Abstract
Precise genetic manipulation of specific cell types or tissues to pinpoint gene function requirement is a critical step in studies aimed at unraveling the intricacies of organismal physiology. Drosophila researchers heavily rely on the UAS/Gal4/Gal80 system for tissue-specific manipulations; however, it is often unclear whether the reported Gal4 expression patterns are indeed specific to the tissue of interest such that experimental results are not confounded by secondary sites of Gal4 expression. Here, we surveyed the expression patterns of commonly used Gal4 drivers in adult Drosophila female tissues under optimal conditions and found that multiple drivers have unreported secondary sites of expression beyond their published cell type/tissue expression pattern. These results underscore the importance of thoroughly characterizing Gal4 tools as part of a rigorous experimental design that avoids potential misinterpretation of results as we strive for understanding how the function of a specific gene/pathway in one tissue contributes to whole-body physiology.
Keywords: Gal4, Drosophila, tissue-specific expression, adult, female
Organismal physiology involves extensive inter-organ communication via circulating factors that are produced and secreted in response to changes in the local, systemic, or external environment. Many organs can sense and communicate such changes by sending signals to other tissues to ensure whole-body homeostasis (Droujinine and Perrimon 2016). For example, growth-blocking peptides produced in the larval fat body (in response to dietary amino acids) activate the epidermal growth factor receptor in inhibitory neurons connected to insulin-producing cells to facilitate insulin secretion (Meschi et al. 2019). Activin-β secreted from enteroendocrine cells in the midgut (in response to a high sugar diet) enhances the response of the fat body to adipokinetic hormone (the Drosophila glucagon analog), resulting in hyperglycemia in larvae (Song et al. 2017). In adults, it was recently shown that ecdysone produced in the ovary stimulates intestinal stem cell (ISC) division in mated females (Ahmed et al. 2020). Oogenesis itself is highly sensitive to changes in physiology and can be modulated by manipulations in peripheral tissues, including the fat body (Armstrong et al. 2014; Matsuoka et al. 2017; Armstrong and Drummond-Barbosa 2018; Weaver and Drummond-Barbosa 2018; Weaver and Drummond-Barbosa 2019), gut (Ameku et al. 2018), and brain (Lafever and Drummond-Barbosa 2005; Sieber and Spradling 2015). Studies aimed at understanding the complex endocrine relationships among organs as organisms respond to physiological or environmental changes require experimental tools that allow cell type/tissue-specific manipulations.
The UAS/Gal4/Gal80 system is commonly used in Drosophila to manipulate a specific cell type or tissue to determine the requirements for genes and pathways either in regulating that same cell type/tissue of interest or in remotely affecting separate tissues (Brand and Perrimon 1993). The UAS/Gal4/Gal80 system employs the yeast transcription factor Gal4 under the control of a “tissue-specific” enhancer/promoter sequence (referred to as the “driver”) in combination with a “responder” that contains an Upstream Activating Sequence composed of Gal4 binding sites upstream of a target gene or sequence of interest (Brand and Perrimon 1993). Gal4 binds to the UAS sequence, thereby inducing tissue-specific expression of the transgene (e.g., fluorescent reporter, hairpin RNA, protein-coding gene, etc). The Gal4 inhibitor Gal80 (Douglas and Hawthorne 1966) can be added to this system for multiple purposes. For example, expression of Gal80 under a tissue-specific promoter can be used to inhibit Gal4 function in a subset of cell types/tissues to generate a more tissue/cell type specific driver (Eliason et al. 2018). Loss of a Gal80 transgene can also be used for the generation of Flp/FRT-induced positively marked loss-of-function clones (expressing a fluorescent reporter driven by Gal4) during genetic mosaic analysis (Lee and Luo 1999). In addition, a temperature-sensitive Gal80 mutant allele can be used to temporally restrict Gal4 activity to specific developmental stages (e.g., larvae or adults) or experimental time windows (McGuire et al. 2003). This system has been instrumental in the use of Drosophila as a model for understanding complex cellular and physiological processes.
A potential caveat to the Gal4/UAS system, however, is that the described cell type- or tissue-specific Gal4 expression patterns can be incomplete, such that published Gal4 lines might have additional unreported sites of expression that could potentially confound the interpretation of experimental results. In fact, when previously assessing published fat body-specific drivers in adult females to identify an adipocyte-specific Gal4, we found that the majority of those drivers were expressed in additional tissues besides the fat body in adult females (Armstrong et al. 2014). As this example illustrates, scientists studying adult female physiology would benefit from having a set of commonly used Gal4 drivers that have been thoroughly analyzed for their expression patterns in adult females, such that their tissue specificity is unequivocal.
In this study, we selected commonly used Gal4 drivers and analyzed their expression patterns in all of the major tissues of the adult Drosophila female. We found that a significant number of Gal4 drivers typically used for the genetic manipulation of specific cell types in the ovary or midgut have previously unreported expression in additional, secondary tissues. By contrast, most of the Gal4 drivers for neuronal subpopulations are indeed specific, as they show their reported pattern without expression in additional tissues. Finally, we highlight techniques commonly used in Drosophila for inhibiting Gal4 expression in secondary tissues, as well as other ways to rule out secondary tissue effects when Gal4 is expressed in multiple tissues.
Materials And Methods
Drosophila strains and culture conditions
Drosophila stocks were maintained at room temperature (22-25°) on standard medium containing cornmeal, molasses, yeast, and agar. Previously described Gal4 lines used in this study are included in Table 1. The nSyb-Gal80 transgene has been previously described (Rubinstein et al. 2010). The UAS-GFP.nls (w1118; P{UAS-GFP.nls}14), UAS-mCD8::GFP (w*; P{10XUAS-IVS-mCD8::GFP}attP2), and UASp-lacZ lines were obtained from the Bloomington Drosophila Stock Center (BDSC; bdsc.indiana.edu/). Additional genetic elements are described in FlyBase (http://www.flybase.org).
Table 1. Full genotypes of Gal4 drivers used in this study.
| Driver | Genotype | Source | Reference |
|---|---|---|---|
| bab1-Gal4 | wa; P{w+mW.hs = GawB}bab1Agal4-5]/TM3, Sb1 | BDSC 6802 | (Cabrera et al. 2002) |
| hh-Gal4MB | sp/CyO; hh-Gal4/TM3 | Michael Buszczak | (Eliazer et al. 2011) |
| hh-Gal4TX | w; hh-Gal4/TM6B | Ting Xie | (Pan et al. 2007) |
| hh-Gal4JF | w1118; P{y+t7.7 w+mC = GMR28E03-GAL4}attP2 | BDSC 45546 | (Jenett et al. 2012) |
| ptc-Gal4 | ptc-Gal4/CyO act-GFP; tub-Gal80ts/TM6B | D.D.-B. Laba | (Forbes et al. 1996) |
| c587-Gal4 | c587-Gal4/FM7i; tub-Gal80ts/CyO, Act-GFP | D.D.-B. Lab | (Hsu and Drummond-Barbosa 2009) |
| tj-Gal4 | tj-Gal4 tub-Gal80ts/CyO twist-gal4.UAS-GFP | D.D.-B. Lab | (Sahai-Hernandez and Nystul 2013) |
| mex1-Gal4 | mex-Gal4/TM6B | Allan Spradling | (Phillips and Thomas 2006) |
| NP3084-Gal4 | wa; P{GawB}NP3084 | Kyoto 113094 | (Hayashi et al. 2002) |
| esg-Gal4 | esg-Gal4; tub-Gal80ts UAS-GFP | Allan Spradling | (Micchelli and Perrimon 2006) |
| dl-Gal4 | y w; tub-Gal80ts/CyO; delta-Gal4/TM3 | Benoit Biteau | (Zeng et al. 2010) |
| Su(H)GBE-Gal4 | y w; GBE Su(H)-Gal4 UAS-GFP/CyO; tub-Gal80ts/TM3 | Benoit Biteau | (Zeng et al. 2010) |
| c42-Gal4 | wa; P{w+mW.hs = GawB}c42 | BDSC 30835 | (Rosay et al. 1997) |
| Uro-Gal4 | wa; P{Uro-GAL4.T}2 | BDSC 44416 | (Terhzaz et al. 2010) |
| mef2-Gal4 | tub-Gal80ts/CyO; mef2-Gal4/TM6B | D.D.-B. Lab | (Ranganayakulu et al. 1998) |
| nSyb.P-Gal4 | y1 w1118; P{y+t7.7 w+mC = nSyb-GAL4.P}attP2 | BDSC 51941 | (Riabinina et al. 2015) |
| nSyb.S-Gal4 | y1 wa; P{w+ma =nSyb-GAL4.S}3 | Mark Wu | (Liu et al. 2012) |
| repo-Gal4 | w1118; P{w+ma =GAL4}repo/TM3, Sb1 | BDSC 7415 | (Sepp et al. 2001) |
| ChAT-Gal4 | w1118; P{w+mC = ChAT-GAL4.7.4}19B/CyO, P{ry+t7.2 = sevRas1.V12}FK1 | BDSC 6798 | (Salvaterra and Kitamoto 2001) |
| pebbled-Gal4 | wa P{w+ma=GAL4}peb | Chris Potter | (Sweeney et al. 2007) |
| Gr5a-Gal4 | pin/CyO; Gr5a-Gal4/TM6b | Chris Potter | (Wang et al. 2004) |
| Gr66a-Gal4 | w-; Gr66a-Gal4; GR93a3 | Chris Potter | (Wang et al. 2004) |
| Ir8a-Gal4 | Ir8a-Gal4/CyO | Chris Potter | (Abuin et al. 2011) |
| Ir25a-Gal4 | Ir25a-Gal4/CyO | Chris Potter | (Abuin et al. 2011) |
| Or83b-Gal4 | w-; Or83b-Gal4/CyO | Chris Potter | (Wang et al. 2004) |
| ppk23-Gal4 | Bl/CyO; ppk23-Gal4/TM6b | Chris Potter | (Wang et al. 2004) |
| tub-Gal4 | y w; tub-Gal80ts; tub-Gal4/TM6B | D.D.-B. lab | (Nabel-Rosen et al. 2002) |
Gal4 lines from D.D.-B. lab were generated by combining Gal4 drivers obtained from the BDSC with tub-Gal80ts through standard genetic crosses.
For tissue- and cell type-specific transgene expression, females of genotypes y w; Gal4*/UAS-transgene or y w; UAS-transgene/+; Gal4*/+ (Gal4* represents Gal4 lines used in this study) were raised at room temperature, and 0-to-2-day-old females were switched to 29° for 7 days to induce transgene expression. For all experiments, standard medium was supplemented with wet yeast paste.
Immunostaining and confocal microscopy
Tissues were dissected in Grace’s insect medium with L-glutamine (Caisson Labs) and fixed in 5.3% formaldehyde (Ted Pella) in Grace’s medium at room temperature. Ovaries were teased apart to separate ovarioles and fixed for 13 min; brains and carcasses were fixed for 20 min; thoraces were fixed for 30 min; and guts with attached Malpighian tubules were fixed for one hour. Samples were rinsed three times and washed three times for 15 min in PBSTx (PBS; 10 mM NaH2PO4/NaHPO4, 175 mM NaCl, pH 7.4, 0.1% Triton X-100), and subsequently incubated for three hours at room temperature in blocking solution consisting of 5% normal goat serum (NGS, MP Biomedicals) and 5% bovine serum albumin (BSA, Sigma-Aldrich) in PBSTx. Samples were incubated at 4° overnight in the following primary antibodies diluted in blocking solution: rabbit anti-GFP (Torrey Pines Biolabs Inc, 1:2500); chicken anti-GFP (Abcam, 1:1000); and mouse anti-β-Galactosidase (Promega, 1:500). Samples were rinsed three times and washed three times for 15 min in PBSTx before incubation for two hours at room temperature in 1:400 Alexa Fluor 488-conjugated goat species-specific secondary antibodies (ThermoFisher Scientific). Samples were rinsed, washed, and mounted in Vectashield with 1.5 μg/mL 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Images were acquired with a Zeiss LSM700 confocal microscope.
Data availability
Drosophila strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results And Discussion
Different hh-Gal4 “niche” drivers have distinct patterns of expression in adult females
Gal4 drivers expressed in subsets of cells in the adult ovary are routinely used for the study of oogenesis (Hudson and Cooley 2014). To determine the degree of cell type/tissue specificity of commonly used ovary Gal4 drivers (Table 2), we carefully examined their expression patterns in adult female tissues using UAS-nucGFP or UAS-mCD8::GFP reporters. (Please note that the expression patterns we report throughout this study were consistently observed with 100% penetrance in the analyzed samples.) We first looked at the expression pattern of terminal filament and cap cell drivers known as hh-Gal4 that were obtained from three independent sources. The first hh-Gal4 line (an enhancer trap Gal4 line obtained from Michael Buszczak and referred hereafter as hh-Gal4MB) (Tanimoto et al. 2000; Eliazer et al. 2011) drove expression of UAS-nucGFP in the cap cells as previously reported (Figure 1A) (Eliazer et al. 2011) but was also expressed in some escort cells [which are somatic cells that envelop and support differentiating germ cells in the anterior portion of germarium prior to the envelopment of 16-cell germline cysts by follicle cells (Margolis and Spradling 1995)] (Figure 1A, yellow arrowheads) and in the hindgut (Figure 2A, yellow arrowhead). Surprisingly, hh-Gal4MB failed to drive expression of UAS-mCD8::GFP in cap cells or escort cells (Figure 1A); however, UAS-mCD8::GFP, like UAS-nucGFP, was also expressed in the hindgut (Figure 2A). The second hh-Gal4 tested (obtained from Ting Xie and referred hereafter as hh-Gal4TX) (Pan et al. 2007) drove expression of UAS-nucGFP in cap cells and a subset of escort cells (Figure 1B, yellow arrowhead) and in the hindgut (Figure 2B, yellow arrowhead). (Please note that we were unable to find any information about how hh-Gal4TX was generated.) Like hh-Gal4MB, however, hh-Gal4TX did not drive expression of UAS-mCD8::GFP in the germarium (Figure 1B), but UAS-mCD8::GFP expression was observed in later stage follicle cells (Figure 1B, white arrowheads) and in some cells in the hindgut (Figure 2B, yellow arrowhead). Lastly, we examined the expression pattern of the Janelia Farm hh-Gal4 driver (referred hereafter as hh-Gal4JF), which was generated by subcloning of the hh regulatory region upstream of Gal4 and site-specific transgene insertion (Jenett et al. 2012). UAS-nucGFP driven by hh-Gal4JF showed robust expression in the terminal filament, cap cells, and escort cells (Figure 1C). UAS-mCD8::GFP driven by hh-Gal4JF, however, was much more strongly expressed in the terminal filament and cap cells than in escort cells (Figure 1C). Both GFP constructs were expressed in follicle cells (Figure 1C, white arrowheads) and in the midgut (Figure 2C) when driven by hh-Gal4JF. These results suggest that different lines termed “hh-Gal4” have distinct patterns of expression that are also in part dependent on the type and insertion site of the UAS reporter transgene. Thus, depending on the hh-Gal4 driver used, some result interpretations might be confounded by additional expression in other tissues and ovarian cell types, and not all UAS transgenes will necessarily be induced in the expected hh-Gal4 pattern.
Table 2. Expression patterns of Gal4 drivers in adult female tissues.
| Reported tissue specificity | Driver | Brain | Muscle | Fat Body | Gut | Ovary | Reference |
|---|---|---|---|---|---|---|---|
| Ovary | bab1-Gal4 | + | — | — | + | + | This study |
| hh-Gal4MB | — | — | — | + | + | This study | |
| hh-Gal4TX | — | — | — | + | + | This study | |
| hh-Gal4JF | — | — | — | + | + | This study | |
| ptc-Gal4 | + | — | — | + | + | This study | |
| c587-Gal4 | + | — | + | — | + | This study | |
| tj-Gal4 | + | — | + | — | + | This study | |
| Gut and Malpighian tubules | mex1-Gal4 | — | — | — | + | — | This study |
| NP3084-Gal4 | + | — | — | + | — | This study | |
| esg-Gal4 | + | — | — | + | — | This study | |
| dl-Gal4 | + | — | — | + | + | This study | |
| Su(H)GBE-Gal4 | + | — | — | + | + | This study | |
| myo31D-Gal4 | + | — | — | + | — | (Weaver and Drummond-Barbosa 2019) | |
| c42-Gal4 | + | — | — | + | — | This study | |
| Uro-Gal4 | — | + | — | — | — | This study | |
| Muscle and brain | MHC-Gal4 | — | + | — | — | — | (Weaver and Drummond-Barbosa 2019) |
| mef2-Gal4 | + | + | — | +a | — | This study | |
| nSyb.P-Gal4 | + | — | — | + | — | This study | |
| nSyb.S-Gal4 | + | — | — | — | — | (Weaver and Drummond-Barbosa 2019) | |
| repo-Gal4 | + | — | — | + | — | This study | |
| ChAT-Gal4 | + | — | — | — | — | This study | |
| Sensory neurons | pebbled-Gal4 | + | — | — | + | + | This study |
| Gr5a-Gal4 | + | — | — | — | — | This study | |
| Gr66a-Gal4 | + | — | — | — | — | This study | |
| Ir8a-Gal4 | + | — | — | — | — | This study | |
| Ir25a-Gal4 | + | — | — | — | — | This study | |
| Or83b-Gal4 | + | — | — | — | — | This study | |
| ppk-Gal4 | + | — | — | — | — | This study | |
| Fat body | adh-Gal4 | + | n.d.b | + | + | + | (Armstrong et al. 2014) |
| cg-Gal4 | + | n.d. | + | — | + | (Armstrong et al. 2014) | |
| FB-Gal4 | — | n.d. | + | + | — | (Armstrong et al. 2014) | |
| 3.1Lsp2-Gal4 | — | n.d. | + | — | — | (Armstrong et al. 2014) | |
| r4-Gal4 | + | n.d. | + | + | + | (Armstrong et al. 2014) | |
| ppl-Gal4 | — | n.d. | + | + | — | (Armstrong et al. 2014) | |
| PromE800-Gal4 | — | — | + | — | — | (Weaver and Drummond-Barbosa 2019) |
Expression in visceral muscle surrounding gut.
n.d., not determined.
Figure 1.
Expression patterns of hh-Gal4 lines in the adult female ovary. Expression of UAS-nucGFP or UAS-mCD8::GFP induced by “niche” drivers hhMB-Gal4 (A), hhTX-Gal4 (B), and hhJF-Gal4 (C). GFP (green); DAPI (blue), nuclei. Scale bars: 10 µm (germarium); 50 µm (ovariole). Arrowheads point to GFP expression in escort cells (yellow) or later follicle cells (white).
Figure 2.
Expression patterns of hh-Gal4 lines in additional adult female tissues. Expression of UAS-nucGFP or UAS-mCD8::GFP induced by “niche” drivers hhMB-Gal4 (A), hhTX-Gal4 (B), and hhJF-Gal4 (C). GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut). Dashed lines separate sections of the gut. Foregut (FG); midgut (MG); hindgut (HG). Yellow arrowheads point to GFP expression in the hindgut.
Ovary Gal4 drivers are expressed in additional tissues in adult females
In addition to hh-Gal4, other Gal4 drivers are used for specific expression in other cell types found in the adult ovary. Of the drivers we tested, almost all showed expression either outside of the ovary or in an additional unreported ovarian cell type (Figures 3 and 4, Table 2). For example, the cap cell and escort cell driver bab1-Gal4 (also known as babAgal4-5) drove robust expression of UAS-nucGFP in the cap cells and escort cells (Figure 3) as reported (Cabrera et al. 2002), but also showed strong GFP expression in the brain and midgut (Figure 4A). Although not tested in our study, an additional bab1-Gal4 line (babPgal4-2) (Cabrera et al. 2002) has also been generated and should be carefully characterized in future studies. The escort cell driver ptc-Gal4 (Forbes et al. 1996) induced GFP in ovarian escort cells as previously reported (Figure 3); however, this driver also showed expression in late stage follicle cells (Figure 3, yellow arrowheads), and in some brain cells (Figure 4A, white arrowhead) and the gut (Figure 4A). The escort cell driver c587-Gal4 (Zhu and Xie 2003; Hsu and Drummond-Barbosa 2009) showed GFP expression in the reported ovarian cell types (Figures 3 and 4B), but showed additional expression in the brain and fat body (Figure 4A), and occasional late stage follicle cells (Figure 3, yellow arrowhead). Finally, the follicle cell driver tj-Gal4 showed GFP expression in the brain and fat body (Figure 4A) in addition to its reported expression in ovarian follicle cells (Figure 3). These results indicate that commonly used ovary drivers have additional sites of expression in multiple tissues in adult females. To determine whether an effect in the ovary is indeed cell type specific, it will be important to rule out potential roles of additional tissues in which these drivers are expressed.
Figure 3.
Expression patterns of commonly used ovary Gal4 drivers in adult female ovaries. Expression of UAS-nucGFP induced by the cap cell and escort cell driver bab1-Gal4, escort cell driver ptc-Gal4, escort cell driver c587-Gal4, or follicle cell driver tj-Gal4. GFP (green); DAPI (blue), nuclei. Scale bars: 10 µm (germarium); 50 µm (ovariole). Yellow arrowheads point to GFP expression driven by ptc-Gal4 or c587-Gal4 in follicle cells.
Figure 4.
Ovary Gal4 driver expression patterns in additional adult female tissues. Expression of UAS-nucGFP induced by the cap cell and escort cell driver bab1-Gal4, escort cell driver ptc-Gal4, escort cell driver c587-Gal4, or follicle cell driver tj-Gal4. GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut). Arrowheads point to some GFP expressing brain cells (white) and adipocytes (yellow). Dashed lines separate sections of the gut. Foregut (FG); midgut (MG); hindgut (HG). (B) Expression of UAS-nucGFP induced by c587-Gal4 in combination with nSyb-Gal80 showing lack of GFP expression in the brain. GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain), 10 µm (germarium).
Gut, muscle, and Malpighian tubule drivers are expressed in multiple tissues in adult females
We previously confirmed that in adult females the myo31DFNP0001-Gal4 driver (Regan et al. 2016) is largely specific for the visceral muscle surrounding the midgut, and showed additional slight expression in the brain (Weaver and Drummond-Barbosa 2019) (Table 2). In addition, NP3084-Gal4 (Hayashi et al. 2002) drove expression of UAS-mCD8::GFP in the gut as reported (Nehme et al. 2007), but also drove expression in the brain (Figure 5A). By contrast, expression of UAS-nucGFP under control of the enterocyte driver mex1-Gal4 (Phillips and Thomas 2006) was restricted to the adult female gut with no GFP expression observed in other tissues (Figure 5A, Table 2). The commonly used ISC/enteroblast driver esg-Gal4 (Micchelli and Perrimon 2006) showed low levels of GFP in a few cells in the brain in addition to its expression in ISCs and enteroblasts (Figure 5A, Table 2), while both the ISC driver dl-Gal4 (Zeng et al. 2010) and the enteroblast driver Su(H)GBE-Gal4 (Zeng et al. 2010) showed expression in the brain and in follicle cells in the ovary in addition to their reported expression in the midgut (Figure 5A, Table 2). Although often overlooked, these additional sites of Gal4 expression are not surprising, given the known expression pattern/function of the genes whose regulatory regions control these Gal4 transgenes (Vässin et al. 1987; Schweisguth and Posakony 1992; Ashraf et al. 1999). For example, Dl was previously shown to be expressed in the follicle cells and the germline throughout oogenesis and is required for fertility (Ruohola et al. 1991). Experiments using these midgut cell type drivers for genetic manipulation of adult females should ideally include additional controls to rule out effects of gene manipulation in the brain or follicle cells. Alternatively, these drivers could be combined with tissue-specific Gal80 expression for suppression of Gal4 activity in the additional cell types that are not of interest to avoid confounding effects.
Figure 5.
Midgut and Malpighian tubule Gal4 driver expression patterns in adult Drosophila females. (A) Expression of UAS-mCD8::GFP induced by the midgut driver NP3084-Gal4. GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain); 50 µm (skeletal muscle); 50 µm (fat body); 250 µm (gut); 50 µm (ovariole). UAS-nucGFP induced by enterocyte driver mex1-Gal4, ISC/enteroblast driver esg-Gal4, ISC driver dl-Gal4, or enteroblast driver Su(H)GBE-Gal4. GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut); 50 µm (ovariole). All rows (except for top row) are shown at the same magnification for corresponding tissues. White arrowhead indicates some GFP expressing cells in the brain. (B) Malpighian tubule drivers c42-Gal4 and Uro-Gal4 expressing UAS-nucGFP. GFP (green); DAPI (blue), nuclei. Scale bars: 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut); 50 µm (ovariole). Yellow arrowheads indicate Malpighian tubules. Dashed lines separate sections of the gut. Midgut (MG); hindgut (HG).
We also examined the expression patterns of two Malpighian tubule drivers and an additional muscle driver (Figure 5B, Figure 6A). The Malpighian tubule driver c42-Gal4 (Rosay et al. 1997) showed high nucGFP levels in both the Malpighian tubules and in parts of the brain, whereas Uro-Gal4 (Terhzaz et al. 2010) showed low expression of GFP in muscles in addition to its strong expression in Malpighian tubules (Figure 5B, Table 2). We previously showed that MHC-Gal4 (Schuster et al. 1996) is specific for adult female skeletal muscle without expression in additional tissues (Weaver and Drummond-Barbosa 2019) (Table 2). Conversely, analysis of the commonly used mef2-Gal4 muscle driver (Ranganayakulu et al. 1998) shows robust expression in the brain in addition to skeletal and visceral (around the gut) muscles (Figure 6A, Table 2). These results suggest that when using drivers for Malpighian tubule-specific manipulation or mef2-Gal4 for muscle-specific experiments, the expression in additional tissues with these drivers should be either blocked with Gal80 or functionally evaluated using other drivers.
Figure 6.
Expression patterns of additional muscle and brain Gal4 drivers in adult females. (A) Expression of UAS-nucGFP induced by the muscle driver mef2-Gal4. The GFP expression observed in the gut of the mef2-Gal4 driver represents the visceral muscle. Arrowheads indicate visceral muscle (pink) and ovariole muscle sheath (white). (B) Expression of UAS-nucGFP induced by the glial cell driver repo-Gal4 and of UAS-mCD8::GFP induced by the cholinergic neuron driver ChAT-Gal4. GFP (green); DAPI (blue), nuclei. Scale bars, 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut); 50 µm (ovariole). Arrowheads indicate Malpighian tubules (yellow). (C) Expression of UAS-mCD8::GFP induced by the neuron drivers nSyb-Gal4.S and nSybGal4.P. GFP (green); DAPI (blue), nuclei. Scale bars, 100 µm (brain, skeletal muscle, fat body, and midgut); 50 µm (ovariole). Dashed lines separate sections of the gut. Foregut (FG); midgut (MG); hindgut (HG).
Commonly used sensory neuron drivers are highly specific in adult females
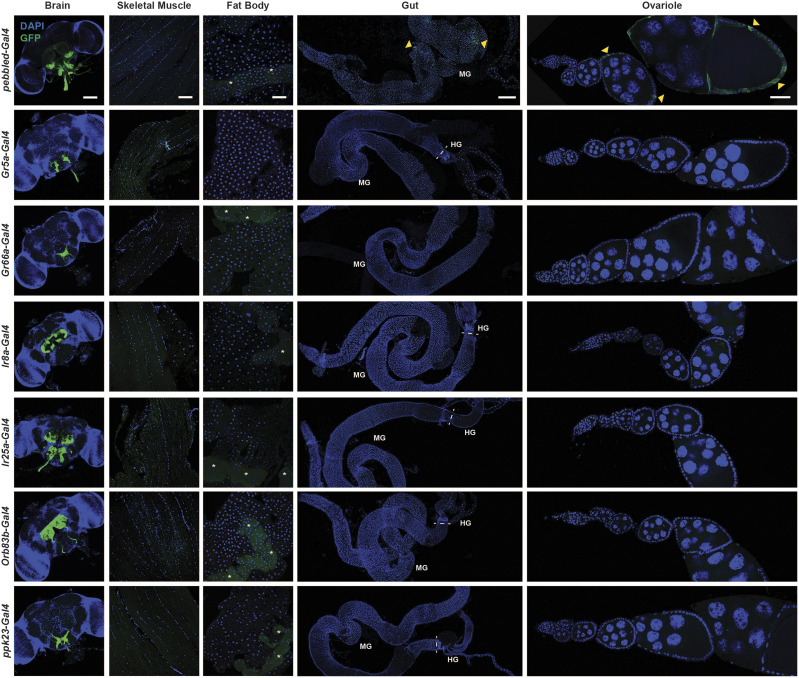
We previously confirmed that in adult females the pan-neuronal driver nSyb-Gal4.S (Pauli et al. 2008) is exclusively expressed in the brain (Weaver and Drummond-Barbosa 2019) (Table 2; also see Figure 6C). Similarly, the cholinergic neuron driver ChAT-Gal4 (Salvaterra and Kitamoto 2001) drives UAS-mCD8::GFP expression only in the brain (Figure 6B, Table 2). By contrast, the glial cell driver repo-Gal4 (Sepp et al. 2001) exhibits some nucGFP expression in the Malpighian tubules in addition to its reported expression in the brain (Figure 6B). In addition to these more broadly expressed brain drivers, we also analyzed multiple sensory neuron drivers using the UAS-mCD8::GFP reporter for their level of specificity (Figure 7). Most of the sensory neuron drivers tested showed highly specific expression in the brain, without additional expression in other tissues. These results are perhaps not surprising given that all sensory neuron drivers we tested are driven by small, gene-specific regulatory regions, ranging in size from 215 bp (Ir8a-Gal4) (Abuin et al. 2011) to 7.4 kb (ChAT-Gal4) (Salvaterra and Kitamoto 2001). The specialized functions of these genes, most of which encode olfactory and gustatory receptors (Chen and Dahanukar 2020), may also contribute to their specificity of expression. One exception was pebbled-Gal4, which showed additional expression in late ovarian follicle cells and in some cells in the gut (Figure 7). However, the expression pattern of pebbled-Gal4 is unsurprising given the known roles of pebbled in promoting the mitotic-to-endocycle switch in follicle cells and follicle cell differentiation (Sun and Deng 2007), and its known expression in the gut (Celniker et al. 2009). Collectively, these results suggest that neuronal drivers in general are more likely to be specifically expressed in neurons, perhaps in part due to the highly specialized nature of these cells. However, additional neuronal drivers still need to be tested to ensure that expression patterns are specific to their neuronal cell population of interest.
Figure 7.
Sensory neuron Gal4 driver expression in adult females. Expression of UAS-mCD8::GFP induced by sensory neuron drivers. GFP (green); DAPI (blue), nuclei. Scale bars, 100 µm (brain); 25 µm (skeletal muscle); 25 µm (fat body); 250 µm (gut); 50 µm (ovariole). The faint green fluorescence observed in the fat body images results from oenocyte autofluorescence (asterisks). Dashed lines separate sections of the gut. Midgut (MG); hindgut (HG).
Conclusions and other considerations for future studies
Many adult tissues produce systemic factors, including peptide hormones, lipids and other types of molecules to modulate the function of multiple tissues within an organism (Droujinine and Perrimon 2016; Castillo‐Armengol et al. 2019; Drummond-Barbosa 2019). Dissecting the complexity of inter-organ signaling networks requires reliable tools for tissue-specific genetic manipulation. This study highlights that many Gal4 drivers commonly used for tissue-specific manipulation of gene function have previously unreported additional sites of expression in adult Drosophila females. These findings are of concern to Drosophila researchers because expression of Gal4 drivers in multiple tissues can confound the interpretation of results aimed at evaluating tissue-specific effects of gene manipulation on a given tissue/biological process.
To ensure that manipulations are indeed tissue-specific, it is crucial to thoroughly test drivers and document their expression patterns broadly across Drosophila tissues according to the specifics of each study. For example, Gal4 expression patterns should be analyzed in specific developmental stages of interest (e.g., larvae vs. adults), in males vs. females, under the specific dietary conditions of the experiment, and in response to any additional physiological conditions considered during the course of a study. Simply put, it would not be wise to assume that the published expression pattern of any given Gal4 driver will remain the same under the specific experimental conditions of a particular study.
While many of the drivers tested are expressed in previously unreported tissues, there are known ways to eliminate expression in secondary tissues by using the Gal4 inhibitor Gal80 (Stoleru et al. 2005; Xie et al. 2018). For example, nSyb-Gal80 is routinely used in combination with Gal4 drivers to inhibit Gal4 specifically in neurons and allow UAS-GFP (or other transgene) expression only in the remaining tissue of interest (Rubinstein et al. 2010). In accordance, we successfully combined c587-Gal4 with nSyb-Gal80 to eliminate the neuronal expression observed in the brain with the c587-Gal4 driver alone, without affecting expression in the ovary (Figure 4B). Analogously, Su(H)GBE-Gal80 is commonly used to inhibit Gal4 in enteroblasts and thus restrict expression of esg-Gal4 to only ISCs (Wang et al. 2014). Evidently, any Gal4 driver could potentially be combined with cell type/tissue-specific Gal80 transgenes to limit Gal4 activity to desired target tissues. However, if a Gal80 transgene is not available for a specific tissue, effects from secondary tissues that express the Gal4 targeting the cell type/tissue of interest could be ruled out by using a separate Gal4 driver specific for that secondary cell type/tissue.
Alternatively, a combinatorial approach commonly used in the Drosophila neuroscience field to generate neuronal type-specific drivers can also be used more broadly to generate cell type/tissue-specific drivers. In this approach, the Gal4 transcription factor is subdivided into its DNA-binding domain (DBD) and its activating domain (AD), and only cells which express both of these components are able to produce a functional Gal4 to induce UAS transgene expression (Xie et al. 2018). By expressing DBD and AD under control of separate enhancers/promoters, it is possible to achieve expression in only the tissues where the expression pattern induced by the two regulatory regions overlap. For example, a truly Malpighian tubule-specific driver could be generated by combining c42-DBD with Uro-AD, since the only tissues in which these two promoters overlap are the Malpighian tubules (Figure 5B). These DBD and AD lines can be generated from existing Gal4 lines using Homology Assisted CRISPR Knock-in (HACK) (Lin and Potter 2016). HACK uses CRISPR-Cas9 technology to induce double-stranded breaks in Gal4 transgenes, which is repaired by a transgenic construct containing Gal4 homologous sequences flanking a cassette (e.g., DBD or AD) to replace the Gal4 transgene. This method has been successfully used to generate TH-AD, TH-DBD, and TH-Gal80 transgenic lines (Xie et al. 2018). Although more labor intensive, having highly specific tools or strategies to rule out effects from other tissues is highly advantageous as we strive for an accurate understanding of complex functional inter-organ relationships.
In addition to the promoter sequence directly upstream of Gal4, the site of the insertion of the Gal4 transgenes can also affect tissue-specific expression. For example, we previously reported that the nSyb-Gal4.S line (Pauli et al. 2008) is expressed only in neurons and in no other tissues (Weaver and Drummond-Barbosa 2019); however, a different nSyb-Gal4 line using the same regulatory sequence but generated by site specific insertion (nSyb-Gal4.P) (Riabinina et al. 2015) has additional expression in the gut (Figure 6C). Therefore, for Gal4 transgenes inserted in different sites along the genome (even under the same regulatory region), it is important to validate each line to ensure that there are no additional sites of expression due to the insertion site.
The UAS responder transgene should also be taken into consideration for tissue-specific manipulations. For example, it is well known that UASt transgenes (referred throughout this study as simply UAS) (Brand and Perrimon 1993) are strongly expressed in somatic cells but show limited, if any, expression in the female germline, whereas the UASp (Rørth 1998) and UASz (Deluca and Spradling 2018) transgenes have been optimized for expression in the female germline. When validating the Gal4 expression pattern of a driver, it would be advisable to use a reporter transgene built using the same UAS vector type as the UAS transgenes intended for experimental manipulations. To illustrate this point, we crossed the ubiquitous tub-Gal4 driver to UASp-lacZ or UAS-mCD8::GFP, which resulted in reporter expression predominantly in germ cells or exclusively in somatic cells in the germarium, respectively (Figure 8). Beyond that, we also documented that even distinct reporter lines built using the same UAS vector can also show differences in expression under control of the same Gal4 driver. For example, expression of UAS-mCD8::GFP with the hh-Gal4JF driver is most strongly expressed in the terminal filament and cap cells of the germarium, with weaker GFP signal in the escort cells (Figure 1). However, uniform expression across all three ovarian cell types was observed using UAS-nucGFP under control of hh-Gal4JF (Figure 1). Furthermore, both hh-Gal4MB and hh-Gal4TX were able to drive expression of UAS-nucGFP, but not of UAS-mCD8::GFP, in cap cells and escort cells (Figure 1), suggesting that these differences are possibly due to reporter insertion site. Indeed, differences in variegation occur due to differences in chromatin accessibility, which have been shown to alter Gal4 expression patterns (Tulin et al. 2002). Therefore, it would be ideal to confirm Gal4 expression patterns with reporters not only made using the same vector, but that also have the same insertion sites.
Figure 8.
Expression of UASp vs. UASt reporters differs in response to the ubiquitous tub-Gal4 driver. Expression of UASp-lacZ and UAS-mCD8::GFP induced by tub-Gal4, illustrating how reporter type can affect recognized Gal4 pattern. β-gal (green); GFP (green); DAPI (blue), nuclei. Scale bar, 10 µm.
Finally, as mentioned above, changes in the external environment or physiology (such as diet, age, infection, temperature, or other stressors) can potentially alter the expression strength or pattern of a driver. For example, expression of the 3.1Lsp2-Gal4 driver (Lazareva et al. 2007) on a yeast-free diet is dramatically reduced compared to that on a yeast-rich diet (Armstrong et al. 2014). In addition, although the UAS/Gal4 system itself shows temperature dependence even in the absence of Gal80ts (Brand et al. 1994), it is also possible that the regulatory regions driving Gal4 might respond in different ways to more subtle changes in temperature than those that activate heat-shock-inducible-Gal4, for instance (Brand et al. 1994). In addition to considering that common manipulations such as changes in diet can alter the expression of Gal4 drivers used for genetic manipulations, one should also evaluate the potential effects of the genetic manipulations themselves on driver expression over the course of the experiment.
Acknowledgments
We thank the Bloomington Stock Center (National Institutes of Health P400D018537), Michael Buszczak, Erika Matunis, Chris Potter, Mark Wu, and Ting Xie for Drosophila stocks. We are grateful to members of the Drummond-Barbosa lab for critical reading of the manuscript. The analyses reported in this paper are described in the main figures and tables. This work was supported by National Institutes of Health (NIH) grants R01 GM069875 (D.D.-B) and R01 GM125121 (D.D.-B.), and Pathway to Independence Award K99 GM127605 (L.N.W.). T.M. was supported by training grant T32 CA009110.
Footnotes
Communicating editor: H. Salz
Literature Cited
- Abuin L., Bargeton B., Ulbrich M. H., Isacoff E. Y., Kellenberger S. et al. , 2011. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69: 44–60. 10.1016/j.neuron.2010.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. M. H., Maldera J. A., Krunic D., Paiva-Silva G. O., Penalva C. et al. , 2020. Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature 584: 415–419. 10.1038/s41586-020-2462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameku T., Yoshinari Y., Texada M. J., Kondo S., Amezawa K. et al. , 2018. Midgut-derived neuropeptide F controls germline stem cell proliferation in a mating-dependent manner. PLoS Biol. 16: e2005004 10.1371/journal.pbio.2005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong A. R., and Drummond-Barbosa D., 2018. Insulin signaling acts in adult adipocytes via GSK-3beta and independently of FOXO to control Drosophila female germline stem cell numbers. Dev. Biol. 440: 31–39. 10.1016/j.ydbio.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong A. R., Laws K. M., and Drummond-Barbosa D., 2014. Adipocyte amino acid sensing controls adult germline stem cell number via the amino acid response pathway and independently of Target of Rapamycin signaling in Drosophila. Development 141: 4479–4488. 10.1242/dev.116467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S. I., Hu X., Roote J., and Ip Y. T., 1999. The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J. 18: 6426–6438. 10.1093/emboj/18.22.6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A.H, A.S. Manoukian, and N. Perrimon, 1994 Ectopic Expression in Drosophila. Methods Cell Biology 44:635–54. DOI: 10.1016/s0091-679x(08)60936-x [DOI] [PubMed] [Google Scholar]
- Brand A. H., and Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Cabrera G. R., Godt D., Fang P. Y., Couderc J. L., and Laski F. A., 2002. Expression pattern of Gal4 enhancer trap insertions into the bric a brac locus generated by P element replacement. Genesis 34: 62–65. 10.1002/gene.10115 [DOI] [PubMed] [Google Scholar]
- Castillo‐Armengol J., Fajas L., and Lopez‐Mejia I. C., 2019. Inter‐organ communication: a gatekeeper for metabolic health. EMBO Rep. 20: e47903 10.15252/embr.201947903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S. et al. , 2009. Unlocking the secrets of the genome. Nature 459: 927–930. 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. D., and Dahanukar A., 2020. Recent advances in the genetic basis of taste detection in Drosophila. Cell. Mol. Life Sci. 77: 1087–1101. 10.1007/s00018-019-03320-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S. Z., and Spradling A. C., 2018. Efficient Expression of Genes in the Drosophila Germline Using a UAS Promoter Free of Interference by Hsp70 piRNAs. Genetics 209: 381–387. 10.1534/genetics.118.300874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., and Hawthorne D. C., 1966. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics 54: 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine I. A., and Perrimon N., 2016. Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu. Rev. Genet. 50: 539–570. 10.1146/annurev-genet-121415-122024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D., 2019. Local and Physiological Control of Germline Stem Cell Lineages in Drosophila melanogaster. Genetics 213: 9–26. 10.1534/genetics.119.300234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliason J., Afify A., Potter C., and Matsumura L., 2018. A GAL80 Collection To Inhibit GAL4 Transgenes in Drosophila Olfactory Sensory Neurons. G3 (Bethesda) 8: 3661–3668. 10.1534/g3.118.200569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S., Shalaby N. A., and Buszczak M., 2011. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 108: 7064–7069. 10.1073/pnas.1015874108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A. J., Spradling A. C., Ingham P. W., and Lin H., 1996. The role of segment polarity genes during early oogenesis in Drosophila. Development 122: 3283–3294. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Ito K., Sado Y., Taniguchi M., Akimoto A. et al. , 2002. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34: 58–61. 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- Hsu H. J., and Drummond-Barbosa D., 2009. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl. Acad. Sci. USA 106: 1117–1121. 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., and Cooley L., 2014. Methods for studying oogenesis. Methods 68: 207–217. 10.1016/j.ymeth.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T. T., Shepherd D., Murphy C. et al. , 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L., and Drummond-Barbosa D., 2005. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309: 1071–1073. 10.1126/science.1111410 [DOI] [PubMed] [Google Scholar]
- Lazareva A. A., Roman G., Mattox W., Hardin P. E., and Dauwalder B., 2007. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 3: e16 10.1371/journal.pgen.0030016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., and Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lin C. C., and Potter C. J., 2016. Editing Transgenic DNA Components by Inducible Gene Replacement in Drosophila melanogaster. Genetics 203: 1613–1628. 10.1534/genetics.116.191783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liu S., Kodama L., Driscoll M. R., and Wu M. N., 2012. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol. 22: 2114–2123. 10.1016/j.cub.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J., and Spradling A., 1995. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121: 3797–3807. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Armstrong A. R., Sampson L. L., Laws K. M., and Drummond-Barbosa D., 2017. Adipocyte Metabolic Pathways Regulated by Diet Control the Female Germline Stem Cell Lineage in Drosophila melanogaster. Genetics 206: 953–971. 10.1534/genetics.117.201921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., and Davis R. L., 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Meschi E., Leopold P. and Delanoue R., 2019. An EGF-Responsive Neural Circuit Couples Insulin Secretion with Nutrition in Drosophila. Dev Cell 48: 76–86.e5. [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., and Perrimon N., 2006. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475–479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H., Volohonsky G., Reuveny A., Zaidel-Bar R., and Volk T., 2002. Two Isoforms of the Drosophila RNA Binding Protein, How, Act in Opposing Directions to Regulate Tendon Cell Differentiation. Dev. Cell 2: 183–193. 10.1016/S1534-5807(01)00118-6 [DOI] [PubMed] [Google Scholar]
- Nehme N. T., Liegeois S., Kele B., Giammarinaro P., Pradel E. et al. , 2007. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3: e173 10.1371/journal.ppat.0030173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Chen S., Weng C., Call G., Zhu D. et al. , 2007. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell 1: 458–469. 10.1016/j.stem.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Pauli A., Althoff F., Oliveira R. A., Heidmann S., Schuldiner O. et al. , 2008. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 14: 239–251. 10.1016/j.devcel.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. D., and Thomas G. H., 2006. Brush border spectrin is required for early endosome recycling in Drosophila. J. Cell Sci. 119: 1361–1370. 10.1242/jcs.02839 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G., Elliott D. A., Harvey R. P., and Olson E. N., 1998. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development 125: 3037–3048. [DOI] [PubMed] [Google Scholar]
- Regan J. C., Khericha M., Dobson A. J., Bolukbasi E., Rattanavirotkul N. et al. , 2016. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife 5: e10956 10.7554/eLife.10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M. N. et al. , 2015. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods 12: 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1998. Gal4 in the Drosophila Female Germline. Mech. Dev. 78: 113–118. 10.1016/S0925-4773(98)00157-9 [DOI] [PubMed] [Google Scholar]
- Rosay P., Davies S. A., Yu Y., Sözen M. A., Kaiser K. et al. , 1997. Cell-type specific calcium signalling in a Drosophila epithelium. J. Cell Sci. 110: 1683–1692. [DOI] [PubMed] [Google Scholar]
- Rubinstein C. D., Rivlin P. K., and Hoy R. R., 2010. Genetic feminization of the thoracic nervous system disrupts courtship song in male Drosophila melanogaster. J. Neurogenet. 24: 234–245. 10.3109/01677063.2010.519805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola H., Bremer K. A., Baker D., Swedlow J. R., Jan L. Y. et al. , 1991. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66: 433–449. 10.1016/0092-8674(81)90008-8 [DOI] [PubMed] [Google Scholar]
- Sahai-Hernandez P., and Nystul T. G., 2013. A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development 140: 4490–4498. 10.1242/dev.098558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaterra P. M., and Kitamoto T., 2001. Drosophila cholinergic neurons and processes visualized with Gal4/UAS–GFP. Gene Expr. Patterns 1: 73–82. 10.1016/S1567-133X(01)00011-4 [DOI] [PubMed] [Google Scholar]
- Schuster C. M., Davis G. W., Fetter R. D., and Goodman C. S., 1996. Genetic Dissection of Structural and Functional Components of Synaptic Plasticity. I. Fasciclin II Controls Synaptic Stabilization and Growth. Neuron 17: 641–654. 10.1016/S0896-6273(00)80197-X [DOI] [PubMed] [Google Scholar]
- Schweisguth F., and Posakony J. W., 1992. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell 69: 1199–1212. 10.1016/0092-8674(92)90641-O [DOI] [PubMed] [Google Scholar]
- Sepp K. J., Schulte J., and Auld V. J., 2001. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev. Biol. 238: 47–63. 10.1006/dbio.2001.0411 [DOI] [PubMed] [Google Scholar]
- Sieber M. H., and Spradling A. C., 2015. Steroid Signaling Establishes a Female Metabolic State and Regulates SREBP to Control Oocyte Lipid Accumulation. Curr. Biol. 25: 993–1004. 10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Owusu-Ansah E., Hu Y., Cheng D., Ni X. et al. , 2017. Activin signaling mediates muscle-to-adipose communication in a mitochondria dysfunction-associated obesity model. Proc. Natl. Acad. Sci. USA 114: 8596–8601. 10.1073/pnas.1708037114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Nawathean P., and Rosbash M., 2005. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438: 238–242. 10.1038/nature04192 [DOI] [PubMed] [Google Scholar]
- Sun J., and Deng W. M., 2007. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12: 431–442. 10.1016/j.devcel.2007.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney L. B., Couto A., Chou Y. H., Berdnik D., Dickson B. J. et al. , 2007. Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron 53: 185–200. 10.1016/j.neuron.2006.12.022 [DOI] [PubMed] [Google Scholar]
- Tanimoto H., Itoh S., ten Dijke P., and Tabata T., 2000. Hedgehog Creates a Gradient of DPP Activity in Drosophila Wing Imaginal Discs. Mol. Cell 5: 59–71. 10.1016/S1097-2765(00)80403-7 [DOI] [PubMed] [Google Scholar]
- Terhzaz S., Cabrero P., Chintapalli V. R., Davies S. A., and Dow J. A., 2010. Mislocalization of mitochondria and compromised renal function and oxidative stress resistance in Drosophila SesB mutants. Physiol. Genomics 41: 33–41. 10.1152/physiolgenomics.00147.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A., Stewart D., and Spradling A. C., 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16: 2108–2119. 10.1101/gad.1003902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vässin H., Bremer K. A., Knust E., and Campos-Ortega J. A., 1987. The neurogenic gene Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. EMBO J. 6: 3431–3440. 10.1002/j.1460-2075.1987.tb02666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zeng X., Ryoo H. D., and Jasper H., 2014. Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet. 10: e1004568 10.1371/journal.pgen.1004568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Singhvi A., Kong P., and Scott K., 2004. Taste representations in the Drosophila brain. Cell 117: 981–991. 10.1016/j.cell.2004.06.011 [DOI] [PubMed] [Google Scholar]
- Weaver L. N., and Drummond-Barbosa D., 2018. Maintenance of Proper Germline Stem Cell Number Requires Adipocyte Collagen in Adult Drosophila Females. Genetics 209: 1155–1166. 10.1534/genetics.118.301137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. N., and Drummond-Barbosa D., 2019. The nuclear receptor seven up functions in adipocytes and oenocytes to control distinct steps of Drosophila oogenesis. Dev. Biol. 456: 179–189. 10.1016/j.ydbio.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Ho M. C. W., Liu Q., Horiuchi W., Lin C. C. et al. , 2018. A Genetic Toolkit for Dissecting Dopamine Circuit Function in Drosophila. Cell Rep. 23: 652–665. 10.1016/j.celrep.2018.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Chauhan C., and Hou S. X., 2010. Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in drosophila. Genesis 48: 607–611. 10.1002/dvg.20661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. H., and Xie T., 2003. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development 130: 2579–2588. 10.1242/dev.00499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Drosophila strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.