA demethylation and as potential therapeutic target, a frequent genomic loss of TET1 has not previously been reported in GBM.
Keywords: Glioblastoma (GBM), Next-generation sequencing (NGS), Copy number variation (CNV), IDH, TET1 deletion, EGFR amplification
Introduction
Glioblastoma multiforme (GBM) is the most common malignant brain tumor in adults with dismal prognosis [1]. Despite a combination of surgical resection, adjuvant radiation and chemotherapy with temozolomide in current standard treatment regimens and ongoing research for targeted therapies using specific drugs (e.g., EGFR kinase inhibitors), GBM mostly remains incurable with a high mortality rate and a 5-year survival of <6% [2]. Among several factors, a high inter- and intratumoral molecular heterogeneity limits confident predictions on drug resistance and therapy response [3,4].
Depending on histopathological and molecular (IDH1/2 status) features, GBM can be classified into IDH-wild-type (wt; ∼90% of cases) and IDH-mutated GBM [5]. Beside IDH mutations, a variety of recurrent genetic aberrations were reported in GBM, including common oncogenic DNA sequence alterations in genes such as EGFR, PTEN, NF1, and TERT promoter (TERTp) as well as large chromosomal aberrations (+7, −10) [6,7]. Moreover, submicroscopic DNA copy number variations (CNV; e.g., at EGFR, MDM2/4, CDK4, PTEN, and CDKN2A/B) were detected as oncogenic aberrations in GBM, ultimately affecting 3 main cell signaling pathways (p53, Rb, and RTK/Ras/PI3K) that initiate and sustain disease progression [8,9].
In addition to key molecular alterations, GBM can be distinguished based on transcriptional and epigenetic differences [6,7]. For example, IDH1-mutated GBM typically harbor other frequent genomic alterations such as mutations of ATRX and TP53 and are characterized by a distinct pattern of DNA hypermethylation via the conversion of α-ketoglutarate (αKG) to 2-hydroxyglutarate (2HG) and subsequent alterations of specific histone marks [10,11]. The production of 2HG inhibits several αKG-dependent oxygenases, including the ten-eleven translocation (TET) enzymes. The TET enzymes act by transcriptional activation or repression of target genes in many cellular processes. For instance, they are the primary mode of active DNA demethylation by catalyzing the re-oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) [12]. Epigenetic alterations initiated by mutations of IDH1/2 and/or TET1/2/3 and subsequent impaired production of 5hmC have been reported in several cancer entities such as myeloid malignancies [13,14].
To identify molecular alterations associated with epigenetic deregulation in IDH-wt GBM, we performed whole exome sequencing and evaluated for the presence of recurrent mutations, chromosomal aberrations and CNVs. Patients’ clinical characteristics were analyzed according to the mutational status. Hereby, we identified TET1 as frequently deleted gene, not previously described in The Cancer Genome Atlas (TCGA) database. Moreover, due to the importance of TET1 for DNA demethylation, we quantified corresponding levels of TET1 mRNA expression and global concentrations of 5hmC and 5mC as a surrogate measure for total DNA (de)methylation. Finally, to address the overall prevalence of this alteration in more detail we comparatively quantified levels of 5mC/5hmC and rates of genomic TET1 deletion in a cohort of 20 patients diagnosed with IDH1-mutated GBM, using PCR-based analysis of STR microsatellites.
Material and Methods
Patient Characteristics and Sample Collection
This study included a total of 55 patients with newly diagnosed IDH-wt GBM who underwent surgical resection at the University Hospital Carl Gustav Carus in Dresden. In addition, 20 patients with IDH1 mutated GBM were included for TET1 deletion mapping. All tumor tissue and blood samples (used as germline control) were collected with written informed consent and after approval by the local ethics committee of the Medical Faculty Carl Gustav Carus Dresden. Tumor tissues were taken intraoperatively and were snap frozen at -80°C. Frozen tumor tissue was sectioned using the Cryostat Jung CM 1800 (Leica, Wetzlar, Germany). To assure a tumor cell content of >80% for DNA extraction, a fresh hematoxylin and eosin-stained reference section was re-reviewed by an experienced local neuropathologist. Genomic DNA was isolated using the QIAmp DNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocols and quantified using NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
IDH and TERT-Promoter Mutation Detection
The IDH1/2 mutational status was determined in tumor tissue prior to exome sequencing by Sanger sequencing, using established primer sets, as published elsewhere [15]. The proximal TERT promoter, covering nucleotides C228 and C250, was amplified using a nested PCR procedure and sequenced on an Ion Torrent S5 NGS system, as described previously [16]. Data were analyzed using the Torrent Suite software v3.2 and the Torrent Variant Caller v.4.0 plugin with default settings and alignment to the hg19 human reference genome from the UCSC Genome Browser (http://genome.ucsc.edu/).
Exome Sequencing, Variant Calling, and Filtering
Library preparation and exome enrichment of paired blood and tumor samples from IDH-wt patients was performed using the SureSelect QXT and SureSelect Human All Exon V6 + COSMIC (Agilent Technologies, Santa Clara, CA, USA) protocols capturing ∼60Mb of exonic targets, according to manufactures protocols. After quantification and quality control using a Qubit 4.0 fluorometer (Life Technologies) and the Agilent 2200 TapeStation (Agilent Technologies), final libraries (mean insert size ∼250bp) were sequenced paired-end (2 × 75bp) on an Illumina HiSeq platform. Between 30 and 60 mio reads were generated per sample. Overall enrichment efficiency was ∼80% (on-target rate) with high coverage uniformity across all chromosomes. Sequencing depth was >10x for over 99% of sequence regions, reflecting the minimum coverage required for SNV analysis. Superior coverage of >30x and >60x was achieved for over 80% and 50% of the sequence regions, respectively. SNV analysis was performed with GATK [17] following the best practices recommended by the GATK team. Briefly, raw paired-end reads were mapped with BWA to human genome hg19 using default parameters [18] and duplicates were tagged using picardtools (http://broadinstitute.github.io/picard/). Insertions/deletions were realigned and base qualities recalibrated using GATK. Variants were identified for individual samples using the HaplotypeCaller tool of GATK in GVCF mode and genotypes were then jointly called using the GenotypeGVCFs tool of GATK. Annotation and filtering of somatic variants in coding regions was performed using tumor/germline pairs with a defined threshold of 10x (read depth) and a cut-off of 10% variant allele frequency using the VariantStudio software (Illumina Inc., San Diego, CA, USA) and the SeattleSeq Annotation 138 software (University of Washington, Seattle, WA, USA).
Copy Number Variation Detection and GISTIC Analysis
CNV analysis was performed with GATK following the best practices recommended by the GATK team. Briefly, raw paired-end reads were mapped with BWA to human genome hg19 using default parameters and duplicates were tagged with picardtools. The next steps were done with tools of the GATK framework. The sequence regions were divided into 1kb bins and counts per bin were obtained. Counts were corrected for GC, converted into log2 copy ratios (using sample median as baseline) and denoised with a panel of normals built from the count data of the blood samples. Additionally, allelic counts were determined for variant sites. Denoised copy ratios as well as allelic counts were then used to model copy ratio segments (i.e., adjacent bins sharing the same copy ratio). For tumor samples, the allelic counts of the matched blood sample were included as well to infer the original nontumor state of the genome. Finally, CN gains and losses were called for the modeled segments. To identify regions of significant amplification or deletion across the set of tumor samples, the GISTIC [19] module was run with the following parameters: -genegistic 1 -smallmem 1 -broad 1 -brlen 0.5 -conf 0.90 -armpeel 1 -savegene 1 -gcm extreme.
Short Tandem Repeat Analysis
To confirm somatic TET1 deletion in tumor/normal samples, loss of heterozygosity (LOH) was mapped using PCR-based analysis of short tandem repeat (STR) loci D10S2480 and GATA121A08 (both in close distance to TET1, 10q21.3), according to standard protocols on an ABI 3130xl (Life Technologies). PCR was carried out using the Qiagen Multiplex PCR Kit (Qiagen, Hilden, Germany) with following Primer sequences: (for D10S2480) 5’-CTGAGTTAGGGTCTTGCTATG-3’ (Forward); 5’-TAAGGAAGACAAACTCATTATTTCC-3’ (Reverse) and (for GATA121A08) 5’-GTTAACAGACTATTACCTGCCTACC-3’ (Forward); 5’-TGAGTATGCCACACTGCA-3’ (Reverse). Additionally, STR analysis was performed in a group of 20 patients diagnosed with IDH1-mutated GBM.
TET1 Expression Profiling
Total RNA from frozen tumor tissue was isolated using the RNeasy Mini Kit (Qiagen) in a QIAcube (Qiagen) according to manufacturer's protocols. Conversion to cDNA was performed using the SuperScript VILO MasterMix with 11.5 µL of RNA for RT (Invitrogen Corporation, Carlsbad, CA, USA). TET1 expression was quantified using a custom TaqMan gene expression assay (Thermo Fisher Scientific, ref: 4331182) with gene specific PCR primers and the TaqMan Universal Master Mix on a 7500 qRT-PCR device (Applied Biosystems, Foster City, CA, USA). Levels of TET1 mRNA were quantified relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Delta-CT values) expression as internal control. The 2^(Delta-Delta-Ct) method was used to calculate relative differences of TET1 mRNA expression between samples, normalized to TET1-wt gene expression.
Quantification of 5-methylcytosine and 5-hydroxymethylcytosine
Global concentrations of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) were quantified on genomic DNA of IDH-wt and IDH-mutated tumors by an ELISA using the MethylFlash Methylated and Hydroxymethylated DNA Quantification Kit (Fluorometric; Epigenetik, Farmingdale, NY, USA) according to the manufacturer's protocol.
Radiological Evaluation and Volumetric Measurements
All patients received a pre- and postoperative MRI with T1, T1 postcontrast, T2, and FLAIR sequences. Radiologic progression was determined using the criteria for Radiologic Assessment in Neuro-Oncology [20]. Additionally, the preoperative T1-weighted postcontrast MR scans were used to measure the enhancing tumor size and obtain volumetric measurements. A combination of manual segmentation using the ITK-SNAP software [21] and intensity filtering based on individually adjusted thresholds was used to select the enhancing tumor tissue with and without the necrotic tumor portion.
Clinical Data and Statistical Analysis
Demographic data, pathological findings and clinical data were retrospectively collected for patients with IDH-wt GBMs. Progression-free survival (PFS) was calculated from the day of first surgery until imaging-based detection of tumor progression (or end of follow-up). Overall survival (OS) was defined as the period from the day of first surgery until death (or the end of follow-up). The statistical association of clinical variables (e.g., age, tumor size, rates of somatic variants, survival) was evaluated using the 2-sided Students t test and Mann-Whitney U test. Survival analysis (PFS and OS) was carried out using the Kaplan-Meier technique. To test for statistical significant differences between survival curves the Mantel-Cox test (Log-rank test) was used. P values <0.05 were considered significant. All calculations were conducted using Prism 5 (GraphPad, La Jolla, CA, USA) and SPSS Statistics 25 (IBM, Armonk, NY, USA).
Results
Demographic Characteristics and Pathological Findings
A total of 55 patients (33 male/22 female) with newly diagnosed IDH-wt GBM were included. Median age at first diagnosis was 58 y (range 23–80 y; interquartile range 44–73 y). IDH-wt classification of tumors was confirmed by Sanger sequencing covering residues R172 (IDH2) and R132 (IDH1) in all patients. Tumors affected the left (n = 22); right (n = 31) or both hemispheres (n = 2) and were mainly localized in the frontal lobe (n = 18), temporal lobe (n = 13), parietal lobe (n = 3), occipital lobe (n = 2), or in multiple lobes (n = 13). Median tumor size was 43.8 cm3 (range 0.53–132.89 cm3) with Ki67 levels ranging between 5% and 90% (median 30%). Methylation of the MGMT promoter was detected in 17 tumors (30.9%). Patient's median OS was 14.4 mo (range 2–66 mo). During the time of follow-up, 47 patients (85%) passed away, 3 patients were lost from follow-up and 5 patients were still alive at the last update of disease progression, conducted in January 2020. Significant associations of clinical variables with outcome were observed using median values of Ki67 proliferation marker (21.7 vs 11.5 mo; P= 0.0002) and tumor size (17.6 vs 12.0 mo; P= 0.020) to dichotomize patients (Figure S1). Moreover, there was a trend towards shorter survival with increasing age (P= 0.071). Demographic characteristics and pathological findings of patients are presented in Supplementary Table S1.
Mutational Landscape of Tumors
Overall, after filtering (≥10fold coverage; ≥10% VAF) and exclusion of frequent polymorphisms (>0.1% population frequency), synonymous variants, variants in noncoding regions (intronic and UTRs) and germline variants (based on tumor/blood subtraction), a total number of 1841 somatic mutations in 1544 different genes were detected in the 55 tumor exomes, corresponding to a mean rate of ∼33 somatic variants per patient. The majority of mutations (93%) were single nucleotide variants (SNVs) (n = 1719), comprising 1616 missense and 103 nonsense SNVs, followed by frame-shift (n = 76) and in-frame InDel mutations (n = 46). Mutations were detected with a median VAF of 31% (range 10%–98%).
As summarized in Figure 1A, we identified a series of frequently mutated genes (grey bars), including well-known GBM driver genes such as TERTp (76%), EGFR (29%), TP53 (22%), PTEN (20%), NF1 (9%), ATRX (5%), PIK3CA (5%), PIK3R1 (4%), and PDGFRA (4%). Other previously reported candidate genes, recurrently mutated in our cohort (in 2 or more patients) were LZTR1 (4%), KIT (4%), and KMT2B (4%). Importantly, we did not detect any TET1 or TET2 mutations in our study group. Beside alterations in common recurrent driver genes, most patients carried one or more additional mutations in cancer associated genes functionally linked to transcriptional regulation (e.g., RB1, RUNX1, CIITA, CNOT3, NCOR1/2, SALL4), histone modification (e.g., SETD2), chromatin-remodeling (e.g., CREBBP and INO80), the SWI/SNF complex (SMARCA1), alternative splicing (e.g., DDX3X and RBM10), or signaling (e.g., MET, MTOR, BRAF, ERBB2; Figure S2). No associations with clinical outcome (PFS/OS) were observed for the mutational status of individual driver genes, including TERTp, EGFR, TP53, PTEN, and NF1 (Figure S3).
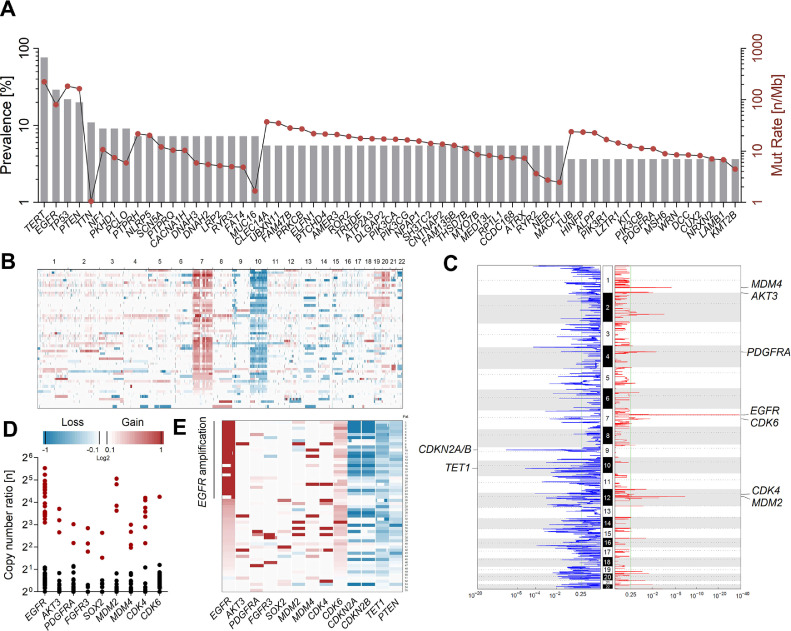
Figure 1.
Somatic genomic alterations in IDH-wt Glioblastoma. (A) Prevalence (%; gray bars) and mutation rate (n Mb−1; red dots) of frequently mutated genes in coding regions from 55 tumor exomes. Filtering of somatic variants was performed using tumor/germline pairs with a defined cut-off of 10% variant allele frequency (VAF). TERT promoter mutations at residues C228 and C250 were analyzed using a targeted NGS procedure (B) Overall genomic copy number variations (CNVs) and chromosomal locations in 55 IDH-wt GBM. (C) Recurrent sites of focal amplification (red) and deletion (blue) determined by the GISTIC module. Most significant focal amplification was detected for EGFR (chr7; q-value = 6.74E−39). The minimal region of allelic loss at chromosome 10 was detected for TET1 (q-value = 3.16E−06). All statistically significant (q-value <0.25) regions are summarized in Supplementary Table S2. (D) Copy number ratios (CNR) of genes frequently affected by focal amplification (CNR >2; red dots). (E) Association of the EGFR amplification status with genomic gains (red) and losses (blue) in relevant oncogenes.
Profiling of Copy Number Variations
NGS coverage-based assessment of copy number ratios (CNR) in exomes from tumor/blood pairs revealed a median frequency of 152 somatic CNVs per patient. Genomic losses accounted for the majority (66%) of detected CNVs (34% gains). Overall, CNV patterns virtually resembled genomic profiles previously reported for IDH-wt GBM, including characteristic gains (CNR 1–2) at chromosome 7p (87%) and/or 7q (82%) as well as partial or complete loss-of-heterozygosity (LOH) at chromosome 10p (78%) and/or 10q (85%) including the tumor suppressor PTEN (10q23; Figure 1B). Large chromosomal gains also affected chromosomes 19 (18%), 20 (27%), and 21q (11%).
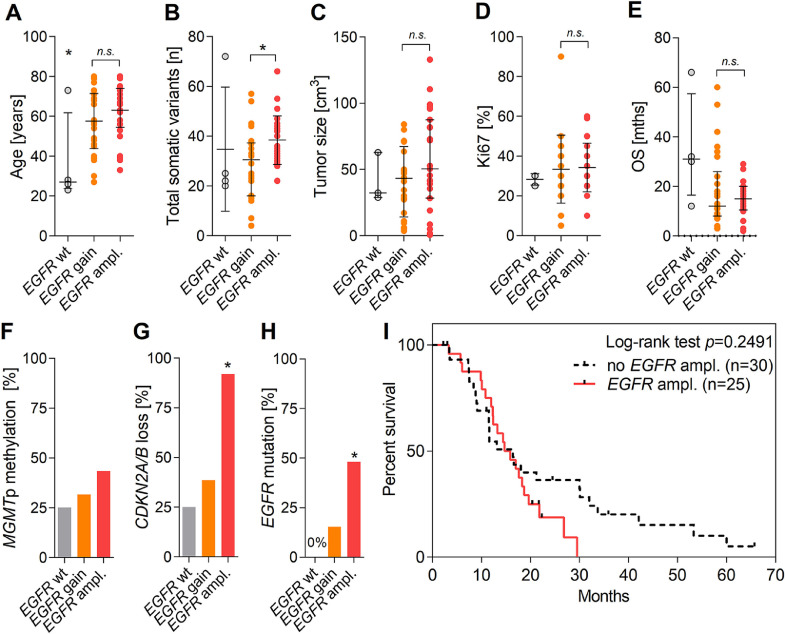
In accordance with the TCGA database, highest rates of focal amplification were detected for EGFR, with CNRs ranging between 8 and 46 fold in roughly half of tumors (45%) (Figure 1C and D). Generally, the EGRF status was not significantly associated with patient's age (Figure 2A). Amplification of the EGFR gene was significantly associated with rates of overall mutational burden (P= 0.0051; Figure 2B), genomic loss of CDKN2A/B at 9p21 (P< 0.0001; Figure 2G) and rates of concomitant EGFR mutations (P< 0.0001; Figure 2H). In addition, EGFR amplification positively correlated with histopathological measures of tumor size (P= 0.0836; Figure 2C), MGMT promoter methylation (P= 0.3706; Figure 2F) and Ki67 staining (P= 0.7910; Figure 2D). Generally, EGFR amplification was not associated with median PFS and OS of patients (P= 0.2283; Figure 2E). However, obviously EGFR amplification correlated with poor long-term survival, irrespective of comparing survival rates at 2 y (33% vs 20%), 3 y (17% vs 0%), or 5 y (7% vs 0%) for patients without or with EGFR amplification (Figure 2I).
Figure 2.
Associations of EGFR amplification in IDH-wt GBM. (A) Age at diagnosis (y), (B) total amount of somatic mutations (n), (C) tumor size (cm3), (D) Ki67 value (%), (E) overall survival (mo), (F) MGMTp methylation (%), (G) CDKN2A/B loss (%) and (H) concomitant EGFR mutations (%) in patients with EGFR-wt (n = 4), EGFR copy number gain (n = 26) and EGFR amplification (n = 25). Error bars indicate median values with interquartile range. (I) Kaplan-Meier analysis showing overall survival (mo) of IDH-wt patients with (n = 25) or without (n = 30) focal amplification of the EGFR gene. Statistical associations were evaluated using the two-sided Students t test, the Mann-Whitney U test and the Mantel-Cox test. P values <0.05 were considered significant.
Almost mutually exclusive to EGFR amplifications, other frequent RTK aberrations were detected at FGFR3 (5%), PDGFRA (4%), AKT3 (5%), and SOX2 (Figure 1E). Furthermore, focal amplifications in other regulatory pathways affected MDM2/4 (both 7%), CDK4 (13%), and CDK6 (2%; Figure 1D). Mutually exclusive to CDK4 amplifications, 65% of patients carried typical deletions of CDKN2A/B with CNRs clearly <0.5 in the majority of cases (∼75%), demonstrating a predominance of homozygous loss at 9p21 (Figure 1C and E). Other cancer associated genes with frequent heterozygous deletions (CNR 0.5–1) were CDK7 (55%), RB1 (25%), and TP53 (13%). The mutational profiles and CNV patterns of IDH-wt exomes are shown in Figure 3. A list of all regions affected by significant genomic deletion and amplification detected using GISTIC analysis is presented in Supplementary Table S2.
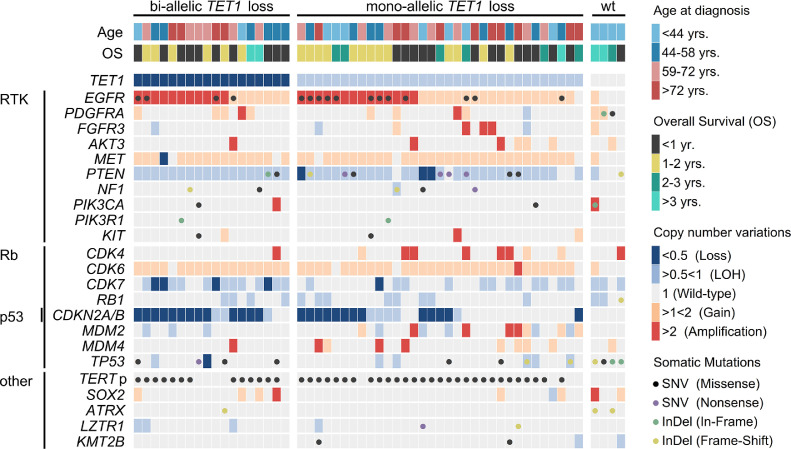
Figure 3.
Somatic profiles of IDH-wt tumors with or without bi-allelic TET1 loss. Distribution of somatic alterations (dots): missense single nucleotide variants (SNV; black), nonsense SNV (purple), in-frame (green) and frame-shift (yellow) insertion/deletion (InDel) mutations. Copy number variations (color coded by type) in relevant oncogenes related to RTK/Rb/p53 signaling pathways and clinical characteristics (age and overall survival) of 55 patients with IDH-wt GBM. TET1 copy number ratios <0.5 are indicative for bi-allelic deletions in at least a subset of tumor clones and were classified “loss.” Associated alterations are shown for patients with bi-allelic TET1 loss, mono-allelic TET1 deletion and TET1 wild type status. The distribution of mutations in other putative oncogenes is presented in Figure S2.
Frequent Copy Number Loss of TET1
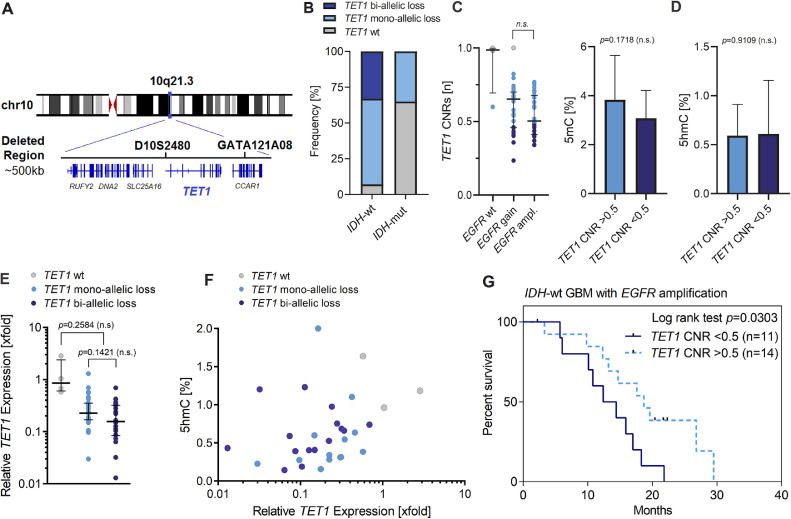
Interestingly, the minimal region of allelic loss at chromosome 10 was detected at 10q21.3, covering a genomic size of ∼500 kb (Figures 1C and 4A). This minimal deleted region in our cohort is not listed in the TCGA datasets and affects a series of genes including RUFY2, DNA2, SLC25A16, CCAR1, and the DNA demethylase TET1 (Tet Methylcytosine Dioxygenase 1). Overall, TET1 deletions were detected in 51 patients (93%) with CNRs ranging between 0.23 and 0.82 (median CNR = 0.58; Figure 3 and Supplementary Table S3). The majority of patients were affected by a heterozygous loss of TET1 due to the presence of LOH at chromosome 10 (53%). Heterozygous TET1 deletions in patients without LOH of chromosome 10 were detected in 7% of GBM patients, indicating the presence of submicroscopic micro-deletions at 10q21.3. In addition, TET1 deletion was detected with CNRs <0.5 in 18 patients (33%) with LOH of chromosome 10, pointing at bi-allelic micro-deletions at 10q21.3 (i.e., not detected at other chr10 regions including genes such as PTEN), inducing complete loss of TET1 activity (Figure 4B). STR-based detection of LOH in tumor/blood pairs, using TET1 microsatellite markers D10S2480 and GATA121A08, confirmed somatic deletion at 10q21.3 in 48 patients with informative results (Supplementary Table S3). LOH detection was not possible in three patients with homozygous STR loci in germline controls. Based on LOH detection using STRs, all bi-allelic TET1 deletions occurred in IDH-wt GBM, whereas only 35% of IDH1-mutated GBM harbored a mono-allelic loss of TET1 (Figure 4B and Supplementary Table S3; P= 0.0011).
Figure 4.
Molecular and clinical associations of genomic TET1 loss. (A) Schematic illustration showing the position of acquired genomic deletion at 10q21.3 (chr10:70066243-70548143) covering a size of ∼500 kb, affected genes (RUFY2, DNA2, SLC25A16, TET1, and CCAR1) and the position of short tandem repeat (STR) microsatellite loci D10S2480 and GATA121A08 used for TET1 deletion mapping in IDH-wt and IDH-mutated GBM. (B) Frequency (%) of TET1 deletion in IDH-wt and IDH-mutated GBM. (C) Association of TET1 copy number ratios (CNRs) with EGFR amplification status. (D) Concentrations (% of gDNA) of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) as measure for global DNA (de)methylation in IDH-wt GBM with mono-allelic (CNR >0.5) and bi-allelic (CNR <0.5) TET1 deletion. (E) The mRNA expression level (2^ddCt) of TET1 measured by qRT-PCR in samples with TET1-wt (n=4), mono-allelic TET1 deletion (n = 33) and bi-allelic TET1 loss (n = 18). Error bars indicate median values with interquartile range. Relative changes to TET1-wt gene expression was analyzed using the Delta-Delta-Ct algorithm. GAPDH expression was used as internal control. (F) Relative TET1 expression [2^ddCt] and corresponding concentrations of global 5hmC (%) in patients with available 5hmC data (n = 31). (G) Kaplan-Meier analysis showing overall survival (mo) of patients affected by EGFR amplification and with (n = 11) or without (n = 14) concomitant bi-allelic loss (CNR <0.5) of the DNA demethylase TET1. Statistical associations were evaluated using the 2-sided Students t-test, the Mann-Whitney U test and the Mantel-Cox test. P values <0.05 were considered significant.
In IDH-wt GBM, bi-allelic TET1 loss (CNR <0.5) was enriched in patients with EGFR amplification (Figures 3 and 4C). No associations were observed with other frequently affected genes such as TP53, CDKN2A/B, or TERTp (Figure 3). Likewise, no significant correlations of bi-allelic TET1 loss with clinical parameters including age, tumor size and Ki67 value were observed. Interestingly, bi-allelic TET1 deletions were not associated with global concentrations of 5hmC (ranging between 0% and 1% of genomic DNA) or 5mC (accounting for 3%–4% of gDNA) in IDH-wt GBM (Figure 4D). Likewise, no significant differences of 5mC/5hmC levels were detected between IDH-wt and IDH-mutated tumors (Fig S4).
However, the genomic loss of TET1 was associated with reduced levels of TET1 mRNA expression in tumors with mono-allelic (relative expression ∼24%) and bi-allelic (relative expression ∼15%) TET1 deletions, compared to control samples (TET1-wt; Figure 4E). In addition, there was a general trend toward higher concentrations of 5hmC with increasing levels of TET1 mRNA expression (Figure 4F). With respect to outcome, genomic TET1 loss and mRNA expression were not independent prognostic factors for PFS and OS in our cohort. However, in patients with EGFR amplification, bi-allelic TET1 loss (CNR <0.5) was significantly (P= 0.0303) associated with decreased OS (median OS = 12.4 mo) compared to patients with TET1 CNRs >0.5 (median OS = 18.7 mo; Figure 4G).
Discussion
Frequent Genomic Loss of TET1 in IDH-wt GBM
In this study, we detected bi-allelic loss of TET1 in 33% of patients in a minimal deleted region at 10q21.3, not previously described for GBM. Correspondingly, genomic TET1 loss was associated with lower levels of TET1 mRNA, demonstrating a reduced expression of TET1 in these tumors. Interestingly, TET1 expression in GBM with bi-allelic TET1 loss was still ∼15% of control samples without TET1 copy number loss (wt), pointing to the presence of micromilieus and intratumoral heterogeneity with TET1 mRNA expression at subclonal levels. Similarly, TET1CNRs of samples with bi-allelic TET1 loss ranged between 0.23 and 0.49, which is indicative for complete genomic TET1 inactivation in a subset of tumor clones. Subsequently, we show that genomic TET1 deletions are frequent alterations primarily associated with IDH-wt GBM. In addition, EGFR amplification and bi-allelic TET1 deletions co-occurred frequently, suggesting a functional interaction indicative of a distinct pathway of genesis in this subset of IDH-wt GBM. More importantly, although the presence of bi-allelic TET1 deletions alone were not an independent prognostic factor, patients with mutual TET1 loss and EGFR amplifications showed significantly worse outcomes compared with patients with only one or none of these alterations. In line, oncogenic EGFR amplifications were recently shown to induce silencing of tumor suppressor genes by repressing the DNA demethylase TET1 in GBM cell lines [22]. Moreover, the latter work reported on a synergistic effect of TET1 in blocking tumor growth by modulating the response to EGFR inhibitors [22].
In addition to EGFR induced repression, the enzymatic inhibition of the TET proteins (TET1/2) via the production of 2-hydroxyglutarate (2HG) is a common mechanism in IDH1-mutated GBM, contributing to the establishment of the CIMP phenotype in proneural GBM [10]. However, while mutations of TET2 are frequently associated with malignant progression in diverse cancers, so far, data on genomic alterations of TET1 in cancer is scarce, except for a frequent DNA copy number loss of TET1 recently reported in prostate cancer [13,14,23]. Notably, as no copy number alterations at 10q21.3 were detected in corresponding blood samples, the presence of germline aberrations or technical errors (e.g., insufficient enrichment or sequencing) can likely be excluded.
In GBM, TET1 was shown to play an important role in the tumorigenicity of GBM cells and to confer recruitment of the CHTOP-methylosome complex by the production of 5hmC [24,25]. Consistently, we show that concentrations of 5hmC were associated with levels of TET1 mRNA expression in IDH-wt GBM. The key role of TET1 for active DNA demethylation in the adult brain via the conversion of 5mC to 5hmC at CpG islands has been previously described [12,26]. Unexpectedly, global concentrations of 5hmC (and 5mC) as measure for TET induced DNA demethylation, were not associated with bi-allelic TET1 loss in our cohort. This is indicative for the presence of alternative mechanisms contributing to overall levels of 5hmC, like the activity of other enzymes involved in active DNA demethylation, namely TET2/3. For example, while TET1 mainly mediates the conversion of 5mC to 5hmC at CpG islands of gene promoters, TET2 and TET3 are involved in DNA demethylation of whole gene bodies (and intronic regions), which is significantly linked to overall cellular production of 5hmC [12,27]. In addition, 5hmC levels are likely affected by passive DNA demethylation in proliferating cells and/or the nuclear exclusion of the TET1 protein, as common feature in glioma cells [28]. Similarly, we did not see any significant differences of 5hmC concentrations in our IDH1-mutated GBM cohort, where genomic loss of TET1 was detected with significantly lower frequencies. To fully understand functional and prognostic consequences of TET1 deletion as potential therapeutic target in IDH-wt GBM, future investigations may specifically profile aberrant DNA methylation at CpG islands in relation to the genomic TET1 loss and expression patterns.
Somatic Genomic Alterations of IDH-wt GBM
Mutations in our cohort which exclusively includes IDH-wt GBM were detected with a mean rate of 33 coding variants per tumor exome, corresponding to a frequency of ∼1.2 somatic mutations per megabase, which is in the range of mutation frequencies previously reported for GBM [6,29]. In line with previous reports [5,6,30], 91% of patients were affected by mutations in classical GBM driver genes TERTp (C228T = 65%; C250T = 11%), EGFR, TP53, PTEN (all in 20%–30%), and NF1 (9%), validating the sensitivity and comparability of our NGS results. Based on the integration of gene size calibrated (n Mb-1) mutation rates (Figure 1A; red dots), elevated numbers of variants in genes such as TTN (11%), PCLO (9%), DNAH2/3 (7%), RYR2 (7%), and MUC16 (7%) detected in our cohort, likely represent random events (passenger mutations), due to the large genomic size of the respective genes [29]. Interestingly, no significant association with PFS or OS was observed for the integration of single recurrent oncogenes with outcome, including TERTp mutations which were previously associated with poor survival in primary GBM [7,30]. However, mutation rates of TERTp were lowest (62%) in younger patients <40 y, confirming an association of TERTp mutations with patients age and clinical performance, respectively [30]. The majority (7 out of 13) of patients without TERTp mutation harbored mutations in TP53 and concomitant inactivating variants in the SWI/SNF complex gene ATRX (in 3 patients), likely representing an alternative mechanism of telomere lengthening in TERTp-wt GBM [6,11,30,31]. In total, 67% of patients carried amplifications in genes associated with RTK signaling (EGFR, PDGFRA, AKT3, SOX2), TP53 induced apoptosis (MDM2/4) and/or cell cycle control via Rb signaling (CDK4/6), representing the 3 key regulatory pathways commonly altered in GBM [6]. EGFR amplification at high copy numbers was the most frequent alteration and closely associated with loss of CDKN2A/B, as described previously [6]. Within the RTK signaling pathway, EGFR amplifications were detected almost mutually exclusive to other frequent RTK lesion (e.g., PDGFRA amplification), reflecting an alternative route of RTK-mediated clonal proliferation [6]. Although EGFR amplification generally confers a more aggressive phenotype, opposing implications were reported for clinical outcome [[6], [7], [8], [9],32]. In our study, EGFR amplification was significantly associated with higher rates of overall mutational burden and histopathological features of tumor aggressiveness, including tumor size, Ki67 values and poor long-term survival (>3y). This points to an increased genomic instability and tumor proliferation potential as a result of upregulated EGFR activity [8,9,32].
Conclusions
Bi-allelic deletions of the DNA demethylase gene TET1 are frequent genomic events that co-occur with EGFR amplifications and are associated with reduced levels of TET1 mRNA expression in IDH-wt GBM. Bi-allelic TET1 loss was not associated with concentrations of 5hmC as measure for global DNA demethylation, suggestive for a site-specific effect of TET1 activity for DNA (de)methylation at specific genomic loci. Although bi-allelic TET1 deletions were not an independent prognostic factor, they are associated with poor outcome in IDH-wt GBM harboring simultaneous EGFR amplification.
Acknowledgments
Authors' Contributions
Conception of the work: T.J., D.K., C.T.; Sample Collection: T.J., S.R., A.Z.; Aquisition/Analysis of Data: S.S., A.P., S.R., A.Z.; Bioinformatic Analysis: S.S., A.P., A.D.; Interpretation of Data: S.S., T.J., G.S., D.K., C.T.; Drafted the manuscript: S.S., T.J.; All Authors have seen and approved the manuscript being submitted.
Acknowledgement
For excellent technical assistance we thank Marita Hartwig, Marika Böhm, Katja Robel and Juliane Blaesche.
Footnotes
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval and consent to participate: All tumor tissue and blood samples (used as germline control) were collected with written informed consent and after approval by the local ethics committee of the Medical Faculty Carl Gustav Carus Dresden.
Data Statement: All data that support the findings of this study are included in this published article and its supplementary information files.
Declaration of Competing Interest: C.T. is CEO and co-owner of AgenDix GmbH, a company performing molecular diagnostics. T.J. received honoraria from Roche and CSL Behring. The remaining authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.10.010.
Appendix. Supplementary materials
References
- 1.Alexander BM, Cloughesy TF. Adult Glioblastoma. J Clin Oncol. 2017;35(21):2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 2.Shergalis A, Bankhead A, 3rd, Luesakul U, Muangsin N, Neamati N. Current challenges and opportunities in treating glioblastoma. Pharmacol Rev. 2018;70(3):412–445. doi: 10.1124/pr.117.014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldape K, Brindle KM, Chesler L, Chopra R, Gajjar A, Gilbert MR, Gottardo N, Gutmann DH, Hargrave D, Holland EC. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–520. doi: 10.1038/s41571-019-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 6.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Chin L. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 8.Mirchia K, Sathe AA, Walker JM, Fudym Y, Galbraith K, Viapiano MS, Corona RJ, Snuderl M, Xing C, Hatanpaa KJ. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7(1):92. doi: 10.1186/s40478-019-0746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz-Hidalgo L, San-Miguel T, Megías J, Monleón D, Navarro L, Roldán P, Cerdá-Nicolás M, López-Ginés C. Somatic copy number alterations are associated with EGFR amplification and shortened survival in patients with primary glioblastoma. Neoplasia. 2020;22(1):10–21. doi: 10.1016/j.neo.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 12.Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi JH, Robertson KD. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15(6):R81. doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150(6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juratli TA, Kirsch M, Robel K, Soucek S, Geiger K, von Kummer R, Schackert G, Krex D. IDH mutations as an early and consistent marker in low-grade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J Neurooncol. 2012;108(3):403–410. doi: 10.1007/s11060-012-0844-1. [DOI] [PubMed] [Google Scholar]
- 16.Stasik S, Salomo K, Heberling U, Froehner M, Sommer U, Baretton GB, Ehninger G, Wirth MP, Thiede C, Fuessel S. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin Biochem. 2019;64:60–63. doi: 10.1016/j.clinbiochem.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 21.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Forloni M, Gupta R, Nagarajan A, Sun LS, Dong Y, Pirazzoli V, Toki M, Wurtz A, Melnick MA, Kobayashi S. Oncogenic EGFR represses the TET1 DNA demethylase to induce silencing of tumor suppressors in cancer cells. Cell Rep. 2016;16(2):457–471. doi: 10.1016/j.celrep.2016.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spans L, Van den Broeck T, Smeets E, Prekovic S, Thienpont B, Lambrechts D, Karnes RJ, Erho N, Alshalalfa M, Davicioni E. Genomic and epigenomic analysis of high-risk prostate cancer reveals changes in hydroxymethylation and TET1. Oncotarget. 2016;7(17):24326–24338. doi: 10.18632/oncotarget.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu R, Ding Y, Luo J, Huang KM, Tang XJ, Li DS, Guo SW. Ten-eleven translocation 1 regulates methylation of autophagy-related genes in human glioma. Neuroreport. 2018;29(9):731–738. doi: 10.1097/WNR.0000000000001024. [DOI] [PubMed] [Google Scholar]
- 25.Takai H, Masuda K, Sato T, Sakaguchi Y, Suzuki T, Koyama-Nasu R, Nasu-Nishimura Y, Katou Y, Ogawa H, Morishita Y. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9(1):48–60. doi: 10.1016/j.celrep.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 26.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26(1):103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller T, Gessi M, Waha A, Isselstein LJ, Luxen D, Freihoff D, Freihoff J, Becker A, Simon M, Hammes J. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol. 2012;181(2):675–683. doi: 10.1016/j.ajpath.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Jr Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Solomon DA, Tihan T. The role of histone modifications and telomere alterations in the pathogenesis of diffuse gliomas in adults and children. J Neurooncol. 2007;132(1):1–11. doi: 10.1007/s11060-016-2349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNulty SN, Cottrell CE, Vigh-Conrad KA, Carter JH, Heusel JW, Ansstas G, Dahiya S. Beyond sequence variation: assessment of copy number variation in adult glioblastoma through targeted tumor somatic profiling. Hum Pathol. 2019;86:170–181. doi: 10.1016/j.humpath.2018.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.