Abstract
The aim of the present study is focused on the decolorization and degradation of azo dyes Ponceau S Red and Methyl Orange by a bacterial strain isolated from the gold mining district of San Martin de Loba, South of Bolivar (Colombia) sediment samples and identified as Franconibacter sp. 1MS (GenBank: MT568543) based on phenotypic and genotypic methods. A higher percentage of decolorization at 100 mg/L concentration, 37 °C, and pH 7 was recorded at 120 h of incubation period for both dyes. The UV–vis, Fourier transform infrared spectroscopy, and gas chromatography–mass spectrometry analysis of the original dyes and their degraded metabolites confirmed that the decolorization was due to degradation. The proposed metabolic pathways for biodegradation of both dyes have been elucidated, which showed the formation of five intermediate metabolites, namely, N,N-dimethylbenzyl-1,4-diamine, sulfonamide, 1,4-diaminobenzene, 2,5-diaminobenzenesulfonic acid, and 1-amino-2-naphthol, which are not only highly toxic but also be able to be converted through metabolic activation into mutagenic, carcinogenic, and/or teratogenic species. The phytotoxicity studies of the original dye and degraded metabolites were tested on Phaseolus vulgaris and divulged that the degraded metabolites have toxic effects. An effective phytostimulation was observed in Ponceau S Red, which could be attributed to its capacity for enrichment of the culture medium with essential nutrients, a favorable environment for the growth of the plant.
Introduction
Environmental pollution has increased in recent years due to the generation of pollutants from different industrial sectors, such as the paper, leather, fertilizer, food, cosmetic, pharmaceutical, and textile industries.1 These industries worldwide discharge large volumes of colored wastewater effluents with high levels of azo dyes (between 100 and 250 mg/L) daily.
The textile sector is the largest generator of contaminated water due to its high consumption and use; it is estimated that approximately 20% of the industrial pollution of water is attributed to it.2 Approximately 10,000 textile dyes are commercially available worldwide, and their annual output is equivalent to 7 × 105 metric tons, of which 10–15% is discharged into water sources during the textile coloring process.3 The World Health Organization (WHO) has established safe limits for the discharge of colored effluents into the environment. However, 2% of these are discharged directly without adequate prior treatment.4 An additional aggravating factor is the incorporation of heavy metals (cadmium, chromium, cobalt, copper, iron, nickel, lead, and mercury) as mordants in industrial processes. According to the Ecological and Toxicological Association of Manufacturers of Organic Dyes and Pigments (ETAD) in the 2017 annual report, the possibility of a new approach for the evaluation of organic metals, pigments, and dyes is recommended, depending on their toxicological profile, biodegradability, and bioaccumulation at relatively low exposure levels.5,6
The detection of dyes and the byproducts of their transformation in wastewater demonstrates the inefficiency of the treatment processes.7 Dyes are detected in concentrations as low as 1 mg/mL, which brings an aesthetic problem; beyond color, it affects the photosynthetic activity of hydrophytes by reducing penetration of light in deep layers, deteriorates water quality, and reduces oxygen solubility, causing acute toxic effects on aquatic flora and fauna.8−10 Harmful effects on human and animal health have also been recognized as dysfunctions of the kidneys, reproductive system, liver, brain, central nervous system,11 tract irritation respiratory, eyes, skin, asthma, sore throat, and allergic contact dermatitis.12,13
Conventional treatment methods used like flocculation, irradiation, adsorption, precipitation, chemical oxidation, coagulation, electrolysis, and photodegradation have presented significant disadvantages such as limited versatility, high cost, low efficiency, mechanical resistance, difficulty in separation, poor resistance to acidic solutions, and interference with other components of wastewater.14 Unlike biological treatments, both aerobic and anaerobic or mixed that are effective, economical, eco-friendly, and efficient for dye degradation, bacteria are the prime focus due to their degradation and even fully mineralized many dyes when used as carbon or energy sources.15 Recent studies report their interest in metal-tolerant bacteria considered potential candidates for ex situ or in situ bioremediation studies due to their ability to accumulate, remove, concentrate, and recover heavy metals from industrial effluents and mine phages due to their ability to develop and adopt different enzymatic detoxification mechanisms, which improve mobility and easy flexibility at discharge sites.16 The biotransformation of some dyes can lead to the formation of molecules with high toxicological potential, such as aromatic amines.17 Studies have shown that azo dyes in clothing undergo biotransformation through the action of various skin bacteria, which leads to the possible release of aromatic amines, which can be absorbed through the skin.18 Products released in the biotransformation process include 40 different aromatic amines derived from approximately 180 azo dyes registered as mutagenic and 14 as carcinogenic.
The aim of the present study is focused on screening of bacteriological decolorization and degradation of azo dyes Ponceau S Red and Methyl Orange by the bacterial strain 1MS. Ponceau S Red and Methyl Orange are dyes containing one and four sulfonate groups, respectively, which means that they have a great capacity to be fully diffused and adsorbed in most environmental conditions.19 This not only evaluates their decolorization efficacy on various concentrations but also optimizes the physiological condition (pH and temperature) for effective decolorization. UV–visible spectrophotometry, Fourier transform infrared spectroscopy (FTIR), and gas chromatography/mass spectrometry (GC–MS) were used to characterize the intermediates of dyes degradation. The test of phytotoxicity allowed us to evaluate the effects of the original dyes and metabolites degraded on plant growth.
Results and Discussion
Isolation, Screening and Identification of Dye Decolorizing Microorganisms
A total of 11 morphologically distinct bacterial colonies were isolated from sediment samples collected in the gold mine located in the district of San Martin de Loba, South of Bolivar (Colombia) using the spread plate method on a nutrient agar supplemented with 100 mg/L of Methyl Orange and Ponceau S Red dyes. The isolated 1MS was selected for further studies due to the fact that it showed a vigorous growth and the highest decolorization ability in both dyes, Methyl Orange (DI: 8.56 cm ± 0.66) and Ponceau S Red (DI: 7.92 cm ± 0.86), within a short period of 24 h incubation, whereas other isolates were not able to form colorless zones, showed difficulties for the growth, or did not grow well above 100 mg/L. A pure culture was maintained on an LB agar and stored at 4 °C until used for further experiments.
The bacterial isolate 1MS was next characterized on the basis of their pheno-genotypic characteristics, which were analyzed according to the standard description of Bergey’s Manual of Determinative Bacteriology and the partial 16S rRNA gene sequence analysis.20 Microscopic examination revealed that the strain 1MS was a Gram-negative, rod-shaped, facultatively anaerobic, motile, and non-spore former. It showed positive response to catalase, urease, Voges–Proskauer, H2S production, and indole utilization and negative response to oxidase, citrate, coagulase, and methyl red. Also, it was able to ferment different carbohydrates like sucrose, d-arabitol and d-sorbitol, whereas it was unable to ferment d-mannitol.
Genotypic characterization of the strain 1MS based on partial 16S rRNA gene sequence analysis was carried out using BLASTn analysis and EzTaxon database, demonstrating that 1MS showed the closest match with various species of the genus Franconibacter. Franconibacter daqui strain AKB4-KU (accession number MN880102) and F. daqui DL503 (accession number NR159235) were the species closest with similarities of 98.97 and 99.44%, respectively. Phylogenetic analysis revealed that strain 1MS was related most closely to F. daqui DL503. On the basis of these results, the isolated 1MS was identified as Franconibacter sp. The nucleotide sequence was deposited in GenBank database under the accession number MT568543 (Figure 1).
Figure 1.
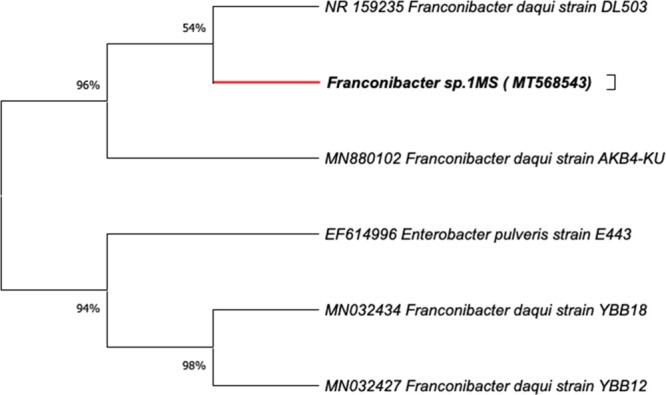
Phylogenetic analysis based on 16S rRNA gene sequence of the strain 1MS (Franconibacter sp.). Neighbor-joining method, bootstrap test (1000 replicates).
The evolutionary distances were computed using the maximum composite likelihood method. Recently, the genus Franconibacter has been described as a new genus of the Enterobacteriaceae family based on the taxonomic reclassification of Enterobacter and Cronobacter species.21 Currently, it comprises three species: F. daqui sp., F. pulveris, and F. helveticus.F. daqui sp., the newest member of the genus, was isolated from a sample of Daqui, which is a saccharifying agent used to initiate fermentation in the production of Chinese liqueurs and vinegars,22 whereas F. helveticus and F. pulveris are recognized as strains of great importance in food contamination isolated from fruit dust and their related environments.23 To the best of our knowledge, there was very little information available on the role of this species in catalytic activities and/or biologicals. Therefore, reported in this study for the first time is the potential of a member of this genus in the decolorization/degradation of dyes.
Decolorization Assay
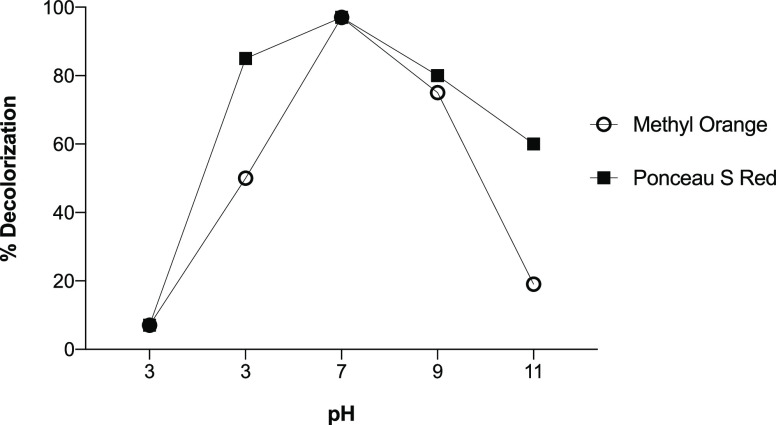
Dye decolorization was studied using a one-factor optimization approach:24 The effects of different pH values, temperatures, time of incubation, and concentrations of Methyl Orange and Ponceau S Red dyes on the rate of decolorization of Franconibacter sp., 1MS were evaluated. In pH assays, a percentage of decolorization of 96% for both dyes was observed at pH 7, whereas at a higher and lower pH, a decrease was observed (Figure 2). Therefore, this pH value was defined as the pH optimum for the activity of decolorization.
Figure 2.
Effect of pH on the decolorization of Methyl Orange and Ponceau S Red dyes using Franconibacter sp., 1MS.
According to Chang et al.,25 the effect of pH may be related to the transport of dye molecules across the cell membrane, which is considered a limiting step in the rate of discoloration. At a pH below 4, H+ ions compete effectively with those dye cations, causing a decrease in color removal efficiency, while at a higher pH above this loading point, the biomass surface is negatively charged, attracting the dye cations that are positively charged through the electrostatic force of attraction. This behavior was observed in our study; at pH 3, the percentage of discoloration was less (6–7%), while at higher pH levels such as pH 5, 9, and 11, they reached values greater than 50%. Interestingly, at pH 1,1 Methyl Orange dye showed a rather marked decrease in the percentage of decolorization (14%).
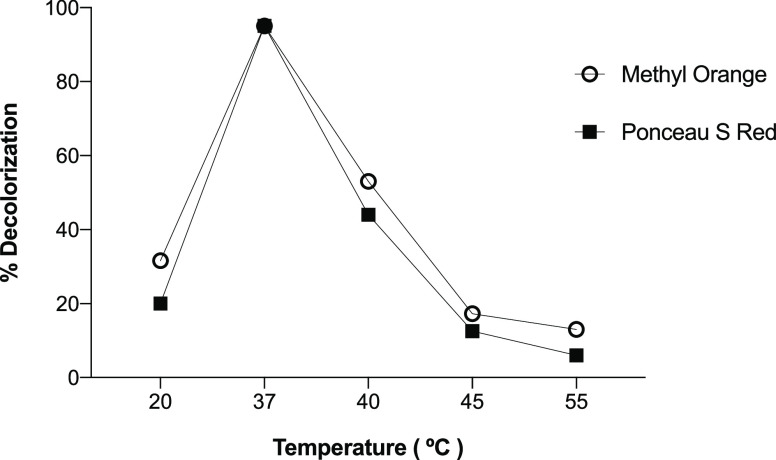
To evaluate the influence of the temperature on the decolorization of the dyes, the experiment was performed under the pH value optimum (pH 7). It was observed that at 37 °C, the percentage of decolorization was higher (94%) than those achieved with the other temperatures tested (20, 40, 45, and 55 °C). However, values above 45 °C led to a significant loss of decolorization, decreasing up to 13% for Methyl Orange and 6% for Ponceau S Red dye (Figure 3). This fact could be due to the high temperatures that can cause unfavorable activity, causing the inactivity of proteins and cellular structures such as the membrane intervening in the redox potential, while the decolorization rate increases with the increase in temperature before reaching the optimum temperature, as described by Zhuang et al.26
Figure 3.
Effect of temperature on the decolorization of Methyl Orange and Ponceau S Red dyes using Franconibacter sp., 1MS.
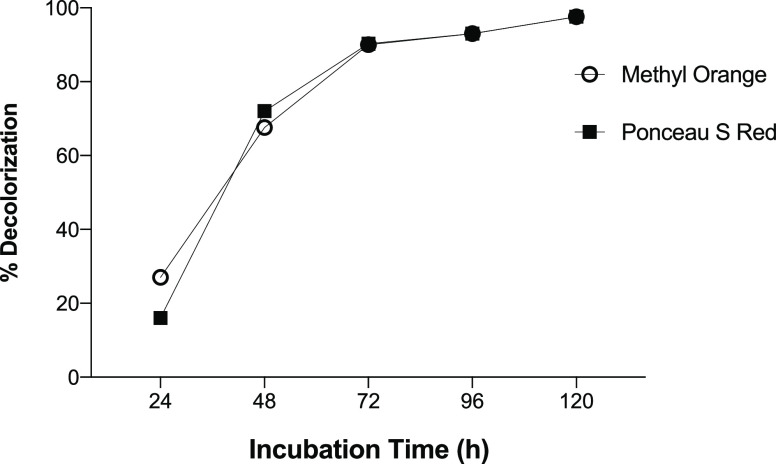
Under the optimal conditions of temperature (37 °C) and pH (7), the effect of incubation time on decolorization of the dyes was carried out at 100 mg/L over a period of 120 h (Figure 4). In this assay, the maximum decolorization was reached only after 120 h for both dyes. However, a percentage of decolorization greater than 80% was observed after 72 h, which can be attributed to a greater metabolic rate and growth of Franconibacter sp. 1MS, which it has helped in fast reduction of dye chromophore.
Figure 4.
Effect of incubation time on the decolorization of Methyl Orange and Ponceau S Red dyes using Franconibacter sp., 1MS.
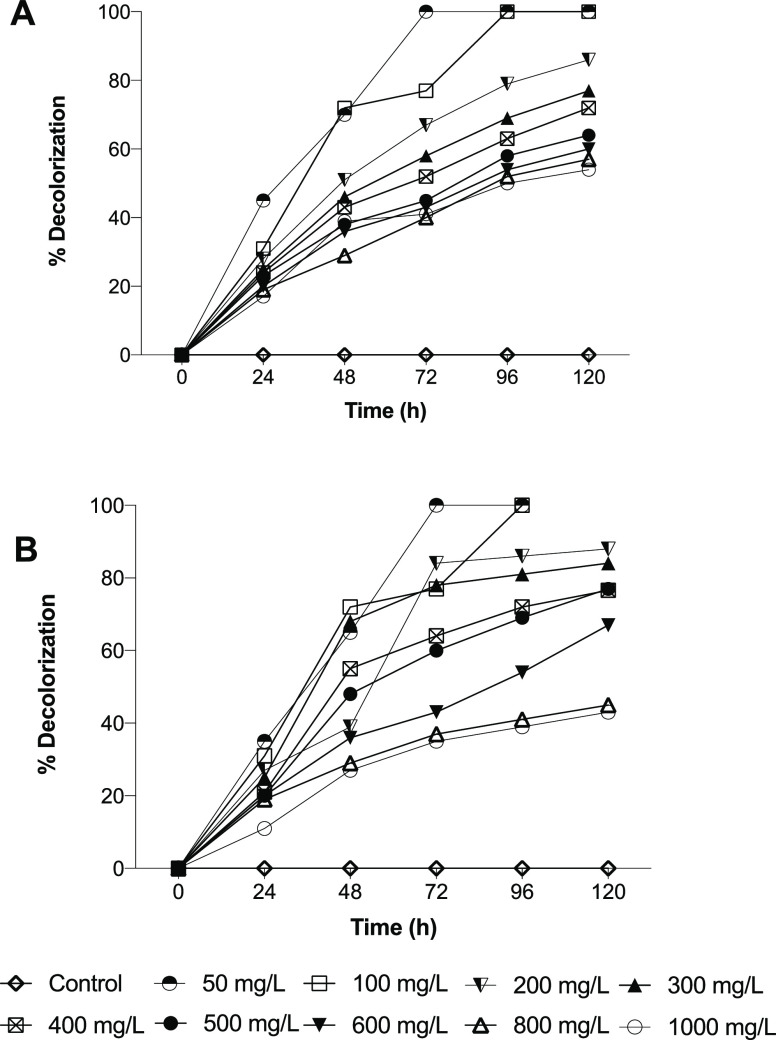
Later, the effect of dye concentration on the decolorization using Franconibacter sp., 1MS was studied in static cultures incubated at 37 °C and pH 7 for 120 h (Figure 5). Franconibacter sp., 1MS evidenced that it is able to decolorize in a wide range of concentrations of 50–1000 mg/L in the two dyes evaluated. However, a decrease in the percentage of decolorization was observed as the dye concentration increased. It is interesting to note that when the concentration of dyes is more than 500 mg/L, the percentage of decolorization is 10 times lower that when 50 mg/L was tested. This decrease may be due possibly to the loss of the cell viability, a lower reproduction rate, a loss of enzyme activity, or toxicity of the dye itself.27,28
Figure 5.
Effect of dyes concentration on the decolorization using Franconibacter sp., 1MS. (A) Methyl Orange dye. (B) Ponceau S red dye. The data in the figure is represented as the mean of three experiments ± standard error.
However, for industrial applications, it is important to know if the microorganism has the ability to withstand high concentrations of the compound since the concentrations of dyes in an industrial effluent can vary between 10 and 50 mg/L.29 In this sense, Franconibacter sp., 1MS meets this characteristic. Decolorization of dyes is a process that is affected by parameters such as pH, temperature, agitation, and incubation time as shown in this study. Other factors, as the complexity of dye, type of enzyme, and use of mediators have been described by several authors. Although the azo dyes used in this study were sulfonated polar dyes, both were efficiently decolorized by Franconibacter sp., 1MS. The percentage of decolorization obtained for Methyl Orange is very similar to other reports carried out with this dye. Zhuang et al.26 reported values of 95% for the degradation using Shewanella ST2 strain, Oceanimonas smirnovii strain ST3, and Enterococcus faecalis strain ST5 evaluated at pH 5–9, setting that the variation in the percentage of decolorization depends on its chemical structure and the intrinsic properties of dyes and bacterial isolate.
On the other hand, in the case of Ponceau S Red, few reports describing decolorization tests using biological methods. Gómez et al.30 reported a degradation greater than 90% using anaerobic sludge. Other studies realized by Meena et al.31 indicated a percentage of decolorization by 99% using photocatalysis, which managed to degrade the dye. In general, the variation in decolorization activity can be associated with the differences in their structure and complexity of the dyes, particularly in the nature and position of substitution groups on aromatic rings and the presence, number, and position of the sulfonated radicals.32 Our results evidence that Ponceau S Red has the highest reduction rate compared to Methyl Orange; this may be due to the presence of the four −SO3Na substituents in its structure unlike the Methyl Orange that only has one, suggesting that the presence of sulfonated groups contribute to a higher degradation dye. In addition, Ponceau S Red dye possesses a hydroxyl group in the ortho position to the azo group, which allows azoreductases to have higher relative activity facilitating the cleavage of the azo bonds. Meanwhile, the presence of methyl groups in the structure of Methyl Orange dye can produce a steric interference and greater difficulty for azoreductase enzymes to form enzymatic substrate complexes with the dye causing the loss of their activity.11
A number of observations were made during the process of decolorization. We observed that the bacterial cells remained colored after 12 h of incubation, indicating that the degradation mechanism would correspond to bio-adsorption; but after 18 h, the cells exposed to the two dyes had no coloration (retention of the original color of the colony), which indicates enzymatic a biodegradation process in both dyes. Analytical studies confirmed the biodegradation of Methyl Orange and Ponceau S Red dyes and the formation of their intermediary metabolites.
Dyes and Degraded Product Analysis
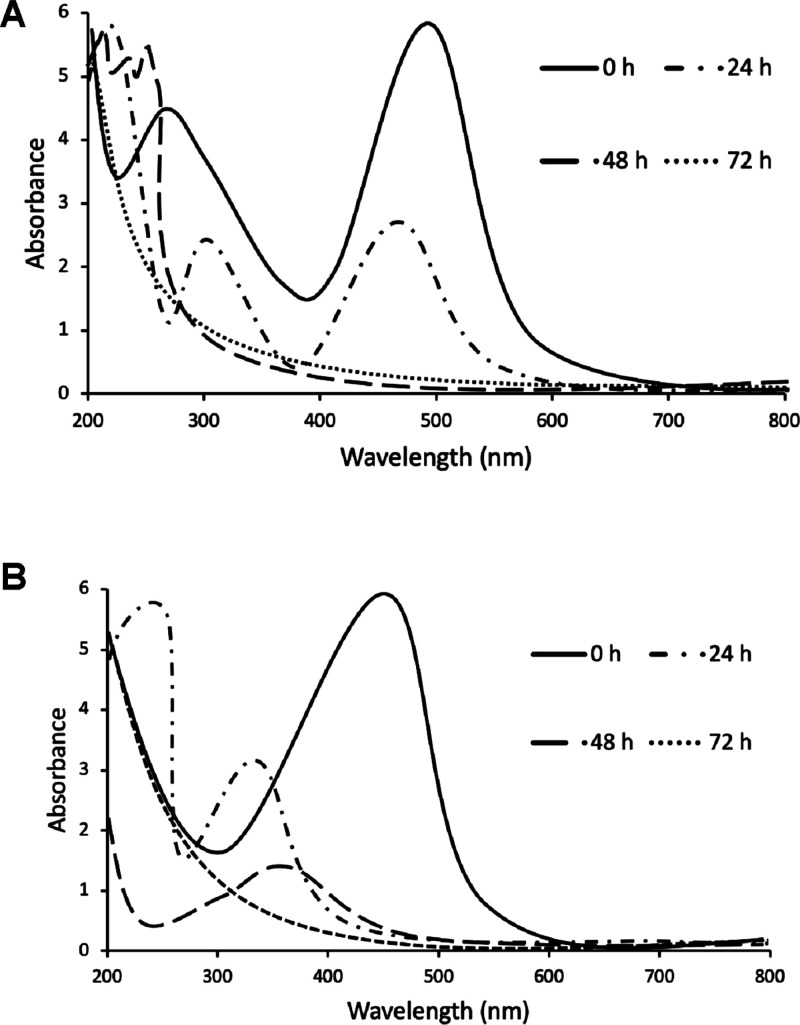
The microbial decolorization process performed by Franconibacter sp., 1MS was analyzed through the spectral changes visualized by UV–vis spectroscopy (Figure 6). During the degradation reaction, the original peak of the absorbance in the region visible to the Methyl Orange was observed at 470 nm at time 0 and decreased without change in λ max. In the same way, it was observed for the case of the Ponceau S red dye at 520 nm. These peaks are attributed to the n → π * transitions of the N=N, C=N, and C=O chromophores. Other peaks at 320 and 200 nm are attributed to π → π * transitions in aromatic rings that affect the aromatic character of dyes.33
Figure 6.
UV–vis spectra of decolorization of (A) Methyl Orange and (B) Ponceau S red dyes at a concentration of 100 mg/L.
The original peak subsequently disappeared, indicating that the chemical bond had been completely broken. The results were consistent with the loss of color in the solution, which indicated that the chromophore group (−N–N−) had degraded. Solid changes in the peaks and the appearance of new peaks in the range of 200–350 nm of the spectrum evidenced that the intermediate products are produced after the chromophores of the dye macromolecules were broken. These results can be attributed to the characteristic peaks of the formation of aromatic amines and sulfonate groups that were absorbed in the region of 260–300 and at 220–230 nm, respectively. However, they were no observed peaks in the visible region (400–700 nm), characteristic of nitro-substituted amines. These differences in the spectral range of 240–350 nm demonstrate the degradation of the original dyes to their metabolic products as reported by Sinha et al.34 and Pinheiro et al.35
Fourier-Transform Infrared (FTIR) Analysis
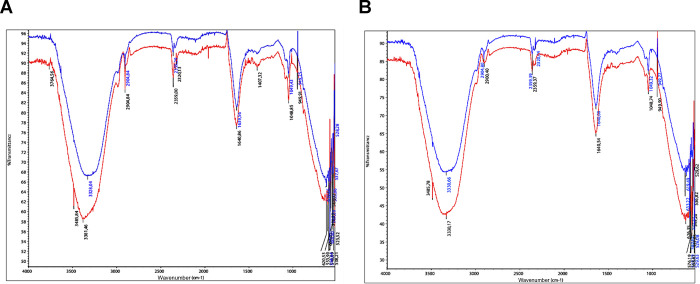
Comparison of FTIR spectrums of original dyes with their products after decolorization for each dye is shown in Figure 7. The spectrum of original dye of Methyl Orange showed a peak at 1407.32 cm–1, which suggests a N–H curve in the azo group, which disappeared in the spectrum of degraded products (Figure 7A). The peak at 2320.13 cm–1 attributed to the C–H stretching was observed only in the original dye. The formation of aromatic amines was attributed to the displacement of the peak at 3381.46 cm–1 in the original dye to 3328.04 cm–1 bands in the spectra of the degradation products, corresponding to the stretching vibration and plane bending vibration of the N–H primary amine bands. In addition, a peak at 1639.56 cm–1 was observed, which is associated with the C=C stretching vibration of the aromatic rings. These results indicated the breakage of azo bonds and the formation of aromatic amines. The peak observed at 1048.85 cm–1 in the original dye represents the stretching of sulfites; the peaks 620.53, 538.21, and 606.05 cm–1 were significantly reduced in the degraded dye, observing the peaks at 619.66, 537.67, and 594.64 cm–1. These changes observed in the IR spectrum of degraded products suggest a degradation of Methyl Orange by Franconibacter sp., 1MS.
Figure 7.
FTIR spectrum of original dyes (red) and their degraded metabolites (blue). (A) Methyl Orange dye. (B) Ponceau S Red dye.
The FTIR spectrum of Ponceau S Red dye and its degraded products is shown in Figure 7B. The peak at 1640.54 cm–1 decreased to 1640.08 cm–1, indicating the reduction of the azo bond. The peaks at 2900.40 and at 2359.37 cm–1 present in the original dye were reduced in the degraded products to 2904.80 and 2359.39 cm–1, respectively; these represent the stretching of the C–H and C–N bonds. A new peak in products degraded at 2320.04 cm–1 was also observed. The presence of aromatic amines was attributed to a shift of the 3485.78 and 3330.17 cm–1 peaks present in the dye to 3330.66 cm–1 in the degraded one. The observed peak at 1048.74 cm–1 was reduced in the degraded products to 1049.32 cm–1, which is associated with the sulfite stretching. Finally, the peaks 626.35, 576.19, and 549.20 cm–1 were reduced significantly in the degraded dye, with peaks being observed at 619.49, 571.57—, and 548.39 cm–1. The results were analyzed and interpreted with the criteria reported by Skong et al.36
Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
The products of the degradation process of Methyl Orange and Ponceau S Red dyes by Franconibacter sp., 1MS were extracted by liquid–liquid extraction and analyzed by gas chromatography coupled to GC–MS mass spectrometry. The chromatogram of the extracted metabolites suggested the formation of multiple degradation products, which is reported based on the values of the mass spectrum and the fragmentation pattern. A peak on the chromatogram at 9.445 min with a charge ion mass standard equal to 136 m/z was present in Methyl Orange and was assigned to the metabolite N,N-dimethylbenzyl-1,4-diamine. In the case of Ponceau S Red, a peak was identified at 8.834 min, and 117 m/z was assigned to the metabolite indole as is depicted in Figure 8. GC–MS analysis revealed the presence of several peaks corresponding to low molecular mass aromatic compounds produced during degradation of both dyes.
Figure 8.
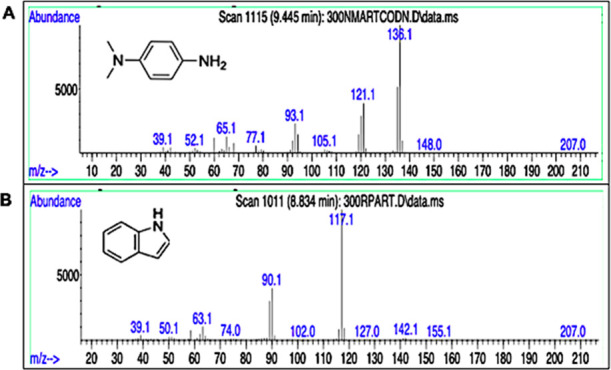
GC–MS spectral data of biodegradation products of Franconibacter sp. (A) Methyl Orange dye. (B) Ponceau S Red dye.
Pathway of Degradation
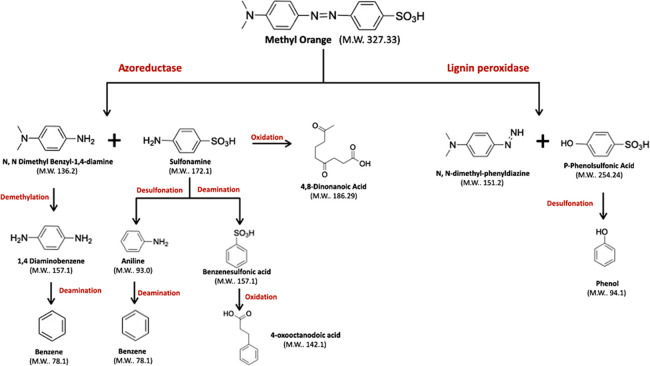
According to the data in this study, the relevant literature and the results found through computational studies using the enzyme-catalyzed reaction pathway prediction server of microorganisms and plants PathPred (https://www.genome.jp /tools/pathpred/),37 a bacterial decolorization/degradation route of Franconibacter sp., 1MS was proposed for Methyl Orange and Ponceau S Red dyes as shown in Figures 9 and 10. For Methyl Orange, Franconibacter sp., 1MS breaks at the azo bond of the dye, which leads to its fragmentation, resulting in the formation of sulfonamide and N,N-dimethylbenzyl-1,4-diamine. This cleavage by reduction of the azo bond, catalyzed by the enzyme azoreductase, is considered the first step in the microbial decolorization process. Then, the sulfonamide can take two routes: (i) a desulfonation that results in aniline or a deamination that results in benzenesulfonic acid or (ii) the sulfonamide is transformed into 4-oxooctanodoic acid. The N,N-dimethylbenzyl-1,4-diamine product could undergo demethylation resulting in formation of 1,4 diaminobenzene (Figure 9).
Figure 9.
Biodegradation pathway proposed for Methyl Orange by Franconibacter sp., 1MS.
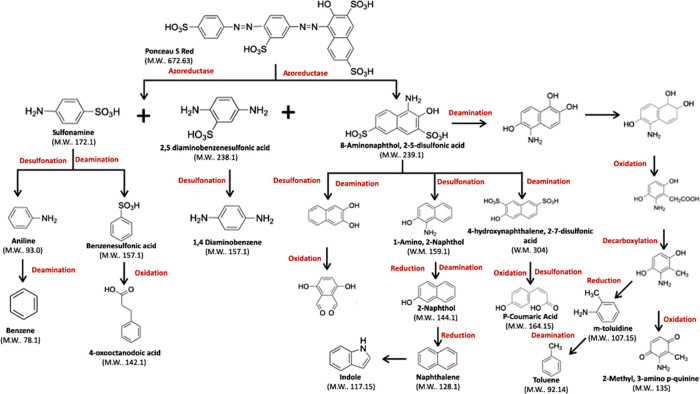
Figure 10.
Biodegradation pathway proposed for Ponceau S by Franconibacter sp., 1MS.
The suggested route for Ponceau S Red dye begins with the breakdown of azo bonds by azoreductase that generates three compounds, sulfonamide, 2,5-diaminobenzenesulfonic acid, and 8-aminonaphthol-2,5-disulfonic acid. Sulfonamide is degraded to the same form as in the Methyl Orange, 2,5-diaminobenzene sulfonic acid undergoes desulfonation resulting in a 1,4-diaminobenzene product, and finally 8-amino naphthol-2,5-disulfonic acid can take four routes as shown in Figure 10.
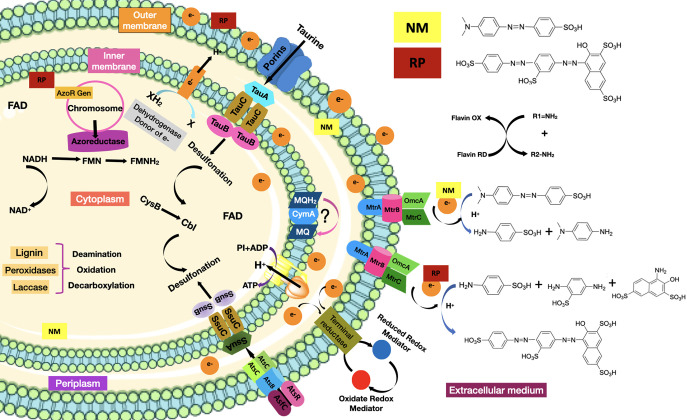
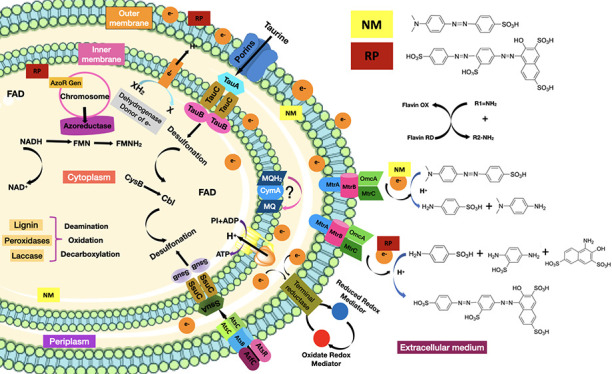
According to the above discussion, we could speculate that Methyl Orange and Ponceau S Red dyes are recognized by the bacterial cell Franconibacter sp., 1MS in the extracellular medium, which induces the transcription and translation of the azoR gene, giving rise to the cytoplasmic enzyme FMN azoreductase dependent on NADH.38 At pH 7 and temperature 37 °C, they penetrate the cell membrane. In the cytoplasm, upon oxidation of NADH, the hydride is transferred to FMN present at the active site of the azoreductase enzyme, which donates the electrons, producing the rupture of azo bonds (−N=N−) and consequently cause the discoloration of the solution, indicating the effective destruction of the chromophore and the generation of the products of the degradation. Subsequently, an oxidative desulfonation and deamination resulting in the synthesis of various intermediate metabolites, whatever they are, are used as substrates in different metabolic pathways, until their final degradation to CO2 and H2O. However, the dyes also could get into the cell although the route-specific transport system for other types of sulfonated substrates known as tauABCD and ssuEADCB39 allowed their uptake (absorption) in the bacterial cell. In the case of the absorption of aromatic sulfonates, it is catalyzed by the AtsBC system. On the other hand, if the extracellular environment is aerobic, then oxygen prevents reduction of the azo dye due to preferential oxidation of the oxygen-reduced redox mediator at the dye site. Also, it has been demonstrated that discoloration is proven to be an extracellular reduction process that requires a multi-component electron transfer pathway consisting of cytoplasmic, periplasmic (MtrA), and outer membrane (OmcB) components,40 which were involved mainly to menaquinones and tetraheme quinol dehydrogenase (CymA) as the crucial components in this pathway, which then branch through a network of periplasmic cytochromes to three cytochromes of the outer membrane. According to Cai et al.,41S. oneidensis MR-1 follows this mechanism for degradation of Methyl Orange dye. Figure 11 shows the proposed mechanism for reducing sulfonated azo dyes using Franconibacter sp., 1MS. Moreover, further studies are needed for the elucidation and verification of these mechanisms.
Figure 11.
Proposed degradation mechanisms for Franconibacter sp., 1MS.
Phytotoxicity Study
Phytotoxicity study was carried out on seeds of Phaseolus vulgaris toward the dye (at a concentration of 100 mg/L) and their degraded products. The toxicity effects were recorded in terms of percentage germination inhibition (GI) and phytotoxicity percentage determined using the lengths of the plumule and radicle of the seeds of P. vulgaris in the water, control dye, and degradation products data shown in Table 1.
Table 1. Phytotoxicities of Dyes and Their Degradation Products by Franconibacter sp., 1MS.
| parametersd | control (water) | Methyl Orange (original dye) | Methyl Orange degradation products | Ponceau S Red (original dye) | Ponceau S Red degradation products |
|---|---|---|---|---|---|
| germination (%) | 100 | 93.3 | 20 | 100 | 26.6 |
| length of radicle (cm) | 3.8 ± 0.75 | 1.96 ± 0.2 | 0b,c | 5 ± 1.4 | 0b,c |
| length of plumule (cm) | 3.5 ± 0.13 | 2.5a ± 0.4 | 0.46b,c ± 0.3 | 3.73a ± 0.2 | 0.26b,c ± 0.3 |
| phytotoxicity (%) | 0 | 49.13 | 100 | 29.31 | 100 |
| germination index (%) | 100 | 47.47 | 0 | 129.31 | 0 |
Radicle and plumule lengths of seeds grown in the original dye are significantly different from those of seeds grown in distilled water by P < 0.001.
Radicle and plumule lengths of seeds grown in degraded products are significantly different from those of seeds grown in distilled water by P < 0.001.
Radicle and plumule lengths of seeds grown in degraded products are significantly different from those of seeds grown in original dye water by P < 0.001
Data are represented as the mean from three sets of experiments ± standard error. Results were analyzed using one-way analysis of variance (ANOVA) with the Tukey–Kramer multiple comparison test.
Phytotoxicity results suggested that degradation products of both dyes were more toxic to the seeds of P. vulgaris. On the contrary, no phytotoxic effect (93–100% germination) was observed for original dyes. A significant variation in the percentage of phytotoxicity with respect to the control in the treatment of P. vulgaris with the degradation products of both dyes was observed (Table 1). Methyl Orange dye showed a significant increase of 50% compared to seeds treated with distilled water (control), while a 30% increase was observed in Ponceau S Red dye.
Original dyes did not show any significant reduction in the germination percentage (GI). According to Nouren et al.,42 GI values less than 50% represent high toxicity, values between 50 and 80% represent moderate toxicity, and values above 80% represent absence of toxicity. In our study, dyes and their degradation products of Methyl Orange were highly phytotoxic. Unlike Ponceau S Red that evidenced a positive effect on germination, according to the parameters described by Lago-Vila et al.,43 which indicate a Gindex: of >110%, these are classified as species with a stimulation effect. Results could be attributed to the use Ponceau S Red as a source of carbon and energy and at the same time to the enrichment of the culture medium with essential nutrients such as potassium, phosphorous, nitrogen, and calcium among others, a favorable environment for the growth of the plant system as reported by Zimmermann et al44 and Bhattacharya et al.45 These positive effects (phytostimulators) were previously reported by Bhattacharya et al.45 in a study that clearly demonstrated the phytostimulatory effects of dyes on the growth of rice seeds.
Results suggest that biodegradation was inefficient in toxicity reduction. Methyl Orange and Ponceau S Red are sulfonated reactive dyes whose degradation products include sulfonated and unsulfonated aromatic amines, which are an important group of environmental pollutants that are toxic in nature.46,47 Our study identified the compounds by GC–MS: N,N-dimethylbenzyl-1,4-diamine, sulfonamide, 1,4-diaminobenzene, 2,5-diaminobenzenesulfonic acid, and 1-amino-2 naphthol as intermediates in the degradation pathway of both dyes, which are not only highly toxic as it is evidenced in Table 2 but also able to be converted through metabolic activation into mutagenic, carcinogenic, and/ or teratogenic species as is indicated by Qu et al.48
Table 2. Toxicity Status of the Chemical Moieties Formed after Biodegradation of Methyl Orange and Ponceau S Red Dye.
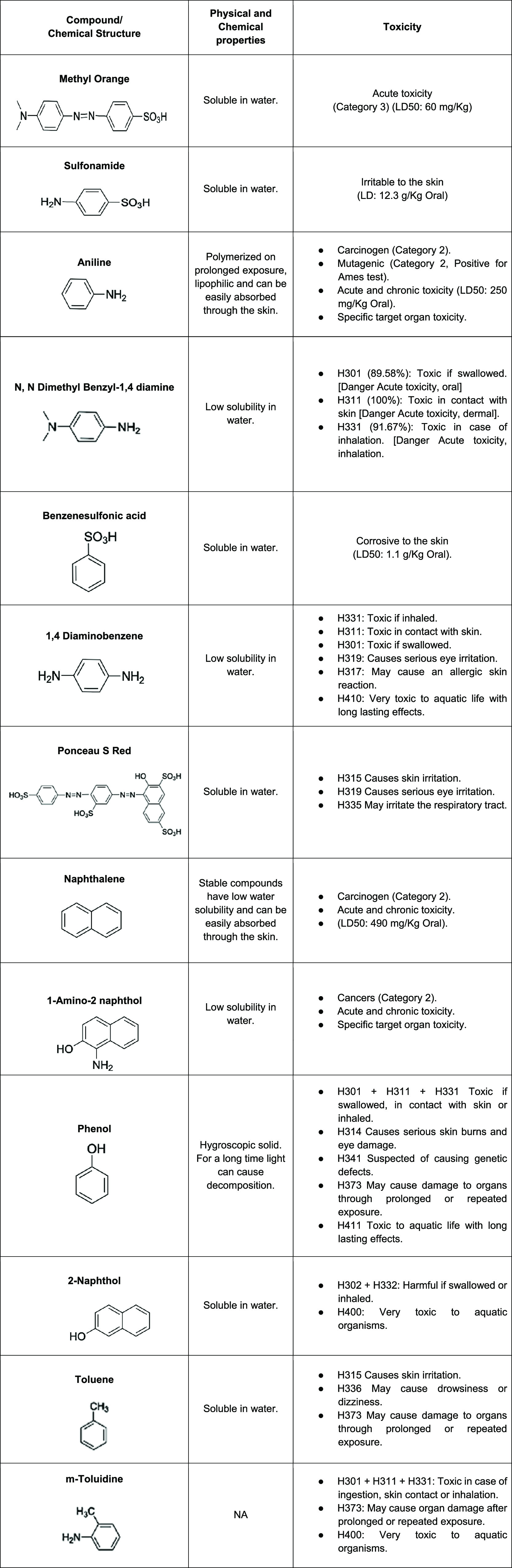
Franconibacter sp., 1MS could be inhibited by these intermediaries and not finish the detoxification, indicating that degradation products were still toxic in some cases of the original dye as emphasized by Rawat et al.49 and Bergsten-Torralba et al.50 According to Kalyani et al.,51 it is of great concern to evaluate the phytotoxicity and microbial toxicity of the dye before and after degradation, given the use of untreated and treated textile effluents that are reported in the agricultural area, and its direct impact on soil fertility. Furthermore, effluents from untreated textiles can cause serious health and environmental risks.
On the other hand, previous reports have shown that azo dye-degrading bacterial strains are sensitive to high salt concentrations, and the metal content in textile effluents can inhibit biodegradation of azo dyes by indigenous bacterial strains due to inhibition of cell growth and their metabolic activities, making treatment times of the process not practical.52,53 Therefore, the use of azo dye-degrading bacteria that are tolerant to extreme environmental conditions remains an effective strategy to improve wastewater treatment systems and improve bioremediation of azo dyes. This study corresponds to the first report of dye degradation in the genus Franconibacter. Based on these results and the genomic potential shown by the F. pulveris DJ34 strain, a member of its genus related to the degradation of crude oil, transport and resistance of metals, reduction of sulfate and nitrate, and synthesis of bio-surfactants among others,54Franconibacter sp., 1MS could be recognized as a species that showed potential characteristics to be studied as a possible candidate in the design of strategies for bioremediation processes.
Conclusions
The bacterial strain was identified as Franconibacter sp., 1MS; this strain can degrade the dyes Methyl Orange and Ponceau S Red as presumably non-toxic; in addition to its discoloration at different dimensions, the degradation of the dyes was not specific because the bacterial strain follows several symmetrical degradation pathways to produce toxic byproducts that cause environmental toxicity at the molecular, cellular, and organismal levels. The strain has potential for discoloration and degradation of dyes under static conditions (pH 7) and temperature (37 °C) through the process of degradation rather than adsorption. However, the strain simply decolorized/degraded the colorants, but did not detoxify them, and generated products such as aromatic amines that are extremely toxic as evidenced in the phytotoxicity study; it is necessary to demonstrate the efficacy of the strain for its application in the treatment of real samples.
Experimental Section
Isolation, Screening, and Identification of Dye Decolorizing Microorganisms
Sediments collected in the gold mine located in the district of San Martin de Loba, South of Bolivar (Colombia) of about 0.1 g was serially diluted in distilled water unto 10–6 dilution. Then, 100 μL of each dilution was inoculated on the nutrient agar plates enriched with 100 mg/L of Ponceau S Red and Methyl Orange by the spread plate technique. The plates were incubated at 37 °C for 72 h. Morphologically distinct colony bacterial isolates with clear zones were selected and repeatedly streaked to get a pure culture on nutrient agar plates. Potential decolorizing bacterial strains were selected based on the decolorization index calculated as the ratio of the total diameter (colony + halo zone)/colony diameter size (cm). Following this, the isolate giving the largest zone of decolorization was characterized through phenotypic and genotypic means. The selected isolate was phenotypically characterized based on its cultural, morphological, and biochemical characteristics by comparing the test results with the standard descriptions given in Bergey’s Manual of Determinative Bacteriology.20 The characterization genotyping was performed based on the partial 16S rRNA gene sequence analysis. DNA genomic was extracted using the DNeasy blood & tissue kit (Qiagen) following the manufacturer’s instructions and used for subsequent polymerase chain reaction (PCR) analysis. A 1.5 Kb 16S rRNA fragment was amplified using the universal primers 27F and 1492R according to the methodology described by Weisburg et al.55 PCR reactions were conducted in 25 μL aliquots containing 50 ng of template DNA, 10 pM of each primer, DreamTaq PCR master mix (2) (Thermo Fisher Scientific, Waltham, MA, USA), and water. The mixture reaction was subjected to denaturation at 95 °C for 5 min followed by 35 cycles consisting of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min and a final extension of 72 °C for 10 min. The PCR products were separated on 1.5% agarose gels in TBE buffer stained with ethidium bromide (0.5 mg/mL) and visualized under UV light. The PCR product was purified and sequenced. The sequences obtained were compared using the BlastN algorithm of NCBI (http://www.ncbi.nih.gov/BLAST/) and EzBioCloud database (https://www.ezbiocloud.net/) for finding the closest homology for the isolate. The phylogenetic reconstruction was performed using the neighbor-joining (NJ) algorithm. The evolutionary distances were computed using the maximum composite likelihood method. The bootstrap testing was done with 1000 replicate samples using MEGA X software.
Decolorization Assay
Dye decolorization was studied using the one-factor optimization approach: The influence of various culture conditions such as pH (3, 5, 7, 9, and 11), temperature (20, 37, 40, 45, and 50 °C), the time of incubation (24, 48, 72, 96, and 120 h), and concentration (50, 100, 200, 300, 400, 500, 600, 800, and 1000 mg/L) of dyes on the rate of decolorization was studied. When investigating one of these factors, all others were kept constant.
Initially, the absorption maxima (kmax) of each dye was checked in a decolorizing medium through scanning the absorption of light within the visible range (200–800 nm) at an interval of 10 nm using a UV–vis spectrophotometer.56 The wavelength reflecting the highest optical density when evaluating the supernatant (10,000 g, 15 min at 4 °C) was regarded as its corresponding maximum wavelength (λ max). The theoretical value of (λ max) reported by the literature for the dyes tested are λ max = 464 for Methyl Orange and λ max = 517–523 nm for Ponceau S Red.
All decolorization experiments were carried out in an LB broth enriched with dye concentration. The solutions of work were prepared containing a bacterial inoculum of 10% (v/v); OD = 1.5 × 108 cells/mL at 600 nM. After 24, 48, 72, 96, and 120 h of incubation, the culture broth was centrifuged (10,000 g, 15 min at 4 °C), and the absorbance of the supernatant was registered at the corresponding kmax of each dye solution. An inoculated dye-free medium was used as a blank. The percentage of decolorization (%D) was calculated according to the formula given by Chen et al.11 All assays were performed in triplicate, and the average values were used in calculations.
| 1 |
where A0 is the absorbance of the dye solution before decolorization, and At is the absorbance of the dye solution after decolorization.
Dyes and Degraded Product Analysis
To analyze the dye degradation by the isolate bacterial 1MS, the culture solutions were harvested at 48 h and then were centrifuged. The supernatants were used for further analysis. The degradation analysis was divided into three experimental parts: (i) measurement of the UV–visible spectrum of the supernatant, (ii) assay of the infrared spectrum of the supernatant by the Fourier transform infrared spectrometer (FTIR), and (iii) identification of degradation metabolites using GC–MS analysis.
UV–Visible Spectrophotometric Analysis
The dye and its degraded metabolites obtained after extraction suspended in water were studied using a UV–visible spectrophotometer to investigate the change in UV–visible spectra before and after decolorization. A supernatant of 2 mL was taken, and an absorption spectrum from 200 to 800 nm was recorded using a UV–visible spectrophotometer.57
Fourier-Transform Infrared (FTIR) Analysis
The dye and its degraded metabolites were subjected to FTIR analysis directly applied to a diamond crystal of ATR, and the resulting spectra of them were corrected for background air absorbance. The spectra were recorded, and samples were measured in the region 4000–400 cm–1. Each spectrum was measured 80 times at resolution 4. The differences in peaks between the dye sample and the degraded dye sample were observed and registered.
Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
Degraded products were centrifuged at 15,000 rpm for 15 min and subsequently filtered through 0.45 mm filters. A liquid–liquid extraction was carried out using acetonitrile and NaCl, which were used in equal volumes. GC–MS was performed with a 7890A mass spectrometer (Agilent Technologies). The injection volume for analysis was 1 mL The analytical column HP-5MS (30 mm × 0.25 mm, 0.25 μm) was used. The initial column temperature range was 80 °C for 1.0 min, then it increased linearly at 5 °C/min to 250 °C and held for 5 min. The helium carrier flow rate was 1 mL/min. The chromatograph was equipped with a reverse-phase glacial acetic acid, methanol, and ultra-pure water. Degradation products were identified by mass spectra and their fragmentation pattern.
Pathway of Degradation
The pathway of azo dyes degradation has been proposed on the basis of results of GC–MS in this study, the relevant literature, and enzyme systems involved using the prediction server of enzyme-catalyzed reaction pathways of microorganisms and plants: PathPred (https://www.genome.jp/tools/pathpred/) and the Integrated Microbial Genomes & Microbiomes system v.5.0 (IMG/M: https://img.jgi.doe.gov/m/). All structures were drawn in the ChemDraw program (https://chemdrawdirect.perkinelmer.cloud/js/sample/index.htlm).
Phytotoxicity Study
The phytotoxicity test was performed to evaluate the toxicity of original dyes and their degradation metabolites (ethyl acetate extracted and dried). This assay was carried out on P. vulgaris following the methodology described by Jadhav et al.58 with few modifications. For this experiment, a set of three experimental groups were prepared: the (i) original dye group, (ii) degradation metabolite group, and (iii) control group (distilled water). Each unit experiment consists of 10 seeds. Seeds of each group were sterilized using 1–5% sodium hypochlorite (15 min) and then washed five times with sterile water. The seeds were placed on different sterile Petri dishes containing a sterile circular filter paper. A 2 mL aliquot of distilled water containing 300 mg/L original dye or its degradation metabolites (300 mg/L) were used to soak the seeds. A control test was carried out using distilled water at the same time. All dishes were kept in laboratory temperature (28 °C). After 5 days, the germination rate differences were recorded, then shoot length and root length were measured and documented. Data are represented as the mean of 10 sprouted seeds from three sets of experiments ± standard error. Germination inhibition (GI), germination index (Gindex), and percentage of phytotoxicity were calculated according to Lago-Vila et al.43 and Roy et al.,59 respectively.
Germination inhibition (GI) was calculated as:
| 2 |
where SRc is the seed germinated average in the control, and SRs is the seed germination average in the sample.
The Gindex was determined according to the equation:
| 3 |
where Gs and Ls are seed germination (%) and root elongation (mm), respectively, while Gc and Lc are their corresponding control values.
The percentage of phytotoxicity was calculated as follows:
| 4 |
where l0 is the length of the root in the control, and lt is the length of the root of the study.
Statistical Analysis
All experiments were performed in triplicates, and results mentioned are the mean average of them. The obtained results were analyzed through one-way analysis of variance (ANOVA) Tukey–Kramer comparison test. Statistical analyses were performed using the statistical package Prism 7.02 GraphPad software. (CA N/A.) p < 0.05 indicates statistical significance.
Acknowledgments
The authors are grateful to University of Cartagena and Comfenalco Technological University Foundation for continuous support to their research group.
The authors declare no competing financial interest.
References
- Ghimpusan M.; Nechifor G.; Nechifor A.-C.; Dima S.-O.; Passeri P. Case studies on the physical-chemical parameters’ variation during three different purification approaches destined to treat wastewaters from food industry. J. Environ. Manage. 2017, 203, 811–816. 10.1016/j.jenvman.2016.07.030. [DOI] [PubMed] [Google Scholar]
- Holkar C. R.; Jadhav A. J.; Pinjari D. V.; Mahamuni N. M.; Pandit A. B. A critical review on textile wastewater treatments: possible approaches. J. Environ. Manage. 2016, 182, 351–366. 10.1016/j.jenvman.2016.07.090. [DOI] [PubMed] [Google Scholar]
- Asad S.; Amoozegar M. A.; Pourbabaee A. A.; Sarbolouki M. N.; Dastgheib S. M. M. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour. Technol. 2007, 98, 2082–2088. 10.1016/j.biortech.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Kono H. Cationic flocculants derived from native cellulose: Preparation, biodegradability, and removal of dyes in aqueous solution. Resour.-Effic. Technol. 2017, 3, 55–63. 10.1016/j.reffit.2016.11.015.. [DOI] [Google Scholar]
- çetin D.; Dönmez S.; Dönmez G. The treatment of textile wastewater including chromium(VI) and reactive dye by sulfate-reducing bacterial enrichment. J. Environ. Manage. 2008, 88, 76–82. 10.1016/j.jenvman.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Haq F.; Butt M.; Ali H.; Chaudhary H. J. Biosorption of cadmium and chromium from water by endophytic Kocuria rhizophila: equilibrium and kinetic studies. Desalin. Water Treat. 2016, 57, 1–19958. 10.1080/19443994.2015.1109561. [DOI] [Google Scholar]
- da Silva Leite L.; de Souza Maselli B.; de Aragão Umbuzeiro G.; Nogueira R. F. P. Monitoring ecotoxicity of disperse red 1 dye during photo-Fenton degradation. Chemosphere 2016, 148, 511–517. 10.1016/j.chemosphere.2016.01.053. [DOI] [PubMed] [Google Scholar]
- Lalnunhlimi S.; Krishnaswamy V. Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Braz. J. Microbiol. 2016, 47, 39–46. 10.1016/j.bjm.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni B. D.; Patel U. D.; Agrawal A.; Ruparelia J. P. Application of BDD and DSA electrodes for the removal of RB 5 in batch and continuous operation. J. Water Process Eng. 2017, 17, 11–21. 10.1016/j.jwpe.2017.01.009. [DOI] [Google Scholar]
- Selvaraj D.; Leena R.; Kamal D. C. Toxicological and histopathological impacts of textile dyeing industry effuent on a selected teleost fish Poecilia reticulata. J Pharmacol. Toxicol. 2015, 3, 26–30. [Google Scholar]
- Chen K. C.; Wu J. Y.; Liou D. J.; Hwang S. C. J. Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 2003, 101, 57–68. 10.1016/S0168-1656(02)00303-6. [DOI] [PubMed] [Google Scholar]
- Gičević A.; Hindija L.; Karačić A.. Toxicity of Azo Dyes in Pharmaceutical Industry. In International Conference on Medical and Biological Engineering; 2019; 581-587, 10.1007/978-3-030-17971-7_88. [DOI] [Google Scholar]
- Kurade M. B.; Waghmode T. R.; Patil S. M.; Jeon B. H.; Govindwar S. P. Monitoring the gradual biodegradation of dyes in a simulated textile effluent and development of a novel triple layered fixed bed reactor using a bacterium- yeast consortium. Chem. Eng. J. 2017, 307, 1026–1036. 10.1016/j.cej.2016.09.028. [DOI] [Google Scholar]
- Li R.; Ning X.; Sun J.; Wang Y.; Liang J.; Lin M.; Zhang Y. Decolorization and biodegradation of the Congo red by Acinetobacter baumannii YNWH 226 and its polymer production’s flocculation and dewatering potential. Bioresour. Technol. 2015, 194, 233–239. 10.1016/j.biortech.2015.06.139. [DOI] [PubMed] [Google Scholar]
- Saleh Y. E.; Hazaa H. A. Decolorization of Congo Red dye by bacterial isolates. J. Ecol. Health Environ. 2017, 5, 41–48. 10.12785/jehe/050201. [DOI] [Google Scholar]
- Nanda M.; Kumar V.; Sharma D. K. Multimetal tolerance mechanisms in bacteria: The resistance strategies acquired by bacteria that can be exploited to ‘clean-up’ heavy metals contaminants from water. Aquat. Toxicol. 2019, 212, 1–10. 10.1016/j.aquatox.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Almeida E. J. R.; Corso C. R. Comparative study of toxicity of azo dye Procion Red MX-5B following biosorption and biodegradation treatments with the fungi Aspergillus niger and Aspergillus terreus. Chemosphere 2014, 112, 317–322. 10.1016/j.chemosphere.2014.04.060. [DOI] [PubMed] [Google Scholar]
- Brüschweiler B. J.; Merlot C. Azo dyes in clothing textiles can be cleaved into a series of mutagenic aromatic amines which are not regulated yet. Regul. Toxicol. Pharmacol. 2017, 88, 214–226. 10.1016/j.yrtph.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Tuttolomondo M. V.; Alvarez G. S.; Desimone M. F.; Diaz L. E. Removal of azo dyes from water by sol–gel immobilized Pseudomonas sp. J. Environ. Chem. Eng. 2014, 2, 131–136. 10.1016/j.jece.2013.12.003. [DOI] [Google Scholar]
- Bergey D. H.Bergey’s manual of determinative bacteriology; 8th ed., Buchanan R. E., Gibbons N. E., Eds.; 1974.
- Iversen C.; Forsythe S. J. Comparison of media for the isolation of Enterobacter sakazakii. Appl. Environ. Microbiol. 2007, 73, 48e52. 10.1128/AEM.01562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.; Su C.; Yang X.; Sun D.; Zeng C.; Chen M.; Hu W.; Zhang C. Franconibacter daqui sp. nov., a facultatively alkaliphilic species isolated from a Daqu sample. Int. J. Syst. Evol. Microbiol. 2017, 67, 4962–4966. 10.1099/ijsem.0.002358. [DOI] [PubMed] [Google Scholar]
- Stephan R.; Grim C. J.; Gopinath G. R.; Mammel M. K.; Sathyamoorthy V.; Trach L. H.; Chase H. R.; Fanning S.; Tall B. D. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2014, 3402. 10.1099/ijs.0.059832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova-Villegas L. G.; Cordova-Villegas A. Y.; Taylor K. E.; Biswas N. Response surface methodology for optimization of enzyme-catalyzed azo dye decolorization. J. Environ. Eng. 2019, 145, 04019013 10.1061/(ASCE)EE.1943-7870.0001513. [DOI] [Google Scholar]
- Chang J. S.; Chou C.; Lin Y. C.; Lin P. J.; Ho J. Y.; Hu T. L. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Water Res. 2001, 35, 2841–2851. 10.1016/S0043-1354(00)00581-9. [DOI] [PubMed] [Google Scholar]
- Zhuang M.; Sanganyado E.; Zhang X.; Xu L.; Zhu J.; Liu W.; Song H. Azo dye degrading bacteria tolerant to extreme conditions inhabit nearshore ecosystems: Optimization and degradation pathways. J. Environ. Manage. 2020, 261, 110222. 10.1016/j.jenvman.2020.110222. [DOI] [PubMed] [Google Scholar]
- Saratale R. G.; Saratale G. D.; Chang J. S.; Govindwar S. P. Bacterial decolorization and degradation of azo dyes: a review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. 10.1016/j.jtice.2010.06.006. [DOI] [Google Scholar]
- Solís M.; Solís A.; Pérez H.; Manjarrez N.; Flores M. Microbial decolouration of azo dyes: a review. Process Biochem. 2012, 47, 1723–1748. 10.1016/j.procbio.2012.08.014. [DOI] [Google Scholar]
- Megha V.; Meenakshi; Rai J. P. N. Optimization of different Parameters on Synthetic Dye Decolorization by Free and Immobilized Mucor hiemalis MV04 (KR078215). Res. J. Chem. Sci. Res. J. Chem. Sci. 2015, 5, 2231–2606. [Google Scholar]
- Gómez S. D. P. A.; Rojas J. A. A.; Duque M. E. G.; Trujillo J. M.; Bermúdez J. M. O.; Martínez G. D. J. R. Degradación del colorante Rojo Punzó por medio de lodos anaerobios. Nova 2010, 8, 229–236. 10.22490/2462944. [DOI] [Google Scholar]
- Meena R. C.; Pachwarya R. B.; Meena V. K.; Arya S. Degradation of textile dyes Ponceau S and Sudan IV using recently developed photocatalyst, immobilized resin Dowex-11. Am. J. Environ. Sci. 2009, 5, 444–450. [Google Scholar]
- Guadie A.; Tizazu S.; Melese M.; Guo W.; Ngo H.; Xia S. Biodecolorization of textile azo dye using Bacillus sp. strain CH12 isolated from alkaline lake. Biotechnol. Rep. 2017, 15, 92–100. 10.1016/j.btre.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W.; Nansheng D.; Helin H. Degradation mechanism of azo dye CI reactive red 2 by iron powder reduction and photooxidation in aqueous solutions. Chemosphere 2000, 41, 1233–1238. 10.1016/S0045-6535(99)00538-X. [DOI] [PubMed] [Google Scholar]
- Sinha A.; Lulu S.; Vino S.; Banerjee S.; Acharjee S.; Osborne W. J. Degradation of reactive green dye and textile effluent by Candida sp. VITJASS isolated from wetland paddy rhizosphere soil. J. Environ. Chem. Eng. 2018, 6, 5150–5159. 10.1016/j.jece.2018.08.004. [DOI] [Google Scholar]
- Pinheiro H. M.; Touraud E.; Thomas O. Aromatic amines from azo dye reduction: status review with emphasis on direct UV spectrophotometric detection in textile industry wastewaters. Dyes Pigm. 2004, 61, 121–139. 10.1016/j.dyepig.2003.10.009. [DOI] [Google Scholar]
- Skong D.A.; Holler F.J.; Crouch S. R.. Principles of instrumental analysis; 6th edn. Brooks/Cole, New York, 2006. [Google Scholar]
- Moriya Y.; Shigemizu D.; Hattori M.; Tokimatsu T.; Kotera M.; Goto S.; Kanehisa M. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010, 38, W138–W143. 10.1093/nar/gkq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed L.; Mahdhi A.; Cheref A.; Bakhrouf A. Decolorization and degradation of azo dye Methyl Red by an isolated Sphingomonas paucimobilis: biotoxicity and metabolites characterization. Desalination 2011, 274, 272–277. 10.1016/j.desal.2011.02.024. [DOI] [Google Scholar]
- Kertesz M. A. Riding the sulfur cycle–metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 2000, 24, 135–175. 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- Hong Y.; Guo J.; Xu Z.; Mo C.; Xu M.; Sun G. Reduction and partial degradation mechanisms of naphthylaminesulfonic azo dye amaranth by Shewanella decolordtionis S12. Appl. Microbiol. Biotechnol. 2007, 75, 647–654. 10.1007/s00253-007-0838-7. [DOI] [PubMed] [Google Scholar]
- Cai P.-J.; Xiao X.; He Y.-R.; Li W.-W.; Chu J.; Wu C.; He M.-X.; Zhang Z.; Sheng G.-P.; Lam M. H.-W.; Xu F.; Yu H.-Q. Anaerobic biodecolorization mechanism of methyl orange by Shewanella oneidensis MR-1. Appl. Microbiol. Biotechnol. 2012, 93, 1769–1776. 10.1007/s00253-011-3508-8. [DOI] [PubMed] [Google Scholar]
- Nouren S.; Bhatti H. N.; Iqbal M.; Bibi I.; Kamal S.; Sadaf S.; Sultan M.; Kausar A.; Safa Y. By-product identification and phytotoxicity of biodegraded Direct Yellow 4 dye. Chemosphere 2017, 169, 474–484. 10.1016/j.chemosphere.2016.11.080. [DOI] [PubMed] [Google Scholar]
- Lago-Vila M.; Rodríguez-Seijo A.; Vega F. A.; Arenas-Lago D. Phytotoxicity assays with hydroxyapatite nanoparticles lead the way to recover firing range soils. Sci. Total Environ. 2019, 690, 1151–1161. 10.1016/j.scitotenv.2019.06.496. [DOI] [PubMed] [Google Scholar]
- Zimmermann T.; Kulla H. G.; Leisinger T. Properties of purified Orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur. J. Biochem. 1982, 129, 197–203. 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya P.; Mallick K.; Ghosh S.; Banerjee P.; Mukhopadhyay A.; Bandyopadhyay S. Algal biomass as potential biosorbent for reduction of organic load in gray water and subsequent reuse: effect on seed germination and enzyme activity. Biorem. J. 2014, 18, 56–70. 10.1080/10889868.2013.847400. [DOI] [Google Scholar]
- Gottlieb A.; Shaw C.; Smith A.; Wheatley A.; Forsythe S. The toxicity of textile reactive azo dyes after hydrolysis and decolourisation. J. Biotechnol. 2003, 101, 49–56. 10.1016/S0168-1656(02)00302-4. [DOI] [PubMed] [Google Scholar]
- Tan N. C. G.; Van Leeuwen A.; Van Voorthuizen E. M.; Slenders P.; Prenafeta-Boldu F. X.; Temmink H.; Field J. A. Fate and biodegradability of sulfonated aromatic amines. Biodegradation 2005, 16, 527–537. 10.1007/s10532-004-6593-x. [DOI] [PubMed] [Google Scholar]
- Qu F.; ElOmari K.; Wagner A.; De Simone A.; Beis K. Desolvation of the substrate-binding protein TauA dictates ligand specificity for the alkanesulfonate ABC importer TauABC. Biochem. J. 2019, 476, 3649–3660. 10.1042/BCJ20190779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat D.; Sharma R. S.; Karmakar S.; Arora L. S.; Mishra V. Ecotoxic potential of a presumably non-toxic azo dye. Ecotoxicol. Environ. Saf. 2018, 148, 528–537. 10.1016/j.ecoenv.2017.10.049. [DOI] [PubMed] [Google Scholar]
- Bergsten-Torralba L. R.; Nishikawa M. M.; Baptista D. F.; Magalhães D. P.; da Silva M. Decolorization of different textile dyes by Penicillium simplicissimum and toxicity evaluation after fungal treatment. Braz. J. Microbiol. 2009, 40, 808–817. 10.1590/S1517-83822009000400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyani D. C.; Patil P. S.; Jadhav J. P.; Govindwar S. P. Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresour. Technol. 2008, 99, 4635–4641. 10.1016/j.biortech.2007.06.058. [DOI] [PubMed] [Google Scholar]
- Cao J.; Sanganyado E.; Liu W.; Zhang W.; Liu Y. Decolorization and detoxification of Direct Blue 2B by indigenous bacterial consortium. J. Environ. Manage. 2019, 242, 229–237. 10.1016/j.jenvman.2019.04.067. [DOI] [PubMed] [Google Scholar]
- Guo G.; Li X.; Tian F.; Liu T.; Yang F.; Ding K.; Liu C.; Chen J.; Wang C. Azo dye decolorization by a halotolerant consortium under microaerophilic conditions. Chemosphere 2020, 244, 125510. 10.1016/j.chemosphere.2019.125510. [DOI] [PubMed] [Google Scholar]
- Pal S.; Kundu A.; Banerjee T. D.; Mohapatra B.; Roy A.; Manna R.; Sar P.; Kazy S. K. Genome analysis of crude oil degrading Franconibacter pulveris strain DJ34 revealed its genetic basis for hydrocarbon degradation and survival in oil contaminated environment. Genomics 2017, 109, 374–382. 10.1016/j.ygeno.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G.; Barns S. M.; Pelletier D. A.; Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M. E.; Dhar K.; Hossain M. T. Decolorization of Textile Reactive Dyes by Bacterial Monoculture and Consortium Screened from Textile Dyeing Effluent. J. Genet. Eng. Biotechnol. 2018, 16, 375–380. 10.1016/j.jgeb.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L.; Ma H.; Zhang L. Degradation process analysis of the azo dyes by catalytic wet air oxidation with catalyst CuO/γ-Al2O3. Chemosphere 2013, 90, 143–149. 10.1016/j.chemosphere.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Jadhav J. P.; Phugare S. S.; Dhanve R. S.; Jadhav S. B. Rapid biodegradation and decolorization of Direct Orange 39 (Orange TGLL) by an isolated bacterium Pseudomonas aeruginosa strain BCH. Biodegradation 2010, 21, 453–463. 10.1007/s10532-009-9315-6. [DOI] [PubMed] [Google Scholar]
- Roy U.; Sengupta S.; Banerjee P.; Das P.; Bhowal A.; Datta S. Assessment on the decolourization of textile dye (Reactive Yellow) using Pseudomonas sp. immobilized on fly ash: Response surface methodology optimization and toxicity evaluation. J. Environ. Manage. 2018, 223, 185–195. 10.1016/j.jenvman.2018.06.026. [DOI] [PubMed] [Google Scholar]