Abstract
BRAF inhibitors were approved for the treatment of BRAF-mutant melanoma. However, most patients acquire the resistance to BRAF inhibitors after several months of treatment. miR-524-5p is considered as a tumor suppressor in many cancers, including melanoma. In this study, we investigated the biological functions of miR-524-5p in melanoma with acquired resistance to BRAF inhibitor and evaluated the endogenous miR-524-5p expression as a biomarker for melanoma. The results showed that the expression of miR-524-5p was 0.481-fold lower in melanoma tissues (n = 117) than in nevus tissues (n = 40). Overexpression of miR-524-5p significantly reduced proliferative, anchorage-independent growth, migratory and invasive abilities of BRAF inhibitor-resistant melanoma cells. Moreover, the introduction of miR-524-5p led to a reduced development of BRAF inhibitor-resistant melanoma in vivo. Remarkably, the MAPK/ERK signaling pathway was decreased after treatment with miR-524-5p. Furthermore, next-generation sequencing analysis implied that the complement system, leukocyte extravasation, liver X receptor/retinoid-X-receptor activation, and cAMP-mediated signaling may be related to miR-524-5p-induced pathways in the resistant cells. The miR-524-5p level was higher on average in complete response and long-term partial response patients than in progressive disease and short-term partial response patients treated with BRAF inhibitors. Our results proposed that miR-524-5p could be considered as a target for treatment BRAF inhibitor-resistant melanoma and a prognostic marker in the response of patients to BRAF inhibitors for melanoma.
Keywords: Melanoma, miRNA, BRAF inhibitor, Resistance, miR-524-5p
Abbreviations: CR, complete response; IHC, immunohistochemistry; IPA, ingenuity pathway analysis, ISH, in situ hybridization; LTPR, long-term partial response; MITF, microphthalmia-associated transcription factor; NGS, next-generation sequencing; PD, progressive disease; RECIST, response evaluation criteria in solid tumors; STPR, short-term partial response
Background
The MAPK/ERK pathway is commonly activated in many cancers, especially in melanoma [1]. It was reported that 50% to 70% of BRAF mutations occur in melanoma [2]. Over 80% of BRAF mutations are caused by the substitution of valine (V) with glutamic acid (E) at codon 600 (BRAFV600E). Therefore, BRAFV600E inhibitors (BRAF inhibitors) are an important therapeutic strategy for the treatment of BRAFV600E melanoma. BRAF inhibitors have the ability to reduce proliferation and induce apoptosis in melanoma through inhibition of BRAFV600E and downstream MAPK/ERK signaling [3]. Two BRAF inhibitors, PLX4032 (vemurafenib), GSK2118436A (dabrafenib), were approved by the U.S. Food and Drug Administration for the treatment of BRAF-mutant melanoma [4,5]. However, despite initially impressive responses, most patients treated with vemurafenib develop acquired resistance after a relatively short period [6]. In 2018, LGX818 (encorafenib) is a new BRAF inhibitor approved by the U.S. Food and Drug Administration for use in combination with binimetinib (MEK inhibitor) [7,8]. Although the combination of dabrafenib and trametinib, a MEK1/2 inhibitor, can increase the overall response rate and progression-free survival, relapse still occurs [9,10]. A few studies have focused on identifying markers of the response to BRAF or MEK inhibitors. The circulating-free DNA of BRAFV600E and MMP9 levels have been considered as monitoring tools for treatment responses to BRAF inhibitors [11], [12], [13]. However, BRAFV600E cell-free DNA was decreased by treatment with a BRAF inhibitor, and MMP9 expression was only observed to be associated with early stages of melanoma patients [13,14]. Hence, there is a clear need for investigation of the effective monitoring and prognostic markers to predict the response to BRAF inhibitors.
The mechanisms of BRAF inhibitor resistance are caused by BRAFV600E amplification or alternative splicing, NRAS mutation, MEK1 or MEK2 mutations, COT upregulation, hyperactive RTK such as PDGFR-β and IGF1-R and activation of the PI3K/AKT pathway [6,[15], [16], [17], [18], [19], [20], [21]]. Several large-scale studies have revealed the roles of miRNAs in melanoma progression. A number of studies have already reported the contribution of miRNAs to the resistance to BRAF inhibitors in melanoma [22], [23], [24], [25]. miR-200c was demonstrated to modulate epithelial–mesenchymal transition, resulting in acquired resistance to BRAF inhibitor, while overexpression of miR-200c reduced the growth of BRAF inhibitor-resistant melanoma cells [22]. miR-125a, miR-204-5p, and miR-211-5p have been shown to enhance BRAF inhibitor resistance through the MAPK and PI3K/AKT pathways [23,24]. Moreover, miR-7 suppressed the resistance to BRAF inhibitors by targeting EGFR/IGFR-1R/CRAF [25]. miR-524-5p has been demonstrated to suppress the growth of not only melanoma but also glioma, gastric cancer, pituitary adenomas, and thyroid cancer [26], [27], [28], [29], [30], [31], [32]. Interestingly, PTEN of the PI3K/AKT pathway and BRAF and ERK2 of the MAPK pathway are direct targets of miR-524-5p [32,33]. Furthermore, it has been reported that miR-524-5p promotes mesenchymal-to-epithelial transition by targeting the EMT-associated genes ZEB2 and SMAD4 [34]. In this study, we investigated the functions of miR-524-5p in BRAF inhibitor resistance, and we assessed whether miR-524-5p could severe as a prognostic marker for the response of melanoma patients to BRAF inhibitors.
Methods
Human specimens
A total of 207 tissues were evaluated in this study. 16 melanoma tissues and 22 nevus tissues were collected from melanoma patients at Landseed Hospital, and 20 melanoma tissues were collected from the patients at Saint Paul's Hospital. Two tissue arrays were purchased from US Biomax (Rockville, MD, USA) and Pantomics, Inc. (San Francisco, CA). ME1004C array contained 61 malignant melanoma, 20 metastatic melanoma, and 18 benign nevi, and MEL961 array as a test group contained 36 tissues of malignant melanoma and 4 benign nevi. Ten tissues of melanoma patients treated with BRAF inhibitor alone or in combination with MEK inhibitor were collected from Kumamoto University Hospital, Japan. The study was agreed by the ethics committees of Landseed Hospital (IRB-14-005-B1), Saint Paul's Hospital (IRB-SPH-10502-01), and Kumamoto University Hospital (IRB-1452).
Cell culture
A375 (BRAFV600E melanoma) and normal human epidermal melanocytes cell lines were purchased from American Type Culture Collection and PromoCell, respectively. SK-Mel-19 cells (BRAFV600E melanoma) were kindly provided by Dr. Neal Rosen (Memorial Sloan-Kettering Cancer Center). Melanoma cells resistant to PLX4032, BRAF inhibitor (named as A375-R and SK-Mel-19-R) were established by treating parental melanoma cells (SK-Mel-19 and A375 cells) with increasing concentrations of the PLX4032. A375 and A375-R cells, and SK-Mel-19 and SK-Mel-19-R cells were maintained in Dulbecco's modified Eagle medium and RPMI-1640 medium (HyClone Lab., Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; HyClone Lab., Logan, UT, USA) and 50 U/mL penicillin and 50 µg/mL streptomycin, respectively. Normal human epidermal melanocytes cells were maintained in Melanocyte media M2 supplemented with 10% FBS and SupplementMix (PromoCell, Heidelberg, Germany). These cells were incubated in a humidified chamber with 5% CO2 at 37 °C and were mycoplasma free.
In situ hybridization and immunohistochemistry
For in situ hybridization (ISH), hsa-miR-524 probes were labeled by double DIG*-labeled miRCURY LNA miRNA (Exiqon, Vedbaek, Denmark). All steps were performed as described previously [35].
For immunohistochemistry (IHC), the slides were deparaffinized with xylene and rehydrated with a decreasing concentration of ethanol. After the inactivation of endogenous peroxidase, the slides were blocked with 2.5% horse serum for 1 h at room temperature. The primary antibody, phospho-MEK1/2 (Cell Signaling Technology, Danvers, MA, USA), incubation was performed at 4 °C overnight. After incubation with secondary antibody from ImmPRESS universal polymer kit (Vector Laboratories, CA, USA) for 1 h at room temperature, ImmPACT VIP substrate kit (Vector Laboratories, CA, USA) was used for antibody detection. The counter was stained with Gill's hematoxylin. Finally, the tissue sections were dehydrated and air-dried.
After staining, images of slides were captured by a light microscope equipped with a camera. The expression of miR-524-5p and phospho-MEK1/2 in the slides was evaluated by 2 pathologists following the Allred scoring system as described previously [35].
Western blot
Protein was extracted by NP-40 lysis buffer and then followed the protocol as described previously [32]. GAPDH, phospho-ERK1/2 (Thr202/Tyr204), total-ERK1/2, phospho-MEK1/2 (Ser221), total-MEK1/2, phospho-RSK (Ser380), phospho-Akt (Ser473), phospho-Akt (Thr308), and total-AKT were purchased from Cell Signaling Technology, and BRAF was purchased from Santa Cruz Biotechnology (CA, USA).
qRT-PCR
Total RNA was extracted by mirVanaTM microRNA Isolation Kit (Ambion, Thermo Fisher Scientific, MA, USA). All steps were followed by the manufacturer's guidance. RNU6 and miR-524-5p TaqMan MicroRNA assays were synthesized from Applied Biosystems. RNU6 was used as a normalized control to quantify the expression of miR-524-5p by ΔΔCt method.
Cell proliferation assay
Cell proliferation was detected by AlamarBlue assay (Invitrogen, Carlsbad, CA). Synergy HT (Biotek, VT, USA) was used to measure the signals at excitation wavelengths of 530 to 560 nm and an emission wavelength of 590 nm.
Soft agar assay
Cells were seeded into 12-well plates containing 2 layers of different agar concentrations (0.6% agar in the bottom layer, 0.3% agar in the top layer). The soft agar assay was performed for 3 to 4 wk and the complete medium was added every 3 d. The colonies were photographed by a microscope.
Wound healing assay
Cells were seeded in 6-well plates inserted a chamber (ibidi) and treated with actinomycin D (10 ng/mL) to suppress cell proliferation. The cell migration was recorded by a microscope at different time points.
Migration and invasion assays
Migration and invasion assays were performed by using 24-well Transwell plates (8 µm pore size, Corning, NY, USA). The cell suspension in the culture medium without FBS was seeded onto the membrane of Transwells and Matrigel-coated Transwells to perform the migration and invasion assays, respectively. The lower chambers were filled with the complete medium containing FBS. After 48 h, the Transwells were incubated in 0.1% crystal violet to stain the cells. Nonmigrated or noninvasive cells on the upper layers of the Transwells were removed by a cotton swab. Stained the migratory and invasive cells located on the bottom layers of the Transwells were scanned by EPSON V750 PRO scanner. ImageJ 1.47 software (NIH, Bethesda, MD, USA) was used to quantify the results.
Xenograft
The experimental protocol was performed under the acceptance of Animal Ethics Committee of Academia Sinica and National Central University (Taiwan). About 5 × 105 A375-R cells were seed in 10-cm culture dishes. After 24 h, the negative control (NC) or miR-524 plasmid were transfected into the cells. Six-week-old male nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (impaired T and B cell lymphocyte development and deficient natural killer cell function) were included in the study. Mice were anaesthetized via intraperitoneal injections. At 48 h of post-transfection, 100 µL of the cell suspension overexpressing the NC or miR-524-5p were injected subcutaneously into the left or the right flanks of the mice (n = 6), respectively. The total 6 mice were kept in a standard ventilated cage in the specific-pathogen-free (SPF) room. When the tumors reached 1000 mm3 (36 d postinjection), the mice were killed by carbon dioxide inhalation followed by the collection of tumors.
Next-generation sequencing and ingenuity pathway analysis
Total RNA was extracted from the cells by mirVanaTM microRNA Isolation Kit (Ambion) and sent to Genomics BioSci & Tech. (Genomics, New Taipei City, Taiwan) for RNA-seq analysis by Illumina. Low qualify reads were filtered out, and the remaining reads were aligned to human genes by using Bowtie2. Gene expression was estimated by RNA-Seq by expectation maximization and recorded in fragments per kilobase of exon per million fragments mapped.
The data of next-generation sequencing (NGS) were analyzed by ingenuity pathway analysis (IPA). The fold change of gene expression of A375-R versus A375 or A375-R-miR-524-5p versus A375-R-NC was calculated. Then, the data were input into IPA, and a fold change of 2 was set as a cut-off. Expression analysis in the core analysis module was selected to analyzed canonical pathways.
Statistical analysis
The data were shown as the mean ± S.D. Student t test (2-tailed) was used to calculate the differences between 2 groups. Mann–Whitney test was used to analyze the difference between groups in vivo experiments. The difference among human tissues was evaluated by χ2 test. The significant differences were considered at P value <0.05. GraphPad Prism software was used to calculate the statistical analysis. Image analysis was performed using ImageJ 1.47 (NIH, Bethesda, MD, USA).
Results
miR-524-5p expression is decreased in the melanoma tissues and related to the response of melanoma patients to BRAF inhibitors
To investigate the endogenous expression of miR-524-5p in normal skin tissue, we analyzed its expression by ISH. It has been reported that miR-524-5p expression is decreased in melanoma cells [32]. The results indicated that miR-524-5p was strongly expressed in normal skin tissue (Figure S1). Furthermore, we asked whether miR-524-5p expression is altered between nevus and melanoma tissues. To address this question, miR-524-5p expression was analyzed in melanoma and nevus tissues from patients in Taiwan and in a commercial tissue array (ME1004C). The staining score was evaluated by 2 pathologists following the Allred scoring system (Figure 1A) [35]. The results showed that miR-524-5p expression was 0.481-fold lower in melanoma tissues than in nevus tissues (Figure 1B). Next, we investigated whether miR-524-5p could be a diagnostic marker of melanoma by receiver operating characteristic curve analysis of miR-524-5p. The results showed that the area under the curve (AUC) values of miR-524-5p were 0.9078 and 0.8378 in the commercial tissue array and Taiwan patients, respectively (Figure 1C). Next, another commercial tissue array (MEL961) was used as the test group. The results showed that the AUC value of the test group reached 0.9757 (95% confidence interval: 0.9228–1.029). miR-524-5p expression by ISH method could detect melanoma from nevus tissues at a cut-off point <2.75 with 86.1% sensitivity and 100% specificity (Figure 1D). phospho-MEK1/2 is a downstream of BRAF. Therefore, the expression of phospho-MEK1/2 was examined by IHC and the results showed that phospho-MEK1/2 had a lower AUC value of 0.7216 in Taiwan patients compared with the AUC values of miR-524-5p (Figure 1C). These findings observed that the staining of miR-524-5p by ISH may be more accurately differentiate between the melanoma and nevus groups than the staining of phospho-MEK1/2.
Figure 1.
miR-524-5p expression is decreased in melanoma tissues and related to the response of melanoma patients to BRAF inhibitors. (A) Staining of miR-524-5p in nevus and melanoma specimens by ISH. (B) Box-whisker plots (left) and the percentage (right) of miR-524-5p expression in 117 melanoma and 40 normal nevus tissues from Taiwan hospitals and a commercial tissue array. ***P value <0.001, χ2 test. (C) The receiver operating characteristic curve of miR-524-5p and phospho-MEK1/2 in Taiwan patients (n = 58; 22 nevus and 36 melanoma tissues) and a commercial tissue array (ME1004C: n = 99; 18 nevus and 81 melanoma tissues). (D) Left panel: The receiver operating characteristic curve of miR-524-5p from the test set (MEL961: n = 40; 4 nevus and 36 melanoma tissues). Right panel: The histogram shows the accuracy of the sensitivity and specificity from the test set. (E) Representative images and the percentage of miR-524-5p and phospho-MEK1/2 expression in melanoma patients before treatment with BRAF inhibitor alone or in combination with MEK inhibitor. Blue and purple colors indicate the staining of miR-524-5p and phospho-MEK1/2, respectively. miR-524-5p and pMEK1/2 signals were observed in the cytoplasm. Brown color indicates melanin by its pigmentation. CR, complete response; LTPR, long-term partial response; PD, progressive disease; STPR, short-term partial response. Bar = 20 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
There is no good prognostic marker for clinical responses to BRAF inhibitor treatment. To examine whether miR-524-5p could serve as a prognostic marker for BRAF inhibitor responses, miR-524-5p and phospho-MEK1/2 expression levels were evaluated in 10 human melanoma clinical samples by ISH and IHC assays, respectively. All patients carried BRAFV600E, and the tissues were collected before the patients were treated with BRAF and MEK inhibitors, or BRAF inhibitor only (Table S1). Ten patients were collected and assessed by response evaluation criteria in solid tumors (RECIST version 1.1). The partial response patients were categorized into 2 groups based on the duration of the response to BRAF inhibitor. The partial response patients with a duration response longer than 12 mo were categorized as having a long-term partial response, and those with a duration response shorter than or equal to 12 mo were categorized as having a short-term partial response. The results showed that the miR-524-5p expression level was higher in the group of complete response and long-term partial response patients than in the group of progressive disease and short-term partial response patients (Figure 1E and Figure S2). However, there was no difference in the expression of phospho-MEK1/2 expression between these groups (Figure 1E and Figure S2). In the limitation of small sample size, these findings proposed that miR-524-5p expression has a possibility to be considered as a prognostic marker for melanoma patients treated with BRAF inhibitors.
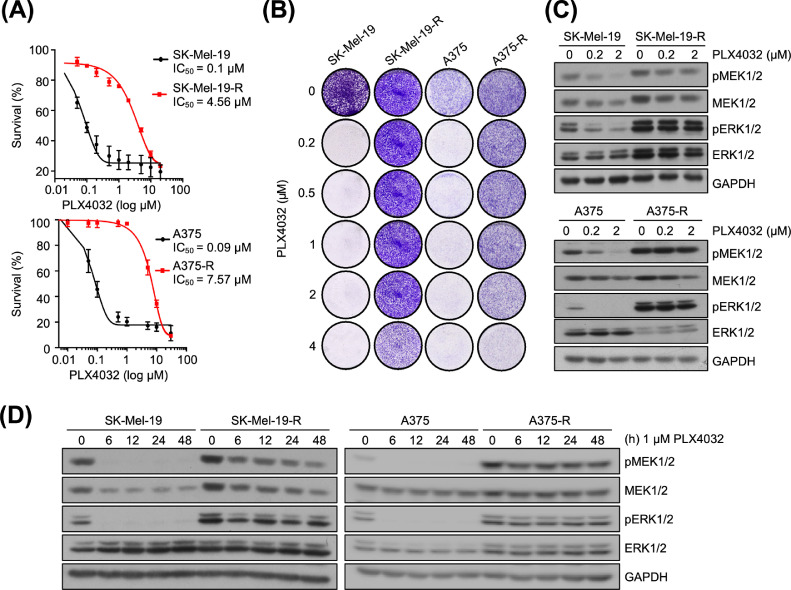
Characterization of PLX4032-resistant melanoma cells
It has been reported that miR-524-5p can reduce melanoma growth and thus the functions of miR-24-5p in BRAF inhibitor-resistant melanoma were further investigated [32]. We first established melanoma cells resistant to PLX4032, a BRAFV600E inhibitor, by treating BRAFV600E mutant melanoma cells (SK-Mel-19 and A375 cells) with increasing concentrations of the PLX4032 (up to 4 µM). To examine the characteristics of the resistance to PLX4032, the cells were treated with PLX4032 at serial concentrations and then analyzed for cell proliferation. The IC50 values were significantly higher in the resistant cells (SK-Mel-19-R cells: IC50 = 4.56 µM; A375-R cells: IC50 = 7.57 µM) than in the parental cells (IC50 = 0.1 µM; Figure 2A). These results indicated that the resistant cells required approximately 45- to 75-fold higher concentrations of PLX4032 were needed to inhibit the growth of the resistant cells. In addition, colony formation assays were performed in the presence of different concentrations of PLX4032 to confirm these resistant cell models. The colonies of the resistant cells were slightly reduced at 4 µM of PLX4032 while the parental cell growth was strongly inhibited at 0.2 µM of PLX4032 (Figure 2B).
Figure 2.
Characterization of PLX4032-resistant melanoma cells. (A) IC50 curves of the parental (SK-Mel-19 and A375 cells) and resistant cells (SK-Mel-19-R and A375-R cells) by treatment with PLX4032 at serial concentrations. After 72 h of PLX4032 treatment, cell survival was measured by AlamarBlue assay. (B) Seven-day colony formation assay of the parental and resistant cells treated with increasing concentrations of PLX4032. (C, D) Western blot of the parental and resistant cells treated with increasing concentrations of PLX4032 for 6 h (C) or treated with PLX4032 at 1 µM for 0, 6, 12, 24, and 48 h (D).
Previous studies indicated that the PLX4032-resistant melanoma displayed persistence of the MAPK/ERK pathway [17,36]. Therefore, the activity of the MAPK/ERK pathway in both resistant cells was analyzed by western blot. As expected, the MAPK/ERK pathway activity in PLX4032-resistant cells was strongly increased compared to that in the parental cells in a dose- and time-dependent manner (Figure 2C, D). The above data suggested that PLX4032-resistant melanoma cells were successfully constructed.
NGS was performed to analyze the transcriptomes of the parental and resistant cells. Genes related to cell proliferation and metastasis including JUN, FOSL1, GLI1, and MMP9 were more highly expressed in the resistant cells than in the parental cells, and similar results have been observed in other studies (Figure S3) [6,37]. Moreover, we further analyzed pathways involved in the resistance to BRAF inhibitor by IPA software. A total of 21 pathways were significantly altered between the resistant and parental cells (P value ≤0.01), of which 11 pathways were reduced and 10 pathways were increased in the resistant cells (Figure S4). Altogether, 21 novel pathways contributed to the resistance to BRAF inhibitor in melanoma.
miR-524-5p decreases proliferation, colony formation, and anchorage-independent growth of PLX4032-resistant melanoma cells
After establishing the resistant cells, we examined the expression of miR-524-5p in the melanocyte, parental, and resistant melanoma cells. As expected, quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed that miR-524-5p expression was reduced in melanoma cells compared to melanocyte cells (Figure 3A). miR-524-5p expression was lower in the resistant cells than in the parental cells, indicating that miR-524-5p may be involved in preventing BRAF inhibitor resistance (Figure 3B). To study the functions of miR-524-5p in the progression of PLX4032-resistant melanoma cells, miR-524-5p mimic or NC was transfected into the resistant cells, and PLX4032 was used as a positive control. The results showed that miR-524-5p overexpression strongly reduced the proliferation in the resistant cells, as demonstrated by AlamarBlue and colony formation assays (Figure 3B, C). Next, the effects of miR-524-5p on anchorage-independent growth of the resistant cells were analyzed by soft agar assay. The number and size of the colonies in the resistant cells were significantly reduced by miR-524-5p in both SK-Mel-19-R and A375-R cells (Figure 3D). Taken together, these results suggested that miR-524-5p reduced the proliferation and anchorage-independent growth of the resistant melanoma cells.
Figure 3.
miR-524-5p reduces the growth of PLX4032-resistant melanoma cells. (A) miR-524-5p expression in melanocyte (NHEM), parental and resistant cells. The expression of miR-524-5p was analyzed by qRT-PCR. (B–D) The parental and resistant cells were treated with PLX4032 (0.5 µM) or transfected with negative control (NC) or miR-524-5p (20 nM in SK-Mel-19-R cells; 40 nM in A375-R cells) to perform the following functional assays. (B) Proliferation was detected by AlamarBlue assay after 72 h. (C) The colony formation assay was performed for 7 d and the purple dots represent cell colonies. (D) Anchorage-independent growth in soft agar was assessed for 3 to 4 wk after transfection (upper panel; bar = 100 µM). The colony size was observed under a microscope and the relative number of colonies was calculated (lower panel). The quantification data are reported as the mean ± SD of 3 independent experiments (n = 3). Student t test: *P value <0.05, **P value <0.01, ***P value <0.001.
miR-524-5p reduces the migration and invasion of PLX4032-resistant melanoma cells
To investigate the effects of miRNA-524-5p on the migratory and invasive ability of PLX4032-resistant melanoma cells, wound healing and Transwell assays were performed to analyze the cell migration and invasion. Overexpression of miR-524-5p strongly reduced the migratory function by decreasing the wound closing at 72 h in SK-Mel-19-R cells and at 24 h in A375-R cells (Figure 4A). To confirm the role of miR-524-5p in the migration of PLX4032-resistant melanoma cells, a Transwell assay was performed. Similarly, the migration of the resistant cells that had migrated to the bottom of the Transwell was highly reduced by approximately 80% to 90% in cells transfected with miR-524-5p compared to the NC cells (Figure 4B). Next, to investigate the effects of miR-524-5p on cell invasion, a Transwell assay was performed in which the Transwell membrane was coated with Matrigel. The resistant cells overexpressing miR-524-5p exhibited decreased the invasion by approximately 85% compared with those expressing the NC (Figure 4C). Collectively, these results suggested that miR-524-5p attenuated the migration and invasion of the resistant melanoma cells.
Figure 4.
miR-524-5p reduces the migration and invasion of PLX4032-resistant melanoma cells. (A) Wound healing assays were performed to assess the cell migration at the indicated time points. (B, C) Representative images of Transwells (upper panel) and the quantification data (lower panel) after 72 h of transfection in cell migration (B) and invasion assays (C). The quantification data are reported as the mean ± SD of 3 independent experiments (n = 3). Student t test: *P value <0.05, **P value <0.01, ***P value <0.001.
miR-524-5p modulates multiple signaling pathways in PLX4032-resistant melanoma cells
As mentioned, activation of the MAPK/ERK and PI3K/AKT pathways is common mechanisms of BRAF inhibitor resistance. Therefore, we examined the influence of miR-524-5p overexpression on the MAPK/ERK and PI3K/AKT pathways in the resistant cells. We observed that overexpression of miR-524-5p led to inactivation of MEK1/2 and ERK1/2 in the MAPK/ERK pathway in both resistant cells (Figure 5). miR-524-5p slightly reduced the phosphorylation of AKT (Ser473) in the PI3K/AKT pathway slightly in SK-Mel-19R cells but not in A375-R cells. These findings indicated that miR-524-5p attenuated the activity of the MAPK/ERK pathway and partially reduced the PI3K/AKT signaling in the resistant melanoma cells.
Figure 5.
miR-524-5p regulates the multiple pathways in PLX4032-resistant melanoma cells. SK-Mel-19 and SK-Mel-19-R cells were treated with PLX4032 for 48 h. A375 and A375-R cells were treated with PLX4032 for 6 h. SK-Mel-19-R cells (A) and A375-R cells (B) were transfected with negative control (NC) or miR-524-5p and the cell lysates were collected at 48 h after transfection for immunoblotting.
Furthermore, gene expression in the resistant cells transfected with miR-524-5p (A375-R-miR-524-5p) or NC (A375-R-NC) was investigated by NGS. Then, the NGS data were analyzed by IPA to elucidate other mechanisms of miR-524-5p in the resistant cells by comparing gene expression and pathways to those of the NC. The results showed that 16 pathways were altered after overexpression of miR-524-5p as compared to the NC (P value ≤0.01; Figure S5). Interestingly, 4 pathways showed significant differences in the fold changes of the z-score between the A375-R versus A375 group and the A375-R-miR-524-5p versus A375-R-NC group (Figures S6 and S7). Notably, decrease of leukocyte extravasation, complement system, and activation of liver X receptor/retinoid-X-receptor (LXR/RXR) activation signaling was found in the resistant cells overexpressing miR-524-5p, while these signaling pathways showed the reverse trend in the resistant cells compared to parental cells. In addition, introduction of miR-524-5p decreased the downregulation of cAMP-mediated signaling. The results indicated that these pathways may be involved in the regulation of miR-524-5p in the resistant cells.
MITFlow has been observed in BRAF inhibitor-resistant melanoma cells. Alteration in the expression of MITF has been linked to melanoma cell plasticity to BRAF inhibitors [38]. The NGS data showed that MITFlow level were detected in the resistant cells compared to the parental cells. In addition, overexpression of miR-524-5p reversed the expression of MITF in the resistant cells. Then, expression of MITF was confirmed by qRT-PCR. The results were similar with the NGS data (Figure S6). This finding indicated that miR-524-5p may contribute to melanoma cell plasticity to BRAF inhibitors.
miR-524-5p reduces the progression of PLX4032-resistant melanoma in vivo
We further determined the effect of miR-524-5p on the progression of PLX4032-resistant melanoma in vivo. A375-R cells overexpressing miR-524-5p or the control plasmid were subcutaneously injected into NOD/SCID mice, followed by the observation of xenograft growth. After 36 d of injection, the mice were sacrificed, and the tumor weight was measured. The results showed that the overexpression of miR-524-5p in A375-R cells led to a decrease in tumor weight (Figure 6A, B). miR-524-5p expression was detected in all tumors by qRT-PCR and ISH analyses (Figure 6C). The average tumor weight was decreased by approximately 40% in the miR-524-5p-expressing group compared to the control group. These results suggested that overexpression of miR-524-5p attenuated the development of PLX4032-resistant melanoma cells in vivo.
Figure 6.
miR-524-5p decreases PLX4032-resistant melanoma growth in vivo. A375-R cells transfected with pre-mir-524-5p or negative control (NC) plasmid were injected into the right or left flanks of NOD/SCID mice (n = 6). After 36 d of injection, the tumors were collected. (A) Images of tumors after sacrifice. (B) The tumor weight was measured. *P value <0.05, Student t test. (C) The expression levels of miR-524-5p in tumors were analyzed by qRT-PCR (left) and in situ hybridization (right). **P value <0.01, Mann–Whitney test. The data are reported as the mean ± SD.
Discussion
Recently, miRNAs have emerged as diagnostic markers for many cancers, such as bladder cancer, oral cancer, and prostate cancer because they are quite stable [39]. About 27.2% and 25% of BRAFV600E were found in nevi and melanoma of Taiwan patients, respectively, but miR-524-5p expression was decreased in melanoma tissue. This indicated miR-524-5p could serve as a potential diagnostic marker of melanoma (Figure S8, right panel and data not shown). Until now, there has been a lack of a potential biomarker for the detection of the BRAF inhibitor response in melanoma. Early detection of the response to BRAF inhibitors can help to choose an appropriate treatment that leads to improved patient survival. Herein, the most interesting observation is the relationship between miR-524-5p and the BRAF inhibitor response. It is possibility to propose that miR-524-5p may be considered as a signature to assess the response of melanoma patients to BRAF inhibitors or the development of BRAF inhibitor resistance (Figure 1E, Table S1 and Figure S8, right panel). Since the number of patients was small in this study, further experiments with larger samples should be included to confirm this observation in the future study.
It is noted that MAPK/ERK pathway was reactivated with higher activity in the resistant cells compared to parental cells (Figure 2C, D). The expression of miR-524-5p is increased when the MAPK/ERK pathway is inhibited but is decreased when MAPK/ERK pathway is reactivated [32]. This study observed that lower miR-524-5p expression in the resistant cells compared to the parental cells is consistent with previous studies (Figure 3A). These results indicated that miR-524-5p expression is response with the activity of MAPK/ERK pathway.
It has been reported that miR-7 and miR-200c expression levels are decreased in BRAF inhibitor-resistant melanoma cells and that overexpression of these miRNAs reversed the resistance [22,25]. However, the clinical correlation between miR-7 and the development of BRAF inhibitor resistance in melanoma patients was not investigated, and the functions of miR-200c in resistant cells in vitro and in vivo were not determined. Our research demonstrated that miR-524-5p overexpression led to reduce the proliferation, anchorage-independent growth, migration, and invasion in the resistant melanoma cells (Figure S8, left panel). miR-524-5p may attenuate the growth of resistant melanoma through the downregulation of the MAPK/ERK and PI3K/AKT pathways partially (Figure 5).
An unknown mechanism of the resistance was reported in more than 40% of tumor samples, although mounting evidence related to mechanisms of BRAF inhibitor resistance has been reported [40]. Herein, NGS data revealed that 21 novel pathways that might be related to BRAF inhibitor resistance (Figure S4). Reactivation of the targeted pathway or activation of alternative pathways is the reasons for drug resistance. This suggests that a therapeutic strategy that inhibits multiple pathways is needed.
In melanoma cells, MITFlow or MITFhigh has been correlated with an invasive or proliferative phenotype [41,42]. The transition from the BRAF inhibitor-sensitive phenotype (MITFhigh) to the BRAF inhibitor-resistant phenotype (MITFlow) is a mechanism to acquire the resistance [43,44]. Similarly, our results showed that loss of MITF expression was shown in the resistant cells compared to the parental cells (Figure S6). NGS and IPA analysis revealed that miR-524-5p alters multiple pathways (Figures S5, S6, and S7). Interestingly, we observed that cAMP-mediated signaling was enhanced in resistant cells transfected with miR-524-5p. MITF is a downstream transcription factor of cAMP-mediated signaling [45,46]. Our results showed that miR-524-5p expression led to increased MITF in the resistance cells (Figure S6). Interestingly, activation of the MAPK/ERK or PI3K/AKT pathway led to increased expression of GLI1 and MMP9 [47,48]. A previous study also reported that GLI1 inhibition can restore the sensitivity of melanoma cells to BRAF inhibitors and was associated with the decreased invasion and MMP2/9 expression [49]. MMP9 promoted the invasion of melanoma, and high expression of MMP9 was observed in the melanoma patients carrying BRAFV600E [50]. Our data showed that high expression of 2 invasion-related genes, GLI1 and MMP9, was shown in the resistant cells compared to the parental cells, and miR-524-5p expression reduced their expression levels (Figure S7 and data not shown). Collectively, these findings indicated that the invasive phenotype was acquired toward BRAF inhibitor resistance in melanoma cells. Enrichment of epithelial–mesenchymal transition, migration, invasion, and mesenchymal signature contributes to BRAF inhibitor resistance [22,44,51]. Better understanding of the molecular mechanisms of resistance will provide more benefits for the identification of novel therapeutic strategies for treating melanoma patients.
RXR and LXR are classified as type II nuclear receptors. Loss of retinoid receptors was shown to be involved in melanoma progression and activation of LXRβ reduced melanoma and the resistance of melanoma to PLX4032 [52,53]. Our data showed that LXR/RXR activation was upregulated by overexpression of miR-524-5p in the resistant melanoma cells. Moreover, it has reported that IL-8 and leukocyte extravasation are highly expressed in melanoma and promote melanoma metastasis [54], [55], [56]. The complement system induces anaphylatoxin, which increases neutrophil extravasation, and the lack of C3aR, the receptor of a key complement protein, led to attenuation of melanoma growth [57,58]. The effects of miR-524-5p expression on decrease of leukocyte extravasation signaling and the complement system have been demonstrated. Recently, tumor-associated macrophages were demonstrated to contribute to BRAF or MEK inhibitor resistance in melanoma [59,60]. These bioinformatic data raised the question whether immune response plays a modulator of the resistance to BRAF inhibitors in melanoma, which may be affected by the introduction of miR-524-5p. In this regard, it will be important to further explore this concept.
In summary, this study reported the novel functions of miR-524-5p in BRAF inhibitor-resistant melanoma. These findings suggested a potential approach for preventing PLX4032-resistant melanoma by miR-524-5p expression. More importantly, this study emphasized the utility of miRNAs not only to develop a new therapeutic strategy for melanoma but also to identify novel and potential biomarkers related to resistance to targeted therapy. mmc1.docx
Data availability statement
NGS data sets were generated and analyzed.
Authors’ contributions
Nianhan Ma: Conceptualization. Mai-Huong Thi Nguyen, Chen-Huan Lin, Szu-Mam Liu, Hsuan Lin, Chi Hou Ng, Jen-Chieh Tsai, and In-Yu Lin: Validation and Investigation. Azusa Miyashita, Satoshi Fukushima, Hironobu Ihn, Ming-Hong Chen, and Mu-Shiun Tsai: Resources. Mai-Huong Thi Nguyen: Writing - Original draft preparation. Azusa Miyashita, Satoshi Fukushima, Shu-Chen Liu, Long-Yuan Li, Jean Lu, and Nianhan Ma: Writing - Reviewing and Editing. Nianhan Ma: Supervision. Satoshi Fukushima, Jean Lu, and Nianhan Ma: Funding acquisition.
Footnotes
Funding: This work was supported by NCU-Landseed International Chronic Disease Research Center (NCU-LSH-108-A-005), Ministry of Science and Technology, Taiwan (MOST-106-2320-B-008-005-MY3 and MOST-108-3114-Y-001-002), Academia Sinica (AS-SUMMIT-109 and AS-KPQ-109-BioMed), and Japan Society for the Promotion of Science (JP18K08301).
Competing Interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.10.009.
Contributor Information
Satoshi Fukushima, Email: satoshi.fukushima.tb@gmail.com.
Jean Lu, Email: jeanlu@gate.sinica.edu.tw.
Nianhan Ma, Email: nianhan.ma@g.ncu.edu.tw.
Appendix. Supplementary materials
References
- 1.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Couselo E., Garcia J.S., Perez-Garcia J.M., Cebrian V.O., Castan J.C. Recent advances in the treatment of melanoma with BRAF and MEK inhibitors. Ann Transl Med. 2015;3:207. doi: 10.3978/j.issn.2305-5839.2015.05.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H., Higgins B., Kolinsky K., Packman K., Go Z., Iyer R., Kolis S., Zhao S., Lee R., Grippo J.F. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70:5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 4.Bollag G., Tsai J., Zhang J., Zhang C., Ibrahim P., Nolop K., Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 5.Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H., Jr., Kaempgen E. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 6.Luebker S.A., Koepsell S.A. Diverse mechanisms of BRAF inhibitor resistance in melanoma identified in clinical and preclinical studies. Front Oncol. 2019;9:268. doi: 10.3389/fonc.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dummer R., Ascierto P.A., Gogas H.J., Arance A., Mandala M., Liszkay G., Garbe C., Schadendorf D., Krajsova I., Gutzmer R. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018;19:1315–1327. doi: 10.1016/S1470-2045(18)30497-2. [DOI] [PubMed] [Google Scholar]
- 8.Patel H., Yacoub N., Mishra R., White A., Long Y., Alanazi S., Garrett J.T. MDPI; 2020. Current Advances in the Treatment of BRAF-Mutant Melanoma Cancers (Basel) p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long G.V., Fung C., Menzies A.M., Pupo G.M., Carlino M.S., Hyman J., Shahheydari H., Tembe V., Thompson J.F., Saw R.P. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 11.Sanmamed M.F., Fernandez-Landazuri S., Rodriguez C., Zarate R., Lozano M.D., Zubiri L., Perez-Gracia J.L., Martin-Algarra S., Gonzalez A. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61:297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]
- 12.Gray E.S., Rizos H., Reid A.L., Boyd S.C., Pereira M.R., Lo J., Tembe V., Freeman J., Lee J.H., Scolyer R.A. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salemi R., Falzone L., Madonna G., Polesel J., Cina D., Mallardo D., Ascierto P.A., Libra M., Candido S. MMP-9 as a candidate marker of response to BRAF inhibitors in melanoma patients with BRAF(V600E) mutation detected in circulating-free DNA. Front Pharmacol. 2018;9:856. doi: 10.3389/fphar.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreuer M., Meersseman G., Van Den Herrewegen S., Jansen Y., Chevolet I., Bott A., Wilgenhof S., Seremet T., Jacobs B., Buyl R. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J Transl Med. 2016;14:95. doi: 10.1186/s12967-016-0852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H., Moriceau G., Kong X., Lee M.K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery C.M., Vijayendran K.G., Zipser M.C., Sawyer A.M., Niu L., Kim J.J., Hatton C., Chopra R., Oberholzer P.A., Karpova M.B. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessen C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.A., Emery C.M., Stransky N., Cogdill A.P., Barretina J. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K., Wubbenhorst B., Xu X., Gimotty P.A., Kee D. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G., Chodon T., Guo R., Johnson D.B., Dahlman K.B. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Tetzlaff M.T., Wang T., Yang R., Xie L., Zhang G., Krepler C., Xiao M., Beqiri M., Xu W. miR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015;28:431–441. doi: 10.1111/pcmr.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koetz-Ploch L., Hanniford D., Dolgalev I., Sokolova E., Zhong J., Diaz-Martinez M., Bernstein E., Darvishian F., Flaherty K.T., Chapman P.B. MicroRNA-125a promotes resistance to BRAF inhibitors through suppression of the intrinsic apoptotic pathway. Pigment Cell Melanoma Res. 2017;30:328–338. doi: 10.1111/pcmr.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz-Martinez M., Benito-Jardon L., Alonso L., Koetz-Ploch L., Hernando E., Teixido J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018;78:1017–1030. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X., Li J., Sun Y., Zhang Y., Dong L., Shen C., Yang L., Yang M., Li Y., Shen G. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7:53558–53570. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G.H., Liu Y.H., Yang Z., Zhu A.L., Zhao C.L. MicroRNA-524-5p suppresses the growth and invasive abilities of gastric cancer cells. Oncol Lett. 2016;11:1926–1932. doi: 10.3892/ol.2016.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Zhang W., Yan W., Han L., Zhang K., Shi Z., Zhang J., Wang Y., Li Y., Yu S. The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis. 2012;33:2276–2282. doi: 10.1093/carcin/bgs261. [DOI] [PubMed] [Google Scholar]
- 28.Xu T., Yu W., Li Q., Li X., Shi Y., Cao B., Zhang Y., Wang S., Zhang Y., Wang T. MicroRNA-524 inhibits the progress of glioma via the direct targeting of NCF2. Am J Transl Res. 2019;11:1605–1615. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Xue X., Hong H., Qin M., Zhou J., Sun Q., Liang H., Gao L. Upregulation of microRNA-524-5p enhances the cisplatin sensitivity of gastric cancer cells by modulating proliferation and metastasis via targeting SOX9. Oncotarget. 2017;8:574–582. doi: 10.18632/oncotarget.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhen W., Qiu D., Zhiyong C., Xin W., Mengyao J., Dimin Z., Chonghui H., Haijun W., Yonghong Z. MicroRNA-524-5p functions as a tumor suppressor in a human pituitary tumor-derived cell line. Horm Metab Res. 2017;49:550–557. doi: 10.1055/s-0043-106437. [DOI] [PubMed] [Google Scholar]
- 31.Zhen Z., Dong F., Shen H., Wang Q.G., Yang L., Hu J. MiR-524 inhibits cell proliferation and induces cell apoptosis in thyroid cancer via targeting SPAG9. Eur Rev Med Pharmacol Sci. 2018;22:3812–3818. doi: 10.26355/eurrev_201806_15265. [DOI] [PubMed] [Google Scholar]
- 32.Liu S.M., Lu J., Lee H.C., Chung F.H., Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5:9444–9459. doi: 10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang M., Qiu X., Cheng D., Zhu C., Chen L. MicroRNA-524 promotes cell proliferation by down-regulating PTEN expression in osteosarcoma. Cancer Cell Int. 2018;18:114. doi: 10.1186/s12935-018-0612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen P.N.N., Choo K.B., Huang C.J., Sugii S., Cheong S.K., Kamarul T. miR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1- and EMT-associated genes to regulate cellular reprogramming. Stem Cell Res Ther. 2017;8:214. doi: 10.1186/s13287-017-0666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S.M., Lin C.H., Lu J., Lin I.Y., Tsai M.S., Chen M.H., Ma N. miR-596 modulates melanoma growth by regulating cell survival and death. J Invest Dermatol. 2018;138:911–921. doi: 10.1016/j.jid.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 36.Su F., Bradley W.D., Wang Q., Yang H., Xu L., Higgins B., Kolinsky K., Packman K., Kim M.J., Trunzer K. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer Res. 2012;72:969–978. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 37.Caporali S., Alvino E., Lacal P.M., Levati L., Giurato G., Memoli D., Caprini E., Antonini Cappellini G.C., D'Atri S. Targeting the PI3K/AKT/mTOR pathway overcomes the stimulating effect of dabrafenib on the invasive behavior of melanoma cells with acquired resistance to the BRAF inhibitor. Int J Oncol. 2016;49:1164–1174. doi: 10.3892/ijo.2016.3594. [DOI] [PubMed] [Google Scholar]
- 38.Ji Z., Erin Chen Y., Kumar R., Taylor M., Jenny Njauw C.N., Miao B., Frederick D.T., Wargo J.A., Flaherty K.T., Jonsson G. MITF modulates therapeutic resistance through EGFR signaling. J Invest Dermatol. 2015;135:1863–1872. doi: 10.1038/jid.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W. MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol Biol. 2017;1617:57–67. doi: 10.1007/978-1-4939-7046-9_4. [DOI] [PubMed] [Google Scholar]
- 40.Johnson D.B., Menzies A.M., Zimmer L., Eroglu Z., Ye F., Zhao S., Rizos H., Sucker A., Scolyer R.A., Gutzmer R. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51:2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arozarena I., Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer. 2019;19:377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 42.Hoek K.S., Eichhoff O.M., Schlegel N.C., Dobbeling U., Kobert N., Schaerer L., Hemmi S., Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 43.Fallahi-Sichani M., Becker V., Izar B., Baker G.J., Lin J.R., Boswell S.A., Shah P., Rotem A., Garraway L.A., Sorger P.K. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol. 2017;13:905. doi: 10.15252/msb.20166796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su Y., Lu X., Li G., Liu C., Kong Y., Lee J.W., Ng R., Wong S., Robert L., Warden C., et al. (2019). Kinetic inference resolves epigenetic mechanism of drug resistance in melanoma. bioRxiv, 724740.
- 45.Johannessen C.M., Johnson L.A., Piccioni F., Townes A., Frederick D.T., Donahue M.K., Narayan R., Flaherty K.T., Wargo J.A., Root D.E. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawakami A., Fisher D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab Invest. 2017;97:649–656. doi: 10.1038/labinvest.2017.9. [DOI] [PubMed] [Google Scholar]
- 47.Pietrobono S., Gagliardi S., Stecca B. Non-canonical hedgehog signaling pathway in cancer: activation of GLI transcription factors beyond smoothened. Front Genet. 2019;10:556. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Napoli S., Scuderi C., Gattuso G., Bella V.D., Candido S., Basile M.S., Libra M., Falzone L. Functional roles of matrix metalloproteinases and their inhibitors in melanoma. Cells. 2020;9(5):1151–1173. doi: 10.3390/cells9051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faiao-Flores F., Alves-Fernandes D.K., Pennacchi P.C., Sandri S., Vicente A.L., Scapulatempo-Neto C., Vazquez V.L., Reis R.M., Chauhan J., Goding C.R. Targeting the hedgehog transcription factors GLI1 and GLI2 restores sensitivity to vemurafenib-resistant human melanoma cells. Oncogene. 2017;36:1849–1861. doi: 10.1038/onc.2016.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guarneri C., Bevelacqua V., Polesel J., Falzone L., Cannavo P.S., Spandidos D.A., Malaponte G., Libra M. NFkappaB inhibition is associated with OPN/MMP9 downregulation in cutaneous melanoma. Oncol Rep. 2017;37:737–746. doi: 10.3892/or.2017.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su Y., Wei W., Robert L., Xue M., Tsoi J., Garcia-Diaz A., Homet Moreno B., Kim J., Ng R.H., Lee J.W. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A. 2017;114:13679–13684. doi: 10.1073/pnas.1712064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chakravarti N., Lotan R., Diwan A.H., Warneke C.L., Johnson M.M., Prieto V.G. Decreased expression of retinoid receptors in melanoma: entailment in tumorigenesis and prognosis. Clin Cancer Res. 2007;13:4817–4824. doi: 10.1158/1078-0432.CCR-06-3026. [DOI] [PubMed] [Google Scholar]
- 53.Pencheva N., Buss C.G., Posada J., Merghoub T., Tavazoie S.F. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell. 2014;156:986–1001. doi: 10.1016/j.cell.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 54.Payne A.S., Cornelius L.A. The role of chemokines in melanoma tumor growth and metastasis. J Invest Dermatol. 2002;118:915–922. doi: 10.1046/j.1523-1747.2002.01725.x. [DOI] [PubMed] [Google Scholar]
- 55.Bar-Eli M. Role of interleukin-8 in tumor growth and metastasis of human melanoma. Pathobiology. 1999;67:12–18. doi: 10.1159/000028045. [DOI] [PubMed] [Google Scholar]
- 56.Slattery M.J., Dong C. Neutrophils influence melanoma adhesion and migration under flow conditions. Int J Cancer. 2003;106:713–722. doi: 10.1002/ijc.11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nabizadeh J.A., Manthey H.D., Steyn F.J., Chen W., Widiapradja A., Md Akhir F.N., Boyle G.M., Taylor S.M., Woodruff T.M., Rolfe B.E. The complement C3a receptor contributes to melanoma tumorigenesis by inhibiting neutrophil and CD4+ T cell responses. J Immunol. 2016;196:4783–4792. doi: 10.4049/jimmunol.1600210. [DOI] [PubMed] [Google Scholar]
- 58.Markiewski M.M., Lambris J.D. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith M.P., Sanchez-Laorden B., O'Brien K., Brunton H., Ferguson J., Young H., Dhomen N., Flaherty K.T., Frederick D.T., Cooper Z.A. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 2014;4:1214–1229. doi: 10.1158/2159-8290.CD-13-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang T., Xiao M., Ge Y., Krepler C., Belser E., Lopez-Coral A., Xu X., Zhang G., Azuma R., Liu Q. BRAF inhibition stimulates melanoma-associated macrophages to drive tumor growth. Clin Cancer Res. 2015;21:1652–1664. doi: 10.1158/1078-0432.CCR-14-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data sets were generated and analyzed.