Abstract
Alteration of the lipid composition of biological membranes interferes with their function and can cause tissue damage by triggering apoptosis. Upon lipid bilayer stress, the endoplasmic reticulum mounts a stress response similar to the unfolded protein response. However, only a few genes are known to regulate lipid bilayer stress. We performed a suppressor screen that combined the auxin-inducible degradation (AID) system with conventional RNAi in C. elegans to identify members of the lipid bilayer stress response. AID-mediated degradation of the mediator MDT-15, a protein required for the upregulation of fatty acid desaturases, induced the activation of lipid bilayer stress-sensitive reporters. We screened through most C. elegans kinases and transcription factors by feeding RNAi. We discovered nine genes that suppressed the lipid bilayer stress response in C. elegans. These suppressor genes included drl-1/MAP3K3, gsk-3/GSK3, let-607/CREB3, ire-1/IRE1, and skn-1/NRF1,2,3. Our candidate suppressor genes suggest a network of transcription factors and the integration of multiple tissues for a centralized lipotoxicity response in the intestine. Thus, we demonstrated proof-of-concept for combining AID and RNAi as a new screening strategy and identified eight conserved genes that had not previously been implicated in the lipid bilayer stress response.
Keywords: Lipid bilayer stress, Lipotoxicity, Unfolded Protein Stress, Auxin-induced degradation, CREB3, NRF2, MDT-15
Biological membranes play an important role in protein folding, signaling, secretion, and the turnover of proteins. Changes in the lipid composition of a membrane alter its properties, and thus, interferes with its function and leads to lipid bilayer stress (LBS) (Covino et al. 2018). Maintaining the membranes’ composition is, therefore, crucial for a cell. High dietary intake of saturated fatty acids leads to a metabolic syndrome referred to as lipotoxicity (Ertunc and Hotamisligil 2016). On a cellular level, elevated levels of saturated fatty acids alter membrane composition. Sensitive for these changes is the endoplasmic reticulum (ER), which is a significant site for protein and lipid synthesis, and the main site of intracellular calcium storage (Schwarz and Blower 2016). Lipid disequilibrium interferes with secretory capacity and renders cells specialized in secretion, such as insulin-producing beta cells, susceptible to cell death (Preston et al. 2009; Acosta-Montaño and García-González 2018). Although LBS has been suggested to play a major part in disease progression, the spectrum of the underlying molecular players sensing LBS remains to be identified.
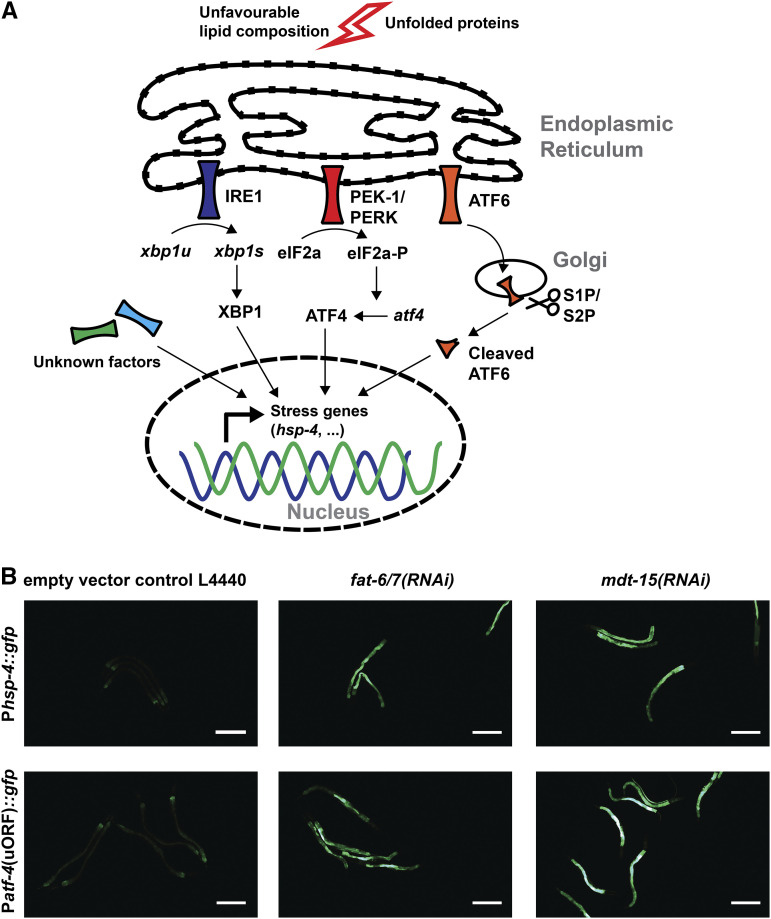
The unfolded protein response (UPR) sensors IRE1, PERK1, and ATF6 are sensitive to changes in membrane fluidity (Koh et al. 2018). On the molecular level, IRE1, PERK1, and ATF6 act in parallel in response to unfolded proteins (Figure 1a). Activated IRE1 splices XBP1 mRNA to stabilize the transcript and allow translation of the spliced XBP1 transcription factor (Figure 1a). PERK1 phosphorylates the initiation factor eIF2alpha, which reduces translation rate and allows preferential translation of genes containing upstream open reading frames (uORFs), such as the transcription factor ATF4 (Harding et al. 2000) (Figure 1a). During ER stress, ATF6 translocates from the ER to the Golgi, where it is cleaved by proteases termed S1P and S2P. Cleaved ATF6 migrates to the nucleus and acts as a transcription factor (Figure 1a). These transcription factors co-regulate many targets, but how the downstream targets of the three arms of the UPR restore membrane homeostasis in detail remains unknown.
Figure 1.
Integrated stress response of C. elegans a) Model of the unfolded protein and lipid bilayer stress response. b) Phsp-4::gfp and Patf-4(uORF)::gfp are activated by fat-6/7(RNAi) and mdt-15(RNAi). Scale bar = 200 μm.
In C. elegans, loss of fatty acid desaturases fat-6 and fat-7 or mdt-15, which is a mediator subunit required for fat-6/7 expression, leads to a higher ratio of saturated fatty acids in the membrane. This activates the ER stress reporter hsp-4::gfp via the IRE-1/XBP-1 axis (Hou et al. 2014). Supplementing C18:1n-9 fatty acid oleate, the product of the FAT-6/7 stearoyl-CoA-desaturases, either partially rescued, in the case of mdt-15 knockdown, or entirely rescued, when fat-6/7 knocked down the induction of ER stress reporter hsp-4::gfp (Hou et al. 2014). The partial rescue of the mdt-15 knockdown may be via other processes or functions that are disrupted in these animals leading to ER stress. The mediator subunit mdt-15 regulates lipid metabolism and is also involved in many other processes, including immunity, stress defense, detoxification, and mitochondrial stress (Taubert et al. 2008; Mao et al. 2019; Lee et al. 2019). Also, activation of the ER stress sensor can also be achieved by depleting the cell’s phosphatidylcholine levels, for example, via knockdown of sams-1/MAT1A, s-adenosyl methionine synthetase (Hou et al. 2014). Curiously, the signature of lipid bilayer stress response is different from the canonical UPR in C. elegans (Hou et al. 2014; Koh et al. 2018). This argues for an additional layer of regulation that fine-tunes the output during activation of the three UPR arms (Figure 1a).
Genetic mutant screening for members of the UPR has been successful (Calfon et al. 2002). However, setting up genetic screens with essential genes that either cause lethality or developmental defects are difficult. RNAi-based forward screens can bypass genes that cause embryonal lethality or developmental defects. However, feeding more than one RNAi simultaneously was previously reported to produce poor results (Min et al. 2010). This suggests a bottleneck for screening strategies where one would like to screen for suppressors of a phenotype caused by a knockdown using an RNAi-mediated screen.
The auxin-inducible degradation (AID) system has been recently introduced to mediate fast and reversible degradation of targeted proteins in C. elegans (Zhang et al. 2015). A protein of interest can be tagged with a short 68 amino acid sequence (degron), which is recognized by the E3 ubiquitin ligase TIR1, derived from Arabidopsis thaliana, in the presence of a small molecule called auxin (Zhang et al. 2015; Martinez et al. 2020). Ubiquitination targets the degron-tagged protein for fast degradation by the proteasome. Depletion times of less than 30 min have been reported for cytosolic proteins after transferring C. elegans co-expressing a degron-tagged protein and TIR1 on plates containing auxin (Zhang et al. 2015). The AID is, therefore, faster and more efficient than RNAi. Since AID initiates protein degradation and RNAi initiates mRNA degradation, these two systems do not compete with each other and can be used in parallel.
Here, we identify suppressors of lipid bilayer stress response using a novel approach combining AID and RNAi-based forward genetic screening. Degradation of MDT-15 by AID was used to induce LBS, which was visualized using the ER-stress reporters Patf-4(uORF)::gfp and Phsp-4::gfp. We screened RNAi libraries targeting kinases and transcription factors. Out of 868 genes, we identified one known and eight novel hits that robustly blocked LBS response upon MDT-15 degradation.
Materials and Methods
C. elegans strains
All strains were maintained at 20° on OP50 Escherichia coli, as described (Stiernagle 2006).
IJ1729: ieSi57 [Peft-3::TIR1::mRuby::unc-54 3′UTR; cb-unc-119] II; yh44 [mdt-15::degron::EmGFP] III. (Lee et al. 2019), SJ4005: zcIs4 [Phsp-4::GFP] V. (Harding et al. 2000), LD1499: [Patf-4(uORF)::GFP::unc-54(3′UTR)], LSD2096: ieSi57 [Peft-3::TIR1::mRuby::unc-54(3′UTR); cb-unc-119] II; yh44 [mdt-15::degron::EmGFP] III; [Patf-4(uORF)::GFP::unc-54(3′UTR)], LSD2102: ieSi57 [Peft-3::TIR1::mRuby::unc-54 3′UTR; cb-unc-119] II; yh44 [mdt-15::degron::EmGFP] III; zcIs4 [Phsp-4::GFP] V.
The screening strain was generated by crossing IJ1729 males with LD1499. 48 F2s were singled out and their offspring were placed onto plates containing 100 μM auxin and the upregulation of the reporter was determined (Figure 2a). In parallel, IJ1729 was crossed to SJ4005.
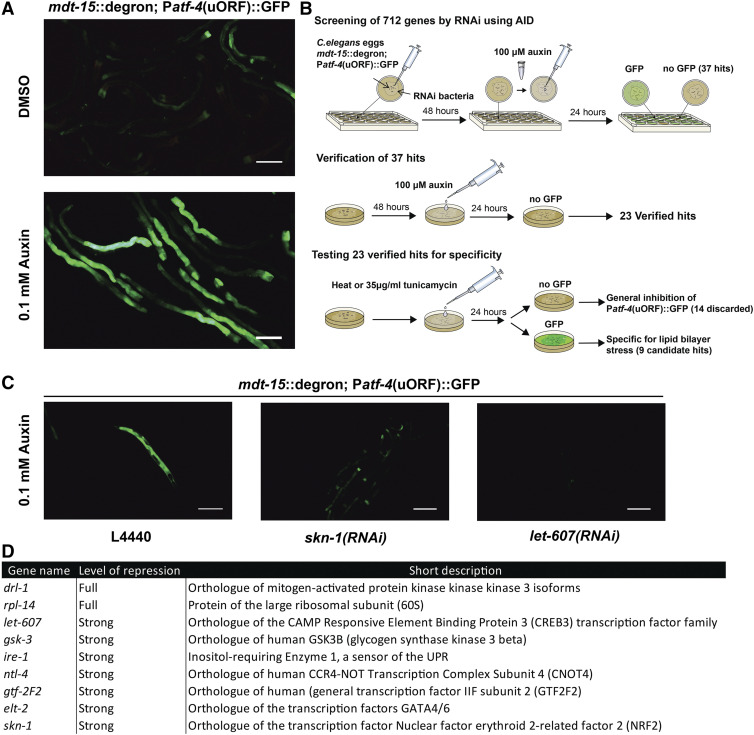
Figure 2.
Suppressor screen of LBS a) Addition of 0.1 mM auxin degrades mdt-15::degron and leads to expression of Patf-4(uORF)::gfp in LSD2096. Pictures were taken 24 h after auxin addition. Scale bar = 200 μm b) Summary of the screening outline. c) Treatment with 0.1 mM auxin to degrade mdt-15::degron after skn-1 and let-607 RNAi represses activation of Patf-4(uORF)::gfp in LSD2096. Pictures were taken 24 h after auxin treatment. Scale bar = 200 μm.
Microscopy
For image acquisition, the animals were placed onto fresh 2% agar pads and anesthetized with 1 mM tetramisole, as previously described (Teuscher and Ewald 2018). Images were obtained using an upright bright field fluorescence microscope (Tritech Research, model BX-51-F) with an attached camera (model DFK 23UX236).
Quantification of GFP fluorescence
GFP fluorescent levels were scored by inspecting transgenic animals with a fluorescent dissecting scope while still on the culturing/treatment condition plates (Ewald et al. 2017). GFP intensity was scored and categorized: 0= none or very low GFP usually corresponding to the untreated control, 1= low, 2= medium, and 3= high GFP fluorescent signal.
Preparation of auxin
70 mg of auxin (3-Indoleacetic acid, Sigma #I3750) was dissolved in 10 mL DMSO to yield a 40 mM stock solution and stored at 4°. The stock was further diluted in M9 to 100 μM before use.
Suppressor screen design
A detailed step-by-step protocol can be found in the supplementary, and a schematic outline is shown in Figure 2b.
Briefly, 24-well plates were filled with Nematode Growth Medium (NGM) containing ampicillin (100 μg/ml), tetracycline (12.5 μg/ml), and 1mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) seeded with 50 μl of freshly grown RNAi bacteria and dried in a sterile laminar flow cabinet. The following day, plates containing gravid LSD2096 adults were washed off and discarded, and the laid eggs were scratched off and collected. Approximately 30-40 eggs were pipetted into each well and incubated at 20°. After 48 hr, the wells were top coated with 50 μl of 100 μM auxin and dried in a sterile laminar flow cabinet at 20° overnight. The following day, the wells were screened for the suppression of the GFP signal. The kinase and transcription factor libraries were screened twice. The preliminary hits had to pass three additional runs on 6 cm plates successfully.
Single and double knockdown by RNA interference for candidate gene validation for LBS
RNAi and double RNAi were performed as described (Ewald et al. 2017).
For single RNAi:
RNAi bacteria cultures were grown overnight in Lysogeny Broth (LB) medium with carbenicillin (100 µg/mL) and tetracycline [12.5 µg/mL], diluted and grown again for 4-6 hr. Bacteria were then concentrated by centrifugation and induced with 1 mM IPTG and spread onto NGM plates containing 1 mM IPTG, tetracycline (12.5 µg/mL) and ampicillin (50 µg/mL). For the empty RNAi vector (EV), plasmid pL4440 was used as a control.
For double RNAi:
bacterial RNAi cultures were grown separately, and after the concentration step, mixed in a 1:1 ratio and seeded onto culture plates, as described above for single RNAi.
Heat-shock and tunicamycin treatment
Animals, RNAi bacteria, and plates were prepared as above, without the addition of auxin. Heat-shock was carried out for 1 hr at 37°, incubated for 5 hr at 25°, and GFP expression was determined in the animals. Plates were top coated with 0.5 ml of 35 μg/ml tunicamycin (Sigma, T7765), incubated for 6 hr at 25°, and GFP expression was again determined.
Data availability
Strains used in this study are either available from CAENORHABDITIS GENETICS CENTER (CGC) or upon request. Supplementary materials are available at figshare: https://doi.org/10.25387/g3.12783044.
Results
Lipid bilayer stress can be induced by knocking down mdt-15 or fat-6/7, resulting in the upregulation of ER-stress reporter Phsp-4::gfp (Hou et al. 2014). We confirmed the induction of Phsp-4::gfp upon RNAi against mdt-15 and fat-6/7 (Figure 1b). We tested a second ER-stress reporter Patf-4(uORF)::gfp, which was also induced upon knockdown of mdt-15 or fat-6/7 RNAi (Figure 1b). The Patf-4(uORF)::gfp reporter contains two upstream open reading frames (uORF) in the 5′ untranslated region (Rousakis et al. 2013). Similar to mammalian ATF4, under unstressed conditions, atf-4 mRNA is not translated but under conditions that lead to a global reduction of protein synthesis (Rousakis et al. 2013; Young and Wek 2016). For screening purposes, we preferred Patf-4(uORF)::gfp over Phsp-4::gfp for its stronger induction of GFP, allowing easier detection in 24- or 96-well plates. Crossing mdt-15(tm2182) mutant with Patf-4(uORF)::gfp led to heterogeneous GFP expression; therefore, it was difficult to use this strain for screening. Hence, we switched to an endogenously degron-tagged mdt-15 strain (Lee et al. 2019). Unstressed MDT-15::degron C. elegans expressed Patf-4(uORF)::gfp only at basal levels at 20°. Incubation with 100 μM auxin for 24 hr increased GFP levels drastically and homogenously throughout the mdt-15::degron; TIR1; Patf-4(uORF)::gfp transgenic animals (Figure 2a), but did not induce GFP fluorescence in wild-type Patf-4(uORF)::gfp (Supplementary Figure 1a, Data Source File 1). Upon treatment with auxin, we also observed typical mdt-15 phenotypes, such as small body size, reduced brood size, and a pale appearance in mdt-15::degron; TIR1; Patf-4(uORF)::gfp animals, consistent with previous reports (Lee et al. 2019). An additional phenotype was observed in our screening strain, the eggs of untreated mdt-15::degron; TIR1; Patf-4(uORF)::gfp animals were sensitive to bleaching, normally used to synchronize C. elegans populations. Either, our mdt-15::degron; TIR1; Patf-4(uORF)::gfp screening strain carried a background mutation or degron-tagged mdt-15 may be partially hypomorph. However, the untreated mdt-15::degron; TIR1; Patf-4(uORF)::gfp screening strain appeared superficially wild type without any induction of GFP. We did not outcross the strain but rather decided to avoid bleach synchronization and continued with our screen by collecting laid eggs off the bacterial lawn. Thus, we established the mdt-15::degron; TIR1; Patf-4(uORF)::gfp strain for screening for LBS suppressors.
Further insight into LBS was gained by taking a targeted RNAi approach. We decided to screen through the majority of C. elegans kinases (382 out of the 438 kinases; (Lehmann et al. 2013)) and about one-third of all transcription factors (330 out of 934 genes; (Reece-Hoyes et al. 2005; Kim et al. 2018)) (Data Source File 1). Our first-pass screening round of the total 712 genes (Data Source File 1) resulted in 6 kinases and 31 transcription factors (Figure 2b). To sort out false positives, we tested the preliminary hits on 6 cm plates, which resulted in 23 verified hits that blocked Patf-4(uORF)::gfp induction upon mdt-15 degradation (Figure 2b, 2c; Supplementary Table 1). To investigate whether these 23 hits were specific for lipid bilayer stress, and not general inhibitors of the unfolded protein response, we heat-shocked the animals and treated them with the N-glycosylation-inhibitor tunicamycin (Figure 2b). Out of the 23 hits, we identified 11 hits specific to LBS. The majority of clones that did not pass this step were positive controls of GFP RNAi from the screening libraries (Supplementary Table 1). Reassuringly, we detected xbp-1, a transcription factor spliced by IRE-1 (Supplementary Table 1). XPB-1 is known to upregulate hsp-4 mRNA during UPR (Calfon et al. 2002). To rule out transgene-specific effects, we crossed Phsp-4::gfp into mdt-15::degron;TIR1 and tested the hits that had passed the previous steps (Figure 2b). Only the weakest hit, ztf-1, did not pass this step (Supplementary Table 1). In addition, knockdown of xbp-1 resulted in the complete absence of any GFP expression even in the control Patf-4(uORF)::gfp strain, suggesting that the transcription factor XBP-1 may be necessary for general expression of atf-4 in C. elegans. Together with XBP-1s importance in transcribing hsp-4, it is difficult to argue whether xbp-1 is necessary for LBS, and further research is required to elucidate this. Thus, we ended up with nine candidate suppressors of LBS from our combined AID with RNAi screen (Figure 2d).
To validate that the nine potential candidates were suppressors of LBS and were not only specific to mdt-15 degradation, we performed single and double RNAi treatment either in combination with fat-6/7(RNAi) or sams-1(RNAi). We found that single knockdown of the candidate genes did not induce Patf-4(uORF)::gfp reporter, except for RNAi against ire-1 or let-607 which induced the Patf-4(uORF)::gfp reporter (Supplementary Figure 1b), suggesting that loss of either these latter two genes may cause ER stress. By contrast, knockdown of ire-1 or let-607 suppressed Patf-4(uORF)::gfp induction upon mdt-15 degradation with 0.1 mM auxin (Figure 2) or with a higher concentration of 1 mM auxin (Supplementary Figure 1c-g). Reassuringly, double RNAi of fat-6/7 with ire-1 or let-607 and double RNAi of sams-1 with ire-1 or let-607 displayed strong suppression of the Patf-4(uORF)::gfp reporter (Supplementary Figure 2; Data Source File 1). Similarly, double RNAi of candidate genes with either fat-6/7 or sams-1 exhibited full suppression of Patf-4(uORF)::gfp reporter, except for elt-2 knockdown (Supplementary Figure 2). Knockdown of elt-2 did not suppress fat-6/7(RNAi) (Supplementary Figure 2a), but suppressed sams-1(RNAi) Patf-4(uORF)::gfp reporter (Supplementary Figure 2b). This suggests a differential genetic activation of LBS from a higher ratio of saturated fatty acids in the membranes compared to depleting the cell’s phosphatidylcholine levels. Thus, our screening approach identified nine high confidence candidates. Eight of these nine candidates have not previously been described to mediate LBS in C. elegans.
Discussion
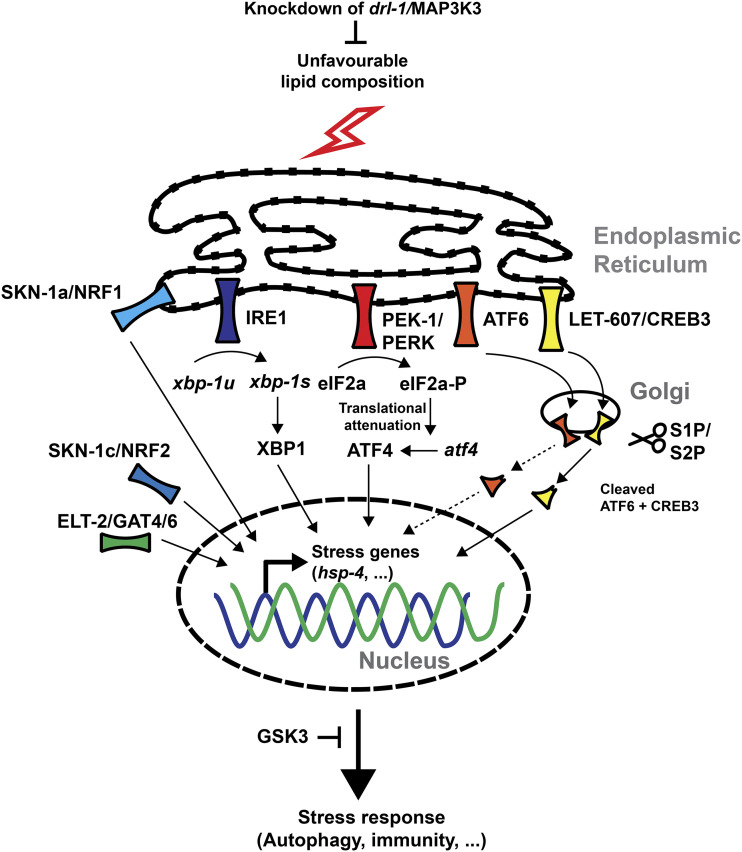
Here we report the first screening approach combining auxin-induced degradation with RNA interference. With our novel approach of combining AID and RNAi screening, we were able to bypass developmental and lethal obstacles caused by the depletion of mdt-15. Our screen revealed a known molecular player (IRE-1) and identified several new genes important to mount a proper LBS response. Thus, our results provide proof-of-concept and support the feasibility of combined AID-RNAi screening approaches. Here we discuss the function of the suppressor genes briefly and propose a hypothetical model for the LBS pathway in C. elegans (Figure 3).
Figure 3.
Hypothetical model of LBS in C. elegans The updated model of LBS in C. elegans indicates a complex network of transcription factors and up- and downstream modulators. 6 out of 9 of our hits are included in this model.
Regulation of LBS by IRE-1 and XBP-1
We unbiasedly detected IRE-1, which was previously proposed as a sensor for LBS in yeast and C. elegans, and its target xbp-1 (Thibault et al. 2012). This confirms the selectivity of our screen. Unfolded proteins in the ER lead to IRE1 oligomerization and the subsequent stimulation of its endoribonuclease activity and splicing of the transcription factor xbp-1. However, monomeric IRE1 still displays RNase activity and splices XBP1 mRNA in HeLa cells during LBS (Kitai et al. 2013; Ho et al. 2020). Thus, we confirmed that IRE-1 branch acts as a major sensor of LBS.
Immunity response network regulates lipid bilayer stress
Knocking down phosphatidylcholine synthesis leads to the activation of genes involved in the immune response (Koh et al. 2018). Many of these transcripts are upregulated in an IRE-1-dependent manner. In addition to ire-1, we detected the NRF1,2,3 homolog skn-1 and the GATA transcription factor elt-2. Both are involved in p38-mediated innate immunity (Block et al. 2015; Ewald 2018). SKN-1 is a major transcription factor for promoting oxidative stress resistance (Blackwell et al. 2015). There are four isoforms of SKN-1: skn-1a, b, c, d (Blackwell et al. 2015). A previous study revealed that IRE-1 had an additional mode-of-action in its monomeric state: elevated levels of reactive oxygen species leads to sulfenylation of cysteine residues in IRE-1 and activates SKN-1a via the p38 MAPK (Hourihan et al. 2016). Isoform skn-1a is similar to mammalian NRF1, which regulates proteostasis and is a transmembrane protein located in the ER (Wang and Chan 2006; Glover-Cutter et al. 2013; Lehrbach and Ruvkun 2019). Curiously, mdt-15 and skn-1c but not skn-1a, co-regulate targets involved in detoxification, such as gst-4 (Goh et al. 2014). This suggests that SKN-1a is activated by loss of mdt-15 and works independently of MDT-15. Knockdown of skn-1 does not only block LBS response but also reverses the small body size and the small number of eggs laid (although these eggs do not hatch as skn-1 knockdown is embryonic lethal; Supplementary Table 1). This suggests that some of the observed phenotypes in mdt-15 mutants or knockdowns are skn-1-dependent. The mammalian SKN-1c ortholog NRF2 has been shown to have protective functions during palmitate-induced lipotoxicity in mammalian cells (Cunha et al. 2016; Park et al. 2015). Taken together, this implies a potential isoform-specific role for skn-1 during LBS.
The GATA transcription factor elt-2 is essential for the mesodermal cell fate and development of the intestine. While the null mutation of elt-2 is embryonic lethal, post-developmental knockdown shortens lifespan, and overexpression extend lifespan (Mann et al. 2016). We observed developmental arrest after elt-2 knockdown. These arrested larvae remained susceptible to heat and tunicamycin treatments, indicating that the UPR was still intact. Like skn-1, elt-2 is recruited to promoters during Pseudomonas aeruginosa infection and co-regulates targets in a p38-mediated fashion (Block et al. 2015). Furthermore, elt-2 and mdt-15 cooperate during heavy metal intoxication (Shomer et al. 2019), supporting the idea of a transcription factor network that cooperatively regulates different stress responses.
Modulators and activators of the LBS response (let-607, gsk-3, and drl-1)
We found three genes, let-607, gsk-3, and drl-1, that are implicated in modulating ER stress responses. RNAi of let-607 suppressed the activation of the atf-4 reporter (Figure 2c). let-607, together with crh-1 and crh-2, was one of the CREB3 orthologs in C. elegans. The mammalian Creb3 family consists of five members (CREB3/Luman, CREB3L1/OASIS, CREB3L2/BBF2H7, CREB3L3/CREBH, and CREB3L4) and is related to ATF6 and SREBP (Sampieri et al. 2019). All are localized in the ER and, like ATF6, are activated by anterograde transport to the Golgi and subsequent cleavage by S1P or S2P. In humans and mice, CREB3L2 upregulates SEC23 and controls secretion load, especially during bone formation (Saito et al. 2009; Tomoishi et al. 2017; Al-Maskari et al. 2018). CREB3 and CREB3L3 are induced after palmitate-induced ER stress, and knockdown of CREB3 by siRNA sensitizes human islet cells to palmitate-induced ER stress (Cnop et al. 2014). CREB3 has been identified in regulating Golgi-stress and activation of ARF4 (Reiling et al. 2013). A previous study in C. elegans links let-607 with the upregulation of sec-23 and other proteins involved in secretion (Weicksel et al. 2016). The let-607 gene has also been identified in a screen for suppressors of PolyQ aggregation and suppresses motility defects caused by mutations in the paramyosin ortholog UNC-15, the basement-membrane protein perlecan UNC-52, the myosin-assembly protein UNC-45, and the myosin heavy chain UNC-54 (Silva et al. 2011). In addition, knockdown of let-607 increased the expression of cytosolic heat-shock proteins. Based on these previous observations and our results, we propose that let-607/CREB3 family is sensing LBS and acts together with the other identified transcription factor encoding genes xbp-1, skn-1, and elt-2 to mount a unique stress response that is different from the canonical UPR.
drl-1/MAP3K3, also known as mekk-3, has been found in a screen for enhancers of dauer formation and extends lifespan by simulating dietary restricted-like conditions (Chamoli et al. 2014). Curiously, loss of drl-1 caused a pale appearance resembling fat-6/7 and mdt-15 mutants, but the mode-of-action appears to be different. The drl-1 promoter is expressed in vulval muscles, body wall muscles, hypodermis, seam cells, some neurons, and tissues lining the pharynx and anus, but not the intestine. Additionally, knockdown in the intestine using tissue-specific RNAi did not extend lifespan (Chamoli et al. 2014). Knockdown of MDT-15 activates Patf-4(uORF)::gfp and Phsp-4::gfp expression mainly in the intestine (Figure 1b). Therefore, knockdown of drl-1 acts in a cell non-autonomous manner. drl-1 decreases fat storage by upregulating fatty acid oxidation (Chamoli et al. 2014). The C. elegans ortholog of the ribonuclease Regnase-1, rege-1, shares many upregulated genes and causes a pale appearance without activation of LBS (Supplementary Table 1; (Habacher et al. 2016)). This suggests a link between drl-1 and rege-1. However, knockdown of rege-1 does not phenocopy loss of drl-1 (Supplementary Table 1). Despite the striking similarities shared by rege-1 and drl-1, only drl-1 modulates LBS. Intriguingly, drl-1 knockdown itself causes ER stress at the L2 stage, and this mounts a protective effect throughout life (Matai et al. 2019). Since drl-1 rewires metabolism by mimicking dietary restriction, we speculate that activation of fatty acid oxidation protects from lipotoxicity. Indeed, during our screen, we observed that starved mdt-15::degron; TIR1; Patf-4(uORF)::gfp transgenic animals in the 24-wells without food failed to upregulate the reporter.
Glycogen synthase kinase-3 (gsk-3) has been described as the busiest of all kinases with over 100 targets known and was found to attenuate palmitate-induced apoptosis (Ibrahim et al. 2011; Beurel et al. 2015). Paradoxically, gsk-3 inhibits skn-1 and stabilizes CREB3, two mode-of-actions contradicting the results of our screen (An et al. 2005; Barbosa et al. 2013). The inhibition of gsk-3 here does not act as previously reported; therefore, we hypothesize other modes of action. One alternative mechanism could be via autophagy. Activation of the lipid bilayer stress activates autophagy via the IRE-1/XBP-1 axis (Ho et al. 2020). Blocking autophagy in this context causes sickness, sterility, and developmental defects. Intriguingly, GSK3 inhibition activates autophagy (Parr et al. 2012). We speculate that prior knockdown of GSK3 leads to an elevated rate of autophagy, which protects from LBS and ameliorates the stress response.
General players in gene expression, but specific for LBS
The last three hits consisted of gtf-2f2, ntl-4, and rpl-14, which are involved in transcription, RNA processing, and translation, respectively. Interestingly, although RNAi against gtf-2f2, ntl-4, and rpl-14 inactivate general processes, the heat- or tunicamycin induced UPR remained functioning and was not affected. This favors the model that UPR and LBS are differentially regulated (Figure 3).
Summary
We demonstrated the feasibility of combining AID and RNAi-based genetic screens. We report the identification of eight novel regulators of the lipid bilayer stress response and grouped them into three categories (Figure 3). skn-1 and elt-2, together with the previously characterized ire-1, are transcription factors involved in immune responses. let-607 may be activated in parallel with the canonical UPR arms, and drl-1 and gsk-3 modulate the ER stress response in our suggested model upstream or downstream, respectively. The last category consists of genes involved in general processes of gene expression. Interestingly, all eight novel candidate genes are well-conserved, suggesting the potential implications of these genes in the mammalian lipid bilayer stress response.
Acknowledgments
We thank Chi Yun and David Ron for the Patf-4(uORF)::gfp strain, Seung-Jae V. Lee for the mdt-15::degron strain, Gary Ruvkun for the kinase and transcription factor RNAi libraries, Keith Blackwell and Ewald Lab members for comments on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Funding from the Swiss National Science Foundation PP00P3_163898 to CYE, Mibelle Biochemistry Research Grant to EJ, and ETH Research Foundation Grant ETH-30 16-2 to RV.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12783044.
Communicating editor: S. Lee
Literature Cited
- Acosta-Montaño P., and García-González V., 2018. Effects of Dietary Fatty Acids in Pancreatic Beta Cell Metabolism, Implications in Homeostasis. Nutrients 10: 393 10.3390/nu10040393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Maskari M., Care M. A., Robinson E., Cocco M., Tooze R. M. et al. , 2018. Site-1 protease function is essential for the generation of antibody secreting cells and reprogramming for secretory activity. Sci. Rep. 8: 14338 10.1038/s41598-018-32705-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. H., Vranas K., Lucke M., Inoue H., Hisamoto N. et al. , 2005. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc. Natl. Acad. Sci. USA 102: 16275–16280. 10.1073/pnas.0508105102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa S., Fasanella G., Carreira S., Llarena M., Fox R. et al. , 2013. An orchestrated program regulating secretory pathway genes and cargos by the transmembrane transcription factor CREB-H. Traffic 14: 382–398. 10.1111/tra.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E., Grieco S. F., and Jope R. S., 2015. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148: 114–131. 10.1016/j.pharmthera.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., and Isik M., 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88: 290–301. 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D. H. S., Twumasi-Boateng K., Kang H. S., Carlisle J. A., Hanganu A. et al. , 2015. The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult C. elegans. PLoS Genet. 11: e1005265 10.1371/journal.pgen.1005265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R. et al. , 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Chamoli M., Singh A., Malik Y., and Mukhopadhyay A., 2014. A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell 13: 641–655. 10.1111/acel.12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnop M., Abdulkarim B., Bottu G., Cunha D. A., Igoillo-Esteve M. et al. , 2014. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 63: 1978–1993. 10.2337/db13-1383 [DOI] [PubMed] [Google Scholar]
- Covino R., Hummer G., and Ernst R., 2018. Integrated Functions of Membrane Property Sensors and a Hidden Side of the Unfolded Protein Response. Mol. Cell 71: 458–467. 10.1016/j.molcel.2018.07.019 [DOI] [PubMed] [Google Scholar]
- Cunha D. A., Cito M., Carlsson P.-O., Vanderwinden J.-M., Molkentin J. D. et al. , 2016. Thrombospondin 1 protects pancreatic β-cells from lipotoxicity via the PERK-NRF2 pathway. Cell Death Differ. 23: 1995–2006. 10.1038/cdd.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertunc M. E., and Hotamisligil G. S., 2016. Lipid signaling and lipotoxicity in metaflammation: indications for metabolic disease pathogenesis and treatment. J. Lipid Res. 57: 2099–2114. 10.1194/jlr.R066514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y., 2018. Redox Signaling of NADPH Oxidases Regulates Oxidative Stress Responses, Immunity and Aging. Antioxidants 7: 130 10.3390/antiox7100130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y., Hourihan J. M., Bland M. S., Obieglo C., Katic I. et al. , 2017. NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. eLife 6: e19493 10.7554/eLife.19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter, K. M., S. Lin, and T. K. Blackwell, 2013 Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet 9: e1003701. 10.1371/journal.pgen.1003701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y. S., Martelli K. L., Parhar K. S., Kwong A. W. L., Wong M. A. et al. , 2014. The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell 13: 70–79. 10.1111/acel.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habacher C., Guo Y., Venz R., Kumari P., Neagu A. et al. , 2016. Ribonuclease-Mediated Control of Body Fat. Dev. Cell 39: 359–369. 10.1016/j.devcel.2016.09.018 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R. et al. , 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6: 1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Ho N., Yap W. S., Xu J., Wu H., Koh J. H. et al. , 2020. Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J. Cell Biol. 219: e201909165 10.1083/jcb.201909165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N. S., Gutschmidt A., Choi D. Y., Pather K., Shi X. et al. , 2014. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc. Natl. Acad. Sci. USA 111: E2271–E2280. 10.1073/pnas.1318262111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourihan J. M., Moronetti Mazzeo L. E., Fernández-Cárdenas L. P., and Blackwell T. K., 2016. Cysteine Sulfenylation Directs IRE-1 to Activate the SKN-1/Nrf2 Antioxidant Response. Mol. Cell 63: 553–566. 10.1016/j.molcel.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S. H., Akazawa Y., Cazanave S. C., Bronk S. F., Elmi N. A. et al. , 2011. Glycogen synthase kinase-3 (GSK-3) inhibition attenuates hepatocyte lipoapoptosis. J. Hepatol. 54: 765–772. 10.1016/j.jhep.2010.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai, Y., H. Ariyama, N. Kono, D. Oikawa, T. Iwawaki et al, 2013 Membrane lipid saturation activates IRE1α without inducing clustering. Genes Cells 18: 798–809. 10.1111/gtc.12074 [DOI] [PubMed] [Google Scholar]
- Kim W., Underwood R. S., Greenwald I., and Shaye D. D., 2018. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics 210: 445–461. 10.1534/genetics.118.301307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J. H., Wang L., Beaudoin-Chabot C., and Thibault G., 2018. Lipid bilayer stress-activated IRE-1 modulates autophagy during endoplasmic reticulum stress. J. Cell Sci. 131: jcs217992 10.1242/jcs.217992 [DOI] [PubMed] [Google Scholar]
- Lee D., An S. W. A., Jung Y., Yamaoka Y., Ryu Y. et al. , 2019. MDT-15/MED15 permits longevity at low temperature via enhancing lipidostasis and proteostasis. PLoS Biol 17: e3000415 10.1371/journal.pbio.3000415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S., Bass J. J., and Szewczyk N. J., 2013. Knockdown of the C. elegans kinome identifies kinases required for normal protein homeostasis, mitochondrial network structure, and sarcomere structure in muscle. Cell Commun. Signal. 11: 71 10.1186/1478-811X-11-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach, N. J., and G. Ruvkun, 2019 Endoplasmic reticulum-associated SKN-1A/Nrf1 mediates a cytoplasmic unfolded protein response and promotes longevity. Elife 8: 516. 10.7554/elife.44425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. G., Van Nostrand E. L., Friedland A. E., Liu X., and Kim S. K., 2016. Deactivation of the GATA Transcription Factor ELT-2 Is a Major Driver of Normal Aging in C. elegans. PLoS Genet 12: e1005956 10.1371/journal.pgen.1005956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Ji F., Breen P., Sewell A., Han M. et al. , 2019. Mitochondrial Dysfunction in C. elegans Activates Mitochondrial Relocalization and Nuclear Hormone Receptor-Dependent Detoxification Genes. Cell Metab. 29: 1182–1191.e4. 10.1016/j.cmet.2019.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M. A. Q., Kinney B. A., Medwig-Kinney T. N., Ashley G., Ragle J. M. et al. , 2020. Rapid Degradation of Caenorhabditis elegans Proteins at Single-Cell Resolution with a Synthetic Auxin. G3 (Bethesda) 10: 267–280. 10.1534/g3.119.400781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matai L., Sarkar G. C., Chamoli M., Malik Y., Kumar S. S. et al. , 2019. Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc. Natl. Acad. Sci. USA 116: 17383–17392. 10.1073/pnas.1900055116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K., Kang J., and Lee J., 2010. A modified feeding RNAi method for simultaneous knock-down of more than one gene in Caenorhabditis elegans. Biotech. 48: 229–232. 10.2144/000113365 [DOI] [PubMed] [Google Scholar]
- Park J. S., Kang D. H., Lee D. H., and Bae S. H., 2015. Concerted action of p62 and Nrf2 protects cells from palmitic acid-induced lipotoxicity. Biochem. Biophys. Res. Commun. 466: 131–137. 10.1016/j.bbrc.2015.08.120 [DOI] [PubMed] [Google Scholar]
- Parr C., Carzaniga R., Gentleman S. M., Van Leuven F., Walter J. et al. , 2012. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-β precursor protein. Mol. Cell. Biol. 32: 4410–4418. 10.1128/MCB.00930-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston A. M., Gurisik E., Bartley C., Laybutt D. R., and Biden T. J., 2009. Reduced endoplasmic reticulum (ER)-to-Golgi protein trafficking contributes to ER stress in lipotoxic mouse beta cells by promoting protein overload. Diabetologia 52: 2369–2373. 10.1007/s00125-009-1506-5 [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes J. S., Deplancke B., Shingles J., Grove C. A., Hope I. A. et al. , 2005. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 6: R110 10.1186/gb-2005-6-13-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J. H., Olive A. J., Sanyal S., Carette J. E., Brummelkamp T. R. et al. , 2013. A CREB3–ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 15: 1473–1485. 10.1038/ncb2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousakis A., Vlassis A., Vlanti A., Patera S., Thireos G. et al. , 2013. The general control nonderepressible-2 kinase mediates stress response and longevity induced by target of rapamycin inactivation in Caenorhabditis elegans. Aging Cell 12: 742–751. 10.1111/acel.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Hino S.-I., Murakami T., Kanemoto S., Kondo S. et al. , 2009. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat. Cell Biol. 11: 1197–1204. 10.1038/ncb1962 [DOI] [PubMed] [Google Scholar]
- Sampieri L., Di Giusto P., and Alvarez C., 2019. CREB3 Transcription Factors: ER-Golgi Stress Transducers as Hubs for Cellular Homeostasis. Front. Cell Dev. Biol. 7: 123 10.3389/fcell.2019.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D. S., and Blower M. D., 2016. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 73: 79–94. 10.1007/s00018-015-2052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomer N., Kadhim A. Z., Grants J. M., Cheng X., Alhusari D. et al. , 2019. Mediator subunit MDT-15/MED15 and Nuclear Receptor HIZR-1/HNF4 cooperate to regulate toxic metal stress responses in Caenorhabditis elegans. PLoS Genet 15: e1008508 10.1371/journal.pgen.1008508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. C., Fox S., Beam M., Thakkar H., Amaral M. D. et al. , 2011. A genetic screening strategy identifies novel regulators of the proteostasis network. PLoS Genet 7: e1002438 10.1371/journal.pgen.1002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans WormBook 11: 1–11. 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Hansen M., Van Gilst M. R., Cooper S. B., and Yamamoto K. R., 2008. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet 4: e1000021 10.1371/journal.pgen.1000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher A. C., and Ewald C. Y., 2018. Overcoming Autofluorescence to Assess GFP Expression During Normal Physiology and Aging in Caenorhabditis elegans. Bio Protoc. 8: e2940 10.21769/BioProtoc.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G., Shui G., Kim W., McAlister G. C., Ismail N. et al. , 2012. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell 48: 16–27. 10.1016/j.molcel.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoishi S., Fukushima S., Shinohara K., Katada T., and Saito K., 2017. CREB3L2-mediated expression of Sec23A/Sec24D is involved in hepatic stellate cell activation through ER-Golgi transport. Sci. Rep. 7: 7992 10.1038/s41598-017-08703-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., and J. Y. Chan, 2006 Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem 281: 19676–19687. 10.1074/jbc.M602802200 [DOI] [PubMed] [Google Scholar]
- Weicksel S. E., Mahadav A., Moyle M., Cipriani P. G., Kudron M. et al. , 2016. A novel small molecule that disrupts a key event during the oocyte-to-embryo transition in C. elegans. Development 143: 3540–3548. 10.1242/dev.140046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. K., and Wek R. C., 2016. Upstream Open Reading Frames Differentially Regulate Gene-specific Translation in the Integrated Stress Response. J. Biol. Chem. 291: 16927–16935. 10.1074/jbc.R116.733899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z., and Dernburg A. F., 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains used in this study are either available from CAENORHABDITIS GENETICS CENTER (CGC) or upon request. Supplementary materials are available at figshare: https://doi.org/10.25387/g3.12783044.