Abstract
During an immune response, cathepsin G (CatG) takes on the role of adaptive and innate immunity and the outcome depends on the localization of CatG. Soluble, cell surface-bound, or intracellular CatG is also responsible for pathophysiology conditions. We applied the activity-based probe MARS116-Bt to mass cytometry by time-of-flight to analyze CatG activity on the cell surface of immune cells. The phosphonate warhead of MARS116-Bt binds covalently to the serine amino acid residue S195 of the catalytic center and thereby CatG activity can be detected. This method contributes to observing the activation or inhibition status of cells during pathogenesis of diseases and enables accurate data acquisition from complex biological samples with a vast panel of cell subset markers in a single-cell resolution.
1. Introduction
Hypertension, cardiac diseases, and diabetes are some of the consequences of cardiac dysfunction. Elevated blood pressure is the result of vasoconstriction, which is triggered by angiotensin II binding to the type 1 angiotensin receptor. Additionally, angiotensin II enhances inflammation and fibrosis.1 How is angiotensin II generated? Angiotensinogen is cleaved by renin to angiotensin I and is further proteolytically digested by the angiotensin-converting enzyme (ACE) to angiotensin II. Importantly, neutrophil-derived cathepsin G (CatG), which can be soluble or bound to the cell surface of neutrophils, and mast cell-derived chymase are also able to convert angiotensin I to angiotensin II.2−5 ACE2 processes angiotensin II to angiotensin 1–7, which is, on the one hand, valuable in lowering blood pressure by vasodilation and provokes the kidneys to excrete sodium and water. On the other hand, it also exhibits an anti-inflammatory capacity. Downregulation of ACE2-expressing cells can cause activation of the renin angiotensin system, which regulates vascular and cardiac physiology and is central to common pathologic conditions such as hypertension and heart failure.6,7
Neutrophils can be identified by a set of cell surface markers, among them are CD11b, CD16, and CD66b, which are reliably expressed across the neutrophilic population. In addition, these cell surface markers are independent from the location or activation status of neutrophils.8 Activated neutrophils, under conditions of both inflammation and homeostasis, express cell surface markers, such as CD62L (L-selectin), CD54 (intracellular adhesion molecule 1, ICAM-1), CD32 (FcγRII), and CD88 (C5a receptor).9 Activation of neutrophils can also be artificially induced using phorbol myristate acetate.10
CatG, a serine protease, together with neutrophil elastase, proteinase 3, and neutrophil serine protease 4 is secreted by activated neutrophils. Cell surface CatG on neutrophils is still proteolytically active,11 although natural serine protease inhibitors, including α1-antitrypsin, are present at the site of inflammation but cannot inhibit cell surface CatG. First, neutrophils release matrix metalloproteases in order to proteolytically inactivate α1-antitrypsin,12 and second, the bulky natural serine protease inhibitor, α1-proteinase inhibitor might not be able to reach the catalytic center of cell surface-bound CatG by steric hindrance.11 CatG possesses a catalytic triad, as with other proteases, containing histidine, aspartate, and serine amino acids within the active center (H57, D102, and S195). The active site cleft is perpendicularly oriented to the two β barrels where the hydroxyl group of S195 nucleophilically attacks the carbonyl carbon of the scissile peptide bond.13,14 Human CatG shows trypsin, chymotrypsin, metase, and lyase activity.15,16 Additionally, it was demonstrated by performing protease profiling that CatG has a preference at the P1 position holding an asparagine (N), at P2 proline (P), and at P3 glutamic acid (E) and at the alternate subsite P1′ isoleucine (I), alanine (A), and serine (S) as well as at the P2′ negatively charged amino acids (aspartic acid, D, and glutamic acid, E).17,18
In our previous work, we analyzed distinct cell populations in the peripheral blood of healthy donors for their cell surface CatG activity by applying the activity-based probe MARS116-Bt in a flow cytometry approach. This method circumvents cell separation from a mixture of cells found in blood or tissue to detect CatG activity by a Western blot-based assay or by enzymatic kinetics.19 Mass cytometry by time-of-flight (CyTOF) is the next generation of flow cytometry to simultaneously analyze a complex panel of cell markers, which is not possible with the classical fluorescence flow cytometer. Hence, a multiplexed profiling of up to 100 surface markers as well as intracellular signaling proteins is possible.20,21 Until now, the detection of cell markers, including proteases, via application of CyTOF has been limited to analysis at the protein level. To this end, we established an approach to determine the proteolytic activity of CatG on the cell surface of neutrophils, NK cells, B cells, and T cells in peripheral blood mononuclear cells (PBMCs) by combining the activity-based probe MARS116-Bt and the anti-biotin-150Nd antibody with CyTOF analysis. The technique outlined here demonstrates that CatG can be detected on CD16+CD66b+ neutrophils and NK cells using the CyTOF methodology. Therefore, MARS116-Bt-anti-biotin-150Nd antibodies are useful to profile a vast panel of different cell subsets in order to evaluate catalytically active CatG, its regulation, or inhibition on the cell surface.
2. Results and Discussion
2.1. Detection of CatG Activity on the Cell Surface of Neutrophils by CyTOF
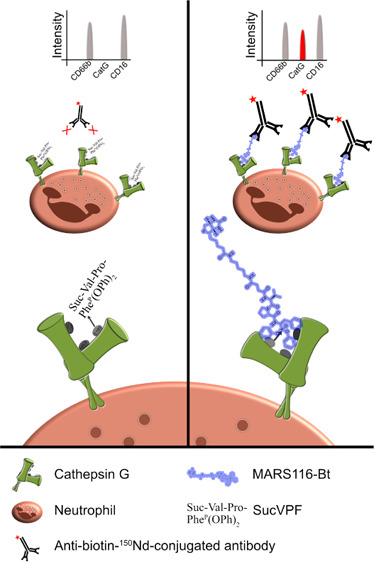
Activity-based probes are applied to detect cysteine and serine protease activity.22−24 MARS116-Bt contains a biotin, spacer, amino acid sequence for specificity, and a warhead. MARS116-Bt, with an incorporated electrophilic phosphonic active site-directed warhead, binds covalently to the oxygen atom of S195 within the catalytic center of CatG.25 PBMCs were incubated with MARS116-Bt with a group of specific antibodies that were conjugated with different isotopes (147Sm-CD20, 154Sm-CD45, 155Gd-CD56, 160Gd-CD14, 162Dy-CD66b, 165Ho-CD16, 168Er-CD8, 170Er-CD3, 173Yb-HLA-DR, and 174Yb-CD4) to determine immune cells and their respective subsets. Furthermore, the groups were treated with different inhibitors to distinguish specificity. One control group was incubated without an inhibitor, two groups were preincubated with the reversible CatG inhibitor (CatGinh.) in a final concentration of 12.5 or 50 μM, and one group was treated with the irreversible CatG inhibitor Suc-Val-Pro-PheP(OPh)2 (SucVPF)26,27 in a final concentration of 50 μM before adding MARS116-Bt. Subsequently, anti-biotin-150Nd antibodies were added to the samples and CatG activity was monitored by CyTOF combined with the respective software (summarized in Figure 1).
Figure 1.
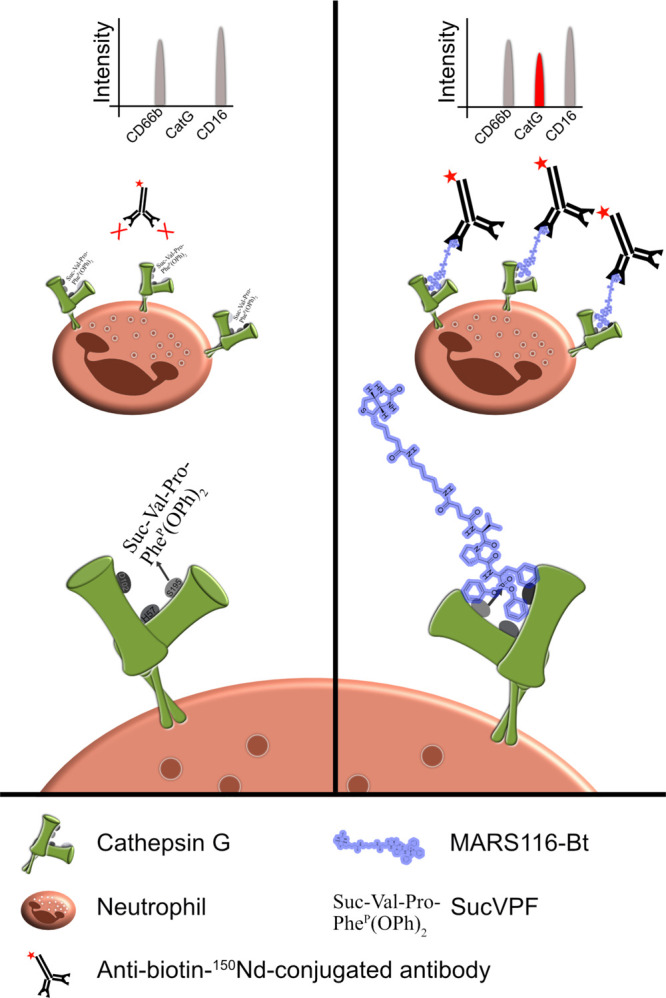
Scheme of MARS116-Bt binding to CatG. Cell surface CatG is drawn with the active-site cleft and the two β barrels. The hydroxyl group of S195 nucleophilically attacks the phosphorus atom of SucVPF and MARS116-Bt resulting in a covalent bond between S195 and the inhibitors. Anti-biotin-150Nd antibodies are added to the cell culture, forming the MARS116-Bt-anti-biotin-150Nd antibody complex, which can be analyzed by mass cytometry by time-of-flight (CyTOF).
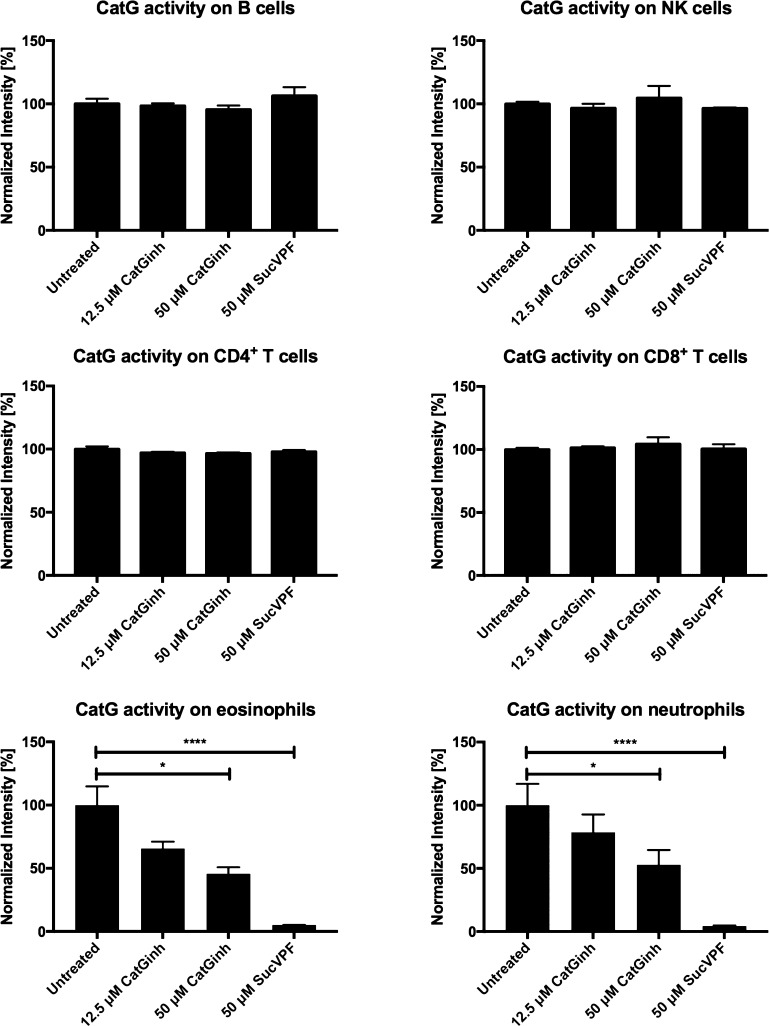
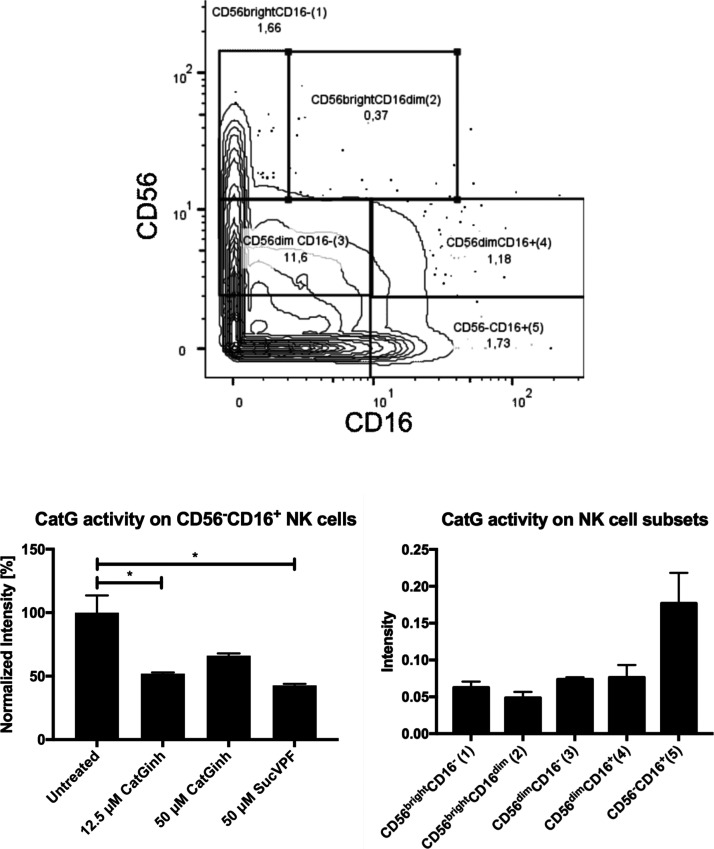
After data acquisition, the FCS files were normalized based on the calibration beads (EQ Four Element Calibration Beads) using the built-in normalizer of the Helios. The beads were gated out of the analysis and the cells were gated for DNA double-positive events (191Ir and 193Ir) to exclude doublets for accurate single-cell identification. CD45 was used as a marker for PBMCs. A further gating procedure was carried out to differentiate between CD4+ T cells (CD3+, CD4+, CD8–, and CD20–), CD8+ T cells (CD3+, CD4–, CD8+, and CD20–), B cells (CD20+ and HLA-DR+), monocytes (CD3–, CD14var, CD16var, CD20–, CD56–, and HLA-DR+), NK cells (CD3–, CD14–, CD16var, CD20–, and CD56var), eosinophils (CD3–, CD16–, CD20–, CD56–, and CD66b+), and neutrophils (CD3–, CD16+, CD20–, CD56–, and CD66b+). NK cells were further subcategorized by gating for the cell surface expression of CD16 and CD56 based on previously established gating procedures of regular flow cytometry.28 In the next step, the 150Nd channel was adjusted for CatG activity selecting CD4+ T cells, CD8+ T cells, B cells, monocytes, NK cells, and their subsets, as well as eosinophils and neutrophils. In contrast to B cells or T cells, eosinophils and neutrophils showed a robust level of proteolytically active CatG (Figure 2 and Supporting Information S1). Additionally, the NK cell subset CD16+CD56– (no. 5) harbors detectable CatG activity, as demonstrated in Figure 3. The specificity of cell surface-active CatG was determined by signal reduction when samples were preincubated with CatG inhibitors (CatGinh. and SucVPF). Thus, the methodology of CyTOF can be used for deep profiling of neutrophils, eosinophils, and NK cells as well as for detection of proteolytically active CatG by applying MARS116-Bt. Furthermore, CyTOF is a valid method to detect not only the presence of specific antigens but also their proteolytic activity.
Figure 2.
Detection of CatG activity on the cell surface of immune cells. PBMCs were stained with the indicated specific antibodies as mentioned in the method section. Cell populations (B cells, NK cells, CD4+ T cells, CD8+ T cells, eosinophils, and neutrophils) were gated by their cell surface expression and CatG activity. Control samples were treated with the CatG inhibitor (CatGinh.), 12.5 or 50 μM, and SucVPF, 50 μM. Values are expressed as normalized intensity ± S.E.M. and significance was calculated by Student’s t-test between two groups (P < 0.1 = * and p < 0.0001 = ****). Technical replicates n = 3 from the same donor.
Figure 3.
NK cell subsets are gated for their CatG activity. NK cell subsets, based on the expression of CD56 and CD16, are classified as CD16–CD56bright (no. 1), CD16dimCD56bright (no. 2), CD16–CD56dim (no. 3), CD16+CD56dim (no. 4), and CD16+CD56- (no. 5); upper panel. NK cell subsets were gated for CatG activity and summarized in a bar diagram (lower, left panel, normalized intensity ± S.E.M.). CD16+CD56– (no. 5) NK cells were treated with CatGinh. or SucVPF (lower, right panel). Values are expressed as intensity ± S.E.M. and significance was calculated (P < 0.1 = *). Technical replicates n = 3 from the same donor.
The next-generation flow cytometry CyTOF setup in combination with MARS116-Bt-anti-biotin-150Nd antibodies allows us to analyze and gate distinct cell subsets and simultaneously determine the proteolytic activity of CatG. This approach can be used to monitor neutrophils for the efficacy of inhibitors to arrest CatG activity as well as disease progression where CatG activity is upregulated. Furthermore, MARS116-Bt-anti-biotin-150Nd antibodies can be added to a vast panel of activation markers in CyTOF analysis to characterize immune cells during inflammation, inducing a pathologic effect, in homeostasis, or when stimulated with different substances. Additionally, different activity-based probes can be applied to CyTOF to detect further serine proteases or even cysteine proteases in a single-cell approach.
In previous work, we detected CatG activity in both NK cell subsets, CD3–CD16–CD56dim and CD3–CD16dimCD56– NK cells,19 by common flow cytometry. Compared to the CyTOF analysis demonstrated here, CatG activity detection was limited to CD16+CD56- NK cells. This gives rise to speculation that a low amount of CatG activity cannot be detected by CyTOF and is certainly a limitation of this technique.
The use of MARS116-Bt-anti-biotin-150Nd antibodies for characterizing the catalytic activity of CatG is not only practical for immune cells from the blood. We also suggest sputum from patients or tissue from animal models because application of the MARS116-Bt-anti-biotin-150Nd antibodies is not restricted to cells from humans. Furthermore, treatment with protease inhibitors to regulate imbalanced proteolytic activity in several diseases is indicated. With respect to serine protease inhibitors, boswellic acids (BAs) inhibit the catalytic activity of CatG.29 Previously, it has been shown that BA interferes with the chemoinvasion of neutrophils, suppresses inflammation, and possesses potential cardioprotective properties.29−31 Inhibition of CatG by BA could be monitored by MARS116-Bt-anti-biotin-150Nd antibodies of a vast panel of cell subsets in a single-cell resolution.
3. Materials and Methods
3.1. PBMC Separation
The content of a buffy coat from one donor (DRK blood donation center Baden–Württemberg–Hessen, Institute Ulm, Germany) was diluted with phosphate-buffered saline (PBS) (pH 7.4, Gibco by Life Technologies, Carlsbad, CA, USA), and PBMCs were separated by density centrifugation (“Ficoll-Pague PLUS”, GE Healthcare, Little Chalfont, UK). After centrifugation (760g for 20 min, without brake), PMBCs were carefully taken from the Ficoll interface and washed three times with PBS. 1 × 107 PBMCs were stored in fetal bovine serum (Gibco by life Technologies, Carlsbad, CA, USA) including 10% dimethyl sulfoxide (DMSO, Serva Electrophoresis GmbH, Heidelberg, Germany) at −80 °C. Notably, neutrophils from PBMCs are low-density neutrophils possessing reduced phagocytic capacity.32 The use of PBMCs for analysis of proteases was approved by the ethics committee at Ulm University (Helmholtzstr. 20, 89069 Ulm, Germany, # 327/14).
3.2. Application of MARS116-Bt in CyTOF
PBMCs from one donor were thawed, washed twice with PBS, and used in a final concentration of 1.5 × 106 PBMCs/staining for both titration of antibodies and the experiments.
The experiment was split into four groups, one control group treated with PBS, two groups that were preincubated with the reversible CatG inhibitor (CatGinh., Calbiochem, Merck Chemicals GmbH, Schwalbach, Germany, Cat. no.: 219372) at two concentrations, 12.5 and 50 μM, and one group treated with 50 μM of the irreversible CatG inhibitor, Suc-Val-Pro-PheP(OPh)2 (SucVPF).26 Preincubation was performed for 15 min at room temperature (RT). Afterward, cells were washed with PBS, centrifuged (300g for 8 min), and incubated with MARS116-Bt, which was synthesized as described previously,25 in a final concentration of 2 μM for 30 min at RT, followed by three wash steps in PBS, and centrifugation at 300g for 8 min. Cells were stained with the following mixture of antibodies, as depicted in Table 1, for 30 min at RT. The cells were washed three times (300g, 8 min) with PBS and fixed with 3.7% of PFA (ThermoFisher, Pierce Methanol-free, Cat# 28906, Markham, Canada) at 4 °C. After two washing steps (600g, 8 min), cells were stained with Ir-Intercalator (191Ir and 193Ir, 125 μM, 1:2000, Fluidigm, Product # 201192A, Markham, Canada) and stored at −80 °C in RPMI supplemented with 10% DMSO until the day of acquisition. The samples were thawed followed by one washing step in PBS with 10% FCS, and three washing steps with MilliQ water (600g, 8 min). Thereafter, cells were acquired at 300 events/second with a Helios CyTOF system (Fluidigm, Markham, Canada). Calibration beads (EQ Four Element Calibration Beads, Fluidigm #201078, Markham, Canada) were used at a concentration of 10%.
Table 1. List of CyTOF Antibodies Used for the Experimentsa.
| metal | antigen | clone | company, product # | |
|---|---|---|---|---|
| 147Sm | CD20 | 2H7 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 150Nd | Biotin | 1d4-C5 | anti-biotin-150Nd-antibodies, Fluidigm, #3150008C | |
| 154Sm | CD45 | HI30 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 155Gd | CD56 | B159 | Fluidigm, #3155008C | |
| 160Gd | CD14 | M5E2 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 162Dy | CD66b | 80H3 | Fluidigm, #3162023C | |
| 165Ho | CD16 | 3G8 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 168Er | CD8 | Sk1 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 170Er | CD3 | UCHT1 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
| 173Yb | HLA-DR | L243 | Fluidigm, #3173005C | |
| 174Yb | CD4 | SK3 | Fluidigm, #201302, Human PB Basic Phenotyping Kit | |
All antibodies were purchased from Fluidigm. Some of them, which are marked, are part of the Human PB Basic Phenotyping Kit.
3.3. Data Analysis
The data were analyzed using manual gating with FlowJo X (FlowJov10.6.2, FlowJo LLC, Ashland, OR, USA) as well as automated clustering approaches using SPADE (Cytobank 7.3.0., implemented version, Santa Clara, CA, USA). Statistical analysis was performed with the commercially available software, GraphPad Prism 6, Inc., San Diego, CA, USA. Data were normalized to the specific untreated control population and expressed as normalized intensity in percent. The ± standard error of the mean (S.E.M.) is shown. Significant differences were considered at p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), or p < 0.0001 (****).
Acknowledgments
FG is a member of the International Graduate School of Molecular Medicine (IGradU) at Ulm University, which is funded by the DFG under the acronym “GSC270”. We gratefully thank Marcin Sienczyk (Faculty of Chemistry, Wroclaw University of Technology, Wroclaw, Poland) for providing MARS-116-Bt, structure, and Suc-VPF. Furthermore, we thank Sarah Warth from the Core Facility of Cytometry (Ulm University, Ulm, Germany) for measuring different samples by CyTOF. T.B. was funded by the Nazarbayev University Faculty-Development Competitive Research Grants Program, reference: 280720FD1907.
Glossary
Abbreviations used
- ACE2
Angiotensin-converting enzyme 2
- BA
boswellic acid
- Cat
cathepsin
- CatG
cathepsin G
- CatGinh
cathepsin G inhibitor
- CyTOF
mass cytometry by time-of-flight
- DMSO
dimethyl sulfoxide
- MMP
matrix metalloprotease
- NK cells
natural killer cells
- PBMCs
peripheral blood mononuclear cells
- PMA
phorbol myristate acetate
- SucVPF
Suc-Val-Pro-PheP(OPh)2
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04092.
CatG activity is plotted as intensity values and cell surface CatG activity, detected on B cells, NK cells, CD4+ T cells, CD8+ T cells, eosinophils, and neutrophils, is summarized in the bar diagram and expressed as intensity (PDF)
Author Contributions
FG performed the experiments, analyzed data, prepared the figures, and wrote the manuscript. UK analyzed the data. MS designed and synthesized MARS116-Bt. TB designed the experiments and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Kessler T.; Schunkert H. Inhibitors of the renin-angiotensin system and SARS-CoV-2 infection. Herz 2020, 45, 323–324. 10.1007/s00059-020-04920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C. A.; Campbell E. J. Angiotensin II generation at the cell surface of activated neutrophils: novel cathepsin G-mediated catalytic activity that is resistant to inhibition. J. Immunol. 1998, 160, 1436–1443. [PubMed] [Google Scholar]
- Klickstein L. B.; Kaempfer C. E.; Wintroub B. U. The granulocyte-angiotensin system. Angiotensin I-converting activity of cathepsin G. J. Biol. Chem. 1982, 257, 15042. [PubMed] [Google Scholar]
- Wintroub B. U.; Klickstein L. B.; Watt K. W. A human neutrophil-dependent pathway for generation of angiotensin II. Purification of the product and identification as angiotensin II. J. Clin. Invest. 1981, 68, 484–490. 10.1172/jci110279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey G. H.; Raymond W. W.; Wolters P. J. Angiotensin II generation by mast cell alpha- and beta-chymases. Biochim. Biophys. Acta 2000, 1480, 245–257. 10.1016/s0167-4838(00)00076-5. [DOI] [PubMed] [Google Scholar]
- Patel V. B.; Zhong J.-C.; Grant M. B.; Oudit G. Y. Role of the ACE2/Angiotensin 1-7 Axis of the Renin-Angiotensin System in Heart Failure. Circ. Res. 2016, 118, 1313–1326. 10.1161/circresaha.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M.; Wang K.; Viveiros A.; Nguyen Q.; Zhong J.-C.; Turner A. J.; Raizada M. K.; Grant M. B.; Oudit G. Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. 10.1161/circresaha.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakschevitz F. S.; Hassanpour S.; Rubin A.; Fine N.; Sun C.; Glogauer M. Identification of neutrophil surface marker changes in health and inflammation using high-throughput screening flow cytometry. Exp. Cell Res. 2016, 342, 200–209. 10.1016/j.yexcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Fortunati E.; Kazemier K. M.; Grutters J. C.; Koenderman L.; Van den Bosch v. J. M. M. Human neutrophils switch to an activated phenotype after homing to the lung irrespective of inflammatory disease. Clin. Exp. Immunol. 2009, 155, 559–566. 10.1111/j.1365-2249.2008.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A.; Nixon J. B.; McPhail L. C. Phorbol myristate acetate induces neutrophil NADPH-oxidase activity by two separate signal transduction pathways: dependent or independent of phosphatidylinositol 3-kinase. J. Leukoc. Biol. 2000, 67, 396–404. 10.1002/jlb.67.3.396. [DOI] [PubMed] [Google Scholar]
- Owen C. A.; Campbell M. A.; Sannes P. L.; Boukedes S. S.; Campbell E. J. Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J. Cell Biol. 1995, 131, 775–789. 10.1083/jcb.131.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers M. C.; George P. M.; Bathurst I. C.; Brennan S. O.; Winterbourn C. C. Cleavage and inactivation of alpha 1-antitrypsin by metalloproteinases released from neutrophils. J. Clin. Invest. 1988, 82, 706–711. 10.1172/jci113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz B.; Horwitz M. S.; Jenne D. E.; Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 2010, 62, 726–759. 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P.; Mayr I.; Huber R.; Korzus E.; Potempa J.; Travis J.; Powers J. C.; Bode W. The 1.8 A crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(OPh)2: a Janus-faced proteinase with two opposite specificities. EMBO J. 1996, 15, 5481–5491. 10.1002/j.1460-2075.1996.tb00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond W. W.; Trivedi N. N.; Makarova A.; Ray M.; Craik C. S.; Caughey G. H. How immune peptidases change specificity: cathepsin G gained tryptic function but lost efficiency during primate evolution. J. Immunol. 2010, 185, 5360–5368. 10.4049/jimmunol.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka M.; Łȩgowska A.; Bulak E.; Jaśkiewicz A.; Miecznikowska H.; Lesner A.; Rolka K. New chromogenic substrates of human neutrophil cathepsin G containing non-natural aromatic amino acid residues in position P(1) selected by combinatorial chemistry methods. Mol. Divers. 2007, 11, 93–99. 10.1007/s11030-007-9063-7. [DOI] [PubMed] [Google Scholar]
- Thorpe M.; Fu Z.; Chahal G.; Akula S.; Kervinen J.; de Garavilla L.; Hellman L. Extended cleavage specificity of human neutrophil cathepsin G: A low activity protease with dual chymase and tryptase-type specificities. PLoS One 2018, 13, e0195077 10.1371/journal.pone.0195077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. T. N.; Shema G.; Zahedi R. P.; Verhelst S. H. L. Protease Specificity Profiling in a Pipet Tip Using “Charge-Synchronized” Proteome-Derived Peptide Libraries. J. Proteome Res. 2018, 17, 1923–1933. 10.1021/acs.jproteome.8b00004. [DOI] [PubMed] [Google Scholar]
- Penczek A.; Sienczyk M.; Wirtz C. R.; Burster T. Cell surface cathepsin G activity differs between human natural killer cell subsets. Immunol. Lett. 2016, 179, 80–84. 10.1016/j.imlet.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Kay A. W.; Strauss-Albee D. M.; Blish C. A. Application of Mass Cytometry (CyTOF) for Functional and Phenotypic Analysis of Natural Killer Cells. Methods Mol. Biol. 2016, 1441, 13–26. 10.1007/978-1-4939-3684-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung R. K.; Utz P. J. Screening: CyTOF-the next generation of cell detection. Nat. Rev. Rheumatol. 2011, 7, 502–503. 10.1038/nrrheum.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach J.; Bartz U.; Gütschow M. Coumarin as a structural component of substrates and probes for serine and cysteine proteases. Biochim. Biophys. Acta Protein Proteonomics 2020, 1868, 140445. 10.1016/j.bbapap.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman L. E.; Bogyo M. Activity-based profiling of proteases. Annu. Rev. Biochem. 2014, 83, 249–273. 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- Chakrabarty S.; Kahler J. P.; van de Plassche M. A. T.; Vanhoutte R.; Verhelst S. H. L. Recent Advances in Activity-Based Protein Profiling of Proteases. Curr. Top. Microbiol. Immunol. 2019, 420, 253–281. 10.1007/82_2018_138. [DOI] [PubMed] [Google Scholar]
- Zou F.; Schmon M.; Sienczyk M.; Grzywa R.; Palesch D.; Boehm B. O.; Sun Z. L.; Watts C.; Schirmbeck R.; Burster T. Application of a novel highly sensitive activity-based probe for detection of cathepsin G. Anal. Biochem. 2012, 421, 667–672. 10.1016/j.ab.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Oleksyszyn J.; Powers J. C. Irreversible inhibition of serine proteases by peptide derivatives of (alpha-aminoalkyl)phosphonate diphenyl esters. Biochemistry 1991, 30, 485–493. 10.1021/bi00216a026. [DOI] [PubMed] [Google Scholar]
- Reich M.; Lesner A.; Łęgowska A.; Sieńczyk M.; Oleksyszyn J.; Boehm B. O.; Burster T. Application of specific cell permeable cathepsin G inhibitors resulted in reduced antigen processing in primary dendritic cells. Mol. Immunol. 2009, 46, 2994–2999. 10.1016/j.molimm.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Poli A.; Michel T.; Thérésine M.; Andrès E.; Hentges F.; Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 2009, 126, 458–465. 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausch L.; Henkel A.; Siemoneit U.; Poeckel D.; Kather N.; Franke L.; Hofmann B.; Schneider G.; Angioni C.; Geisslinger G.; Skarke C.; Holtmeier W.; Beckhaus T.; Karas M.; Jauch J.; Werz O. Identification of human cathepsin G as a functional target of boswellic acids from the anti-inflammatory remedy frankincense. J. Immunol. 2009, 183, 3433–3442. 10.4049/jimmunol.0803574. [DOI] [PubMed] [Google Scholar]
- Abdel-Tawab M.; Werz O.; Schubert-Zsilavecz M. Boswellia serrata: an overall assessment of in vitro, preclinical, pharmacokinetic and clinical data. Clin. Pharmacokinet. 2011, 50, 349–369. 10.2165/11586800-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Efferth T.; Oesch F. Anti-inflammatory and anti-cancer activities of frankincense: Targets, treatments and toxicities. Semin. Canc. Biol. 2020, 10.1016/j.semcancer.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Sagiv J. Y.; Michaeli J.; Assi S.; Mishalian I.; Kisos H.; Levy L.; Damti P.; Lumbroso D.; Polyansky L.; Sionov R. V.; Ariel A.; Hovav A.-H.; Henke E.; Fridlender Z. G.; Granot Z. Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Rep. 2015, 10, 562–573. 10.1016/j.celrep.2014.12.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.