Abstract
Satellite DNAs (satDNAs) are a ubiquitous feature of eukaryotic genomes and are usually the major components of constitutive heterochromatin. The 1.688 satDNA, also known as the 359 bp satellite, is one of the most abundant repetitive sequences in Drosophila melanogaster and has been linked to several different biological functions. We investigated the presence and evolution of the 1.688 satDNA in 16 Drosophila genomes. We find that the 1.688 satDNA family is much more ancient than previously appreciated, being shared among part of the melanogaster group that diverged from a common ancestor ∼27 Mya. We found that the 1.688 satDNA family has two major subfamilies spread throughout Drosophila phylogeny (∼360 bp and ∼190 bp). Phylogenetic analysis of ∼10,000 repeats extracted from 14 of the species revealed that the 1.688 satDNA family is present within heterochromatin and euchromatin. A high number of euchromatic repeats are gene proximal, suggesting the potential for local gene regulation. Notably, heterochromatic copies display concerted evolution and a species-specific pattern, whereas euchromatic repeats display a more typical evolutionary pattern, suggesting that chromatin domains may influence the evolution of these sequences. Overall, our data indicate the 1.688 satDNA as the most perduring satDNA family described in Drosophila phylogeny to date. Our study provides a strong foundation for future work on the functional roles of 1.688 satDNA across many Drosophila species.
Keywords: Satellite DNA, 1.688, 359 bp satellite, Drosophila melanogaster group, heterochromatin, concerted evolution
Satellite DNAs (satDNAs) consist of tandem repeat sequences typically organized in large arrays in the heterochromatic regions of eukaryotic chromosomes (reviewed by Plohl et al. 2012). Regarded as one of the fastest evolving components of the eukaryotic genome (Strachan et al. 1985), closely related species may differ dramatically in the number, abundance, and genomic distribution of a given satDNA (Plohl et al. 2012; Altemose et al. 2014). There are several examples showing that satDNAs are often subject to concerted evolution because the repeat sequences display both a high degree of similarity within a species and a high degree of divergence between species (e.g., Bachmann and Sperlich 1993; Palomeque and Lorite 2008; Pérez-Gutiérrez et al. 2012; de Lima et al. 2017).
SatDNAs were initially identified in Drosophila by their buoyant densities (in g/ml) on cesium chloride gradients to characterize the most abundant sequences in D. virilis, D. melanogaster and D. hydei genomes (Gall et al. 1974; Renkawitz 1979). In the past decades, satDNAs have been mostly studied from a small sample of cloned repeats isolated from a limited number of species and individuals. (Brutlag et al. 1977; Waring and Pollack 1987; Bachmann and Sperlich 1993) More recently, the usage of genomic approaches has provided novel interesting insights on the origin, organization and evolution of satDNA in Drosophila (Larracuente 2014; Dias et al. 2014; Khost et al. 2017; McGurk and Barbash 2018; Sproul and Khost et al. 2020).
In several Drosophila species, satDNAs account for more than 30% of the genome. Loss and amplification events of distinct satDNA families are important factors in shaping the architecture and size of the Drosophila genome (Bosco et al. 2007). Several studies in Drosophila point to a biological role for satDNAs, usually related to centromere structure (Sun et al. 1997; Henikoff et al. 2001) and chromatin organization and modulation (Usakin et al. 2007; Gallach 2014). For example, transcripts of the 1.688 satDNA family in D. melanogaster contribute to the safeguard mechanism that is required for proper kinetochore complex assembly and accurate chromosomal segregation (Rosic et al. 2014). Other findings indicate that siRNAs generated from 1.688 satDNA X-linked euchromatic arrays contribute to the recruitment of the MSL complex and seem to play an important role in the dosage compensation mechanisms in D. melanogaster (Menon et al. 2014). Despite their importance for genome organization, function and evolution, studies focusing on satDNA evolution have been absent in Drosophila species with sequenced genomes and in eukaryotes in general.
In D. melanogaster, one of the most well-understood satDNAs is the 1.688 g/ml satellite (herein referred to as 1.688 satDNA). This satDNA is also known as 359 bp because the bulk of this satellite is composed of 359 bp repeats present in the heterochromatic portion of the X chromosome (Brutlag 1980); less abundant repeat variants, also known as subfamilies, of 260 bp and 353-356 bp are present on chromosomes 2 and 3, respectively (Losada and Villasante 1996; Abad et al. 2000). The 1.688 satDNA is part of the genome of Drosophila species from the melanogaster subgroup, but its relative abundance may vary more than 40-fold between species (Barnes 1978; Strachan et al. 1985; Lohe and Roberts 1988). In addition to the long arrays in heterochromatin, short arrays are present in euchromatic regions of the X chromosome of D. melanogaster, D. simulans, D. sechellia and D. mauritiana (Dibartolomeis et al.1992; Sproul and Khost et al. 2020).
Even though the 1.688 satDNA is one of the best-studied in Drosophila melanogaster, only limited information exists for this satDNA in other species from the melanogaster subgroup, and none from species outside the subgroup. To rectify this, we took advantage of the wealth of genomic data in several Drosophila species to study the 1.688 satDNA in more detail and identify new facets of its evolution, including its origins and evolutionary patterns in heterochromatin and euchromatin. Taken together, our in-depth genomic analysis of 14 Drosophila species from both inside and outside the melanogaster subgroup is the most comprehensive evaluation of the 1.688 satDNA to date. Our work (1) provides strong evidence that the 1.688 satDNA family is the most broadly maintained satDNA in Drosophila phylogeny to date (2) exposes how chromatin domains can impact the sequence evolution of satDNA repeats and (3) suggests potential new functions of this satDNA family.
Materials and Methods
Data mining and sequence analysis
1.688 satDNA repeats were mined from several sequenced species of Drosophila by performing BLASTN searches in FlyBase (http://flybase.org/blast) with a collection of D. melanogaster subgroup 1.688 satDNA consensus sequences described by Strachan et al. (1985). BLASTN searches were restricted to assembled genomes due to the high single-pass error-rates (11–15% for PacBio, similar for Nanopore) (Tørresen et al. 2019). The majority of these errors consist of insertion and deletions (indels), leading to misalignments and possible misleading evolutionary analyses.
The Tandem Repeat Finder (Benson 1999) program was used to bolster the data-mining efforts by confirming the monomer size present in each array. The 1.688-like satDNA arrays found in files representing chromosomes, scaffolds or contigs were manually curated by a dot-plot using Dotlet applet (Junier & Pagni 2000; https://dotlet.vital-it.ch/), one by one, to determine the start and end of each repeat, using the 1.688 satDNA consensus sequences of each species or the closest related species as parameters. Sequences were designated as “full repeats” if they had the same start and end relative to our reference 1.688 satDNA repeats. We cannot exclude the possibility that other sequences with homology to 1.688 satDNA exist in the genome of Drosophila species but were not retrieved through this methodology.
Aiming to identify new variants of 1.688 satDNA repeats in different species, we used the short sequence cluster method RepeatExplorer (http://www.RepeatExplorer.org/) (Novák et al. 2013). RepeatExplorer identifies repetitive elements de novo using a graph-based method to group reads into discrete clusters based on all-by-all blast similarity. This analysis is unbiased and uses a large repertoire of sequences, resulting in a higher quality analysis and more variability of sequences due to its overall sequence clusterization. All reads used in this analysis were generated by Illumina whole-genome shotgun reads (Supp. Table 1). Reads were trimmed to 100 bp and sequencing adapters were removed. We excluded reads that were more than 10% below our quality cut-off value of 30. The quality filtering approaches used herein have been described as sufficient to minimize downstream analysis artifacts (Minoche et al. 2011). The clusterization threshold was explicitly set to 90% sequence similarity spanning at least 65% of the read length. Only clusters with genomic proportion equal or higher to 0.01% were analyzed in this study, as recommended by the developers of RepeatExplorer (Novák et al. 2013).
Table 1. List of species-specific sets of primers used to amplify 1.688 satDNA tandem repeated arrays in nine Drosophila species.
| SPECIES | FWD | REV |
|---|---|---|
| D. MELANOGASTER | 5`-cgttagcactggtaattagctgc-3` | 5`-cgatccctattactttttgaagg-3` |
| D. SIMULANS/D. SECHELLIA | 5`-gtttgtttcttaaatcccaatcg-3 | 5`-ctcaacgaggtatgacattcc-3` |
| D. ERECTA | 5`-gccggatgttttaggaggtt-3` | 5`-aggtatggcattccactcttggac-3` |
| D. YAKUBA | 5′-ccatacctcgttgaattcg-3` | 5`-cattccactttggcaac-3` |
| D. EUGRACILIS | 5`-tcatacatcgatgaactcgt-3` | 5`-cattccatagtccgacaa-3` |
| D. BIARMIPES | 5`-ccaataaattggcatcaa-3` | 5`-cgagctcagcaaggtatgaca-3` |
| D. TAKAHASHII | 5`-cttaattctcaatcgatttgc-3` | 5`- ctacgagctcaacaaggta-3` |
| D. SUZUKII | 5`-ggctaaacaacgactga-3` | 5`-aatcaaggcgtacagctaa-3` |
Assignment of repeats to heterochromatin and euchromatin
Our 1.688-like satDNA repeats were assigned to heterochromatin or euchromatin based on analyses of the flanking sequences within 0.5 to 2 kb, as previously described (Kuhn et al. 2012). We searched for flanking sequences using BLASTN against each species genome and manually checked them in GBrowse (http://flybase.org/cgi-bin/gbrowse2). The presence of transcriptionally active genes in genomic previously described as euchromatic in D. melanogaster and their orthologs in D. simulans, D. sechellia, D. erecta and D. yakuba was taken as supporting evidence of euchromatic location of repeats, and similarly for the melanogaster subgroup using features in FlyBase (http://flybase.org). Phylogenetic clusters were used to additionally separate the groups of sequences based on similarity to those previously described within euchromatic regions. 1.688 satDNA repeats derived from heterochromatin and euchromatin differ by ∼30% and tend to form distinct phylogenetic clusters (Kuhn et al. 2012; de Lima et al. 2017), therefore for species that lack annotated genomes we relied solely on phylogenetic clusterization to assign arrays. Multiple sequence alignments were performed using Muscle 4.0 (Edgar 2004) with default options. Incomplete, partial or truncated monomers (< 75% of the expected monomer size) were not used in the interspecific analyses. We identified the best substitution model with jModelTest2 (Darriba et al. 2012) for each alignment, which was subsequently used for phylogenies and distance matrices. The optimal model tended to be Kimura 2-parameters/T93 models with gamma distribution. The MEGA software version 7.1 (Tamura et al. 2014, Kumar et al. 2016) was used for the calculation of genetic distances and construction of Neighbor-Joining (NJ) dendrograms and Tajima’s D test calculations.
The identification of conserved motifs in 1.688 satDNA derived from heterochromatin was carried out using the software MEME (http://meme.sdsc.edu) (Bailey et al. 2009). We searched for motifs of 10 - 200 bp that had statistical values lower than 10−6.
Genomic distribution on assembled genomes of D. melanogaster, D. simulans and D. yakuba
To map the distribution of 1.688 satDNA arrays in euchromatin, we retrieved assembled files of chromosomes 2L, 2R, 3L, 3R and X from D. melanogaster, D. simulans and D. yakuba present on FlyBase (www.flybase.org). We derived a chromosome-specific consensus sequence and used each one as a query for local BLASTN searches (Altschul et al. 1990) with the corresponding chromosome and species files. To calculate density, the position of the hit and the amount of 1.688 satDNA (in bps) was plotted in non-overlapping 100 kb intervals. Data were plotted using software R version 3.2.2 (R Core Team 2015).
To identify the presence of 1.688 satDNA sequences in the vicinity of genes we used the Table Browser genomic tool implemented in the UCSC GenomeBrowser (www.genome.ucsc.edu) for the annotated genomes of D. melanogaster, D. simulans, D. sechellia, D. erecta and D. yakuba. We isolated genic and intronic sequences, along with 5 kb up and downstream for all annotated genes in all five species. Each of these data sets was independently queried with local BLASTN searches using the species-specific 1.688 satDNA consensus sequence. We used as a cutoff level 65% of identity, 70% of coverage and e-value <10−05 in order to prevent non-specific results. With the exception of D. yakuba, we did not have redundancies in our queries despite Drosophila genomes having a high gene density (Hou et al. 2012): of the 792 initial hits in D. yakuba, only ten hits were found in both the upstream and downstream query results (all intronic hits were unique).
Fly stocks:
All fly stocks were raised on standard Bloomington medium at 25°, and male and female third instar wandering larvae were used. The following fly stocks were used: D. melanogaster (Oregon R), D. simulans w501 (DSSC#14021-0251.195), D. sechellia (DSSC#14021-0248.01), D. erecta (DSSC#14021-0224.01), D. yakuba (DSSC#14021-0261.01), D. eugracilis (DSSC#14026-0451.02), D. biarmipes (DSSC#14023-0361.06), D. takahashii (DSSC#14022-0311.10). D. suzukii specimens were identified and collected by Prof. Jennifer Gleason in Missouri and Kansas locations during the years of 2017 and 2018. D. suzukii flies were cultivated by Prof. Jennifer Gleason at the Ecology and Evolutionary Biology Department at University of Kansas, Lawrence, in accordance with FDA regulations.
Genomic DNA was extracted from 30 frozen flies using the Promega Maxwell 16 robot protocol and its respective extraction kit (www.promega.com/-/media/files/resources/protocols/technical-manuals/0/maxwell-16-instrument-as1000-operating-manual.pdf). Species-specific primer sets were designed to amplify highly conserved regions identified from alignment data (Table 1). PCR reactions consisted of an initial denaturation step of 94° for 3 min, followed by 30 cycles of 94° for 60 sec, 55° for 60 sec, and 68° for 60 sec, and then a final extension at 68° for 10 min. PCR products were excised from 1% agarose gels and purified with the IBI Gel Extraction Kit. Probes were labeled with 16-dUTP-biotin by nick-translation according to the Enzo Life Sciences’ Nick Translation DNA Labeling System.
Preparation, DNA staining and FISH on mitotic and polytene chromosome spreads
Mitotic chromosome spreads from larval brain tissue were prepared as previously described in detail (Hanlon et al. 2018). The brain from a single third-instar larvae was dissected in 0.7% sodium chloride and moved to a fresh 50 µl drop of 0.5% sodium citrate for hypotonic treatment for 5 min, followed by fixation for 4 min in 2 ml of fixative solution (45% acetic acid, 2.5% paraformaldehyde). After fixation, the brain was transferred to a 3 µl drop of 45% acetic acid on an 18-mm × 18-mm siliconized coverslip, gently squashed with an inverted microscope slide, then heavily squashed for 2 min using a hand clamp. The slide was then immediately placed into liquid nitrogen for at least 5 min, after which the coverslip was popped off the slide using a razor blade, dehydrated in 70% ethanol for at least 10 min at −20°, then transferred to 100% ethanol at −20° for at least 10 min. Each slide was air-dried and 21 µl of the FISH solution (20 ul 50% formamide, 10% dextran sulfate, 2× SSC plus 1 ul of biotinylated probe, see above) was applied directly to the sample area and a clean 22 mm × 22 mm glass coverslip was placed on top. The slide was heated to 95° on a heat block for 5 min in darkness, then transferred to a hybridization chamber consisting of a container lined with damp paper towels (to maintain humidity and prevent the samples from drying out) at 30° for overnight (16–24 h).
After the incubation, slides were washed three times in 0.1x SSC for 15 min each, then put in blocking solution (4x SSC, 3% BSA, 0.1% Tween-20) at 37° for 30 min. Excess blocking solution was wiped from around the sample area, followed by application of the secondary solution (40 μl 4x SSC, 1% BSA, 0.1% Tween-20 plus 0.8 μl avidin 488 (Roche) antibody). Each slide was covered with a 22 mm × 22 mm coverslip then put into a hybridization chamber at 37° for 30 min. Slides were washed three times in SSCT (4x SSC, 0.1% Tween-20), followed by three washes in 0.1x SSC; each wash was five minutes. Excess liquid was wiped from around the sample area, and each slide was mounted with 5 μl Vectashield+DAPI and a 22 mm × 22 mm coverslip. Coverslip edges were sealed with nail polish and imaged immediately.
Polytene chromosomes FISH experiments were conducted as described in de Lima et al. (2017). Briefly, salivary glands from third stage larvae were dissected on 0.7% NaCl solution and transferred immediately to 1X PBS solution. Next, the salivary glands were fixed in Carnoy solution (3:1 ethanol/acetic acid) and then moved to a 50-µl drop of 1:1 lactic acid/acetic acid for 5 min and squashed. The slide was incubated at room temperature for two hours and then immediately placed into liquid nitrogen for at least 3 min. Immediately after removal, the coverslip was popped off the slide using a razor blade, and dehydrated at room temperature in 100% ethanol for at least 10 min. For FISH, each dried slide was fixed with paraformaldehyde 4% solution for 30 min and then transferred to 100% ethanol. Slides were then incubated in 2xSSC at 65° for 30 min and dehydrated in 70% and 96% ethanol for 10 min each. Chromosomes were denatured in 0.07 M NaOH solution for 30 sec and immediately incubated in 2XSSC solution for 10 min. Slides were then dehydrated in two consecutive 2 min incubations of 70% and 96% ethanol. For each slide, 20 µl of the FISH solution (50% formamide, 10% dextran sulfate, 2× SSC, 100 ng fluorescence-labeled probe) was applied directly to the dried slide and a clean Parafilm 30 mm × 30 mm coverslip was placed on top. Slides were incubated at 37° overnight (16–24 hr), and washed twice in 2XSSC at 37° for 5 min. The probes were detected with avidin 488 (Roche) then mounted by applying 5 µl Vectashield with DAPI to a clean 22-mm × 22-mm no. 1.5 glass coverslip.
Microscopy and image processing
Mitotic chromosome images were acquired with a DeltaVision microscopy system (GE Healthcare, Piscataway, NY) consisting of a 1 × 70 inverted microscope with a high-resolution charge-coupled device (CCD) camera. Polytene chromosome images were acquired with an Inverted Zeiss LSM 780 confocal microscope. All imaging used a 63x objective for polytene and 100x objective with 1.6 auxiliary magnification for mitotic chromosomes. Stacks of deconvolved images were combined in a z-projection showing maximum intensity, cropped to the region of interest, recolored, and adjusted for brightness and contrast in FIJI/ImageJ.
Data availability
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1549. All stocks (except D. suzukii) and reagents available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.12952253.
Results
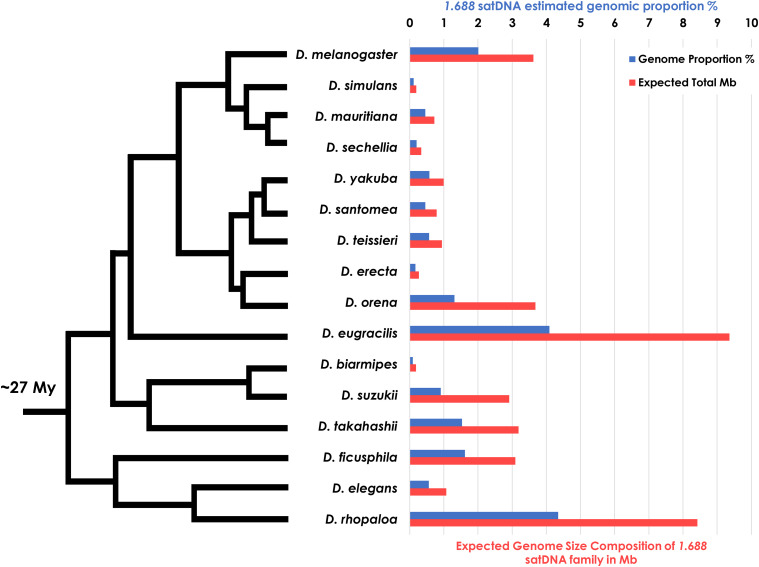
To broadly identify satDNA sequences across the Drosophila phylogeny belonging to the 1.688 satDNA family, we performed BLAST searches using a collection of D. melanogaster 1.688 consensus sequences (Strachan et al. 1985) against all available Drosophila sequenced genomes to date. We found 16 species that carried 1.688 satDNA repeats, 14 of which have fully assembled genomes (Figure 1). As expected, we found 1.688 satDNA repeats present in all five sequenced species from the melanogaster subgroup, i.e., D. melanogaster, D. simulans, D. sechellia, D. erecta and D. yakuba, but absent in the genomes of other species from the melanogaster group including D. kikkawai, D. ananassae and D. bipectinata, and several other more phylogenetically distant species such as D. pseudoobscura, D. miranda and D. persimilis (obscura group), D. mojavensis (repleta group) and D. virilis (virilis group). The usage of the RepeatExplorer short sequence de novo cluster method in this study aimed to minimize the underrepresentation of previous estimations of satDNAs and other repetitive families calculated in Drosophila by means of genome assemblies (Drosophila 12 genomes Consortium et al. 2007). After applying a stringent quality cutoff, the reads used in the overall de novo clusterization had genome coverage of at least 0.55-fold (Table S1), suggesting that most, if not all, repetitive sequences were clustered and analyzed in this study (See Methods). We retrieved a total of 10,855 monomers of 1.688 satDNA repeats from the assembled genomes of 14 species with evidence for significant abundance in the genomes of D. melanogaster subgroup species as well as D. eugracilis, D. suzukii, D. biarmipes, D. takahashii, D. ficusphila, D. elegans, and D. rhopaloa (Table 2). These species shared a common ancestor ∼27 Mya, making the 1.688 satDNA the most broadly maintained satDNA sequence in Drosophila phylogeny described to date. The genomic contribution varied significantly among the species, especially in D. elegans and D. rhopaloa, which show a greater genomic proportion when compared to the D. melanogaster subgroup (Figure 1). Notably, we did not observe any satDNA family with similar characteristics in D. kikkawai or any other more phylogenetically distant species (D. leontia, D. ananassae and D. bipectinata), either in assembled genomes or RepeatExplorer outputs -that were analyzed with a similar genomic coverage (Table S1). Therefore, the presence of 1.688 satDNA sequences in the rhopaloa subgroup and absence in D. kikkawai, D. leontia (montium subgroup) and D. ananassae and D. bipectinata (ananassae subgroup) suggests that the 1.688 satDNA family either originated or expanded before the D. melanogaster-D. rhopaloa clade diversification that occurred approximately 27 Mya.
Figure 1.
Variation in the genomic proportion of the 1.688 satellite DNA family throughout Drosophila phylogeny. Bars indicate the estimated genomic proportion (blue) and the total amount of DNA in Mb (red) comprised by the 1.688 satDNA family based on the RepeatExplorer analysis for each of the 16 species of Drosophila analyzed. Phylogeny adapted from Russo et al. (2013).
Table 2. Number of 1.688 satDNA copies analyzed in each Drosophila genome and characterized as heterochromatic or euchromatic (see Methods).
| Heterochromatic copies | Euchromatic copies | 1.688 satDNA subfamilies (bp) | |
|---|---|---|---|
| D. melanogaster | 1860 | 168 | 360 /353 /257 |
| D. simulans | 1568 | 129 | 360 /199 |
| D. sechellia | 951 | 144 | 361 /198 |
| D. mauritiana | 829 | 102 | 361 /198 |
| D. erecta | 297 | 595 | 361 /198 |
| D. orena | 1473 | 42a | 360 /198 |
| D. santomeab | — | — | 191 |
| D. teissierib | — | — | 191 |
| D. yakuba | 77 | 930 | 360 /191 |
| D. eugracilis | 95 | 63 | 358 |
| D. biarmipes | 121 | 73 | 360 |
| D. suzukii | 244 | 35 | 350 /198 |
| D. takahashii | 59 | 47 | 337 |
| D. ficusphila | 61 | 18 | 197 |
| D. elegans | 52 | 45 | 390 |
| D. rhopaloa | 347 | 51 | 365 /189 |
D. orena euchromatic copies identified were mostly truncated or partial when compared to heterochromatic copies.
D. santomea and D. teissieri do not have assembled genomes. 1.688 satDNA sequences analyzed were retrieved from assembled contigs generated as output of RepeatExplorer pipeline.
A new 1.688 subfamily of ∼190 bp is present in 11 species of Drosophila
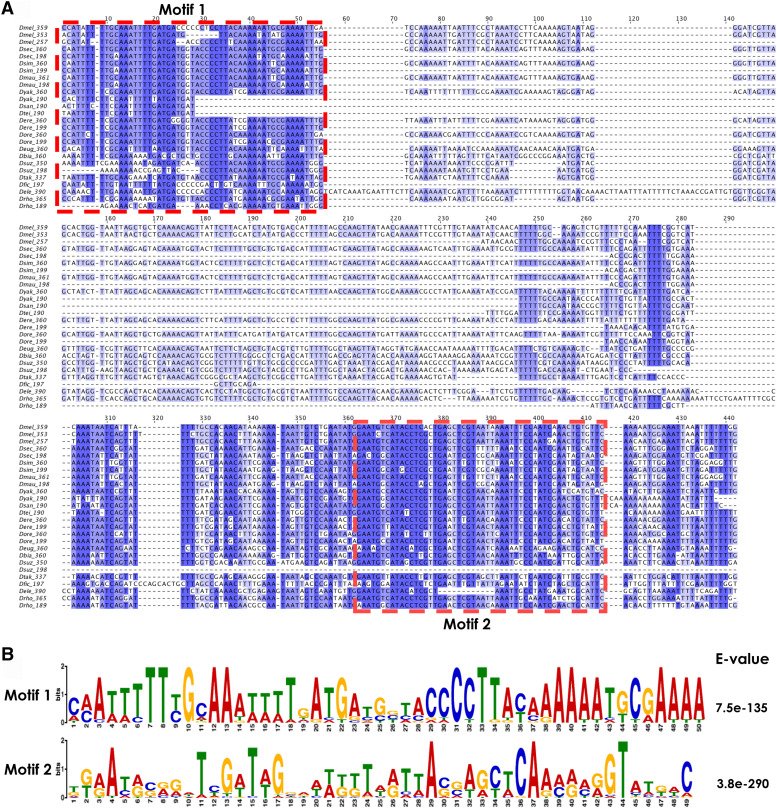
The most common monomer size for the 1.688 satDNA repeat in most species is ∼360 bp, but in our sequence analysis we identified a new variant (subfamily) of ∼190 bp present throughout Drosophila phylogeny. Previous studies have described two chromosome-specific subfamilies of ∼260 bp and 353 bp on chromosomes 2 and 3, respectively, in D. melanogaster (Losada and Villasante 1996; Abad et al. 2000), and a 196 bp monomer has been previously described by Strachan et al. (1985) in D. teissieri, indicating that 1.688 satDNA monomer size is not restricted to ∼360 bp. In fact, the 190 bp subfamily is a common 1.688 satDNA variant present in 11 of the 16 species we analyzed, similar to the range of species in which the 360 bp subfamily is found (Table 2). Also, like the 360 bp variant, the number of ∼190 bp monomers was variable in each species (Table S2). When the monomers from the 190 bp subfamily were aligned alongside the 360 bp subfamily, two striking features emerge. First, monomers from the 190 bp subfamily share a common deletion of ∼170 bp in the central portion when compared to the 360 bp consensus (Figure 2A and Figure S1). Second, both subfamilies share two well-conserved motifs, referred to as Motif 1 and Motif 2 (Figure 2B). The presence of these two conserved regions shared among 1.688 satDNA sequences from different species may indicate functional constraints, possibly related to protein-binding sites (Ugarkovic 2005). Evolutionary conservation can be selected based on primary sequence or the ability to form non-canonical secondary or tertiary structures needed for functional interactions. Further experimentation will be required to fully understand the significance of the motifs. D. suzukii is notable because the 190 bp subfamily is still ∼190 bps, but does not share the same deletion compared to the other species (Figure 2 and Figure S1). The D. suzukii 198 bp monomer has Motif 1 and most of the central region that is conserved in the 360 bp subfamily monomers but does not carry Motif 2 and its surrounding sequence (Figure 2). Based on these findings we hypothesize that the 190 bp subfamily is a derivative of the 360 bp subfamily, produced from an event that occurred independently prior to the D.melanogaster-D.rhopaloa clade diversification (see Discussion).
Figure 2.
1.688 satDNA sequence analysis for repeats derived from heterochromatin. A. Consensus sequences for subfamilies from each of the 16 species were aligned by species. The subfamily description is denoted on the left for each monomer. Dark blue indicates regions with high sequence conservation whereas white regions indicate no conservation among the sequences. B. Two conserved motifs within the 360 bp subfamily were identified by MEME and are shared by all 16 species of Drosophila.
Concerted evolution of 1.688 satDNA in heterochromatin and enrichment on the X chromosome
We identified 10,855 copies of 1.688 satDNA repeats (Table 2). Since 1.688 satDNA has been shown to be present in both heterochromatin and euchromatin, we used two approaches to computationally assign each individual copy as being heterochromatic or euchromatic following established protocols (Kuhn et al. 2012). Analyses of flanking sequences within 0.5 to 2 kb, when available, together with the methodology of phylogenetic tree clusterization of heterochromatic and euchromatic copies enabled us to assign 8,034 repeats as heterochromatic, while the other 2,821copies were inferred as euchromatic (see Methods). As a result of this analysis, we determined the proportion of heterochromatic and euchromatic copies among the 14 species we studied with assembled genomes: D. melanogaster, D. simulans, D. sechellia, D. mauritiana, D. orena, D. erecta, D. yakuba, D. eugracilis, D. biarmipes, D. suzukii. D. takahashii, D. ficusphila, D. elegans and D. rhopaloa. (Figure S2). With the exception of D. erecta and D. yakuba, each of the species we analyzed have a higher abundance of heterochromatic copies than euchromatic copies. The overall abundance of 1.688 satDNA sequences identified and characterized may differ among the species analyzed due to differences in genomic assembly efforts. However, the pattern observed is consistent with previous models that predict an increased accumulation of repetitive sequences in chromosomal regions with low recombination, such as heterochromatin (Charlesworth et al. 2005).
To better understand the evolutionary relationships among 1.688 satDNA family monomers, we ran a Neighbor-Joining (NJ) tree constructed with 4,111 full-length heterochromatic 1.688 satDNA repeats extracted from the sequencing reads of the 14 species with assembled genomes. We restricted the analysis to full-length copies to avoid biased alignments originating from partial or truncated sequences (see Methods). The clusterization produced trees with branches that contained repeats from a single species (e.g., D. melanogaster) or phylogenetically very closely related species (e.g., D. simulans, D. mauritiana and D. sechellia) (Figure 3). This preferential clusterization in a species-specific pattern indicates a concerted mode of evolution, i.e., repeats within each species are more similar to each other than to repeats between species. Our results also recapitulated the chromosome-specific clustering pattern for 1.688 satDNA in D. melanogaster present in chromosomes X, 2 and 3, (Kunh et al. 2012; Khost et al. 2017) (Figure S3A), indicating intrachromosomal concerted evolution. A similar pattern was observed in D. erecta, D. suzukii, D. elegans and D. rhopaloa suggesting chromosomal- or array-specific evolution of 1.688 satDNA in these species (Figures S3B-D). In contrast, D. simulans, D. mauritiana and D. sechellia formed a single large branch that lacked species-specific clusters. This may be the result of the close phylogenetic relationship of these species (Garrigan et al. 2012), reinforcing the idea that concerted evolution is a time-dependent mechanism (Strachan et al. 1985).
Figure 3.
Clusterization reveals species specificity, indicative of concerted evolution in 1.688 satDNA heterochromatic arrays. An unrooted phylogenetic tree using 4111 full-length 1.688-like monomers derived from heterochromatin from 14 species of Drosophila shows a species-specific pattern throughout the melanogaster group phylogeny. The evolutionary history was inferred using the Neighbor-Joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions with less than 85% site coverage were eliminated, i.e., fewer than 15% alignment gaps, missing data, and ambiguous bases were allowed at any position.
The outcome of the NJ analysis is congruent with the species-specific nucleotide divergence observed in the phylogenetic analysis (Table 3). The interspecific variation among 1.688 satDNA repeats exceeds 21% on average, except between D. simulans, D. mauritiana and D. sechellia that diverge only ∼8% from each other, and D. orena and D. melanogaster in which the copies show an 18.2% nucleotide divergence. Intraspecific values trended much lower than interspecific values and varied from 2% in D. yakuba to 31% in D. eugracilis (Table 3). In summary, the evolutionary pattern of the 1.688 satDNA family is incongruent with the phylogenetic relationships for the melanogaster group (Figure 3; Figure S4), consistent with the idea that satDNA families are good taxonomic markers (Picariello et al. 2002), but do not reflect the phylogenetic history of the D. melanogaster-D. rhopaloa clade.
Table 3. Intra- and interspecific nucleotide divergence (p-distance) of 4,111 1.688 satDNA heterochromatic copies retrieved from 14 species of Drosophila.
| D. mel | D. sim | D. sec | D. mau | D. ere | D. yak | D. ore | D. eug | D. bia | D. suz | D. tak | D. fic | D. ele | D. rho | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. mel | 0.100 | |||||||||||||
| D. sim | 0.261 | 0.080 | ||||||||||||
| D. sec | 0.251 | 0.084 | 0.080 | |||||||||||
| D. mau | 0.257 | 0.079 | 0.079 | 0.070 | ||||||||||
| D. ere | 0.264 | 0.306 | 0.300 | 0.301 | 0.130 | |||||||||
| D. yak | 0.264 | 0.299 | 0.297 | 0.293 | 0.202 | 0.020 | ||||||||
| D. ore | 0.182 | 0.260 | 0.256 | 0.257 | 0.280 | 0.268 | 0.070 | |||||||
| D. eug | 0.324 | 0.317 | 0.316 | 0.316 | 0.380 | 0.372 | 0.341 | 0.310 | ||||||
| D. bia | 0.304 | 0.343 | 0.333 | 0.334 | 0.369 | 0.343 | 0.323 | 0.392 | 0.150 | |||||
| D. suz | 0.349 | 0.419 | 0.415 | 0.410 | 0.366 | 0.394 | 0.330 | 0.434 | 0.400 | 0.100 | ||||
| D. tak | 0.307 | 0.341 | 0.335 | 0.332 | 0.340 | 0.329 | 0.314 | 0.399 | 0.332 | 0.276 | 0.190 | |||
| D. fic | 0.223 | 0.241 | 0.237 | 0.239 | 0.262 | 0.246 | 0.256 | 0.359 | 0.270 | 0.356 | 0.285 | 0.140 | ||
| D. ele | 0.286 | 0.306 | 0.300 | 0.299 | 0.349 | 0.357 | 0.302 | 0.392 | 0.373 | 0.399 | 0.338 | 0.312 | 0.200 | |
| D. rho | 0.258 | 0.351 | 0.353 | 0.356 | 0.353 | 0.372 | 0.293 | 0.390 | 0.348 | 0.402 | 0.392 | 0.310 | 0.356 | 0.21 |
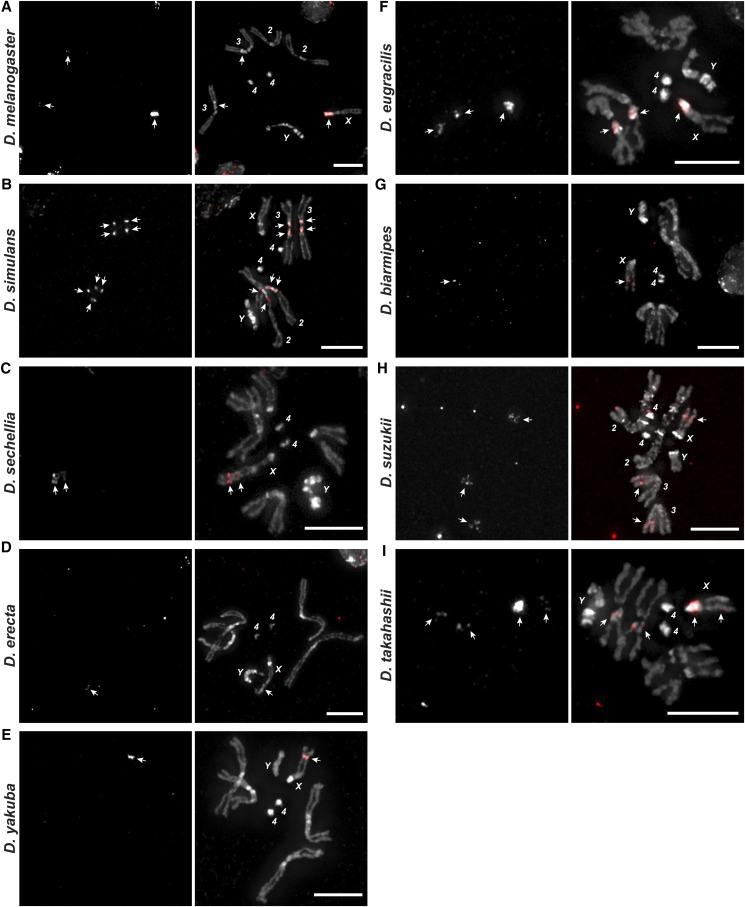
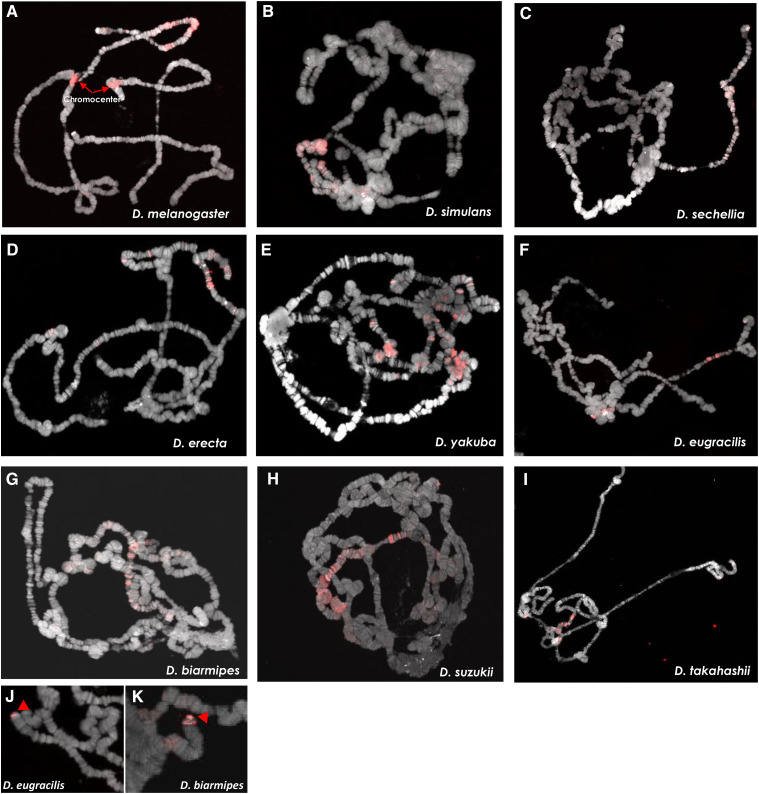
To confirm and extend our computational analysis, we experimentally mapped the distribution of the 1.688 satDNA family throughout Drosophila species by performing fluorescent in-situ hybridization (FISH) experiments using species-specific probes on the mitotic chromosomes of squashed larval brains from D. melanogaster, D. simulans, D. sechellia, D. erecta, D. yakuba, D. eugracilis, D. biarmipes, D. suzukii and D. takahashii (Figure 4). We observed dramatic variation in abundance that corroborates the predictions from RepeatExplorer (Figure 1). All species except D. simulans had 1.688 satDNA repeats on the X chromosome. Additionally, we observed a signal on the distal portion of the X chromosome in D. sechellia, D. erecta, D. yakuba, D. biarmipes, D. suzukii, and D. takahashii that had a similar chromosomal location but differed in intensity. This distal X chromosome signal was the only detectable signal observed in D. sechellia, D. erecta, D. yakuba, D. biarmipes, suggesting that the region could be the initial point of amplification of this satDNA family. 1.688 satDNA arrays were also present at different sites and on different chromosomes (Figure 5), although the precise determination of autosome (second or third) for D. eugracilis and D. takahashii was not possible (Figures 4 and 5). Taken together, our data provide striking evidence that, despite being conserved for ∼27 My, the 1.688 satDNA sequences show significant differences in abundance, sequence evolution, and chromosomal distribution in Drosophila species.
Figure 4.
Differential and shared locations of 1.688 arrays in heterochromatin visualized in nine Drosophila species from the melanogaster group. FISH was performed on neuroblast chromosome spreads from D. melanogaster, D. simulans, D. sechellia, D. erecta, D. yakuba, D. eugracilis. D. biarmipes, D suzukii and D. takahashii (A-I, respectively). The panels show species-specific probes for 1.688 satDNA (red) labeled with Bio-16-dUTP combined with DAPI staining (gray). The X and Y sex chromosomes and autosomal labels (when known) are identified in each panel. Note that the X chromosome from all species except D. simulans has a clear signal. Bar = 5 µm. White arrows indicate signals of hybridization.
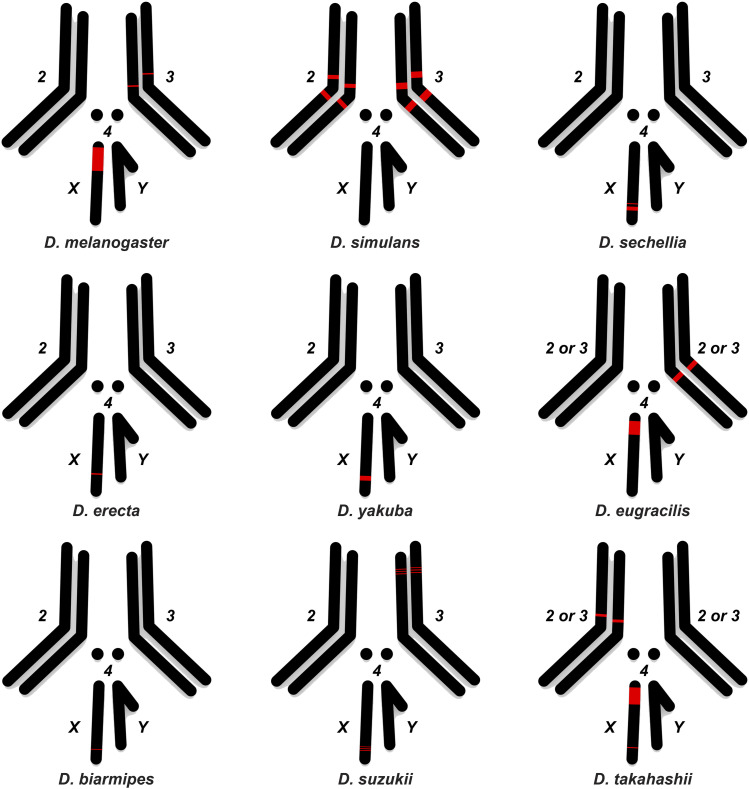
Figure 5.
Ideogram showing chromosomal location of 1.688 heterochromatic arrays mapped by FISH in nine Drosophila species from the melanogaster group. Simplified karyotypes (black) from D. melanogaster, D. simulans, D. sechellia, D. erecta, D. yakuba, D. eugracilis. D. biarmipes, D suzukii and D. takahashii are shown along with the approximate location and relative abundance of 1.688 satDNA (red) for each species. Note that although chromosomes may have different sizes in each species, the ideograms are similarly sized for the sake of simplicity. The 1.688 satDNA arrays on chromosome 2L in D. melanogaster described by Abad et al. (2000) were not detected with the probes in this study (see Table 1).
1.688 satDNA is spread throughout euchromatic regions in the D. melanogaster-D. rhopaloa clade
With our methodology, we identified 2,821 putative euchromatic copies of 1.688 satDNA in the 14 species with assembled genomes. This is the first report of 1.688-like satDNA copies in the euchromatin of D. erecta, D. yakuba, D. eugracilis, D. biarmipes, D. suzukii, D. takahashii, D. ficusphila, D. elegans and D. rhopaloa. (Figure S2 and Table 2). These euchromatic copies are difficult to detect cytologically on mitotic chromosomes due to their lower copy number and lack of compaction as compared to copies within heterochromatin. Therefore, to confirm the presence of 1.688 satDNA arrays in euchromatic regions, FISH experiments were performed on polytene chromosomes from the salivary glands of nine species (Figure 6). Polytene chromosomes undergo several rounds of endoreduplication in their euchromatic regions, but the centromeric heterochromatin does not endoreduplicate and instead forms a dense central mass known as the chromocenter (Riddle et al. 2011). Therefore, FISH on polytene chromosomes enables both the detection of low-copy number targets due to their amplification as well as the confirmation that they are located within the euchromatic regions of the chromosomes. Our FISH analysis showed several hybridization signals throughout the euchromatic regions for all nine species (Figure 6). We detected the presence of 1.688 satDNA repeats within euchromatic regions of the X, 2nd and 3rd chromosomes in different species, but no signals were detected on the 4th and Y chromosomes. Notably, we detected strong signals on the telomeric portions of D. eugracilis and D. biarmipes chromosomes (Figure 6 J-K, respectively), confirming and expanding previous findings by Saint-Leandre et al. (2019).
Figure 6.
Localization of 1.688 arrays in euchromatin of polytene chromosomes for nine Drosophila species from the melanogaster group. FISH using species specific probes was performed on salivary gland polytene chromosome from D. melanogaster, D. simulans, D. sechellia, D. erecta, D. yakuba, D. eugracilis. D. biarmipes, D suzukii and D. takahashii (A-I, respectively). The panels show probes (red) labeled with Bio-16-dUTP combined with DAPI staining (gray). J-K. FISH indicating telomeric/subtelomeric 1.688 satDNA signals of hybridization (red arrowheads) on D. eugracilis and D. biarmipes chromosomes.
To identify possible preferential euchromatic locations, we ran BLAST searches using a species-specific euchromatic consensus sequence as a query in the available genome assemblies of D. melanogaster, D. simulans and D. yakuba. The locations of the euchromatic 1.688 satDNA varied significantly throughout the chromosomes of the three species and, consistent with our FISH analyses, we did not detect copies on the fourth and Y chromosomes. D. yakuba showed the highest presence of euchromatic arrays, followed by D. melanogaster and D. simulans. (Table S3). Furthermore, euchromatic repeats are enriched on the X chromosome in all three species (Table S3). In D. melanogaster and D. simulans the majority of euchromatic arrays are on the X chromosome (83% and 80%, respectively). In contrast, D. yakuba has only 61% of 1.688 satDNA arrays localized on the X chromosome (Table S3). To further understand the genomic distribution of the euchromatic repeats in these three species, we analyzed their density throughout each chromosome and found that each species showed a unique pattern of localization (see Methods; Figures S5-9). Altogether, 1.688-like satDNA sequences in euchromatic regions is a common feature of Drosophila genomes within the D. melanogaster-D. rhopaloa clade. We suspect the species-specific distribution pattern occurred during the initial phases of the process of speciation, with differential expansions through recurrent events of mobilization (Sproul and Khost et al. 2020).
Euchromatic repeats display high nucleotide divergence in intra- or interspecific comparisons
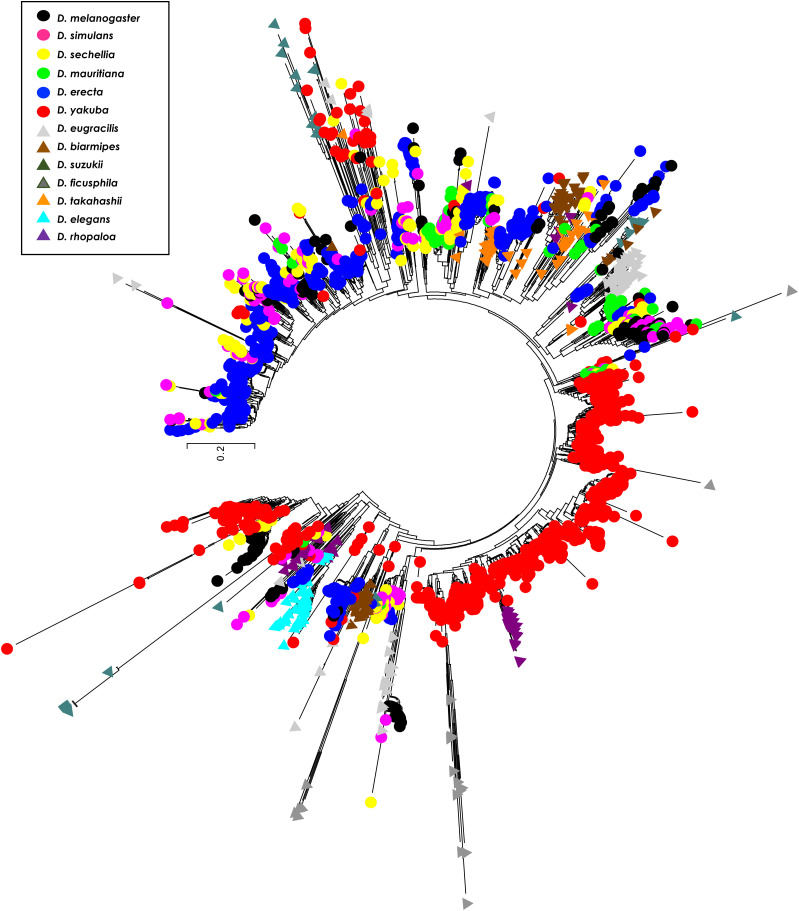
Similar to our analysis of heterochromatic repeats, we ran an NJ phylogenetic analysis with all 2,393 full-length repeats retrieved from 13 species. We excluded D. orena from this analysis because the euchromatic copies we identified were mostly truncated or partial when compared to heterochromatic copies. The tree topology showed large branch lengths with three major groups (Figure 7). One branch is composed almost exclusively of monomers from the 190 bp satDNA subfamily in D. yakuba, indicating a high degree of sequence divergence of this variant within this species. The two other branches do not show any consistent phylogenetic grouping, and sequences derived from the same arrays do not cluster predominantly together. Further, a species-specific analysis of the euchromatic arrays of D. melanogaster, D. simulans, D. sechellia, D. erecta and D. yakuba failed to show a higher frequency of clusterization of 1.688 satDNA sequences positioned on the same chromosome when compared to arrays located on non-homologous chromosomes, although some branches appear to show array-specific or species-specific patterns. Our analysis of the intra- and interspecific nucleotide divergence of the euchromatic 1.688 satDNA copies showed a broad range of variation (Table 4). Intraspecific nucleotide divergence ranged from 14.2% in D. elegans to 43.3% in D. ficusphila, and overall, we observed a higher degree of divergence for 1.688 satDNA euchromatic copies (23.7% on average) than heterochromatic copies (13.2% on average), indicating euchromatic copies tend to be highly divergent within a species. Together, the patterns of clusterization and nucleotide divergence indicate that 1.688 satDNA euchromatic and heterochromatic sequences experience different evolutionary pressures.
Figure 7.
Lack of species-specific clusterization (except in D. yakuba) of euchromatic copies of 1.688 satDNA indicates broad interspecies divergence. An unrooted phylogenetic tree using 2393 full-length 1.688-like monomers derived from euchromatin from 13 species of Drosophila shows intermingling throughout melanogaster group phylogeny. D. yakuba is a notable exception in which the species-specific branch indicates a recent expansion of 190 bp repeats in this species. The evolutionary history was inferred using methods and parameters described in Figure 3.
Table 4. Intra- and interspecific nucleotide divergence (p-distance) of 2,393 1.688 satDNA euchromatic copies retrieved from 13 species of Drosophila.
| D. mel | D. sim | D. sec | D. mau | D.ere | D.yak | D.eug | D.bia | D.suz | D.tak | D.fic | D.ele | D.rho |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.274 | ||||||||||||
| 0.273 | 0.233 | |||||||||||
| 0.276 | 0.24 | 0.239 | ||||||||||
| 0.261 | 0.236 | 0.247 | 0.185 | |||||||||
| 0.269 | 0.229 | 0.229 | 0.242 | 0.198 | ||||||||
| 0.284 | 0.252 | 0.253 | 0.231 | 0.239 | 0.176 | |||||||
| 0.325 | 0.303 | 0.309 | 0.297 | 0.29 | 0.297 | 0.282 | ||||||
| 0.303 | 0.279 | 0.284 | 0.265 | 0.27 | 0.274 | 0.319 | 0.18 | |||||
| 0.397 | 0.377 | 0.378 | 0.377 | 0.361 | 0.361 | 0.408 | 0.375 | 0.369 | ||||
| 0.261 | 0.239 | 0.245 | 0.217 | 0.227 | 0.233 | 0.285 | 0.233 | 0.369 | 0.18 | |||
| 0.409 | 0.387 | 0.389 | 0.388 | 0.379 | 0.373 | 0.443 | 0.423 | 0.46 | 0.378 | 0.433 | ||
| 0.335 | 0.314 | 0.318 | 0.3 | 0.312 | 0.3 | 0.358 | 0.297 | 0.398 | 0.286 | 0.411 | 0.142 | |
| 0.285 | 0.267 | 0.268 | 0.25 | 0.249 | 0.235 | 0.316 | 0.269 | 0.364 | 0.238 | 0.381 | 0.286 | 0.196 |
1.688-Like elements are present in the vicinity of genes of melanogaster subgroup species
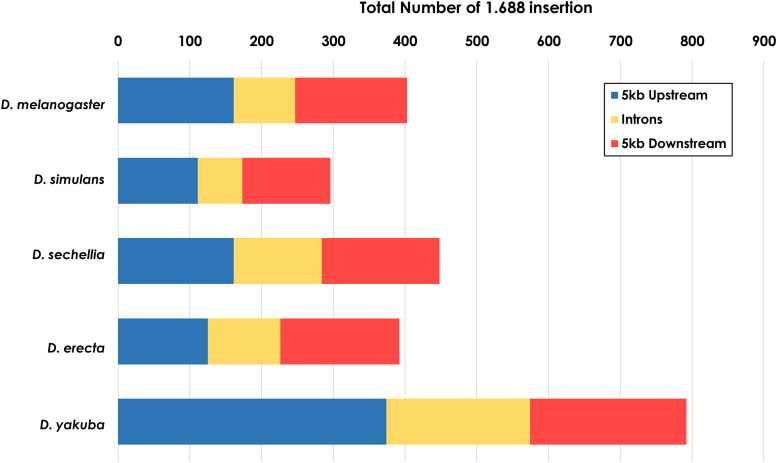
Due to the pervasive localization of 1.688 satDNA arrays in euchromatin, we analyzed whether 1.688 satDNA insertion sites are present near gene regions. We ran species-specific BLASTN searches in the promoter/upstream, coding, intronic and downstream regions of all genes annotated in the UCSC GenomeBrowser (www.genome.ucsc.edu) for the five species of the melanogaster subgroup that have annotated genomes (D. melanogaster, D. simulans, D. sechellia, D. erecta and D. yakuba). Using stringent query parameters to prevent non-specific hits, we identified both full and partial 1.688 satDNA arrays in proximity to genic regions (maximum of 5 kb) or within introns (See Methods). From all five species, we found 921 insertions in the promoter/upstream regions, 825 insertions in downstream regions, and 572 insertions in intronic regions (Supp. Mat. 3).
The number of insertions varied in the different types of regions and are not well conserved among the species (Figure 8). D. yakuba has the largest number of insertions with 374, 218 and 200 instances in upstream, downstream and intronic regions, respectively. In D. simulans, we identified 111 1.688 satDNA sequences present in upstream regions, 123 in downstream regions and 62 in intronic regions. D. sechellia and D. erecta showed similar distribution patterns in the three regions (Figure 8). D. melanogaster has the second lowest number of gene proximal insertions in all five species with 149, 156 and 86 insertions in upstream, downstream and intronic regions, respectively. Altogether, we find that 1.688 satDNA sequences are not only present in euchromatic regions but can be in close proximity to genes, with each species displaying a unique and dynamic pattern throughout euchromatin.
Figure 8.
Insertions of 1.688 satDNA sequences in the vicinity of genes. Species-specific BLASTN searches in the upstream, intronic and downstream regions of all annotated genes from the UCSC GenomeBrowser (www.genome.ucsc.edu) was performed for the five species of the melanogaster subgroup with annotated genomes.
The intronic insertion of 1.688 satDNA within Netrin paralogs is evolutionarily conserved
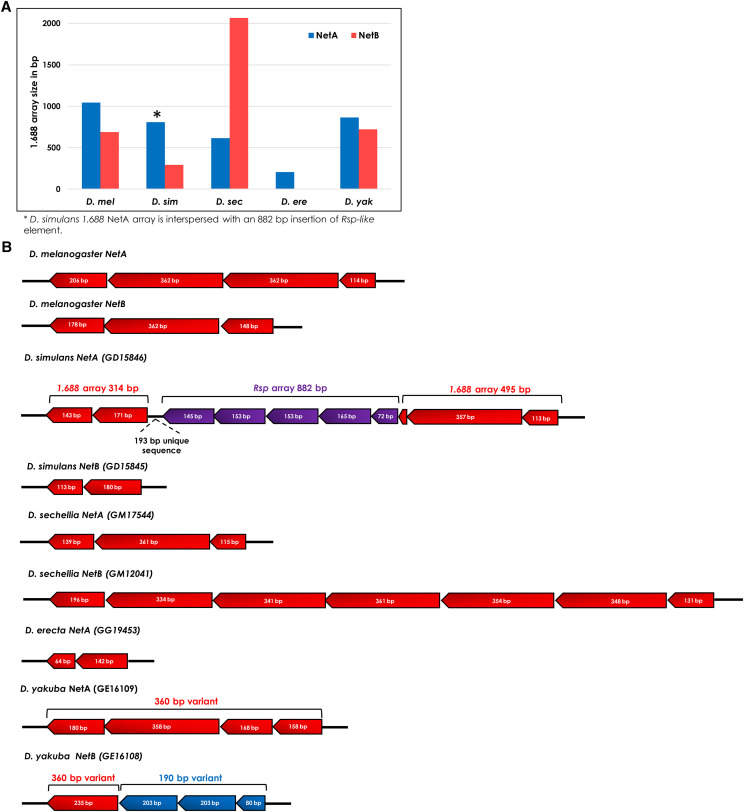
A striking example of an evolutionarily conserved insertion site is the intronic regions of the duplicated gene Netrin. The products of NetA (Dmel: FBgn0015773) and NetB (Dmel: FBgn0015774) are crucial instructive cues for target recognition at the fly neuromuscular junction (Orr et al. 2014). The genes are in tandem on the X chromosome and contain a 1.688 satDNA array in the intronic region in all five species with annotated genomes except D. erecta, which has the shortest 1.688 satDNA array in NetA and no array in NetB (Figure 9A). Surprisingly, in D. yakuba the NetA array carries the 360 bp variant whereas NetB array carries the 190 bp variant as identified by its deletion signature (Figure S9B). In other species, only the 360 bp variant is present in the NetA and NetB introns. This pattern of evolutionary conservation suggests that the 1.688 satDNA insertion likely occurred prior to the gene duplication event, which was before the speciation of the melanogaster subgroup. Interestingly, the 1.688 satDNA monomeric sequences present in the NetA and NetB introns do not evolve neutrally. Although the NetA coding sequence appear to have evolved under neutral selection (D=-0.09, Tajima’s test), the 1.688 satDNA sequences inserted in NetA and B introns, and the NetB coding sequence, all show signs of positive selection (D=-0.53, -0.43, -0.74, respectively). This finding suggests that 1.688 satDNA euchromatic insertions in the vicinity of genes could play a role as a cis-acting gene regulation element.
Figure 9.
Differential pattern of 1.688satDNA insertions in the intronic regions of duplicated genes NetA and NetB in five species of the melanogaster subgroup. A. Histogram representation of 1.688 array size insertion in the intronic regions of NetA and NetB genes, respectively, for five species of melanogaster subgroup. B. Schematic representation of 1.688 insertion in the intronic regions of NetA and NetB genes in five species of melanogaster subgroup evidencing a species-specific pattern of 1.688 array size and organization. The 360 bp subfamily is represented in red, whereas the 190 bp subfamily is in blue. Responder-like arrays are represented in purple. Note that repeat units sizes identified here are based on the monomeric sequence designed for each species.
Additionally, we found an insertion of 882 bp of Responder-like satDNA exclusively inside the NetA 1.688 satDNA array in D. simulans (Figures 9B and S10A). This insertion also carries a 193 bp sequence highly similar to the intronic region of the Flotillin 2 gene (Dsim: FBgn0068599) and an adjacent 1.688 satDNA array(Figure S10). The same 193 bp sequence is observed in the orthologous sites of closely related species from the melanogaster subgroup (Figure S10A). In D. simulans, the high sequence identity between the 193 bp sequence present at Flotillin 2 and NetA (98.4% of identity), together with the nearly identical 233 bp flanking fragment of 1.688 on both gene introns (96.2% of identity) (Figure S10B) and the absence of this insertion in the close related species D. sechellia suggests a recent transposition event of a Responder-like element from Flotillin 2 to NetA in which a partial sequence of the 1.688 satDNA array was carried during this element mobilization. Finally, the Responder-like element transposition from one 1.688 satDNA array to another reinforces the hypothesis that the evolution and proliferation of both satDNA families in euchromatin are co-dependent (Sproul and Khost et al. 2020).
Discussion
In the present study we performed a large-scale analysis of the 1.688 satDNA family in genomes from 16 species in the melanogaster group utilizing a combination of assembled genomes, de novo repeat identification, similarity estimation methods and molecular cytogenetic analyses. Our report greatly expands the knowledge regarding the prevalence and evolution of 1.688 satDNA sequences in the melanogaster group of species (Figure 1; Figure 4). Additionally, our data are consistent with previous observations that 1.688 satDNA sequences are not confined to heterochromatic regions but are also present in euchromatin, which we demonstrate for the first time in D. erecta, D. yakuba, D. eugracilis, D. biarmipes, D. suzukii and D. takahashii (Figure 6) and predict in D. ficusphila, D. elegans and D. rhopaloa (Figure S2). Our data suggest that 1.688 satDNA amplification probably occurred in the ancestral lineage of the D. melanogaster-D. rhopaloa clade after the divergence from the common ancestor shared with the montium subgroup of species ∼27 Mya. Alternatively, the absence of 1.688 satDNA copies in D. kikkawai and D. leontia may derive from stochastic processes or concerted evolution that have led to significant nucleotide divergence, which may hinder identification in these species. The maintenance of conserved satDNA sequences can indicate functional constraints, such as the centromeric satDNA in primates (Alkan et al. 2007). However, in Drosophila the long-term evolutionary maintenance of satDNA is rare. Previous analyses have described the maintenance of the satDNA family pvB370 for a period of about 20 My in the D. virilis group species, with varying abundance in each species (Heikkinen et al. 1995; Biessmann et al. 2000). To our knowledge, this was the most broadly maintained satDNA family until our description of 1.688 satDNA as a genomic feature of 16 species in melanogaster group. Although 1.688 satDNA has been maintained for an exceptionally long period of time, it also exhibits great variability in genomic proportions, varying from 0.091% in D. biarmipes to 4.345% in D. rhopaloa (Figure 1). Recurrent turnover events of different satDNA families, such as Rsp, dodeca, and simple satellite DNAs, along with heterogeneity in chromosomal localization of satDNA families has been described in Drosophila species (Larracuente 2014; Carmena et al. 1993; Jagannathan et al. 2017; Wei et al. 2018), reinforcing that satDNA evolution is dynamic and can impact the genomic landscape of a species It is important to highlight that the two satDNA families with broader coexistence patterns throughout Drosophila phylogeny, pvB370 and 1.688, were previously described on both domains of chromatin (Biessmann et al. 2000 and herein). satDNA in heterochromatin turns over faster than in euchromatin, so its presence in euchromatin may aid its evolutionary maintenance.
Computational and experimental evidence both support differences in the abundance and location of the 1.688 satDNA family throughout the melanogaster group phylogeny (Figures 4 and 5). However, the presence of 1.688 satDNA arrays on the telomere proximal region of chromosome X in six out of nine species analyzed (Figure 4 and 5) suggests that this 1.688 satDNA block may be evolutionarily conserved and leads us to speculate that this particular location may be the ancestral position. We demonstrate that 1.688 heterochromatic arrays are present at different locations and on different chromosomes, even in closely related species (Figure 5), indicating the spread of heterochromatic 1.688 satDNA may have followed unique trajectories in each lineage. A striking example is shown by the dramatic differences in chromosome localization of the 1.688 satDNA heterochromatic arrays in D. simulans and D. sechellia (Figure 5), which is in spite of the monomers lacking a clear species-specific cluster or significant nucleotide divergence (Figure 3; Table 3). These findings suggest that even though satDNAs have a high rate of evolution in eukaryotic genomes (Charlesworth 1994), 1.688 satDNA sequences do not necessarily evolve as rapidly as their location changes, at least in the heterochromatin of these species. Overall, our findings highlight a paradoxical evolutionary characteristic of the 1.688 satDNA family: persistence over a long evolutionary period with species-specific patterns of location and sequence variation.
In addition to heterochromatic arrays, 1.688 satDNA euchromatic copies can contribute significantly to the genome content of the melanogaster group, especially in D. yakuba (Figure S2). Notably, 1.688 satDNA euchromatic arrays are enriched on, but not restricted to, the X chromosome (Figure 6, Table S3). The enrichment of 1.688 satDNA in the euchromatic portions of the X chromosome may be related to specific sex-chromosome characteristics (Gallach 2014) or to functional constraints, as X-linked transcripts derived from 1.688 satDNA are reported to act as siRNAs that enhance the dosage compensation by MSL complex proteins (Menon et al. 2014). The exact mechanisms of colonization of different heterochromatic/euchromatic regions is still unknown. However, a recent study suggests that Rsp-dependent transposition by extrachromosomal DNA may mobilize and spread 1.688 satDNA sequences (Sproul and Khost et al. 2020). Therefore, we speculate that 1.688 satDNA repeats were established in euchromatin early in the evolution of the melanogaster group and subsequent unique mobilization and diversification events contributed to the species-specific patterns observed.
The species-specific patterns of 1.688 satDNA repeats in heterochromatin (Figure 3) contrasts with the lack of array-specific or chromosome clusterization for most repeats in euchromatin (Figure 7). Taken together, these observations suggest the operation of different molecular forces in the two types of chromatin. The concerted evolution of repeats in heterochromatin indicates molecular drive mechanisms (Dover 1986). Conversely, recombination in repeated regions may be repressed in euchromatin since unequal exchanges may have negative consequences such as gene deletions (Feliciello et al. 2006). Our evidence that 1.688 satDNA sequences display different evolutionary signatures depending on the chromatin context suggests that they may be subject to different molecular processes, leading us to speculate that the differential location of 1.688 satDNA sequences in heterochromatin and euchromatin may shape the sequence landscape of the 1.688 satDNA family in each species.
Although the most prevalent 1.688 satDNA monomer size is ∼360 bp (Carlson and Brutlag 1977), the 190 bp variant is present throughout D. melanogaster group phylogeny (Table 2). First described in the D. teissieri genome (Strachan et al. 1985), our data indicate that this variant is pervasive and can comprise the majority of copies in some species, such as in D. ficusphila and D. suzukii. The prevalence of the 360 bp and 190 bp variants throughout the D. melanogaster-D.rhopaloa clade as described here leads us to conclude that both were likely present in the last common ancestor ∼27 Mya, and while it is impossible to know which variant is the most ancestral, we favor a model where the 360 bp variant gave rise to the 190 bp variant through some recombination event during the evolutionary history of this clade. Notably, the 190 bp variant in D. suzukii does not share the same structure observed in the rest of the species (Figure 2). This result suggests that 190 bp subfamilies either passed through a species-specific homogenization event, or that an independent event generated the 190 bp variant, as we hypothesize for the D. suzukii 190 bp variant. Interestingly, the length of both 1.688 satDNA subfamilies (190 and 360 bp) corresponds to the length of DNA wrapped around 1 or 2 nucleosomes, respectively (Henikoff et al. 2001), suggesting that the maintenance of both monomer sizes could be linked to this genomic architecture feature.
D. yakuba has the largest number of euchromatic 1.688 satDNA copies out of all 14 species we analyzed (Figure S2, Table 2). Due to the species-specific pattern of their clustering (Figure 7), we speculate there was a relatively recent expansion by lineage-specific events such as rolling-circle amplification and insertion (Sproul and Khost et al. 2020). It is noteworthy that different mechanisms, such as gene conversion, may also perform important roles in the expansion of the 190 bp subfamily, as observed in NetB repeats (Figure S10). Intriguingly, D. yakuba also has the largest number (28) of fixed chromosomal inversions identified in Drosophila (Ranz et al. 2007), the breakpoint of which are described as hotspots for repeat insertions (Casals et al. 2006). Therefore, we suggest that the high number of 1.688 satDNA euchromatic insertions in D. yakuba may reflect genomic instability events that occurred during the evolution of this species.
1.688 satDNA arrays are present at telomeric regions in D. biarmipes and D. eugracilis genomes, suggesting that this satDNA family may have been co-opted to perform the telomeric function in these species. Drosophila telomeric regions are composed of transposable elements (Het-A/TART) responsible for maintaining telomeric sequences and a protein complex (HOAP) required for telomere stability (Silva-Sousa and Casacuberta 2012). A recent study by Saint-Leandre et al. (2019) failed to identify the expected telomere-specialized elements in the D. biarmipes genome and suggested that telomeric function relies on euchromatic D. melanogaster 1.688 satDNA repeats (also known as SAR2 sequences) and Helitron-like transposable elements. Previous studies have shown that D. melanogaster satDNA sequences interact with recombinant HOAP complex proteins (Shareef et al. 2001). Therefore, the presence of 1.688 satDNA in the telomeric regions of D. biarmipes and D. eugracilis combined with the apparent absence of Het-A and TART sequences (in D. biarmipes) suggests that 1.688 satDNA may serve a telomeric function in these two species.
Finally, we found evidence for insertions of 1.688-like satDNA elements in gene proximal regions in melanogaster subgroup species (Figure 8). This distribution indicates that at least at some positions, gene-proximal insertions are tolerated (Feliciello et al. 2015). Furthermore, the insertions in the intronic regions of NetA and NetB genes show evidence of positive selection. Previous studies proposed that 1.688 satDNA sequences may act as cis-regulatory elements, such as promoters, insulators or transcription factor binding sites (Menon and Meller 2015). Thus, the presence of 921 1.688 satDNA hits located on the 5 kb upstream regions of genes for the five species analyzed suggests that 1.688 satDNA repeats might act as local modulators of transcription, as observed for transposable elements inserted immediately upstream of protein coding genes (Faulkner et al. 2009). The transcriptional potential of 1.688 satDNA (Menon et al. 2014), as well its distribution near protein-coding genes, supports the hypothesis that 1.688 satDNA may represent a rich source for the assembly of gene regulatory systems (Menon and Meller 2015). Moreover, Ekhteraei-Tousi et al. (2020) have recently shown that the depletion of the 1.688 satDNA block within chromosome X pericentromeric regions may influence the differential expression of genes involved in eggshell formation, and that the transcripts from these regions may act in trans. Much remains to be done to understand the influence of 1.688 satDNA sequences on the transcriptional landscape. However, it is tempting to speculate that the disparate pattern of 1.688 satDNA insertions close to protein-coding genes, along with its unique diversification in Drosophila species, could contribute to the establishment of lineage-specific or species-specific patterns of gene expression.
In summary, our work represents the most comprehensive analysis of 1.688 satDNA evolution to date, demonstrating that this satDNA family likely originated after the divergence of D. melanogaster-D. rhopaloa and montium group species (27 Mya). The locations of 1.688 satDNA in genomes throughout Drosophila phylogeny, and the presence of conserved sequence motifs, suggests 1.688 satDNA may provide a “multi-use tool” function for chromosome biology, consistent with roles at centromeres, telomeres, and in regulating gene expression. Our findings lay a strong foundation for future studies aimed to better understand the biological roles of 1.688 satDNA sequences, and satDNAs in general, during the evolutionary history of Drosophila species.
Acknowledgments
We thank Dr. Jennifer Gleason (Ecology and Evolutionary Biology Department at University of Kansas, Lawrence) for generously providing D. eugracilis, D. takahashii, D. biarmipes and D. suzukii fly stocks. SLH is supported by the NICHD Pathway to Independence Award number K99HD09927.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12952253.
Communicating editor: E. Betran
Literature Cited
- Abad J. P., Agudo M., Molina I., Losada A., Ripoll P. et al. , 2000. Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol. Gen. Genet. 264: 371–377. 10.1007/s004380000331 [DOI] [PubMed] [Google Scholar]
- Alkan C., Ventura M., Archidiacono N., Rocchi M., Sahinalp S. C. et al. , 2007. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLOS Comput. Biol. 3: 1807–1818. 10.1371/journal.pcbi.0030181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altemose N., Miga K. H., Maggioni M., and Willard H. F., 2014. Genomic characterization of large heterochromatic gaps in the human genome assembly. PLOS Comput. Biol. 10: e1003628 10.1371/journal.pcbi.1003628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bachmann L., and Sperlich D., 1993. Gradual evolution of a specific satellite DNA family in Drosophila ambigua, D. tristis, and D. obscura. Mol. Biol. Evol. 10: 647–659. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E. et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S. R., Webb D. A., and Dover G., 1978. The distribution of satellite and main-band DNA components in the melanogaster species subgroup of Drosophila. Chromosoma 67: 341–363. 10.1007/BF00285965 [DOI] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Zurovcova M., Yao J. G., Lozovskaya E., and Walter M. F., 2000. A telomeric satellite in Drosophila virilis and its sibling species. Chromosoma 109: 372–380. 10.1007/s004120000094 [DOI] [PubMed] [Google Scholar]
- Bosco G., Campbell P., Leiva-Neto J. T., and Markow T. A., 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177: 1277–1290. 10.1534/genetics.107.075069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Appels R., Dennis E. S., and Peacock W. J., 1977. Highly repeated DNA in Drosophila melanogaster. J. Mol. Bio. 112: 31–47. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., 1980. Molecular arrangement and evolution of heterochromatic DNA. Annu. Rev. Genet. 14: 121–144. 10.1146/annurev.ge.14.120180.001005 [DOI] [PubMed] [Google Scholar]
- Carmena M., Abad J. P., Villasante A., and Gonzalez C., 1993. The Drosophila melanogaster dodecasatellite sequence is closely linked to the centromere and can form connections between sister chromatids during mitosis. J. Cell Sci. 105: 41–50. [DOI] [PubMed] [Google Scholar]
- Carlson M., and Brutlag D., 1977. Cloning and characterization of a complex satellite DNA from Drosophila melanogaster. Cell 11: 371–381. 10.1016/0092-8674(77)90054-X [DOI] [PubMed] [Google Scholar]
- Casals F., González J., and Ruiz A., 2006. Abundance and chromosomal distribution of six Drosophila buzzatii transposons: BuT1, BuT2, BuT3, BuT4, BuT5, and BuT6. Chromosoma 115: 403–412. 10.1007/s00412-006-0071-7 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227. 10.1017/S0016672300032365 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., and Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. 10.1038/sj.hdy.6800697 [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., and Posada D., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9: 772 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima L. G., Svartman M., and Kuhn G. C., 2017. Dissecting the Satellite DNA Landscape in Three Cactophilic Drosophila Sequenced Genomes. G3 (Bethesda) 7: 2831–2843. 10.1534/g3.117.042093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias G. B., Svartman M., Delprat A., Ruiz A., Kuhn G. C., 2014. Tetris is a foldback transposon that provided the building blocks for an emerging satellite DNA of Drosophila virilis. Genome biology and evolution, 6: 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBartolomeis S. M., Tartof K. D., and Jackson F. R., 1992. A superfamily of Drosophila satellite related (SR) DNA repeats restricted to the X chromosome euchromatin. Nucleic Acids Res. 20: 1113–1116. 10.1093/nar/20.5.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A., 1986. Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet. 2: 159–165. 10.1016/0168-9525(86)90211-8 [DOI] [Google Scholar]
- Dover G., 1982. Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. 10.1038/299111a0 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium; Clark A. G, Eisen M. B., Smith D. R., Bergman C. M. et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekhteraei-Tousi S., Lewerentz J., and Larsson J., 2020. Painting of Fourth and the X–Linked 1.688 Satellite in D. melanogaster is Involved in Chromosome-Wide Gene Regulation. Cells 9: 323 10.3390/cells9020323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G. J., Kimura Y., Daub C. O., Wani S., Plessy C. et al. , 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Feliciello I., Picariello O., and Chinali G., 2006. Intra-specific variability and unusual organization of the repetitive units in a satellite DNA from Rana dalmatina: molecular evidence of a new mechanism of DNA repair acting on satellite DNA. Gene 383: 81–92. 10.1016/j.gene.2006.07.016 [DOI] [PubMed] [Google Scholar]
- Feliciello, I., Akrap, I., and Ugarković, Đ. 2015 Satellite DNA modulates gene expression in the beetle Tribolium castaneum after heat stress. PLoS Genet. 11: e1005466. doi: 10.1371/journal.pgen.1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., and Atherton D. D., 1974. The satellite DNAs of Drosophila virilis. In Cold Spring Harbor Symposia on Quantitative Biology (Vol. 38, pp. 417–421). Cold Spring Harbor Laboratory Press. [DOI] [PubMed] [Google Scholar]
- Gallach M., 2014. Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol. Evol. 6: 1279–1286. 10.1093/gbe/evu104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Andolfatto P., Clark A. G. et al. , 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22: 1499–1511. 10.1101/gr.130922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon S. L., Miller D. E., Eche S., and Hawley R. S., 2018. Origin, Composition, and Structure of the Supernumerary B Chromosome of Drosophila melanogaster. Genetics 210: 1197–1212. 10.1534/genetics.118.301478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen E., Launonen V., Müller E., and Bachmann L., 1995. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J. Mol. Evol. 41: 604–614. 10.1007/BF00175819 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., and Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- Hou C., Li L., Qin Z. S., and Corces V. G., 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48: 471–484. 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M., Warsinger-Pepe N., Watase G. J., and Yamashita Y. M., 2017. Comparative Analysis of Satellite DNA in the Drosophila melanogaster Species Complex. G3 (Bethesda) 7: 693–704. 10.1534/g3.116.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T., and Pagni M., 2000. Dotlet: diagonal plots in a web browser. Bioinformatics 16: 178–179. 10.1093/bioinformatics/16.2.178 [DOI] [PubMed] [Google Scholar]
- Kim, M., S. Ekhteraei-Tousi, J. Lewerentz, and J. Larsson, 2018 The X–Linked 1.688 Satellite in Drosophila melanogaster Promotes Specific Targeting by Painting of Fourth. Genetics 208: 623–632. 10.1534/genetics.117.300581 [DOI] [PMC free article] [PubMed]
- Khost D. E., Eickbush D. G., and Larracuente A. M., 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 27: 709–721. 10.1101/gr.213512.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn G. C. S., Küttler H., Moreira-Filho O., and Heslop-Harrison J. S.. 2012. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol. Biol. Evol. 29: 7–11. 10.1093/molbev/msr173 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., and Tamura K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., 2014. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol. Biol. 14: 233 10.1186/s12862-014-0233-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., and Roberts P. A., 1988, pp. 148–186 in Evolution of satellite DNA sequences in Drosophila. Heterochromatin: molecular and structural aspects. Cambridge: Cambridge University Press, University Press, New York. [Google Scholar]
- Losada A., and Villasante A., 1996. Autosomal location of a new subtype of 1.688 satellite DNA of Drosophila melanogaster. Chromosome Res. 4: 372–383. 10.1007/BF02257273 [DOI] [PubMed] [Google Scholar]
- McGurk M. P., and Barbash D. A., 2018. Double insertion of transposable elements provides a substrate for the evolution of satellite DNA. Genome Res. 28: 714–725. 10.1101/gr.231472.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon D. U., Coarfa C., Xiao W., Gunaratne P. H., and Meller V. H., 2014. siRNAs from an X-linked satellite repeat promote X-chromosome recognition in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 111: 16460–16465. 10.1073/pnas.1410534111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon D. U., and Meller V. H., 2015. Identification of the Drosophila X chromosome: The long and short of it. RNA Biol. 12: 1088–1093. 10.1080/15476286.2015.1086864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche A. E., Dohm J. C., and Himmelbauer H., 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol. 12: R112 10.1186/gb-2011-12-11-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P., Neumann P., Pech J., Steinhaisl J., and Macas J., 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. 10.1093/bioinformatics/btt054 [DOI] [PubMed] [Google Scholar]
- Orr B. O., Borgen M. A., Caruccio P. M., and Murphey R. K., 2014. Netrin and Frazzled regulate presynaptic gap junctions at a Drosophila giant synapse. J. Neurosci. 34: 5416–5430. 10.1523/JNEUROSCI.3183-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomeque T., and Lorite P., 2008. Satellite DNA in insects: a review. Heredity 100: 564–573. 10.1038/hdy.2008.24 [DOI] [PubMed] [Google Scholar]
- Pérez-Gutiérrez M. A., Suárez-Santiago V. N., López-Flores I., Romero A. T., and Garrido-Ramos M. A., 2012. Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Mol. Biol. 78: 19–29. 10.1007/s11103-011-9848-z [DOI] [PubMed] [Google Scholar]
- Picariello O., Feliciello I., Bellinello R., and Chinali G., 2002. S1 satellite DNA as a taxonomic marker in brown frogs: molecular evidence that Rana graeca graeca and Rana graeca italica are different species. Genome 45: 63–70. 10.1139/g01-125 [DOI] [PubMed] [Google Scholar]
- Plohl M., Meštrović N., and Mravinac B., 2012. Satellite DNA evolution, in Repetitive DNA (ed. Schmid, M.) 126–152. [DOI] [PubMed] [Google Scholar]
- R Core Team 2015 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
- Ranz J. M., Maurin D., Chan Y. S., Von Grotthuss M., Hillier L. W. et al. , 2007. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 5: e152 10.1371/journal.pbio.0050152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., 1979. Isolation of twelve satellite DNAs from Drosophila hydei. Int. J. Biol. Macromol. 1: 133–136. 10.1016/0141-8130(79)90029-1 [DOI] [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B. et al. , 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163. 10.1101/gr.110098.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rošić S., Köhler F., S.and Erhardt, 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Bio. 207: 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo C. A., Mello B., Frazão A., and Voloch C. M., 2013. Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool. J. Linn. Soc. 169: 765–775. 10.1111/zoj.12062 [DOI] [Google Scholar]
- Saint-Leandre B., Nguyen S. C., and Levine M. T., 2019. Diversification and collapse of a telomere elongation mechanism. Genome Res. 29: 920–931. 10.1101/gr.245001.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef M. M., King C., Damaj M., Badagu R., Huang D. W. et al. , 2001. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol. Biol. Cell 12: 1671–1685. 10.1091/mbc.12.6.1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Sousa R., and Casacuberta E., 2012. Drosophila telomeres: an example of co-evolution with transposable elements. Genome Dyn. 7: 46–67. 10.1159/000337127 [DOI] [PubMed] [Google Scholar]
- Sproul J. S., Khost D. E., Eickbush D. G., Negm S., Wei X., et al. 2020. Dynamic evolution of euchromatic satellites on the X chromosome in Drosophila melanogaster and the simulans clade. Molecular Biology and Evolution 37: 2241–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T., Webb D., and Dover G., 1985. Transition stages of molecular drive in multiplecopy DNA families in Drosophila. EMBO J. 4: 1701–1708. 10.1002/j.1460-2075.1985.tb03839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wahlstrom J., and Karpen G., 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007–1019. 10.1016/S0092-8674(00)80491-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tørresen O. K., Star B., Mier P., Andrade-Navarro M. A., Bateman A. et al. , 2019. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 47: 10994–11006. 10.1093/nar/gkz841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., and Salzberg S. L., 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13: 36–46. 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic D., 2005. Functional elements residing within satellite DNAs. EMBO Rep. 6: 1035–1039. 10.1038/sj.embor.7400558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usakin L., Abad J., Vagin V. V., de Pablos B., Villasante A. et al. , 2007. Transcription of the 1.688 satellite DNA family is under the control of RNA interference machinery in Drosophila melanogaster ovaries. Genetics 176: 1343–1349. 10.1534/genetics.107.071720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring G. L., and Pollack J. C., 1987. Cloning and characterization of a dispersed, multicopy, X chromosome sequence in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84: 2843–2847. 10.1073/pnas.84.9.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. H. C., Lower S. E., Caldas I. V., Sless T. J., Barbash D. A. et al. , 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Mol. Biol. Evol. 35: 925–941. 10.1093/molbev/msy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1549. All stocks (except D. suzukii) and reagents available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.12952253.