The single-crystal structures of three salts of bedaquiline, a drug used for the treatment of drug-resistant tuberculosis (TB), are described.
Keywords: bedaquiline, bedaqulinium salt, drug-resistant tuberculosis, crystal structure
Abstract
Bedaquiline [systematic name: 1-(6-bromo-2-methoxyquinolin-3-yl)-4-(dimethylamino)-2-(naphthalen-1-yl)-1-phenylbutan-2-ol, C32H31BrN2O2] is one of two important new drugs for the treatment of drug-resistant tuberculosis (TB). It is marketed in the US as its fumarate salt {systematic name: [4-(6-bromo-2-methoxyquinolin-3-yl)-3-hydroxy-3-(naphthalen-1-yl)-4-phenylbutyl]dimethylazanium 3-carboxyprop-2-enoate, C32H32BrN2O2 +·C4H3O4 −}, and about a dozen other salts of bedaquiline have been described in patent literature, but none have so far been structurally described. In a first communication, we present the crystal structure of bedaquilinium fumarate and of two new benzoate salts, as well as that of a degradation product of the reaction of bedaquilinium fumarate with sodium ethoxide, 3-benzyl-6-bromo-2-methoxyquinoline, C17H14BrNO. The fumarate and benzoate salts both feature cations monoprotonated at the dimethylamino group. The much less basic quinoline N atom remains unprotonated. Both salts feature a 1:1 cation-to-anion ratio, with the fumarate being present as monoanionic hydrofumarate. The conformations of the cations are compared to that of free base bedaquiline and with each other. The flexible backbone of the bedaquiline structure leads to a landscape of conformations with little commonalities between the bedaquiline entities in the various structures. The conformations are distinctively different for the two independent molecules of the free base, the two independent molecules of the hydrofumarate salt, and the one unique cation of the benzoate salt. Packing of the salts is dominated by hydrogen bonding. Hydrogen-bonding motifs, as well as the larger hydrogen-bonded entities within the salts, are quite similar for the salts, despite the vastly differing conformations of the cations, and both the hydrofumarate and the benzoate structure feature chains of hydrogen-bonded anions that are surrounded by and hydrogen bonded to the larger bedaquilinium cations, leading to infinite broad ribbons of anions, cations, and (for the benzoate salt) water molecules. The benzoate salt was isolated in two forms: as a 1.17-hydrate (C32H32BrN2O2 +·C7H5O2 −·1.166H2O), obtained from acetone or propanol solution, with one fully occupied water molecule tightly integrated into the hydrogen-bonding network of anions and cations, and one partially occupied water molecule [refined occupancy 16.6 (7)%], only loosely hydrogen bonded to the quinoline N atom. The second form is an acetonitrile solvate (C32H32BrN2O2 +·C7H5O2 −·0.742CH3CN·H2O), in which the partially occupied water molecule is replaced by a 74.2 (7)%-occupied acetonitrile molecule. The partial occupancy induces disorder for the benzoate phenyl ring. The acetonitrile solvate is unstable in atmosphere and converts into a form not distinguishable by powder XRD from the 1.17-hydrate.
Introduction
Bedaquiline, 1, is one of two important new drugs for the treatment of drug-resistant tuberculosis (TB). Bedaquiline is marketed in the US as the fumarate salt (2) with the trade name Sirturo (Brigden et al., 2015 ▸). The fumarate salt is described in US Patent 8 546 428 (Hegyi et al., 2013 ▸). The citrate, sulfate, phosphate, and tartrate salts are described in two other patents (Zvatora, Dammer, Krejcik et al., 2016 ▸; Zvatora, Dammer, Ridvan et al., 2016 ▸). However, none of these salts has been structurally described in detail. For the fumarate, as well as one each of the two sulfate and citrate salts, well-resolved powder X-ray patterns have been reported, but the structures were not solved and no single-crystal data are reported. For the remaining salts (the phosphate and tartrate salts, and the second sulfate and citrate polymorphs), the powder patterns indicate the samples to have either extremely small particle distributions or to be entirely amorphous. Detailed structural data are reported solely for the free base form of bedaquiline (Petit et al., 2007 ▸).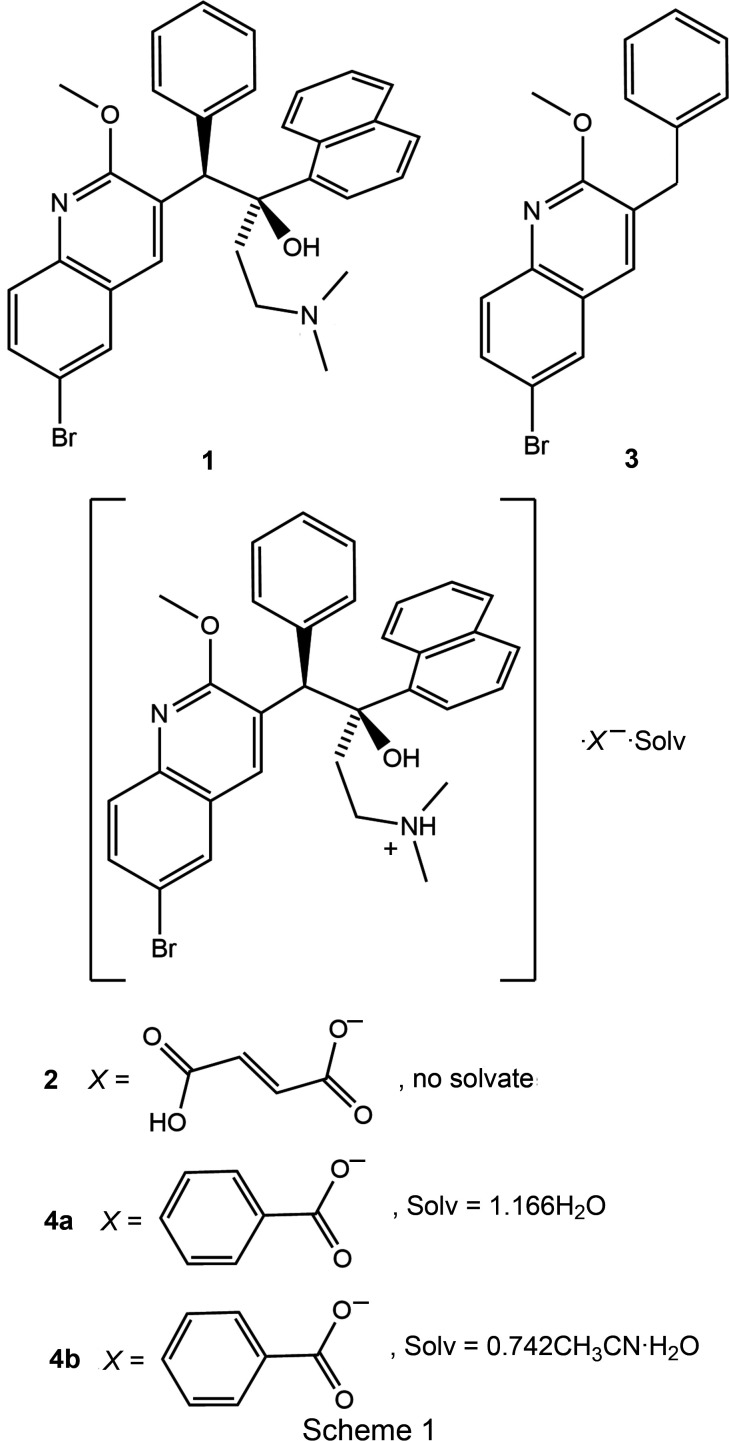
Bedaquiline features two basic N atoms that are amenable to protonation, i.e. the tertiary amine appended to the dangling ethylene group and the pyridine N atom. The two sites have distinctly different basicities and selective protonation of only the more basic amine site should be possible. Salts of both mono- and dicationic bedaquilinium ions can thus be formed, depending on the strength and amount of acid used for salt formation. The formation of cocrystals (with no or incomplete proton transfer) can also be imagined.
The NMR data reported in the patent publications indicate bedaquilinium fumarate to have a 1:1 anion-to-cation ratio. Whether the bedaquiline is protonated once or twice (and the fumarate deprotonated once or twice) had not been disclosed. For the sulfate salts, a 1:1 molar ratio of bedaquiline-to-sulfuric acid was used, but the anion-to-cation ratio in the salt was not determined. The given reaction yields, assuming a 1:1 salt, are around 33%, which would allow formation of a 1:2, a 1:1, or a 2:1 salt. The patent specifically states a wide range for the molar ratio of bedaquiline to sulfuric acid: ‘The molar ratio of bedaquiline:sulfuric acid may be in the range of 10:1 to 1:3, preferably 1:1, 2:1, and 1:2’. Similar statements have been made for the tartrate salts, one of the citrate salts, and the phosphate salt. No elemental analysis data are given to support any of the possible ratios, thus leaving the stoichiometry and the overall nature of the presented salts in question. The possibility of hydrate or solvate formation was also not properly addressed in the patent claims.
This lack of structural knowledge and even of basic chemical composition frustrates the understanding of the chemical, physical, and physiological properties of bedaquiline and its derivatives. To reduce this paucity of information on the bedaquiline system, there is interest in developing additional salts of bedaquiline and obtaining detailed analysis and structural data for these compounds, to better understand and possibly improve their properties, such as solubility, which in turn affect pharmacokinetics and dosage. Additional reasons for this study include finding a bedaquiline salt with improved stability and hygroscopicity.
Experimental
Melting points were determined using a Thomas Hoover Capillary Melting Point apparatus and are uncorrected. NMR data were collected in acetonitrile-d 3 (ACN-d 3) using a Bruker DRX-500 spectrometer and were referenced against the residual nondeuterated solvent peak.
Benzoic acid was purchased from Mallinckrodt, acetone from Fischer Chemicals, and acetonitrile from VWR Chemicals. Hydrochloride in methanol 1.25 M was obtained from Fluka. Bedaquiline fumarate was obtained from Johnson & Johnson. All chemicals were used as received without further purification.
Polarized light microscopy images were collected using an Olympus Series BX51TRF (Olympus America Inc., Melville, NY) polarized light microscope equipped with 12 V/100 W illumination; an Achromat 0.9 NA polarized-light condenser; Olympus Series UPlanFL N objectives: 40X/0.75 NA, 20X/0.50 NA, 10X/0.30 NA, and 4X/0.13 NA; an intermediate tube with variable position analyzer and compensator; and a trinocular viewing head with a Lumenera Series Infinity X (Teledyne Lumenera, Ottawa, Ontario, Canada) digital camera using Infinity (Version 6.5.6) and Infinity Analyze software (Version 7.0.2.930, Build date 01-Feb-2020). A small portion of sample was placed on a cleaned microscope slide and a No. 1 1/2-cover glass placed over the sample. Mineral oil, USP (CAS: 8042-47-5), was allowed to cover the sample by capillarity. Images were acquired as a collection of three: (i) plane polarized light, (ii) crossed polarized light, and (iii) crossed polarized light with a first-order red compensator. Microscopy observations revealed crystal habits for bedaquilinium benzoate powders as birefringent platy anhedral agglomerates that are softly bound and easily dispersed under light pressure from a tungsten needle on the cover glass. A representative collection of images is given in the supporting information.
IR microspectroscopy experiments were conducted using a Smiths Detection (Danbury, CT) IlluminatIR 1.5 IR Microspectrometer accessory on an Olympus Series BX41TF polarized-light microscope (Olympus America Inc., Melville, NY), which provided the base optical platform. The IlluminatIR 1.5 is equipped with a gray-body ceramic IR source, a 60° Michelson Interferometer with a zinc–selenide (ZnSe) beam splitter, a 4 wavenumber (cm−1) spectral resolution, and a 0.25 × 0.25 mm liquid-nitrogen-cooled mercury cadmium telluride (MCT) photoconductive detector, and the sample area was defined using a fixed circular 100 µm aperture. The IlluminatIR 1.5 is computer-interfaced using universal serial bus (USB) communications with Smiths Detection QualID App (Version 2.51, 2005) software. Advanced data processing was conducted using either Thermo Galactic spectral analysis software packages GRAMS/AI and SpectralID, or Thermo Fisher Scientific OMNIC software (Version 9.11.706, 2020). IR analyses were performed by reflection/absorption (R/A) using an all-reflecting objective (ARO, 15X, 0.88 NA). A small amount of sample was transferred to a low-E microscope slide (Smiths Detection P/N: 006-4013) and dispersed to a thin layer. IR microprobe analyses were conducted on what appeared microscopically to be a single crystal. An FT–IR spectral background was collected immediately prior to each sample spectral analysis.
Powder X-ray diffraction (XRD) data were collected in focusing mode on a PANalytical Empyrean X-ray diffractometer equipped with Bragg–Brentano HD optics, a sealed-tube copper X-ray source (λ = 1.54178 Å), Soller slits on both the incident and receiving optics sides, and a PixCel3D Medipix detector. Samples were hand ground using an agate mortar and pestle, and packed into a silicon single-crystal zero-background sample holder, 16 mm wide and 0.25 mm deep. Antiscatter slits and divergence slits, as well as the mask, were chosen based on sample area and starting θ angle. Data were collected between 4 and 40° in 2θ under ambient conditions using the PANalytical Data Collector software (PANalytical, 2015 ▸). Rietveld refinements were performed against the 150 K models of the single-crystal structure data sets using HighScore (PANalytical, 2015 ▸) software. Refinement of preferred orientation was included using a spherical harmonics model. Plots of Rietveld fits for all compounds are given in the supporting information.
Synthesis and crystallization
Free base bedaquiline (1)
The free base used during synthesis was prepared by extracting a CH2Cl2 solution of the fumarate three times with saturated NaHCO3 solution (Rombouts et al., 2016 ▸). The identity and purity of the free base thus afforded from the material supplied by Johnson & Johnson was verified using NMR spectroscopy [m.p. 175–176 °C; literature value 181°C (Zvatora, Dammer, Ridvan et al., 2016 ▸)]. 1H NMR (500 MHz, ACN-d 3): δ 8.82 (s, 1H), 8.66 (d, J = 8.7 Hz, 1H), 8.03 (s, 1H), 8.02 (d, J = 7.3 Hz, 1H), 7.87 (d, J = 8.0 Hz, 1H), 7.66 (t, J = 7.7 Hz, 4H), 7.49 (t, J = 7.7 Hz, 1H), 7.30 (m, 3H), 6.87 (m, 3H), 5.88 (s, 1H), 4.20 (s, 3H), 2.52 (d, J = 14.6 Hz, 1H), 2.01 (m, 2H), 1.89 (m, 7H).
Single crystals were grown by dissolving bedaquiline (30 mg, 0.054 mmol) in acetone (1 ml) in a 5 ml scintillation vial and the solution was allowed to evaporate slowly to obtain medium-sized plate-shaped crystals of 1.
Decomposition of bedaquiline fumarate by sodium ethoxide
Sodium ethoxide (1.5 g, 22.0 mmol) was dissolved in EtOH (20 ml). The resulting solution was added to a solution of bedaquiline fumarate (5 g, 7.44 mmol) in ACN/EtOH (50 ml, 1:1 v/v). After 1 h, water was added slowly and the resulting mixture extracted with EtOAc. The combined organic layers were dried (MgSO4) and then concentrated to provide a colorless crystalline material that was found by IR and NMR spectroscopies to not match free base bedaquiline. Individual crystals were identified as 3-benzyl-6-bromo-2-methoxyquinoline (3) by single-crystal X-ray diffraction, and no further analyses were performed.
Bedaquilinium fumarate (2)
Bedaquiline (30 mg, 0.054 mmol) was mixed with fumaric acid (6.3 mg, 0.054 mmol) dissolved in acetone (1 ml) in a 10 ml scintillation vial. Propyl alcohol (5 ml) was then added and the mixture was allowed to evaporate slowly to obtain large colorless block-shaped crystals that were analyzed by single-crystal and powder X-ray diffraction. 1H NMR (500 MHz, ACN-d 3): δ 8.68 (d, J = 8.3 Hz, 1H), 8.55 (s, 1H), 8.05 (d, J = 7.2 Hz, 1H), 7.97 (s, 1H), 7.88 (d, J = 7.9 Hz, 1H), 7.66 (m, 4H), 7.51 (t, J = 7.2 Hz, 1H), 7.32 (m, 3H), 6.89 (m, 3H), 6.32 (s, 2H), 5.89 (s, 1H), 4.21 (s, 3H), 3.02 (m, 1H), 2.69 (m, 1H), 2.24 (s, 7H), 2.09 (m, 2H).
Bedaquilinium benzoates 4a and 4b
Bedaquiline (30 mg, 0.054 mmol) was mixed with benzoic acid (6.7 mg, 0.055 mmol). The mixture was dissolved in acetone (2 ml) in a 5 ml scintillation vial and was allowed to evaporate. The 1.17-hydrate 4a was obtained in the form of colorless rod-shaped crystals (m.p. 127–129 °C). 1H NMR (500 MHz, ACN-d 3): δ 8.75 (d, J = 8.4 Hz, 1H), 8.67 (s, 1H), 8.12 (d, J = 7.1 Hz, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.86 (m, 3H), 7.75 (d, J = 8.0 Hz, 2H), 7.56 (m, 4H), 7.38 (m, 5H), 6.93 (m, 3H), 5.95 (s, 1H), 4.26 (s, 3H), 3.05 (m, 1H), 2.96 (m, 1H), 2.25 (m, 7H), 1.96 (m, 2H). An identical material with the same water content was obtained when crystallization was carried out from 2-propanol instead of acetone.
Bedaquiline (30 mg, 0.054 mmol) was mixed with benzoic acid (6.8 mg, 0.056 mmol) in acetonitrile (10 ml) and was allowed to evaporate slowly. The acetonitrile solvate monohydrate 4b was obtained in the form of thin colorless needles (m.p. 127–129 °C).
Refinement
Crystal data, data collection, and structure refinement details are summarized in Table 1 ▸. The two benzoate salt structures 4a and 4b are isomorphous, differing from each other only in the nature of part of the solvent molecules and some slight shifts to other atoms, and they were refined against a common structural model, with the structure of 4b being solved by isomorphous replacement from that of 4a. The atom-naming scheme for the published bedaquiline free base structure (Petit et al., 2007 ▸), as deposited in the Cambridge Structural Database (CSD; Groom et al., 2016 ▸; refcode KIDWAW), was used for the remeasured 150 K data of free base bedaquiline 1 and was also adopted for the bedaquilinium cations in the two benzoate salts 4a and 4b, and fumarate salt 2.
Table 1. Experimental details.
Experiments were carried out at 150 K. Absorption was corrected for by multi-scan methods (SADABS2016; Krause et al., 2015 ▸).
| 1 | 2 | 3 | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C32H31BrN2O2 | C32H32BrN2O2 +·C4H3O4 − | C17H14BrNO |
| M r | 555.50 | 671.57 | 328.20 |
| Crystal system, space group | Orthorhombic, P212121 | Monoclinic, P21 | Orthorhombic, P212121 |
| a, b, c (Å) | 11.1584 (8), 13.6425 (14), 36.061 (4) | 16.4556 (6), 10.3205 (3), 20.1636 (8) | 4.3606 (6), 10.820 (2), 29.886 (11) |
| α, β, γ (°) | 90, 90, 90 | 90, 109.1832 (15), 90 | 90, 90, 90 |
| V (Å3) | 5489.5 (9) | 3234.2 (2) | 1410.1 (6) |
| Z | 8 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 1.53 | 1.32 | 2.91 |
| Crystal size (mm) | 0.21 × 0.13 × 0.05 | 0.45 × 0.37 × 0.17 | 0.41 × 0.06 × 0.05 |
| Data collection | |||
| Diffractometer | Bruker D8 Quest diffractometer with PhotonII charge-integrating pixel array detector (CPAD) | Bruker D8 Quest diffractometer with PhotonII charge-integrating pixel array detector (CPAD) | Bruker D8 Quest diffractometer with PhotonII charge-integrating pixel array detector (CPAD) |
| T min, T max | 0.603, 0.747 | 0.438, 0.495 | 0.658, 0.747 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 66520, 17893, 12296 | 115858, 24622, 18572 | 28030, 5125, 4504 |
| R int | 0.052 | 0.040 | 0.037 |
| (sin θ/λ)max (Å−1) | 0.770 | 0.771 | 0.768 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.044, 0.111, 1.03 | 0.043, 0.117, 1.06 | 0.023, 0.057, 1.05 |
| No. of reflections | 17893 | 24622 | 5125 |
| No. of parameters | 675 | 837 | 196 |
| No. of restraints | 0 | 1 | 0 |
| H-atom treatment | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement | Only H-atom displacement parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.48, −0.58 | 1.23, −1.28 | 0.28, −0.38 |
| Absolute structure | Flack x determined using 4397 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) | Flack x determined using 7327 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) | Flack x determined using 1685 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.034 (3) | −0.0144 (14) | −0.011 (3) |
| 4a | 4b | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C32H32BrN2O2 +·C7H5O2 −·1.166H2O | C32H32BrN2O2 +·C7H5O2 −·0.742C2H3N·H2O |
| M r | 698.70 | 726.10 |
| Crystal system, space group | Monoclinic, P21 | Monoclinic, P21 |
| a, b, c (Å) | 12.6384 (5), 7.9259 (3), 17.5249 (8) | 12.8661 (8), 8.0386 (5), 17.4704 (10) |
| α, β, γ (°) | 90, 99.8450 (17), 90 | 90, 101.093 (3), 90 |
| V (Å3) | 1729.63 (12) | 1773.13 (19) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Cu Kα |
| μ (mm−1) | 1.24 | 1.97 |
| Crystal size (mm) | 0.55 × 0.21 × 0.13 | 0.31 × 0.05 × 0.05 |
| Data collection | ||
| Diffractometer | Bruker D8 Quest diffractometer with PhotonII charge-integrating pixel array detector (CPAD) | Bruker D8 Quest diffractometer with PhotonIII_C14 charge-integrating and photon counting pixel array detector |
| T min, T max | 0.638, 0.746 | 0.599, 0.754 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 80228, 13080, 10456 | 39739, 7360, 6750 |
| R int | 0.049 | 0.060 |
| (sin θ/λ)max (Å−1) | 0.770 | 0.639 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.032, 0.073, 1.03 | 0.035, 0.085, 1.06 |
| No. of reflections | 13080 | 7360 |
| No. of parameters | 445 | 515 |
| No. of restraints | 5 | 195 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.36, −0.48 | 0.40, −0.50 |
| Absolute structure | Flack x determined using 4051 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) | Flack x determined using 2778 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.006 (3) | 0.004 (8) |
For powder X-ray data collection and refinement, see the Experimental (§2).
H-atom treatment
C-bound H atoms were added in calculated positions and refined using a riding model. C—H bond distances were constrained to 0.95 Å for aromatic C—H moieties, and to 1.00, 0.99, and 0.98 Å for aliphatic C—H, CH2, and CH3 moieties, respectively. Alcohol O—H and ammonium NR 3H N—H bond distances were either freely refined (for 2) or were constrained to 0.84 and 1.00 Å, respectively. Methyl CH3 and hydroxy H atoms were allowed to rotate but not to tip to best fit the experimental electron density. The positions of the carboxylate H atoms (in 2) were refined freely. The positions of the fully occupied water H atoms were refined freely and O—H distances were restrained to 0.84 (2) Å. The H atoms of the partially occupied water molecule in 4a were initially refined and O—H and H⋯H distances were restrained to 0.84 (2) and 1.36 (2) Å, respectively, while a damping factor was applied. The position of water atom H6E (in 4a) was further restrained based on hydrogen-bonding considerations, i.e. to be hydrogen bonded to the pyridine H atom, with the H⋯N distance restrained to 2.35 (2) Å. In the final refinement cycles, the damping factor was removed and the H atoms were set to ride on the parent O atom. For all structures, the U iso(H) values were set to a multiple of U eq(C), being 1.5 for CH3 and OH, and 1.2 for C—H, CH2, and N—H groups, respectively.
Disorder modeling
In the structure of 4a, one fully occupied and one partially occupied water molecule are present in the lattice. The occupancy ratio refined to 0.166 (7). In 4b, the partially occupied water molecule is replaced by an approximately three-quarter-occupied acetonitrile molecule. In the absence of the acetonitrile molecule, the neighboring benzene ring of the benzoate anion tilts slightly to move towards the void left by the absent solvent molecule. The two disordered benzene moieties were restrained to have similar geometries. The Uij components of the anisotropic displacement parameters (ADPs) for disordered atoms closer to each other than 2.0 Å were restrained to be similar. The ADPs of the ipso C atoms, which occupy nearly identical positions, were constrained to be identical. Subject to these conditions, the occupancy ratio refined to 0.742 (7):0.258 (7) in favor of the acetonitrile molecule being present.
Salt screening and methods
Salt screening is a complex and challenging endeavor involving potentially millions of experiments. For bases like bedaquiline, these experiments can involve up to 50 commonly used salt formers in various ratios, as well as crystallizations from up to 60 different solvents by varying temperature, concentration, agitation, pH, and other factors. Further mixtures of these solvents can be used. Antisolvent crystallization using these solvents is also of interest and introduces even more variables.
For bedaquiline, the first step in screening for additional salts involved recovering bedaquiline free base from the commercially available bedaquilinium fumarate. This was first attempted by deprotonation of the bedaquilinium cation of the fumarate salt using the base sodium ethoxide. However, the alkoxide, when used in excess, proved to be too strong a base and led to fragmentation of the bedaquiline molecule. One of the products of the decomposition reaction was isolated by crystallization and identified, by single-crystal X-ray diffraction, as 3-benzyl-6-bromo-2-methoxyquinoline (3) (Fig. 1 ▸) and its structure will be described below. The other half of the decomposition reaction was not recovered or identified, but is assumed to be the ketone elimination product of the remaining bedaquiline fragment, 4-(dimethylamino)-1-(naphthalen-1-yl)butan-2-one. The reaction most likely proceeds through initial deprotonation of all acidic groups by the ethoxide, including the central alcohol of bedaquiline. The tertiary alkoxide thus formed can undergo a reverse Grignard reaction (Zook et al., 1959 ▸), under elimination of the ketone and the carbanion of 3. Using much less basic sodium bicarbonate as the neutralizing agent avoids this decomposition reaction. Bedaquiline free base could be recovered that way from the fumarate salt, following the procedure described by Rombouts et al. (2016 ▸), thus allowing us to proceed to use the free base in salt screening experiments (Fig. 1 ▸).
Figure 1.
Isolation of free base bedaquiline (1) from commercially available bedaquilinium fumarate (2), and decomposition to 3.
Because the fumarate, citrate, sulfate, phosphate, and tartrate salts were known, salt formation screening focused on the crystallization of salts such as acetate, benzoate, benzenesulfonate, hydrobromide, succinate (1:1 and 1:2), hydrochloride, tartrate (1:1 and 1:2), lactate, maleate (1:1 and 1:2), malate (1:1 and 1:2), and mesylate. In general, the crystallizations involved mixing stoichiometric amounts of bedaquiline with the acids at either 1:1 or 1:2 molar ratios in acetone, acetonitrile, tetrahydrofuran, and ethyl acetate, either with or without the antisolvents water and hexane. The solvents were evaporated either slowly or rapidly, and materials were typically first screened using polarized-light microscopy (PLM) to ensure that a crystalline material had formed, and that the sample was uniform. Samples that passed the first screening step were submitted for further analysis. Crystals were analyzed by NMR (dissolved in an appropriate solvent) to confirm the presence of both components in the material. In the next step, materials were further screened using IR microspectroscopy, to confirm that the material was indeed a new substance (a salt or a cocrystal), and not just a mixture of the two components. Although some investigators have advanced the theory that Raman spectroscopy is the best method for analysis and determination of salt formation (e.g. Kojima et al., 2010 ▸), we found IR microspectroscopy had better specificity than Raman microscopy for the bedaquiline free base and salts; therefore, screening materials via Raman methods was abandoned. IR microspectroscopy proved to be a superior method to determine the formation of bedaquilinium salts. Materials that passed these screening steps (PLM, NMR, and IR spectroscopy) were then analyzed by powder X-ray diffraction. Rietveld refinement was used to identify known crystal phases. For samples for which suitable crystals could be obtained, single-crystal X-ray diffraction was used to determine the structures of phases not yet structurally described.
Example IR spectra comparing bedaquilinium benzoate and free base bedaquiline are given in Fig. 2 ▸; see Fig. 3 ▸ for the synthesis. The spectra are distinctly different, indicating transformation of the free base into a material containing both benzoate and bedaquiline fragments. A range of bands in the fingerprint region indicate the presence of a bedaquiline component in both compounds. A shoulder near 1700 cm−1 in the bedaquiline benzoate spectrum can be assigned to the C=O stretch of benzoate, confirming the formation of the salt. Further evidence for the formation of a salt, rather than a simple mixture of the two starting materials, is provided by the absence of bands in the range 2830–2760 cm−1. Tertiary amines (of which the free base is one) have a characteristic N—CH2 in-phase stretch that occurs in this range (Colthup et al., 1990 ▸). The bands in this range of the free base spectrum are not present in the bedaquiline benzoate spectrum, suggesting the formation of a salt. Note: the free base spectrum contains some spectral features due to ethanol, which was used in the synthesis process.
Figure 2.
IR spectra of bedaquilinium benzoate (4a) (blue) and bedaquiline free base (1) (red).
Figure 3.
Synthesis of bedaquilinium benzoate (4).
In the course of our investigations, we had been so far able to determine the single-crystal structures of bedaquilinium fumarate (2), the commercially available form of bedaquiline, as well as isolate and characterize two other previously unknown bedaquilinium salts: the mono-benzoate salt, in the form of its 1.17-hydrate (4a), and a mixed hydrate acetonitrile solvate (4b). Their structures, as well as that of the degradation product from reaction of bedaquilinium fumarate with sodium ethoxide, 3-benzyl-6-bromo-2-methoxyquinoline (3), will be described below. The structure of free base bedaquiline (1) was re-recorded at 150 K for easier comparison with the low-temperature data of 2, 4a, and 4b. Implications for the larger bedaquiline system will be discussed.
Results and discussion
3-Benzyl-6-bromo-2-methoxyquinoline (3), the solvolysis product from reaction of bedaquiline with sodium ethoxide, lacks an easy-to-identify NMR signature and was identified by single-crystal X-ray diffraction. It crystallized from acetonitrile in the orthorhombic and chiral space group P212121 (Fig. 4 ▸). The starting bedaquilinium fumarate is a chiral compound and an enantiopure sample was used. This chiral information and all chiral centers are, however, lost in the degradation reaction to 3-benzyl-6-bromo-2-methoxyquinoline (3). In the solid state, the molecule does, however, exhibit chirality, and the crystal analyzed was found to be enantiopure, with a Flack parameter of −0.011 (3). In solution, the material is expected to be a rapidly interconverting racemic mixture, as simple rotation of the benzyl group to the other side of the mean plane of the molecule creates the inversion-related enantiomer. Molecules of 3 are divided into two planar fragments: the benzyl group and the 6-bromo-2-methoxyquinoline moiety. Both fragments are close to ideally flat, with r.m.s. deviations from planarity of only 0.0052 and 0.0194 Å, respectively. The methoxy group is thus ideally coplanar with the remainder of the bromoquinoline fragment. It points away from the benzyl CH2 group to avoid steric interactions. The torsion angle between the two mean planes is 73.01 (4)°.
Figure 4.
The structure of decomposition product 3 (50% probability displacement ellipsoids).
The structures of the three bedaquilinium salts, i.e. fumarate 2, and benzoates 4a and 4b, are substantially more complicated (Figs. 5 ▸ and 6 ▸), but they share some commonalities. In all three salt structures, the bedaquilinium cation is singly protonated at the dimethylamino fragment, with 1:1 anion-to-cation ratios. In the structure of 2, the fumarate anions remain singly protonated hydrofumarate(1−) anions, thus being monoanionic, as are the benzoate anions. The quinoline N atoms remain unprotonated, even though there are additional acidic protons available in the structure of 2. At first glance, this might be surprising, since many pyridinium salts of both benzoic and fumaric acids have been reported [973 pyridinium benzoate derivatives and 44 pyridinium fumarate salts are reported in the CSD (Groom et al., 2016 ▸), accessed August 2020]. The behaviour is, however, in agreement with the pKa values of the acids and with the reduced basicity of the quinoline N atom of bedaquiline, compared to ordinary pyridine. The first pKa of fumaric acid is 3.053, the second is 4.494, and that of benzoic acid is 4.202 (Martell & Smith, 1976 ▸), which are easily sufficient to protonate the amine moiety of bedaquiline [the pKa of trialkylammonium ions are around 10–11 (Bioquest, 2020 ▸)]. The pKa of the conjugated acid of bedaquiline protonated at the quinoline N atom is not reported but can be estimated from the known values for quinoline, pyridine, and 2-methoxypyridine, which are 4.9, 5.23, and 3.06, respectively (Bioquest, 2020 ▸). Methoxy substitution in the 2-position to the N atom substantially reduces the basicity of the N atom (the pKa of the conjugated acid drops by 2.17 between pyridine and 2-methoxypyridine). Assuming other effects to be negligible yields a pKa value of 2.73 for 2-methoxyquinolinium. The quinoline N atom of bedaquiline is thus not basic enough to be protonated by medium-strength acids, such as benzoic or fumaric acid, in agreement with the findings from the crystal structures for 2, 4a, and 4b. Stronger acids, such as mineral acids or maleic acid (first pKa is 1.94; Bioquest, 2020 ▸), might be able to double protonate bedaquiline if used in sufficient excess. Experiments in this direction are ongoing in our laboratories.
Figure 5.
Single-crystal structure of fumarate salt 2 (50% probability displacement ellipsoids).
Figure 6.
Single-crystal structures of benzoate salts of 4a (left) and 4b (right) (50% probability displacement ellipsoids). Hydrogen bonds are shown as turquoise dashed lines.
All three salts do crystallize in the chiral monoclinic space group P21. The core of the bedaquilinium cation in the three structures is formed by the ethylene moiety of atoms C1 and C2, from which the four major substituents radiate off: the phenyl ring and the bromo(methoxy)quinoline group from C1, and the naphthyl and (dimethylamino)ethyl fragments from C2. The hydroxy group is also attached to C2, while C1 also carries a single H atom. Atoms C1 and C2 are also the chiral centers of the bedaquilinium cation, which were modeled to have R and S chiralities, respectively, in agreement with the reported absolute structure for free base bedaquiline (Petit et al., 2007 ▸). The Flack parameters refined to −0.014 (1) for 2, and to 0.006 (3) and 0.004 (8) for 4a and 4b, respectively, confirming that the crystals were enantiopure.
The arrangement of anions and cations, and packing interactions, however, vary substantially between the fumarate and the two benzoate salts. The fumarate salt crystallized in an anhydrous and unsolvated form. Two crystallographically unique ion pairs are present in the lattice (i.e. Z′ = 2 for compound 2). The structure obtained agrees with the reported powder patterns of commercially available bedaquilinium fumarate (see Fig. 7 ▸ for a Rietveld refinement plot).
Figure 7.
Powder XRD pattern (ambient temperature) of bedaquilinium fumarate (Johnson & Johnson) with a Rietveld refinement fit against the single-crystal structure of 2. The room-temperature unit-cell parameters refined to a = 16.5879 (2), b = 10.4952 (8), c = 20.183 (2) Å, β = 109.238 (2)° and V = 3317.4 (4) Å3. Rietveld fits for 4a and 4b are given in the supporting information.
The two newly isolated benzoate salts are distinctly different from 2, both being solvates, but they are very similar to each other, and are indeed isomorphous (the acetonitrile solvate was solved by isomorphous replacement from the hydrate). Both structures feature one tightly bound water molecule (atom O5). A second interstitial site does, however, differ between the two phases. In 4a, it is occupied by a second water molecule, which is partially occupied. In 4b, on the other hand, this site is partially occupied by a disordered acetonitrile molecule, which in turn induces disorder in the phenyl ring of the benzoate anion [see Refinement (§2.2.2) for disorder details].
The ethane backbone of the bedaquiline core gives the cations a three-dimensional (3D) shape, but the individual aromatic fragments remain planar. Similar to 3, the 6-bromo-2-methoxyquinoline moiety is planar, with the methoxy group pointing away from the core of the cation. The r.m.s. deviations from planarity are 0.1127 and 0.1363 for the two cations in 2 (Z′ = 2), 0.1019 Å in 4a, and 0.0922 Å in 4b.
The ethane backbone and the malleable ethylamine fragment gives the bedaquilinium cations a high degree of conformational flexibility. Differing packing arrangements, induced by the presence (or absence) of varying anions and solvent molecules, led to a landscape of conformations observed for the cations in 2, 4a, and 4b, as well as free base bedaquiline 1. The dihedral angles between the mean planes of the 6-bromo-2-methoxy-quinoline fragment (plane 1), the phenyl ring (plane 2), and the naphthyl group (plane 3), as well as the torsion angles along the ethane backbone and the ethylamine fragment, are given in Table 2 ▸. Besides the obvious similarities between the values for isomorphous 4a and 4b, no general trend is observed. The conformations vary not only between the four structures, but even between independent molecules within the same structure (both free base 1 and fumarate 2 are Z′ = 2 structures). The two C1—C2—C3—C4 torsion angles in fumarate salt 2, for example, are −63.8 (3) and 174.92 (19)°, which are distinctly different from each other. However, some similarities can be observed: the interplanar angles between the phenyl and 6-bromo-2-methoxyquinoline planes are between 70 and 90° in all structures, and the torsion angles involving the ethane backbone and the ipso phenyl atom (C17—C1—C2—C3) are close to antiperiplanar (‘trans’) in all the compounds. No other similarities common to all four structures can be found and the overall trend is one of pronounced flexibility and variability.
Table 2. Selected torsion angles (°) for free base bedaquiline 1, fumarate salt 2, and benzoate salts 4a and 4b .
| 1 a,b | 2 b | 4a a | 4b a | |
|---|---|---|---|---|
| τ plane 1 versus plane 2c | 73.29 (7), 81.37 (7) | 85.33 (9), 86.02 (8) | 77.31 (6) | 76.2 (1) |
| τ plane 2 versus plane 3c | 86.13 (8) 76.95 (7) | 89.7 (1), 89.74 (9) | 45.62 (6) | 44.2 (1) |
| τ plane 1 versus plane 3c | 14.0 (1), 8.16 (9) | 8.71 (3), 16.67 (4) | 37.50 (6) | 36.7 (1) |
| τ C1—C2—C3—C4 | 175.7 (3), 177.0 (3) | −63.8 (3), 174.92 (19) | 166.84 (14) | 165.1 (3) |
| τ C2—C3—C4—N1 | 58.7 (4), 63.0 (4) | 137.2 (2), 133.7 (2) | 164.04 (15) | 165.5 (3) |
| τ C17—C1—C2—C3 | 169.0 (3), 178.8 (3) | 169.7 (2), 171.38 (19) | 174.44 (14) | 174.1 (3) |
Notes: (a) measured at 150 K; (b) Z′ = 2; (c) plane 1 = 6-bromo-2-methoxyquinoline, plane 2 = phenyl, and plane 3 = naphthyl.
While the geometries and conformations in the four bedaquiline structures do not follow any general trend, there are some differences between the geometries of free base bedaquiline and its salts that can be rationalized. Directional interactions that differ between the free base and the salts play a major role. In bedaquiline, only one actual hydrogen bond is present, and this is an intramolecular O—H⋯N hydrogen bond (Table 3 ▸). It induces the amine N atoms to turn towards the hydroxy group, thus enforcing a gauche geometry of the (dimethylamino)ethyl group (see torsion angle C2—C3—C4—N1 in Table 2 ▸). Intermolecular interactions between molecules in 1 are limited to weaker and less directional interactions, specifically Br⋯Br interactions and π-stacking, which had been discussed in detail by Petit et al. (2007 ▸). In the fumarate and benzoate salts 2 and 4, the opposite is observed. The amine moiety, being protonated, is unable to act as the acceptor for an O—H⋯N hydrogen bond, and it takes up the role of a hydrogen-bond donor towards the benzoate or fumarate anions. This releases the amine from the gauche conformation enforced by the intramolecular O—H⋯N hydrogen bond in 1, and the ethylamine instead extends into a more relaxed conformation, approaching anti in 4 and close to eclipsed in 2 (see torsion angle C2—C3—C4—N1 in Table 2 ▸). The hydroxy groups in the salts are freed up to also form intermolecular hydrogen bonds. In 4, they hydrogen bond with the fully occupied water molecule, and in 2 with a fumarate carboxylate group. The water molecule in 4 in turn extends hydrogen bonds to the O atoms of two neighboring benzoate anions, and the fumarate anions in 2 are involved in a series of hydrogen bonds among each other, forming an infinite chain of hydrogen-bonded anions along the b-axis direction (Fig. 8 ▸). Thus, one intramolecular hydrogen bond in free base bedaquiline 1 is converted into a whole series of strong intermolecular hydrogen bonds (Tables 4 ▸, 5 ▸ and 6 ▸), creating for the salts a much stronger cohesion between neighboring structural entities than what was observed in the free base structure. Weaker interactions that dominate for bedaquiline, such as Br⋯Br halogen bonds and π-stacking, are not observed for the salts, but the strong hydrogen bonds are augmented by a range of other not-as-strong directional interactions, such as C—H⋯O and C—H⋯Br hydrogen bonds (see hydrogen-bond Tables 4 ▸, 5 ▸, and 6 ▸ for a full listing).
Table 3. Hydrogen-bond geometry (Å, °) for 1 .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯N1 | 0.84 | 1.94 | 2.696 (4) | 150 |
| C1—H1⋯O2 | 1.00 | 2.24 | 2.763 (4) | 111 |
| O3—H3O⋯N3 | 0.84 | 1.93 | 2.685 (4) | 149 |
| C33—H33⋯O4 | 1.00 | 2.23 | 2.773 (4) | 112 |
Figure 8.
Hydrogen-bonding pattern in 2. (Top) Chains along b created by hydrofumarate anions. For clarity, only the 3-dimethylazaniumyl-1-hydroxypropyl fragments of the cations are shown, and H atoms not involved in hydrogen bonds have been omitted. (Bottom) Segment of an entire ribbon, including full cations. Color coding: O, N, and Br atoms are shown with 50% probability displacement ellipsoids, with all other atoms in capped-stick mode and color coded by the symmetry equivalence of their anion and cation (A and B cations in light and dark green, and A and B anions in orange and red). Hydrogen bonds are shown as turquoise and red dashed lines.
Table 4. Hydrogen-bond geometry (Å, °) for 2 .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1A—H1AB⋯O4A | 0.86 (4) | 1.88 (4) | 2.699 (3) | 159 (4) |
| O5A—H5A⋯O4B | 1.04 (4) | 1.58 (4) | 2.603 (3) | 169 (4) |
| N1A—H1AN⋯O3A | 0.98 (4) | 1.68 (4) | 2.641 (3) | 163 (3) |
| C1A—H1A⋯O2A | 1.00 | 2.23 | 2.767 (3) | 112 |
| C3A—H3AB⋯Br1B i | 0.99 | 3.01 | 3.850 (3) | 143 |
| C5A—H5AA⋯O1A ii | 0.98 | 2.58 | 3.501 (3) | 157 |
| C5A—H5AC⋯Br1B i | 0.98 | 2.82 | 3.612 (3) | 139 |
| C26A—H26A⋯O5B iii | 0.95 | 2.54 | 3.459 (4) | 163 |
| C34A—H34A⋯O5B iii | 0.95 | 2.52 | 3.379 (3) | 151 |
| O1B—H1B⋯O4B iv | 0.89 (4) | 1.88 (4) | 2.741 (3) | 160 (4) |
| O6B—H6B⋯O4A v | 0.86 (5) | 1.80 (5) | 2.625 (3) | 159 (4) |
| N1B—H1BN⋯O3B iv | 0.85 (4) | 1.83 (4) | 2.632 (3) | 158 (4) |
| C5B—H5BA⋯O1B vi | 0.98 | 2.42 | 3.337 (4) | 156 |
| C5B—H5BB⋯Br1A | 0.98 | 2.88 | 3.468 (3) | 119 |
| C5B—H5BB⋯O6A | 0.98 | 2.42 | 3.287 (4) | 148 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  .
.
Table 5. Hydrogen-bond geometry (Å, °) for 4a .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1B⋯O5 | 0.84 | 1.87 | 2.681 (2) | 164 |
| O5—H5D⋯O4i | 0.90 (4) | 1.85 (4) | 2.724 (2) | 163 (3) |
| O5—H5E⋯O3 | 0.77 (3) | 1.96 (3) | 2.724 (2) | 172 (4) |
| O6—H6E⋯N2 | 0.87 | 2.38 | 3.148 (13) | 147 |
| N1—H1⋯O4i | 1.00 | 1.64 | 2.643 (2) | 178 |
| C5—H5C⋯Br1ii | 0.98 | 3.09 | 3.910 (2) | 142 |
| C6—H6A⋯O3 | 0.98 | 2.53 | 3.465 (3) | 161 |
| C6—H6B⋯O6iii | 0.98 | 2.28 | 3.249 (13) | 169 |
| C28—H28⋯O5ii | 0.95 | 2.59 | 3.421 (3) | 146 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Table 6. Hydrogen-bond geometry (Å, °) for 4b .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1B⋯O5 | 0.77 | 1.94 | 2.691 (4) | 163 |
| O5—H5D⋯O4i | 0.86 (3) | 1.89 (3) | 2.738 (4) | 168 (6) |
| O5—H5E⋯O3 | 0.82 (3) | 1.92 (3) | 2.734 (4) | 172 (6) |
| N1—H1⋯O4i | 1.00 | 1.62 | 2.619 (4) | 178 |
| C5—H5C⋯Br1ii | 0.98 | 3.13 | 3.964 (4) | 144 |
| C6—H6A⋯O3 | 0.98 | 2.63 | 3.562 (6) | 159 |
| C28—H28⋯O5ii | 0.95 | 2.66 | 3.498 (5) | 148 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
In the acetonitrile solvate 4b, an H atom of the naphthyl group interacts in a C—H⋯N hydrogen-like bond with the solvent molecule, when present. In its absence, the phenyl ring of the benzoate anion relaxes towards the void created, inducing disorder for the anion [see Refinement (§2.2.2) for details]. For the 1.17-hydrate 4a, the partially occupied water molecule is hydrogen bonded to the pyridine N atom and acts as an acceptor for a C—H⋯O interaction. There is, however, no second possible hydrogen-bond acceptor near the partially occupied water molecule, and the second water H atom is not involved in any recognizable interaction. This is energetically unfavorable, which helps to explain why this position is occupied less than 20% of the time [the refined value is 16.6 (7)%], while the other solvent water molecule, tightly hydrogen bonded on all sides, is fully occupied. Lack of space appears to be no issue, as the larger acetonitrile molecule in 4b has a higher occupancy of around three-quarters [refined value 74.2 (7)%]. Kinetic factors during crystal growth (a lack of water molecules during crystallization from mostly dry solvents for 4a, but no lack of acetonitrile molecules for 4b) seem to dominate. The presence or absence of either water or acetonitrile in this void does not appear to hinder continuation of crystal growth, while this cannot be said for the tightly incorporated O5 water molecule present in both structures, which appears to be essential for the formation of this structure. The larger size of acetonitrile versus water, and the low prevalence of the second water molecule in 4a, lead to a slightly larger unit-cell volume for 4b compared to 4a, i.e. 1773.13 (19) versus 1729.63 (12) Å3. The visually most obvious difference between the two structures is the difference in the β angle, which is expanded by slightly more than 1° in 4b, leading to a significant offset between atoms in the two structures when the unit cells are overlaid (Fig. 9 ▸). The same is evident when comparing the powder patterns of 4a and 4b simulated from the 150 K single-crystal data, which are distinctively different (see supporting information).
Figure 9.
Least-squares overlay of the structures of 4a (red) and 4b (blue) using the non-H atoms for one of the two ion pairs of the asymmetric unit (r.m.s. fit value = 0.152), ignoring water and solvent molecules, and disorder. When extending the fit to the atoms of both ion pairs of the unit cell, the r.m.s. value increases to 0.195, showing the lattice mismatch between 4a and 4b.
Observations based on powder XRD data indicate that the samples of 4b quickly desolvate, even under ambient conditions. The powder patterns of 4b more closely match the parameters of hydrate 4a than would be expected for acetonitrile solvate 4b, even if the data were collected on samples exposed to air only for a few minutes prior to data collection (Table 7 ▸). Rietveld refinements of samples of 4b yield β angles that match those of 4a (at both room temperature and 150 K), and the room-temperature volume of 4b is actually slightly smaller than that of 4a, in agreement with the assumption that all the acetonitrile in the structure of 4b is lost when exposed to the atmosphere, while the solvent water molecules of 4a, being hydrogen bound, do remain in the lattice under these conditions. These observations will be further investigated in a more detailed upcoming study, focusing on a larger series of bedaquilinium salts, including their solvation properties, thermal stability, and responses to ambient moisture.
Table 7. 150 K and room-temperature (RT) unit-cell dimensions for 1, 2, 4a, and 4b .
RT data were obtained from powder XRD patterns via Rietveld refinement (see supporting information for Rietveld plots).
| 1, 150 K | 1, RT | 2, 150 K | 2, RT | ||
|---|---|---|---|---|---|
| a (Å) | 11.1584 (8) | 11.230 (1) | 16.4556 (6) | 16.5879 (2) | |
| b (Å) | 13.6425 (14) | 13.766 (1) | 10.3205 (3) | 10.4952 (8) | |
| c (Å) | 36.061 (4) | 36.455 (3) | 20.1636 (8) | 20.183 (2) | |
| β (°) | 90 | 90 | 109.1832 (15) | 109.238 (2) | |
| V (Å3) | 5489.5 (9) | 5636 (1) | 3234.2 (2) | 3317.4 (4) | |
| ρ (Mg m−3) | 1.344 | 1.309 | 1.379 | 1.344 | |
| R wp (%) | 19.99 | 20.48 | |||
| R exp (%) | 15.67 | 13.27 | |||
| S | 1.27 | 1.54 | |||
| 4a, 150 K | 4a, RT | 4b, 150 K | 4b, RTa | ||
| a (Å) | 12.6384 (5) | 12.7267 (4) | 12.8661 (8) | 12.720 (1) | 12.7242 (8) |
| b (Å) | 7.9259 (3) | 8.0157 (3) | 8.0386 (5) | 8.0094 (5) | 8.0116 (6) |
| c (Å) | 17.5249 (8) | 17.6438 (7) | 17.4704 (10) | 17.626 (2) | 17.632 (1) |
| β (°) | 99.8450 (17) | 99.7928 (6) | 101.093 (3) | 99.836 (2) | 99.813 (1) |
| V (Å3) | 1729.63 (12) | 1773.7 (1) | 1773.13 (19) | 1769.4 (3) | 1771.2 (2) |
| ρ (Mg m−3) | 1.342 | 1.309 | 1.360 | 1.311 | 1.361 |
| R wp (%) | 5.57 | 30.65 | 28.65 | ||
| R exp (%) | 10.78 | 10.50 | 14.26 | ||
| S | 1.94 | 2.12 | 2.01 | ||
Note: (a) data from two independent samples and Rietveld refinements.
The structure of 2, on the other hand, is solvent free. Like 4, the main intermolecular interactions are strong hydrogen bonds, already briefly discussed, with the bedaquilinium cation always acting as a hydrogen-bond donor, and the hydrofumarate anions acting as both H-atom acceptors and donors. The quinoline N atom does not act as an acceptor for a hydrogen bond, in agreement with its reduced basicity, already discussed. Originating from the cation are N—H⋯O hydrogen bonds, formed by the ammonium cations, and O—H⋯O hydrogen bonds, originating from the alcohol moieties. The N—H⋯O and O—H⋯O hydrogen bonds from one cation are towards the two O atoms of the same fully deprotonated carboxylate group, yielding an  (10) hydrogen-bonding graph-set motif. This motif is identical for the two crystallographically independent ion pairs in 2. The remaining strong hydrogen bonds are between the hydrofumarate anions. The hydrogen-bond donors are the carboxylic acid groups of each hydrofumarate anion, while the hydrogen-bond acceptor is always the O atom also hydrogen bonded to the bedaquilinium alcohol group, thus tying fumarate anions together in a head-to-tail fashion, forming infinite chains that extend along the b-axis direction (Fig. 8 ▸). The individual hydrogen-bonding motifs for these hydrogen bonds are D(2). The hydrofumarate anion chains bridge between bedaquilinium cations connecting anions and cations into ribbons that extend along the b-axis direction.
(10) hydrogen-bonding graph-set motif. This motif is identical for the two crystallographically independent ion pairs in 2. The remaining strong hydrogen bonds are between the hydrofumarate anions. The hydrogen-bond donors are the carboxylic acid groups of each hydrofumarate anion, while the hydrogen-bond acceptor is always the O atom also hydrogen bonded to the bedaquilinium alcohol group, thus tying fumarate anions together in a head-to-tail fashion, forming infinite chains that extend along the b-axis direction (Fig. 8 ▸). The individual hydrogen-bonding motifs for these hydrogen bonds are D(2). The hydrofumarate anion chains bridge between bedaquilinium cations connecting anions and cations into ribbons that extend along the b-axis direction.
These strong hydrogen bonds in 2 are again augmented by a series of other not-as-strong directional interactions, such as C—H⋯O and C—H⋯Br hydrogen bonds (Table 4 ▸) that interconnect between ribbons to create a fully 3D network and structure. Segments of neighboring ribbons do also interdigitate with each other, further stabilizing the overall structure.
The arrangement of anions and cations in 4a and 4b, and their connection via hydrogen-bonding interactions, follows a similar pattern to that in 2, but it is augmented by the solvate water molecules, which play a similar role as the protonated fumarate carboxylic acid groups do in 2 in connecting anions and cations with each other into infinite ribbons (Fig. 10 ▸). Bedaquilinium cations and water molecules act as hydrogen-bond donors and the benzoate anions act as hydrogen-bond acceptors. The N—H⋯O hydrogen bond from the cation is towards one O atom of the benzoate carboxylate group and the O—H⋯O hydrogen bond is towards the water molecule, which in turn is hydrogen bonded to the same benzoate O atom as the ammonium fragment. The  (10) graph-set motif in 2 is thus converted in 4 into an
(10) graph-set motif in 2 is thus converted in 4 into an  (10) motif, but otherwise fulfils the same function as in 2, connecting the hydroxy and ammonium segments of one cation to the same carboxylate group. The fully occupied water molecules in 4 assume the role of the carboxylic acid groups in 2, acting as bridges between anions [
(10) motif, but otherwise fulfils the same function as in 2, connecting the hydroxy and ammonium segments of one cation to the same carboxylate group. The fully occupied water molecules in 4 assume the role of the carboxylic acid groups in 2, acting as bridges between anions [ (6) hydrogen-bonding motif], creating infinite benzoate–water chains that extend along the b-axis direction. Wrapped around these chains, and connected to them via O—H⋯O and N—H⋯O hydrogen bonds, are the bedaquiline cations, thus creating wider ribbons of cations, anions, and solvent water molecules (Fig. 10 ▸), emulating the pattern already observed for 2. The partially occupied water molecules are located on the outer rim of the ribbon, hydrogen bonded to the quinoline N atom but not bridging or connecting between any structural entities.
(6) hydrogen-bonding motif], creating infinite benzoate–water chains that extend along the b-axis direction. Wrapped around these chains, and connected to them via O—H⋯O and N—H⋯O hydrogen bonds, are the bedaquiline cations, thus creating wider ribbons of cations, anions, and solvent water molecules (Fig. 10 ▸), emulating the pattern already observed for 2. The partially occupied water molecules are located on the outer rim of the ribbon, hydrogen bonded to the quinoline N atom but not bridging or connecting between any structural entities.
Figure 10.
Hydrogen-bonding pattern in 4a. (Top) Chains along b created by benzoate anions and water molecules. For clarity, only the 3-dimethylazaniumyl-1-hydroxypropyl fragments of the cations are shown, and H atoms not involved in hydrogen bonds have been omitted. (Bottom) Segment of an entire ribbon, including cations and partially occupied water molecules. Color coding: O, N, and Br atoms are shown with 50% probability displacement ellipsoids, and all other atoms are in capped-stick mode and color coded by the symmetry equivalence of their anion and cation (cations in dark green and benzoate anions in orange). Hydrogen bonds are shown as turquoise and red dashed lines.
The strong intermolecular interactions present in 4a and 4b do compensate for the presence of the only partially or not at all filled voids present, and the packing efficiency for the salts of 4 is comparable to that of free base bedaquiline. Indeed, the density for the 1.17-hydrate is, at 1.342 Mg m−3, virtually identical to that of the free base of 1.344 Mg m−3 (both measured at 150 K), and the acetonitrile solvate is, at 1.360 Mg m−3, even denser than the solvent-free base. The best packing efficiency is, however, observed for fumarate salt 2, featuring a multitude of strong hydrogen bonds, and having neither incorporated solvent molecules nor unoccupied void space. Its density, at 150 K, is 1.379 Mg m−3 (Table 7 ▸).
Supplementary Material
Crystal structure: contains datablock(s) 1, 2, 3, 4a, 4b, global. DOI: 10.1107/S2053229620013455/ov3143sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2053229620013455/ov31431sup2.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2053229620013455/ov31432sup3.hkl
Structure factors: contains datablock(s) 3. DOI: 10.1107/S2053229620013455/ov31433sup4.hkl
Structure factors: contains datablock(s) 4a. DOI: 10.1107/S2053229620013455/ov31434asup5.hkl
Structure factors: contains datablock(s) 4b. DOI: 10.1107/S2053229620013455/ov31434bsup6.hkl
Supporting information file. DOI: 10.1107/S2053229620013455/ov31431sup7.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31432sup8.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31433sup9.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31434asup10.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31434bsup11.cml
Polarized light microscopy images and PXRD parretns. DOI: 10.1107/S2053229620013455/ov3143sup12.pdf
Acknowledgments
This material is based on work supported by the National Science Foundation (NSF) through the Major Research Instrumentation Program, which provided funding for the single-crystal X-ray diffractometer.
Funding Statement
This work was funded by National Science Foundation, Directorate for Mathematical and Physical Sciences grant 1625543 to M. Zeller. Bill and Melinda Gates Foundation grant INV-017799 to S. R. Byrn.
References
- Bioquest (2020). AAT Bioquest pKa and pKb Reference Table, https://www.aatbio.com/data-sets/pka-and-pkb-reference-table, accessed August 21, 2020.
- Brigden, G., Hewison, C. & Varaine, F. (2015). Infect. Drug Resist. 8, 367–378. [DOI] [PMC free article] [PubMed]
- Bruker (2019). APEX3 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Colthup, N. B., Daly, L. H. & Wiberley, S. E. (1990). In Introduction to Infrared and Raman Spectroscopy, 3rd ed. New York: Academic Press Inc.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hegyi, J. F. A. L., Aelterman, W. A. A., Lang, Y. L., Stokbroekx, S. C. M., Leys, C., Van Remoortere, P. J. M. & Faure, A. (2013). US Patent 8 546 428, Janssen Pharmaceuticals, USA.
- Hübschle, C. B., Sheldrick, G. M. & Dittrich, B. (2011). J. Appl. Cryst. 44, 1281–1284. [DOI] [PMC free article] [PubMed]
- Kojima, T., Tsutsumi, S., Yamamoto, K., Ikeda, Y. & Moriwaki, T. (2010). Int. J. Pharm. 399(1–2), 52–59. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Martell, A. E. & Smith, R. M. (1976). In Critical Stability Constants, Vols. 1–4. New York: Plenum Press.
- PANalytical (2015). Data Collector (XRD data collection software, Version 5.3.0.62) and HighScore (Version 4.5). PANalytical BV, Almelo, The Netherlands.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Petit, S., Coquerel, G., Meyer, C. & Guillemont, J. (2007). J. Mol. Struct. 837, 252–256.
- Rombouts, J. A., Veenboer, R. P., Villellas, C., Lu, P., Ehlers, A. W., Andries, K., Koul, A., Lill, H., Ruijter, E., Orru, R. V. A., Lammertsma, K., Bald, D. & Slootweg, J. C. (2016). RSC Adv. 6, 108708–108716.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zook, H. D., March, J. & Smith, D. F. (1959). J. Am. Chem. Soc. 81, 1617–1620.
- Zvatora, P., Dammer, O., Krejcik, L., Zvonicek, V. & Hert, J. (2016). Int. Patent WO/2016/198031A1, Zentiva, Czech Republic.
- Zvatora, P., Dammer, O., Ridvan, L., Lustig, P., Pekarek, T., Stefco, M., Krejcik, L. & Tkadlecova, M. (2016). Int. Patent WO/2016/058564, Zentiva, Czech Republic.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, 2, 3, 4a, 4b, global. DOI: 10.1107/S2053229620013455/ov3143sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2053229620013455/ov31431sup2.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2053229620013455/ov31432sup3.hkl
Structure factors: contains datablock(s) 3. DOI: 10.1107/S2053229620013455/ov31433sup4.hkl
Structure factors: contains datablock(s) 4a. DOI: 10.1107/S2053229620013455/ov31434asup5.hkl
Structure factors: contains datablock(s) 4b. DOI: 10.1107/S2053229620013455/ov31434bsup6.hkl
Supporting information file. DOI: 10.1107/S2053229620013455/ov31431sup7.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31432sup8.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31433sup9.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31434asup10.cml
Supporting information file. DOI: 10.1107/S2053229620013455/ov31434bsup11.cml
Polarized light microscopy images and PXRD parretns. DOI: 10.1107/S2053229620013455/ov3143sup12.pdf












