Abstract

Microbial nucleic acids in the extracellular milieu are recognized in vertebrates by Toll-like receptors (TLRs), one of the most important families of innate immune receptors. TLR9 recognizes single-stranded unmethylated CpG DNA in endosomes. DNA binding induces TLR9 dimerization and activation of a potent inflammatory response. To provide insights on how DNA ligands induce TLR9 dimerization, we developed a detailed theoretical framework for equilibrium ligand binding, modeling the binding of the ssDNA at the two main sites on the TLR9 ectodomain. Light scattering and fluorescence anisotropy assays performed with recombinant TLR9 ectodomain and a panel of agonistic and antagonistic DNA ligands provide data that restrain the binding parameters, identify the likely ligand binding intermediates, and suggest cooperative modes of binding. This work brings us one step closer to establishing a rigorous biochemical understanding of how TLRs are activated by their ligands.
Vertebrates rely on the innate immune system as their first line of defense against pathogens.1 Innate immune receptors detect pathogen-associated molecular patterns (PAMPs) that are common and conserved in microbes, but absent in the host. A major family of innate immune receptors is the Toll-like receptors (TLRs).2 TLR3, TLR7, TLR8, and TLR9 are found in endosomes and recognize nucleic acid PAMPs.3−7 TLR9 recognizes single-stranded DNA (ssDNA) containing unmethylated CG nucleotide sequence motifs (CpGs).7 CpGs are more prevalent in bacteria and viruses than in the mammalian genome, in which most CG sequences are methylated.8,9
The crystal structures of TLR9 ectodomain fragments from mouse, horse, and cow have been determined without ligand (apo), bound to antagonistic ligands 4084 and iSUPER, and bound to 1668-12mer,10 a truncated version of the activating oligonucleotide ligand 1668.11 These structures provided the structural basis for the CpG specificity of TLR9 ligand recognition. The apo-TLR9 structure was monomeric, but the TLR9 ectodomain formed a dimer with bound 1668-12mer, suggesting a model of TLR9 signal activation through dimerization. Full-length membrane-inserted TLR9 is thought to form loosely assembled inactive homodimers prior to binding ssDNA, with ligand binding inducing a conformational rearrangement and tightening of the dimer assembly necessary to activate signaling.12 The two TLR9 ectodomains assemble around two 1668-12mer oligonucleotides to form a 2:2 TLR9:oligonucleotide complex.10 The oligonucleotides, sandwiched between the two ectodomains, function as “molecular glue” between the two TLR9 subunits.10 Each ligand in the dimer interacts with two distinct binding surfaces on TLR9, near the N- and C-terminal ends of the ectodomain, respectively.10 An additional binding site in TLR9 was recently identified in the central region of the ectodomain, with specificity for short ssDNA oligonucleotides containing the motif 5′-xCx,13 which function as auxiliary ligands to enhance signaling.14 Auxiliary ligands with analogous functions in signal augmentation have been identified for TLR7 and TLR8.15,16
Although structural studies have shed light on how TLR9 recognizes ssDNA ligands, key open questions remain concerning the signaling mechanism of TLR9. A reductionist approach to determine the minimal sequence requirements for an oligonucleotide to maximally activate TLR9 identified a length of between 23 and 29 nucleotides as the optimal length for mouse TLR9 agonists, with a 5′-TCC motif and CpG motif located 5–7 nucleotides from the 5′ end.17,18 It remains unclear why extending the length of the ligand beyond the 12 nucleotides observed in the TLR9:1668-12mer structure enhances signaling. Moreover, modeling of ssDNA binding has been limited by the use of either a 1:1 binding model (rather than a 2:2 model) or of the Hill equation.10,13,19,20 Although it is known that the ligand-saturated state of a signaling-competent TLR9-ssDNA complex is a 2:2 dimer, the set of assembly intermediates through which this complex assembles is unknown. TLR9 dimerization upon ligand binding can theoretically occur if two TLR9 ectodomains first form 1:1 protein:ligand complexes and then come together to form 2:2 dimers, or alternatively if a single TLR9 first binds two oligonucleotides (one at each binding site), and this 2:1 complex then recruits a free TLR9. Determining the most prevalent intermediates in TLR9 dimerization and measuring binding cooperativity would provide key missing links in our understanding of TLR9 activation. Here, we propose an equilibrium binding model for ligand-dependent dimerization of TLR9 ectodomain, providing a theoretical framework for different possible modes of binding. We support and refine our model with biochemical and biophysical analyses of ligand binding. Our work brings us one step closer to establishing a detailed and rigorous understanding of the assembly intermediates and energy landscape of DNA-dependent TLR9 activation.
Materials and Methods
Purification of Mouse TLR9 Ectodomain (mTLR9-ECD)
A pMT plasmid encoding mTLR9-ECD with a secretion signal and a C-terminal protein A tag was cotransfected with pCoBlast at a 10:1 pMT:pCoBlast molar ratio into S2 insect cells. Stable cell lines were selected with 100 μg/mL blasticidin. Protein expression was induced with 0.5 mM copper sulfate. Five days postinduction, the culture media was concentrated by tangential-flow filtration on a 30 kDa cutoff membrane (Merck). mTLR9-ECD was purified by protein A-affinity chromatography with IgG Sepharose 6 Fast Flow resin (Cytiva) in PBS. mTLR9-ECD was eluted with 0.1 M glycine–HCl pH 3.5 and 0.15 M NaCl and immediately neutralized with 1/20 (v/v) 1 M Tris pH 8. mTLR9-ECD was further purified on a MonoS 4.6/100 PE ion-exchange column (Cytiva) in 10 mM MES pH 6.0, 0.06–1 M NaCl. Protein eluting at 0.25–0.32 M NaCl was pooled, cleaved, and further purified on a Superdex 200 10/300 size-exclusion column (Cytiva) in 10 mM Tris pH 7.4, 0.15 M NaCl. Uncleaved protein eluted as a mixture of monomer and dimer. To remove the tag and proteolytically activate mTLR9-ECD, the protein was incubated with 1/20–1/50 (w/w) GluC protease (NEB) for 24–48 h at 4 °C. GluC was removed with Benzamidine Sepharose 4 Fast Flow resin (Cytiva). Cleaved mTLR9-ECD eluted as a monomer.
Dynamic Light Scattering (DLS)
mTLR9-ECD (2 μM) was incubated with 2 μM oligonucleotide (Sigma-Aldrich) in 10 mM MES pH 6, 0.15 M NaCl, for 1 h at room temperature. After spin-filtering through a 0.22 μm membrane (Costar), 30 μL samples were loaded into black, clear-bottomed, 384-well plates (Corning). Data were collected on a Wyatt Technologies DynaPro II plate reader at 25 °C. Five acquisitions were collected for each sample, with five measurements per acquisition.
Size-Exclusion Chromatography Coupled to Multiangle Light Scattering (SEC-MALS)
mTLR9-ECD (8 μM) was incubated with 20 μM oligonucleotide 1668. The mixture was loaded onto a Superdex 200 10/300 column in 10 mM MES pH 6.0, 0.15 M NaCl, with a flow rate of 0.5 mL min–1 at 293 K. Protein was detected with a UV detector at 280 nm (Agilent Technology 1260), a quasielastic light scattering module (DAWN-8+, Wyatt Technology), and a differential refractometer (Optilab T-rEX, Wyatt Technology). Molar masses of peaks in the elution profile were calculated from the light scattering and protein concentration, quantified using the differential refractive index of the peak, assuming dn/dc = 0.186, with ASTRA6 (Wyatt Technology).
Relative Fluorescence Anisotropy
mTLR9-ECD was titrated into a solution of oligonucleotide labeled at the 5′ end with Alexa 488 (Sigma-Aldrich). The oligonucleotide concentration used was 2 or 5 nM. Protein was added to a maximum concentration of 100 nM. 30 μL samples were assayed in 384-well black, clear-bottomed plates (Corning) with a ClarioSTAR plate reader (BMG Labtech) using a 482/530 nm filter.
Data were fitted with a 1:1 binding model that accounted for ligand depletion,21 using the following equation:
where Af denotes the anisotropy of the free ligand, Ab the anisotropy of the bound ligand, L the total ligand concentration, and R the total protein concentration. L was fixed, and Ab, Af, and Kd were fitted using the known values of R and A. The fit was performed with Prism8 (GraphPad).
For competition assays where fluorescent oligonucleotide was displacing unlabeled oligonucleotide (Figure S1A), 2 μM oligonucleotide 4084 was briefly incubated at room temperature with increasing amounts of mTLR9-ECD. Fluorescent oligonucleotide (5 nM) was then added. For competition assays where unlabeled oligonucleotide was displacing bound fluorescent oligonucleotide (Figure 3E,F, Figure S1B), 100 nM mTLR9-ECD and 2 nM fluorescent oligonucleotide were preincubated for 30 min at room temperature in 10 mM MES pH 6, 0.15 M NaCl. Unlabeled oligonucleotide was titrated in, and measurements were taken at 1.5 and 4.5 h. The inhibitor constant, Ki, was calculated with the Cheng–Prusoff equation:
Figure 3.
Apparent binding affinities of mTLR9-ECD for various ligands measured by relative fluorescence anisotropy. (A–D) The equilibrium binding affinities of different Alexa 488-labeled oligonucleotides for proteolytically activated mTLR9 were calculated from fitting to a 1:1 binding model. The average Kd’s from replicate experiments are shown. Numbers in parentheses indicate 95% confidence intervals. (E, F) Competition fluorescence anisotropy experiments reveal slow oligonucleotide dissociation. (E) Change of anisotropy over time after unlabeled 4084 was added to a solution containing 100 nM mTLR9-ECD preincubated with 5 nM fluorescent 1668-12mer. (F) Anisotropy as a function of unlabeled 4084 concentration 1.5 h (black) and 4.5 h (green) after addition to mTLR9-ECD bound to 1668-12mer. The red dotted lines mark the anisotropy of the fluorescent 1668-12mer, which is the expected anisotropy if all of the fluorescent oligonucleotide has been competed off the protein. (A–F) The data shown are representative of one to four independent experiments (see Data Set S1).
Anisotropy Simulations
A custom script was written in Python 3.7.3 to solve a system of equations as defined by the relationships between KA, KB, and the components of the system, P, D, PD, PD2, and P2D2. A similar script was used for simulation with a simplified model using only K1 and K4 and for fitting K1 and K4 to data. The scripts are available upon request or at https://github.com/StevieReikine/TLR9_Anisotropy.
Mass Photometry
100 nM uncleaved mTLR9-ECD was incubated with 100 nM 1668 oligo or buffer control. 10 μL of the protein was applied to 10 μL of buffer on a coverslip on a RE-FEYN OneMP mass photometry system (RE-FEYN). Movies were acquired for 60 s. The mass was calculated using a standard protein calibration curve.
Results
Equilibrium Ligand Binding Model for TLR9
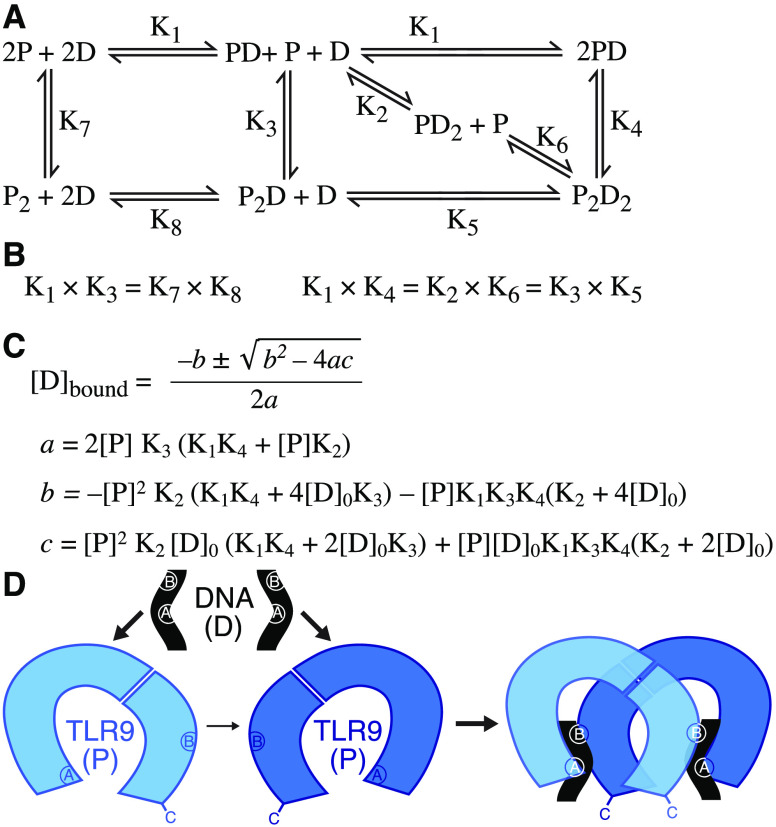
To generate a quantitative description of ligand-induced dimerization of TLR9, we first need to establish a model of complex formation that can be tested experimentally. A stoichiometric binding equilibrium model representing the possible intermediate states in TLR9 dimerization is presented in Figure 1A. The model allows for assembly of the 2:2 active TLR9:DNA complex via TLR9 dimerization upon binding two ligands, a single ligand, or no ligands. The term [PD] represents the apparent binding of a DNA oligonucleotide (D) to TLR9 (P). Since TLR9 has two oligonucleotide binding sites, [PD] is the sum of [PDA] and [PDB], representing the oligonucleotide bound at site A or site B, respectively. Hence, the macroscopic equilibrium constants K1, K2, and K3 are each composed of at least two microscopic binding constants, which describe the equilibria between [PDA] and [PDB] and the previous or subsequent state. These macroscopic binding constants will also include other microscopic constants if binding induces a conformational change or is cooperative. This model notably describes the interactions between activating ligands and the TLR9 ectodomain at the two primary sites and does not take into account potential contributions of the auxiliary 5′-xCx binding site, the transmembrane and TIR domains, or membrane-tethering to complex assembly.
Figure 1.
Proposed general equilibrium model for TLR9-ECD agonist binding. (A) Stoichiometric representation of the possible species in equilibrium as TLR9 (P) binds an activating ssDNA ligand (D) and dimerizes. The macroscopic equilibrium binding constants are labeled. (B) Relationships between the macroscopic constants. (C) Solution for the concentration of bound ssDNA, [D]bound. (D) Schematic of DNA ligand-dependent TLR9 ectodomain dimerization. The two TLR9-DNA interaction sites, sites A and B, are labeled on the protein and ligand. The line width of the arrows represents the approximate frequency of the indicated interaction in solution.
Our model has implications for the relationships between the macroscopic binding constants (Figure 1B). Since mTLR9 ectodomain (mTLR9-ECD) remains predominantly monomeric at high protein concentrations,10 our model predicts that K7 is large relative to K1. Moreover, given that K1K3 = K7K8, K3 would thus be very large. Similarly, if we assume that in the presence of DNA TLR9 preferentially forms 2:2 TLR-DNA dimers, even at low protein concentrations, K1K4 would be relatively small, which would imply that K5 is very small, since K1K4 = K3K5, and K3 is very large. This would lead to the interesting hypothesis that the 2:1 TLR9:DNA dimer species (P2D) rarely occurs. Our general model of equilibrium binding can be solved for the concentration of bound ssDNA, [D]bound, accounting for mass action (Figure 1C). [D]bound can be measured experimentally in a ligand binding assay. A complete solution of all macroscopic constants is not readily accessible experimentally, but numerical solutions or simulations could in principle be used to identify possible values for each constant.
Our equilibrium binding model is expressed in terms of macroscopic binding constants, but considering its implications for the microscopic constants is also informative. First, we considered the scenario where the two ssDNA binding sites are independent and not cooperative. The microscopic binding constants for K1 are KA and KB, describing DNA binding to site A and site B, respectively. Writing K1 in terms of the microscopic constants, we obtain the following:
For K2, the microscopic binding constants are also KA and KB, provided ligand binding at one site does not alter the binding affinity at the second site, for example, through a conformational change in TLR9 or other allosteric mechanism. Writing K2 in terms of the microscopic constants, K2 = KA + KB. The macroscopic constants K1 and K2 are therefore related as follows:
Since the constants cannot be negative, K2 > K1. This analysis suggests that the most common intermediates in ligand-induced TLR9 dimerization are 1:1 protein:DNA (PD) complexes with one of the binding sites saturated with ligand, which then assemble into 2:2 dimers (P2D2) (Figure 1D). However, this analysis assumes that the two binding sites are independent. If binding of the second ligand is cooperative, K2 could be smaller than K1.
The microscopic binding constants for K3 are more complex than for K1 and K2. The binding affinity of a free protein to an ssDNA that is part of a protein:DNA complex is different than its binding affinity to free DNA. Additionally, protein:protein interactions may promote the K3 transition. In summary, our theoretical analysis of TLR9 ligand binding based on a specific set of assumptions makes testable predictions, specifically K3 > K1 and K2 > K1, and provides a framework for experimental characterization of ligand-induced TLR9 dimerization.
Agonists Induce TLR9 Dimerization
We set out to test our equilibrium binding model and measure key parameters experimentally with recombinant mTLR9-ECD and selected ligands. Oligonucleotides 1668 and 1668-12mer were shown previously to induce dimerization of mTLR9.10 Other agonistic ligands are thought to activate TLR9 in the same manner,22 but a systematic comparison of the effect of different ligands on the oligomeric state of TLR9 has not been performed. Hence, we measured the oligomeric state of recombinant mTLR9-ECD in the presence of five oligonucleotides (Table 1) by dynamic light scattering (DLS). Our panel of ligands included the prototypical agonists 1668 and 2006;11,23 the 1668-12mer oligonucleotide used in the structural studies;10 minM, identified in cell-based assays as the minimal DNA sequence required for potent activation of mouse TLR9;17,18 and antagonistic oligonucleotide 4084, as a control for binding without dimerization.10
Table 1. Sequences and Properties of TLR9 DNA ssDNA Ligands Used in This Study.
As expected, the hydrodynamic radii and molecular masses calculated from DLS indicated that mTLR9-ECD formed a 1:1 complex with the antagonist 4084, and 2:2 complexes with all four agonistic oligonucleotides (Figure 2A,B). The experimentally determined molecular diameters of the complexes were slightly larger than expected and the molecular masses correspondingly smaller, because the DLS data were fitted to a globular model whereas mTLR9-ECD has a nonglobular horseshoe shape.
Figure 2.
Ligand binding assays with mTLR9 ectodomain (mTLR9-ECD) in the presence of various ssDNA ligands. (A) Molecular diameter histograms from dynamic light scattering (DLS). The data shown are representative of at least two independent experiments (see Data Set S1). (B) Table of experimental and theoretical molecular diameters and masses calculated from DLS data. The polydispersity of each sample, related to the peak width in part A, is listed. The theoretical diameters were calculated as twice the radius of gyration, Rg, of monomeric or dimeric TLR9 from the crystal structures10 divided by 0.775, to convert to diameter of hydration, Rh (assuming Rh = Rg/0.775). (C) SEC-MALS of 8 μM mTLR9-ECD with 20 μM oligonucleotide 1668. (D) Masses, Rh, and Rg determined from SEC-MALS data or calculated from the crystal structure.
To obtain a more direct measure of the mass of a TLR9 dimer, size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) was performed on mTLR9 bound to oligonucleotide 1668. As expected, the measured mass of 232 kDa was consistent with a 2:2 dimer (Figure 2C). We note that the experimental hydrodynamic radii (Rh) determined from SEC-MALS and DLS (5.3–5.6 nm) were approximately 10% larger than the theoretical radius predicted from the TLR9:1668-12mer crystal structure (4.9 nm; Figure 2D). This slight discrepancy could be due to the method used to calculate Rh (which was based on the root-mean-square distance from the center of mass), or to the eight additional nucleotides in 1668 versus 1668-12mer, which were not taken into account.
Agonist Binding Assays Are Not Accurately Fitted by a 1:1 Model
To further investigate the binding modes of TLR9 ligands, relative fluorescence anisotropy ligand binding assays were performed. mTLR9-ECD was titrated into 2 nM solutions of oligonucleotides 1668, 1668-12mer, and 4084 labeled with Alexa 488. The binding curves were fitted with a 1:1 ligand binding model, accounting for receptor depletion (Figure 3A–D). The apparent mTLR9 binding affinities were 29 nM for 4084 (95% confidence interval (CI) 18–48 nM, n = 1); 11 nM for 1668-12mer (95% CI 8–15 nM, n = 4); 2.8 nM for 1668 (95% CI 1.7–4.4 nM, n = 3); and 3.2 nM for minM (95% CI 1.7–5.9 nM, n = 1). These values are consistent with expectations, since minM is the most potent ligand, and 1668-12mer has a shorter than optimal sequence. The anisotropy data fitted the 1:1 binding model well for oligonucleotide 4084, which does not induce dimerization. For the agonistic ligands, the data points follow a steeper sigmoidal trajectory than the 1:1 model curve. The 1:1 binding model, while yielding reasonable overall binding curve fits, consistently fails to capture the full cooperativity of the 2:2 dimer complex assembly observed in the data. Without additional data constraining some of the parameters, the 2:2 model in Figure 1 contains too many variables to produce a fit to the anisotropy data with a single well-constrained solution.
Competition Assays Suggest Slow Oligonucleotide Dissociation
The binding sites of the 1668-12mer agonist and 4048 antagonist oligonucleotides partially overlap.10 To establish whether these two oligonucleotides bind competitively, a competition experiment was performed by titrating in mTLR9-ECD preincubated with a molar excess of unlabeled 4048 oligonucleotide into a solution containing Alexa 488-labeled 1668-12mer. No binding of 1668-12mer was observed, indicating that binding of 1668 and 4084 is competitive (Figure S1A). We examined the equilibrium dynamics of this competition by preincubating mTLR9 with Alexa 488-labeled 1668-12mer, titrating in a molar excess of unlabeled 4084 and monitoring displacement of 1668-12mer over time. Unexpectedly, the competition experiments took several hours to reach equilibrium (Figure 3E,F). This was also true when unlabeled 1668-12mer was used instead of 4084 as the competing oligonucleotide (Figure S1B). We conclude that oligonucleotides dissociate from the dimer very slowly, on the time scale of hours.
Deconvolution of the Two Binding Sites Reveals Cooperativity
To deconvolute the contributions of the two ligand binding sites in mTLR9, two key residues involved in ligand binding at site B were mutated. The mutations, E617A and H642A, are predicted to inhibit ligand binding at site B. Since the H642A mutation alone abolished TLR9-dependent signaling in a cell-based assay,10 we also predicted that these mutations would inhibit dimerization. Indeed, Alexa 488-labeled 1668 oligonucleotide bound mTLR9-E617A/H642A with an anisotropy response curve fitting a 1:1 binding model similar to the binding curve for uncleaved mTLR9 (Figure 4A), suggesting that the mutations in site B prevent dimerization. The binding affinity of mTLR9-E617A/H642A for 1668 was 40 nM (95% CI 23–53 nM, n = 2). This provides the affinity of oligonucleotide 1668 for site A, which corresponds to the microscopic constant KA.
Figure 4.
Experimental and simulated ligand binding assays with mTLR9-ECD variants show evidence of cooperativity between the two oligonucleotide binding sites. (A) Affinity of Alexa Fluor 488-labeled oligonucleotide 1668 for mTLR9-ECD mutated at site B. (B) Equilibrium binding affinity of oligonucleotide 1668 to mTLR9-ECD mutated at site A. Despite the mutations, this TLR9 variant remained partially competent for dimerization (see Figure S2). (C) Affinity of oligonucleotide 1668 for mTLR9-ECD without proteolytic activation. Simulated curves calculated using different values of KA and KB are shown alongside the experimental data. (A–C) The data shown are representative of two independent experiments (see Data Set S1). The average Kd’s of the replicates are shown. Numbers in parentheses indicate 95% confidence intervals. (D) A simplified model fitting K1 and K4 alone produced a similar curve (pink) as the 1:1 binding model for 1668 (black). The fitted value for K1 (8.5 nM) is consistent with the observed Kd for uncleaved mTLR9 binding to 1668 (panel C), which is the same as K1 if K2 = 0. The experimental data in panel D are the same as those in Figure 3D.
To similarly determine the microscopic constant KB, mutations predicted to disrupt ligand binding to site A, W96A and Y132A,10 were introduced, and the affinity for oligonucleotide 1668 was measured. The anisotropy data for mTLR9-W96A/Y132A binding to Alexa 488-labeled 1668 yielded a binding affinity for KB of 141 nM (95% CI 40–169 nM, n = 2), calculated from fitting to a 1:1 binding model (Figure 4B). The curve did not fit the 1:1 binding model as well as the site B mutant, however, suggesting that the W96A/Y132A mutations weakened but did not abolish binding of the ligand to site A. Indeed, mass photometry analysis showed that a fraction of mTLR9-W96A/Y132A dimerized in the presence of ligand (Figure S2). The number of contact sites between the protein and ligand at site A is greater than at site B,10 and it may not be possible to fully inhibit binding to site A without destabilizing the fold of mTLR9-ECD.
Cleavage of the ectodomain by an endosomal protease is necessary for dimerization but not for ligand binding.10,24,25 To confirm this in our system, fluorescence anisotropy was measured with uncleaved mTLR9-ECD and Alexa 488-labeled 1668. The binding affinity was 12 nM (95% CI 6–24 nM, n = 2), with a relatively good fit to a 1:1 model curve, consistent with the expected inability of the uncleaved ectodomain to dimerize (Figure 4C). Moreover, since uncleaved TLR9 cannot dimerize, this binding affinity reports on only two of the macroscopic equilibrium constants defined in Figure 1, K1 and K2. The deviation in the data from the theoretical fit (Figure 4C) is likely due to the presence of two ligand binding sites on TLR9, sites A and B, which the crystal structure suggests have different binding affinities.10 Hence, early in the titration the ligand will primarily bind the high-affinity site (site A), with the low-affinity site (site B) becoming saturated with ligand last.
To evaluate whether the experimental binding curves could be accurately predicted, computer simulations of binding curves for 1668 to uncleaved mTLR9-ECD were performed with microscopic constants KA and KB set to values within the ranges determined in Figure 4A,B. Simulations assuming no cooperativity between sites A and B generated binding curves that were shallower than the experimental binding curve (Figure 4C). Lower values of KA (10–20 nM) improved the fit to the experimental data, with KA = 10 nM and KB = 50 nM producing the best-fitting simulated curve, but all simulated curves were less sigmoidal than the data. This suggests that there is cooperativity between the two binding sites, which is unexpected given their physical separation.
We hypothesize above that the most common intermediates in ligand-induced TLR9 dimerization are 1:1 protein:DNA (PD) complexes with one of the binding sites saturated with ligand, which then assemble into 2:2 dimers (P2D2). If the two sites are independent, then modeling the binding as taking place only through the pathways described by the macroscopic constants K1 and K4 should be a good approximation (Figure 1D). To test this, simulations were performed on mTLR9-ECD and 1668 data from Figure 3D using a simplified model in which only K1 and K4 were fitted. This simulation produced a similar curve as the 1:1 model for 1668 and failed to fully capture the sigmoidal shape of the data (Figure 4D). This simulation provides further indirect support that there is cooperativity between the two sites. Binding cooperativity between sites A and B would explain the more sigmoidal shape of all fluorescence anisotropy binding curves presented in this study relative to theoretical or simulated curves plotted assuming the sites were independent.
Discussion
Here, we present a robust theoretical equilibrium binding model for TLR9 binding to DNA ligands complemented by in vitro biophysical data on mTLR9-ECD binding to ligands. Our ligand binding assays confirm that agonistic oligonucleotides induce dimerization of proteolytically activated mTLR9-ECD, whereas it remained monomeric in the presence of antagonistic ligand 4084. All ligands bound TLR9 tightly with overall apparent Kd values in the low nanomolar range. More importantly, binding of ligands that induced TLR9 dimerization did not fit a 1:1 binding model, consistent with a more complex binding mode. The shape of the fluorescence anisotropy binding curves is more sigmoidal than predicted from a 1:1 binding model, suggesting that binding of DNA to sites A and B is cooperative and involves two or more binding events. Moreover, the unexpectedly long time that it took ligand competition experiments to reach equilibrium (several hours) revealed that dissociation of the dimeric complex (P2D2) is very slow, despite the rate of dimer assembly being relatively rapid.
Binding assays with a TLR9 variant containing mutations at site B provided clear evidence that both oligonucleotide binding sites are required for dimerization and provided the microscopic constant for ligand binding to site A, KA (28 nM). Together, these experiments and the DLS data for apo-TLR9 provide experimental evidence that K3 > K1. Computational modeling of uncleaved mTLR9-ECD binding to two ligands with a range of KA and KB values further supports that the two sites are cooperative. With both KA and KB known, ligand binding curves for uncleaved TLR9, an obligate monomer, could be fitted to our 2:2 binding model to further verify whether sites A and B are cooperative, in contrast to what is suggested by the crystal structure.
We present a theoretical model which can be tested experimentally, as advances in biophysics will allow more sensitive equilibrium measurements. The model could be further constrained, for example, by determining whether the species PD2 can be observed in equilibrium conditions containing excess DNA, thus constraining K2 and K4.
Our ligand binding studies were performed with a soluble ectodomain fragment in the absence of auxiliary oligonucleotides (5′-xCx), which were recently shown to augment signaling. The purpose of this study was to develop an accurate model for TLR9-ECD binding to agonistic oligonucleotides and including auxiliary oligonucleotides would have complicated interpretation of ligand binding data. However, the role of auxiliary oligonucleotides is an important area for further study. In particular, it will be important to examine whether 5′-xCx oligonucleotide binding at the auxiliary site is independent of ligand binding at sites A and B, and to determine the mechanism through which auxiliary ligands promote dimerization. Future studies with full-length membrane-inserted TLR9 are also required to understand how the transmembrane and TIR domains may contribute to complex formation.
A complete model for TLR9-DNA binding is presented, and while there are many solutions for the macroscopic equilibrium constants a priori, the experimental data presented narrow the relationships between the macroscopic binding constants. To obtain a unique solution for the complex 2:2 binding model of TLR9 to its ligands, further experimental and numerical analyses are required. Given the structural and mechanistic similarities to other TLRs, most notably TLR7 and TLR8, this work will help establish a more general model for TLR activation and guide future efforts to design TLR9 agonists or antagonists.
Acknowledgments
We thank Christopher M. Johnson (MRC-LMB Biophysics) for guidance in binding assay setup. We thank Prof. Toshiyuki Shimizu (Univ. of Tokyo) for kindly providing TLR9 cDNAs. We thank members of the Modis lab for insightful discussions.
Glossary
Abbreviations
- CpG
unmethylated CG DNA nucleotide sequence motif
- mTLR9-ECD
mouse Toll-like receptor 9 ectodomain
- ssDNA
single-stranded DNA
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00466.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by NIH grant R01-GM102869 and Wellcome Trust Senior Research Fellowships 101908/Z/13/Z and 217191/Z/19/Z to Y.M.
The authors declare no competing financial interest.
Supplementary Material
References
- Beck G.; Habicht G. S. (1996) Immunity and the invertebrates. Sci. Am. 275, 60–66. 10.1038/scientificamerican1196-60. [DOI] [PubMed] [Google Scholar]
- Medzhitov R.; Preston-Hurlburt P.; Janeway C. A. Jr. (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397. 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Blasius A. L.; Beutler B. (2010) Intracellular toll-like receptors. Immunity 32, 305–315. 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L.; Holt A. C.; Medzhitov R.; Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738. 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Diebold S. S.; Kaisho T.; Hemmi H.; Akira S.; Reis e Sousa C. (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531. 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Heil F.; Hemmi H.; Hochrein H.; Ampenberger F.; Kirschning C.; Akira S.; Lipford G.; Wagner H.; Bauer S. (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529. 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hemmi H.; Takeuchi O.; Kawai T.; Kaisho T.; Sato S.; Sanjo H.; Matsumoto M.; Hoshino K.; Wagner H.; Takeda K.; Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745. 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Leleux J. A.; Pradhan P.; Roy K. (2017) Biophysical Attributes of CpG Presentation Control TLR9 Signaling to Differentially Polarize Systemic Immune Responses. Cell Rep. 18, 700–710. 10.1016/j.celrep.2016.12.073. [DOI] [PubMed] [Google Scholar]
- Krieg A. M. (2002) CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20, 709–760. 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- Ohto U.; Shibata T.; Tanji H.; Ishida H.; Krayukhina E.; Uchiyama S.; Miyake K.; Shimizu T. (2015) Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature 520, 702–705. 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- Whitmore M. M.; Li S.; Falo L. Jr.; Huang L. (2001) Systemic administration of LPD prepared with CpG oligonucleotides inhibits the growth of established pulmonary metastases by stimulating innate and acquired antitumor immune responses. Cancer Immunol. Immunother. 50, 503–514. 10.1007/s002620100227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E.; Verma A.; Visintin A.; Gong M.; Sirois C. M.; Klein D. C.; Monks B. G.; McKnight C. J.; Lamphier M. S.; Duprex W. P.; Espevik T.; Golenbock D. T. (2007) Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 8, 772–779. 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- Ohto U.; Ishida H.; Shibata T.; Sato R.; Miyake K.; Shimizu T. (2018) Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation. Immunity 48, 649–658. 10.1016/j.immuni.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Pohar J.; Lainscek D.; Ivicak-Kocjan K.; Cajnko M. M.; Jerala R.; Bencina M. (2017) Short single-stranded DNA degradation products augment the activation of Toll-like receptor 9. Nat. Commun. 8, 15363. 10.1038/ncomms15363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Ohto U.; Shibata T.; Krayukhina E.; Taoka M.; Yamauchi Y.; Tanji H.; Isobe T.; Uchiyama S.; Miyake K.; Shimizu T. (2016) Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 45, 737–748. 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Tanji H.; Ohto U.; Shibata T.; Taoka M.; Yamauchi Y.; Isobe T.; Miyake K.; Shimizu T. (2015) Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 22, 109–115. 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- Pohar J.; Lainscek D.; Fukui R.; Yamamoto C.; Miyake K.; Jerala R.; Bencina M. (2015) Species-Specific Minimal Sequence Motif for Oligodeoxyribonucleotides Activating Mouse TLR9. J. Immunol. 195, 4396–4405. 10.4049/jimmunol.1500600. [DOI] [PubMed] [Google Scholar]
- Pohar J.; Kuznik Krajnik A.; Jerala R.; Bencina M. (2015) Minimal sequence requirements for oligodeoxyribonucleotides activating human TLR9. J. Immunol. 194, 3901–3908. 10.4049/jimmunol.1402755. [DOI] [PubMed] [Google Scholar]
- Weiss J. N. (1997) The Hill equation revisited: uses and misuses. FASEB J. 11, 835–841. 10.1096/fasebj.11.11.9285481. [DOI] [PubMed] [Google Scholar]
- Li Y.; Berke I. C.; Modis Y. (2012) DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J. 31, 919–931. 10.1038/emboj.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favicchio R.; Dragan A. I.; Kneale G. G.; Read C. M. (2009) Fluorescence spectroscopy and anisotropy in the analysis of DNA-protein interactions. Methods Mol. Biol. 543, 589–611. 10.1007/978-1-60327-015-1_35. [DOI] [PubMed] [Google Scholar]
- Vollmer J.; Krieg A. M. (2009) Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv. Drug Delivery Rev. 61, 195–204. 10.1016/j.addr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hartmann G.; Weeratna R. D.; Ballas Z. K.; Payette P.; Blackwell S.; Suparto I.; Rasmussen W. L.; Waldschmidt M.; Sajuthi D.; Purcell R. H.; Davis H. L.; Krieg A. M. (2000) Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J. Immunol. 164, 1617–1624. 10.4049/jimmunol.164.3.1617. [DOI] [PubMed] [Google Scholar]
- Ewald S. E.; Lee B. L.; Lau L.; Wickliffe K. E.; Shi G. P.; Chapman H. A.; Barton G. M. (2008) The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662. 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.; Brinkmann M. M.; Spooner E.; Lee C. C.; Kim Y. M.; Ploegh H. L. (2008) Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9, 1407–1414. 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.