Abstract
Mammalian pluripotent stem cells (PSCs) have distinct molecular and biological characteristics among species, but to date we lack a comprehensive understanding of regulatory network evolution in mammals. Here, we carried out a comparative genetic analysis of 134 genes constituting the pluripotency gene regulatory network across 48 mammalian species covering all the major taxonomic groups. We report that mammalian genes in the pluripotency regulatory network show a remarkably high degree of evolutionary stasis, suggesting the conservation of fundamental biological process of mammalian PSCs across species. Nevertheless, despite the overall conservation of the regulatory network, we discovered rapid evolution of the downstream targets of the core regulatory elements and specific amino acid residues that have undergone positive selection. Our data indicate development of lineage-specific pluripotency regulating networks that may explain observed variations in some characteristics of mammalian PSCs. We further revealed that positively selected genes could be associated with species’ unique adaptive characteristics that were not dedicated to regulation of PSCs. These results provide important insight into the evolution of the pluripotency gene regulatory network underlying variations in characteristics of mammalian PSCs.
Keywords: adaptation, embryonic development, gene network, Mammalia, natural selection, pluripotent stem cells
Significance
Pluripotent stem cells (PSCs) have emerged as advanced cellular and genetic material for mammalian research. Despite their interspecies similarities related to their ability to differentiate into other somatic cells and to divide into make more stem cells with maintenance of the undifferentiated state, PSCs from different species show variations in some of their characteristics such as pluripotent state and gene expression. Various studies have shown molecular mechanisms underlying the maintenance of PSC characteristics that constitute a complex gene regulatory network. Unfortunately, such knowledge is largely limited to primates and rodents. This study provides comparative analyses involving pluripotency-regulating genes across mammalian taxa. As such, it helps the understanding of the genetic basis for the evolution of the mammalian PSCs.
Introduction
Pluripotent stem cells (PSCs) are undifferentiated cells that exhibit unlimited self-renewability and pluripotency, the potential to give rise to cells from all three embryonic germ layers. Pluripotent embryonic stem cells are isolated from the inner cell mass of developing preimplantation mouse or human blastocysts (Evans and Kaufman 1981; Martin 1981; Thomson 1998). Although PSCs of rare animal species represent powerful resources for research and conservation, little equivalent derivations have been attempted with these species for ethical and technical reasons.
Recent advances in somatic cell reprogramming into induced pluripotent stem cells (iPSCs) have broadened the possibility of obtaining PSCs from variety of mammals, including endangered species (Ben-Nun et al. 2011). Derivation of iPSCs has been reported from a wide range of taxonomic groups, including Carnivora (Shimada et al. 2009; Verma et al. 2012, 2013; Menzorov et al. 2015), Cetartiodactyla (Ezashi et al. 2009; Han et al. 2011; Liu et al. 2012), Chiroptera (Mo et al. 2014), Lagomorpha (Osteil et al. 2013), Metatheria (Weeratunga et al. 2018), Monotremata (Whitworth et al. 2019), Perissodactyla (Ben-Nun et al. 2011; Breton et al. 2013), Rodentia (Takahashi and Yamanaka 2006; Liao et al. 2009; Miyawaki et al. 2016; Lee et al. 2017), and Primates (Takahashi et al. 2007; Tomioka et al. 2010; Marchetto et al. 2013; Wunderlich et al. 2014; Ramaswamy et al. 2015). However, questions remain about species variation in the properties of the PSCs, such as pluripotent state, reprogramming efficiency, and optimal culture conditions.
A highly interconnected pluripotency gene regulatory network (PGRN) controls the characteristics of PSCs (Li and Belmonte 2017). Mammals seem to possess evolutionarily conserved PGRN because iPSCs from different taxonomic groups have been derived using human genetic sequences of identical sets of transcription factors (OCT4, SOX2, KLF4, and MYC; collectively referred to as OSKM). However, various pluripotent states and configurations are observed across different species (Ezashi et al. 2016; Weinberger et al. 2016; Paterson et al. 2018), indicating diversity and specificity of the PGRN among species. However, our understandings of the PGRN are narrow, largely limited to primates and rodents (Manor et al. 2015), and to date the evolutionary history of the PGRN across different mammalian taxonomic groups has not been studied in depth.
Comparative approaches can provide insights into the evolution of gene regulatory processes by examining the conservation and divergence of networks (Thompson et al. 2015). Evolutionary conservation and adaptations can be inferred by detecting purifying and positive selection (Nielsen et al. 2007). We have recently described genetic signatures for phenotypic adaptations in cetacean lipid metabolism (Endo et al. 2018), and numerous studies have revealed the effects of natural selection in the development of adaptive characteristics of animals and plants (Lenski 2017). Importantly, effects of changes on a regulatory process depends on the hierarchical position of the changes within the regulatory network (Erwin and Davidson 2009) so an holistic approach is recommended. Evolutionary pattern of the PGRN within the Class Mammalia and how natural selection acts on the genes involved in the PGRN, however, are yet to be described.
In this study, we aimed to investigate whether any evolutionary pattern among mammalian taxa could be the result of selection of genes involved in the PGRN. To achieve this we conducted a comparative selection analyses of the PGRN genes across mammals covering all the major taxonomic groups. 1) To assess the evolutionary conservation profiles of the mammalian PGRN, we estimated trends in the stringency of purifying selection. 2) To identify rapidly evolving subcircuits of the PGRN, we compared the evolutionary rate based on the ratio of substitution rates at nonsynonymous and synonymous sites (dN/dS). 3) To detect phylogenetic inference of the variations in the PGRN, we identified genes under positive selection for lineages and investigated concordance in functional regions of the proteins. This paper presents the evolutionary history underlying the conservation and variations in mammalian PGRN, and provides the genetic basis for the evolution of the PGRN in mammals.
Materials and Methods
Data Sampling
Target genes were selected to include all 138 genes involved with “Signaling pathways regulating pluripotency of stem cells” (hsa04550) from KEGG pathway database (Kanehisa et al. 2017). Human coding DNA sequences (CDS) of those genes were used as references and blasted against publicly available genomes of 61 mammals, covering all major taxonomic groups. Gallus gallus, Xenopus tropicalis, and Danio rerio were used as an outgroup. The set of genes was tested for orthologs using OMA stand alone v2.3.0 (Altenhoff et al. 2018). When a CDS was not inferred as an ortholog, another CDS was selected based on its annotation and tested with OMA again. Because the genomes were of varying quality, mammalian species with the same or lower number of orthologs than G. gallus were removed. Genes in which orthologs were found in less than 80% of the study species were also excluded from the further analyses. Orthologs were aligned with using PRANK v.140603 (Löytynoja and Goldman 2005) and the aligned sequences were further tested for their quality with GUIDANCE2 v2.02 (Sela et al. 2015) using the default settings. For the chicken genes, NANOG (Gene ID: 100272166, NP_001139614.1) was included in analyses even though it was not determined as ortholog by OMA because it is well studied and known ortholog of human NANOG (Fuet and Pain 2017). A consensus tree was prepared based on commonly accepted mammalian phylogenies (Meredith et al. 2011; Perelman et al. 2011) and applied for later natural selection analyses. The gene ontology (GO) and the KEGG orthology (KO) were determined according to the Gene Ontology Annotation and KEGG, respectively.
Testing the Stringency of Natural Selection
Natural selection was tested for each gene using RELAX (Wertheim et al. 2015) in HyPhy (Pond et al. 2005). RELAX is a general hypothesis testing framework that determines whether selective strength, distinguishing purifying or positive selection, was relaxed or intensified in the subsets of interest (Wertheim et al. 2015). RELAX estimates the selection intensity parameter, k, where k ≥ 1 indicates intensification and k < 1 indicates relaxation of natural selection, where ω > 1 represents positive selection and ω < 1 represents purifying selection. Mammalian lineages were set as a test branch and nonmammalian vertebrates as a reference branch. Because RELAX requires reference branches, genes without nonmammalian orthologs were excluded. P values were corrected for false discovery rate (FDR) using the Benjamini–Hochberg method (Benjamini and Hochberg 1995) with a cutoff of 10%.
Evolutionary Rate of Each Gene
Evolutionary rate of each gene was estimated by using the model M0 in CodeML (PAMLv.4.8) (Yang 2007). The synonymous substitution rate (dS), the nonsynonymous substitution rate (dN), and their ratio, dN/dS (also noted as ω) were calculated, where dN/dS < 1 indicates purifying selection, dN/dS = 1 indicates neutral, and dN/dS > 1 indicates positive selection. The dN/dS ratio was estimated for each gene using mammalian orthologs.
Comparison of Conservation Patterns among Subcircuits
The categories of subcircuits were determined according to KEGG descriptions in the “Pathway” category, including Core, JAK–STAT, MAPK, OCT4 (POU5F1), SOX2, and NANOG (OSN) activated, OSN suppressed, PI3K–Akt, TGFβ, and Wnt signaling pathways. As for the Core subcircuits, we included the genes in the core transcriptional network, OSN and the genes which interact with the OSN inside of nucleus. When one gene was tagged in multiple subcircuits, it was grouped in all the related pathways. Because subcircuits contain unequal sample sizes, multiple paired comparisons for the means of dN/dS ratios were statistically tested using ANOVA followed by the Tukey–Kramer post-test with P value of 0.05.
Lineage-Specific Positive Selection
Positively selected genes were identified using the branch-site model (Zhang et al. 2005) implemented in CodeML (PAMLv.4.8) (Yang 2007). The modified model A as an alternative hypothesis was compared with the model A1 as the null. P values were evaluated under the likelihood ratio test by comparing 2Δl of the two models to the 1:1 mixture of 0 and χ2 distribution. Lineage-specific positive selection was tested by setting each contemporary species and ancestral branch as a priori specified target separately and corrected for FDR using the Benjamini–Hochberg method (Benjamini and Hochberg 1995) with a cutoff of 10%.
Functional Impact
Positively selected sites were determined using the Bayes empirical Bayes (BEB) implemented in the branch-site model (Yang et al. 2005) with 95% posterior probability. Lineage-specific amino acid substitutions were explored in positively selected genes identified in ancestral branches as indicative of significant changes in protein function (Tian et al. 2013). The lineage-specific amino acid substitutions were identified manually by looking at a conversion of residue that occurred and fixed in more than 90% of the descendant contemporary species Functional domain structures and regions were referred to UniPort and Pfam using human protein as a reference.
Results
Summary of Sample Data
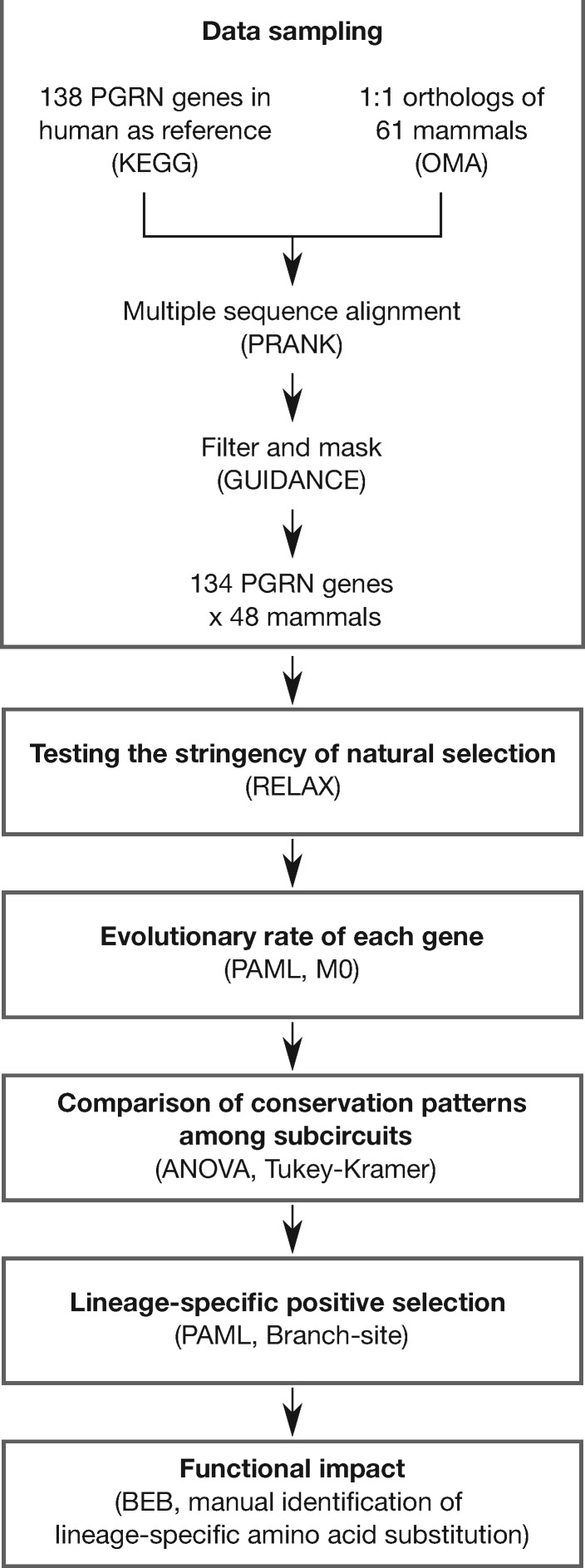
To investigate the evolutionary pattern of the gene regulatory network, the PGRN, that maintains pluripotency and self-renewability of PSCs, we performed comparative genetic analyses across mammalian species (fig. 1). We collected the protein coding sequences of PGRN-related genes covering all the major taxonomic groups. A total of 61 mammalian species were used in this study including Classes Primates, Rodentia, Cetartiodactyla, Chiroptera, Carnivora, Insectivora, Afrotheria, and Metatheria, with at least two species from each. For each species, we searched orthologs of the 138 genes assigned to functions associated with signaling pathways regulating pluripotency of stem cells. Our filtering strategy retained 134 gene sets from 48 mammalian species for later analyses (see Materials and Methods section, supplementary table S1, Supplementary Material online).
Fig. 1.
Flow diagram detailing the strategy to investigate the evolution of the PGRN across mammals. Boxes represent steps of methods in processing order. Key approaches used for each step are indicated in parenthesis.
Evolutionarily Conserved Profiles across Mammals
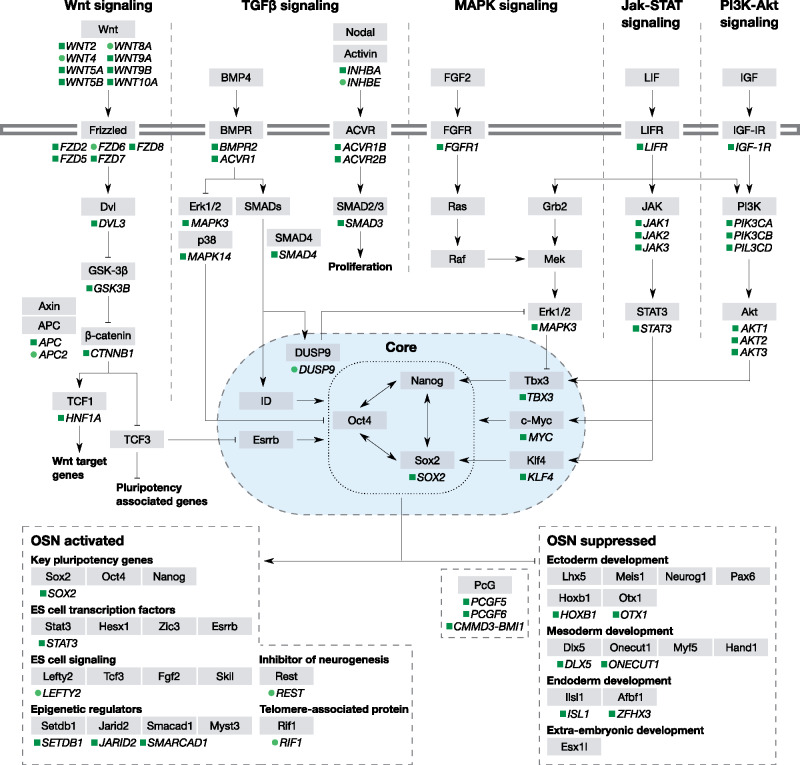
To assess evolutionary constraint and reduction of selective strength in the mammalian PGRN, we examined the level of purifying selection with each gene. Of the 127 orthologous genes analyzed, we observed intensification of purifying selection in 53 genes (41.7%) as well as relaxation in 9 genes (7.1%) at P < 0.05 FDR <10%, revealing that intensification of purifying selection was more common among the PGRN (fig. 2, supplementary table S2, Supplementary Material online). For the commonly used reprogramming factor, OSKM, we found high degrees of genetic conservation in SOX2, KLF4, and MYC across mammals. We observed relaxation of purifying selection in a number of genes involved in the molecular mechanisms that determine the pluripotent states (APC2, DUSP9, INHBE, LEFTY2, and RIF1) and in a number of oncogenes and tumor suppressor genes across mammals (APC2, FZD6, INHBE, REST, RIF1, WNT4, and WNT8A). Intensification of purifying selection was observed across the PGRN, whereas relaxation appeared to be limited in particular subcircuits. This led to the question whether any conservation patterns among the PGRN subcircuits.
Fig. 2.
Trends in the stringency of purifying selection on the PGRN genes. Schematic representation of the PGRN according to KEGG is shown. Only genes with significant shifts in the stringency of purifying selection at 10% FDR or less are indicated: Green rectangle, intensification; lime circle, relaxation. Dashed lines indicate the subcircuits of the PGRN. Horizontal double line indicates the cellular envelop. Blue oval represents the nucleus. The genes under each subcircuit are simplified, see supplementary table S4, Supplementary Material online.
Conservation Patterns among the PGRN Subcircuits
We then asked if there are differences in degree of conservation among subcircuits constituting the PGRN. In the purifying selection analysis identified above, we found relatively higher number of genes with intensified purifying selection in JAK–STAT, OSN suppressed, and PI3K–Akt signaling pathways (52.9%, 50%, and 44.8%, respectively), whereas more relaxed genes were found in OSN activated and Wnt signaling pathways (20% and 10%, respectively) (table 1).
Table 1.
Comparison of Selective Pressure among the Subcircuits
| No. Gene with Significant Shifts in Strength of Purifying Selection |
|||
|---|---|---|---|
| Subcircuits | No. gene | Intensified (%) | Relaxed (%) |
| Core | 11 | 4 (36.4) | 1 (9) |
| JAK–STAT | 17 | 9 (52.9) | 0 |
| MAPK | 26 | 8 (30.8) | 1 (3.8) |
| OSNa activated | 15 | 4 (26.7) | 3 (20) |
| OSNa suppressed | 18 | 9 (50) | 0 |
| PI3K–Akt | 29 | 13 (44.8) | 0 |
| TGFβ | 27 | 9 (33.3) | 2 (7.4) |
| Wnt | 40 | 16 (40) | 4 (10) |
OCT4 (POU5F1), SOX2, and NANOG.
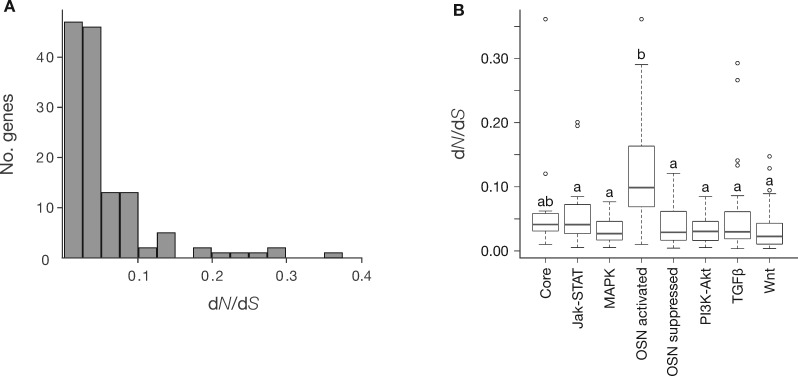
To gain insights into the main force that has shaped the evolution of mammalian PGRN, we estimated the evolutionary rates, dN/dS, of each gene. The estimated dN/dS values ranged from 0.0036 to 0.3614. All the genes showed dN/dS values lower than 1 and 95.5% lower than 0.2 (fig. 3A, supplementary table S3, Supplementary Material online). Mean dN/dS values of each subcircuit were generally low suggesting high degree of evolutionary stasis at level of protein-coding sequence (supplementary table S4, Supplementary Material online). ANOVA indicated that evolutionary rates differ among subcircuits (supplementary table S5, Supplementary Material online), and the following Tukey–Kramer showed that the OSN activated subcircuits, the downstream of the core transcriptional network, had significantly higher evolutionary rate compared with other subcircuits with P < 0.05 (fig. 3B and supplementary table S5, Supplementary Material online).
Fig. 3.
Conservation patterns of the PGRN. (A) Histogram of gene counts for dN/dS values showing distribution of evolutionary rates for the PGRN genes. (B) Box plots of the mean dN/dS values for the PGRN subcircuits. Multiple paired comparisons were tested with the Tukey–Kramer method, where subcircuits not assigned by the same latter are significantly different with P value of 0.05; error bars, 95% confidence interval. OSN represents OCT4 (POU5F1), SOX2, and NANOG.
Lineage-Specific Positive Selection
If genes in the OSN activated subdivision experience lower evolutionary constraint across mammals, some species may have developed unique characteristics through changes in these genes over evolutionary time periods. To test this, we performed positive selection analysis for the genes in the OSN activated subdivision on all the ancestral branches and contemporary species in the mammalian taxonomic tree containing our study species. Of the 15 OSN activated genes, we identified 8 positively selected genes in 4 ancestral branches and 22 contemporary species with 10% FDR correction (fig. 4 and supplementary table S6, Supplementary Material online). The four ancestral branches where positively selected genes were found include the common ancestor of Eutheria, Primatomorpha (Primates and the flying lemur), Megachiroptera, and Pan. The number of positive selection events was the highest in the gene TCF3, which is an integral component of the core regulatory circuitry of PSCs (Cole et al. 2008) (one ancestral branch and eight contemporary species). Although three of the four tested epigenetic regulators (KO: 03036) have shown to be under intensified purifying selection across mammals (fig. 2), a relatively high number of positive selection events with KAT6A, SETDB1, and JARID2 were observed in Primates (five events over nine total events in Primates and over 11 events with these genes across mammals).
Fig. 4.
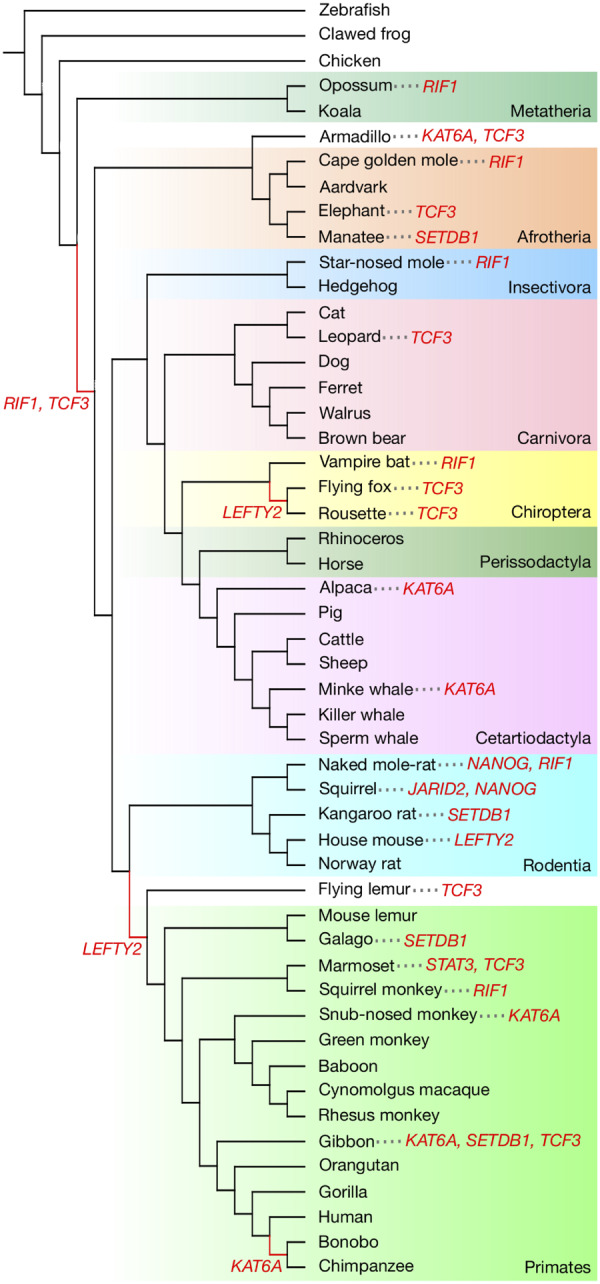
Mapping of positively selected genes identified in ancestral branches and contemporary species of mammals. Positively selected genes at 10% FDR or less (red) are indicated on a commonly accepted phylogeny of mammals. Ancestral branches with which positively selected genes were identified are shown in red. Colored boxes represent major taxonomic groups referred to the NCBI taxonomy.
Functional Impact of Positively Selected Genes at Nucleotide Level
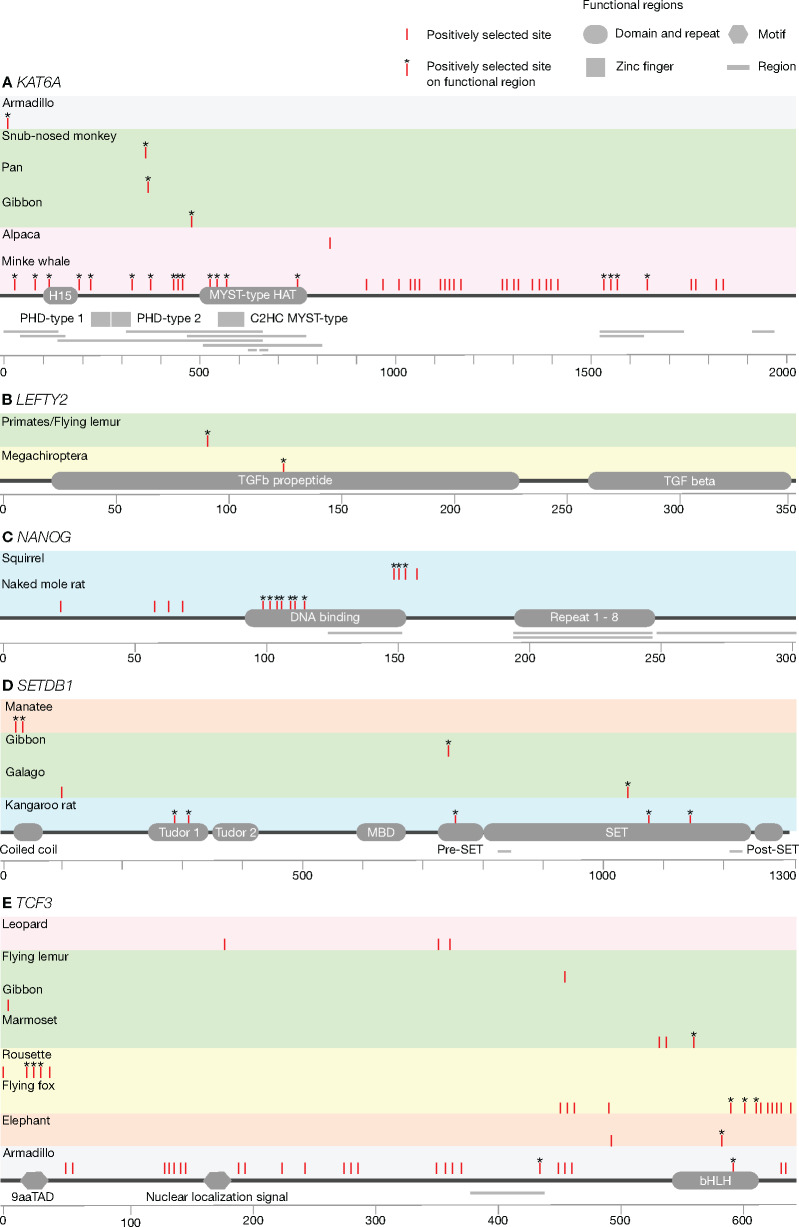
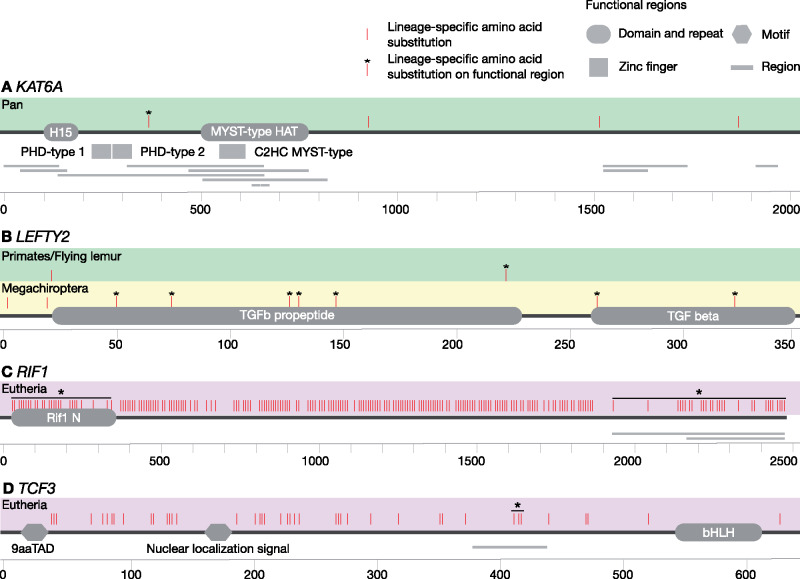
To assess whether the identified positively selected genes have undergone functional modification, we investigated the positively selected sites of the eight positively selected genes for each selected lineage (supplementary table S7, Supplementary Material online). The positions of positively selected sites were compared with protein functional regions using humans as reference. Although the majority of the positively selected sites were found outside of known functional regions, we found sites under positive selection in functional regions of five proteins encoded by KAT6A, LEFTY2, NANOG, SETDB1, and TCF3 (fig. 5). We also found lineage-specific amino acid substitutions in functional regions of the four positively selected genes at ancestral branches, including KAT6A, LEFTY2, RIF1, and TCF3 (fig. 6 and supplementary table S8, Supplementary Material online). These lineage-specific amino acid substitutions were unique to and shared among the descendent contemporary species, indicating functional importance of these genetic mutations (Tian et al. 2013).
Fig. 5.
Positively selected sites of the OSN activated genes: (A) KAT6A, (B) LEFTY2, (C) NANOG, (D) SETDB1, and (E) TCF3. Positively selected lineages and their positively selected sites with 95% posterior probability are mapped on the coded protein with the arrangements of functionally important regions. Only genes with significant positively selected sites in at least one lineage and on at least one functional region are shown. The functional region represented with bar is a region of interest that cannot be described in other subsections according to UniPort. Colored boxes represent major taxonomic groups [Afrotheria {orange}, Carnivora {red}, Cetartiodactyla {fuchsia}, Chiroptera {yellow}, Primates/flying lemur {lime}, Rodentia {aqua}, and other {gray}].
Fig. 6.
Lineage-specific amino acid substitutions in positively selected genes at ancestral branch: (A) KAT6A, (B) LEFTY2, (C) RIF1, and (D) TCF3. Positively selected lineages and their specific amino acid substitutions are mapped on the coded protein with the arrangements of functionally important regions. The functional region represented with bar is a region of interest that cannot be described in other subsections according to UniPort. Colored boxes represent major taxonomic groups [Eutheria {purple}, Chiroptera {yellow}, and Primates/flying lemur {lime}].
Discussion
Genetic Conservation of the PGRN
The relatively higher number and prevalence of evolutionary conserved genes across the PGRN indicates overall comparability of the regulating mechanisms for pluripotency and self-renewability of mammalian PSCs (fig. 2). In context of the reprogramming somatic cells into iPSCs, although human sequences are effective (Ben-Nun et al. 2011), the efficient combination of reprogramming factors may differ among species (Tomioka et al. 2010; Verma et al. 2013; Mo et al. 2014; Weeratunga et al. 2018). Evidence of evolutionary conservation in the three commonly used reprogramming factors explains the efficiency of human genetic sequences for reprogramming somatic cells from variety of species (Ezashi et al. 2016). However, low success rate and inefficiency of reprogramming by human OSKM are reported in some species (Verma et al. 2013; Mo et al. 2014; Whitworth et al. 2019). For increasing reprogramming efficiency, additional expression of other pluripotency-associated genes have been shown to enhance reprogramming rate (Takahashi and Yamanaka 2016). We detected significant evidence of purifying selection across mammals for TBX3, which improves reprogramming efficiency in mouse (Han et al. 2010), implying that TBX3 might be a first candidate enhancer for derivation of mammalian iPSCs with low reprogramming efficiency.
The characteristics of mammalian PSCs appear to differ among species (Ezashi et al. 2016; Weinberger et al. 2016; Paterson et al. 2018) and reduction in selective constraint highlights evolutionary flexibility and innovation (Lahti et al. 2009; Moczek 2010; Hunt et al. 2011). For example, pluripotent state of PSCs can be described with multiple types such as naive or primed states, and the amenabilities of the pluripotent state in vitro differ between species (Boroviak et al. 2015). Although naive state PSCs are cultured with dual inhibition (2i) of MEK and GSK3 and leukemia inhibitory factor (LIF), human and mouse embryonic stem cells show distinct transcriptional responses to the 2i/LIF (Huang et al. 2014). Our findings of relaxation of purifying selection on genes, such as RIF1, may explain the diversity of the amenable pluripotent state of the species. RIF1 is involved in cellular response to LIF (GO: 1990830). INHBE and LEFTY2 participate in the regulation of MAPK cascade (GO: 0043408), the target of MEK inhibitor, and the protein encoded by DUSP9 is the essential regulator of MAPK (GO: 0000165, 0000187, 0000188). APC2 is known to form a protein complex with the scaffold Axin and the kinases GSK3 and CK1 (Pronobis et al. 2015). Mammals lack correlation between body size or life span and cancer risk which is referred to as “Peto's paradox” (Peto et al. 1975; Caulin and Maley 2011; Peto 2015). Our findings of the relaxation of selective strength for oncogenes and tumor suppressor genes may imply evolutional flexibility in mammalian cancer resistance.
Variable Evolutionary Conservation Patterns among PGRN Subcircuits
The patterns of genetic conservation among the PGRN subcircuits provide insight into the conservation and variations of the characteristics of mammalian PSCs (table 1 and fig. 3B). Multiple upstream signaling pathways serve to maintain the self-renewability and pluripotency of PSCs. We found relatively high degree of conservation in JAK–STAT, OSN-suppressed, and PI3K–Akt signaling pathways, suggesting conservation of fundamental biological characteristics and homeostasis in mammalian PSCs. JAK–STAT signaling pathway is stimulated by LIF and essential for self-renewal (Niwa et al. 1998). PI3K signaling pathway plays a crucial role for proliferation in mouse embryonic stem cells (Takahashi et al. 2005) and equivalency of its function in mouse and primate ES cells has been reported (Watanabe et al. 2006). Furthermore, the role of PI3K signaling pathway in the proliferation of PSCs has been demonstrated in the horse (Whitworth et al. 2014). Our findings support the common regulatory mechanisms observed among phylogenetically distant taxonomic groups. However, we observed the relatively higher number of relaxed genes in the Wnt signaling pathway, the downstream target of which, ESRRB, is necessary and sufficient to mediate self-renewal independently of JAK–STAT signaling pathway (Martello et al. 2012), that may suggest evolutionary flexibility in the parallel pathways supporting self-renewal.
The core transcriptional network genes, OSN, orchestrate a cascade of regulatory events involving an autoregulatory loop involving the other pluripotency regulators (Cole et al. 2008; Li and Belmonte 2017). We observed relatively high evolutionary rates with the genes activated by OSN suggesting diversity of subsequent regulating mechanisms among mammals. Importantly, changes in the proximal targets of the master regulators could have profound influence on the downstream circuitry (Erwin and Davidson 2009), our observations of relatively high evolutionary rates with the OSN activated subcircuit may reflect the variations of some characteristics among mammalian PSCs. In addition, understanding mammalian diversity in the downstream targets of the core transcriptional network may help elucidate the mechanisms of reprogramming toward pluripotency and increase the reprogramming efficiency (Takahashi and Yamanaka 2016). Thus, we focused our later analyses on the genes in the OSN activated subcircuit for exploring variations in the mammalian PGRN.
Development of Lineage-Specific PGRN
Evidence for positive selection on PGRN genes has implications for development of lineage-specific regulatory networks (fig. 4). We observed evidence of frequent episodes of positive selection with TCF3, implying that alternation of the PGRN have occurred multiple times during mammalian evolution. TCF3 is the integral component of the core regulatory circuitry of PSCs and encodes a member of the E protein family of helix-loop-helix transcription factors and plays a crucial role by binding to the component of the core transcription factors (Cole et al. 2008; Yi et al. 2008). We found that the common ancestor of Eutheria has been positively selected with fixed lineage-specific amino acid substitutions with TCF3, which targets and controls the expression of ESRRB (Yi et al. 2008; Martello et al. 2012). ESRRB has been proposed to be recruited to the Eutheria PGRN after the divergence of marsupials and Eutheria because neither Tasmanian devil nor platypus iPSCs express ESRRB (Weeratunga et al. 2018; Whitworth et al. 2019). Importantly, ESRRB not only regulates self-renewal in PSCs (Martello et al. 2012) but also plays a key role in trophoblast, which forms placenta (Luo et al. 1997; Latos et al. 2015). Our findings imply that functional modification in TCF3 may have caused rewiring Eutherian PGRN to incorporate ESRRB that subsequently led to the acquisition of placenta. We also report genetic evidence of positive selections with TCF3 and its another target LEFTY2 (Cole et al. 2008) in Megachiroptera. Positive selection has happened in the ancestral branch of Megachiroptera with LEFTY2 and the two descendant species, the large flying fox (Pteropus vampyrus) and the rousette (Rousettus aegyptiacus) in TCF3, highlighting the evolutionary history of pre- and postdivergence of these taxa. Possible implications from this observation will be discussed in the following section. TCF3 also regulates the expression of NANOG (Pereira et al. 2006; Cole et al. 2008). The reprogramming efficiency of NANOG varies among species; whereas overexpression of NANOG increases the reprogramming rate in felids (Verma et al. 2012, 2013), it does not increase the reprogramming rate in marmosets (Tomioka et al. 2010). We found that both the leopard and the marmoset have positively selected sites in their TCF3, but at different positions (fig. 5E), which may reflect the variations of response to NANOG. This could have implications in development of lineage-specific PGRNs in these taxa.
Potential Drivers of Mammalian PGRN Variation
Our findings of lineage-specific positively selected genes among the PGRN genes provide insight into the influence of species adaptation on the PGRN and the characteristics of mammalian stem cells. The architecture of developmental gene regulatory networks, such as the PGRN, is composed of diverse components. Certain subcircuits of regulatory networks are not dedicated to a particular biological process, but are also used for diverse functions that might have led to species adaptations. The naked mole-rat (Heterocephalus glaber) exhibits extraordinary longevity and cancer resistance (Buffenstein 2008; Delaney et al. 2013). The iPSCs derived from the naked mole-rat have shown to resist tumor formation through the expression of ARF (Miyawaki et al. 2016), in which the Arf/p53 pathway has a protective role from cancer and aging (Matheu et al. 2008). Consistent with this finding, we found NANOG and RIF1 to be positively selected in this species. RIF1 encodes a protein that regulates DNA replication and damage, interacting with 53BP1 (Kumar and Cheok 2014) which enhances p53-dependent transcriptional responses (Cuella-Martin et al. 2016). NANOG is not only one of the core transcriptional network genes, but also exhibits tumorigenic activity through interaction with p53 (Cuella-Martin et al. 2016; Kim et al. 2016). Our finding of positively selected sites on the DNA-binding motifs of NANOG further suggests development of cancer resistance with p53 pathway in addition to ARF (fig. 5C).
Alternatively, the evidence of positive selection on RIF1 may indicate convergent evolution of species adaptation for perception. We identified that RIF1 has been positively selected in the naked mole-rat, the Cape golden mole (Chrysochloris asiatica), the star-nosed mole (Condylura cristata), and the vampire bat (Desmodus rotundus). RIF1 plays an important role in DNA replication and damage (Kumar and Cheok 2014), and these responses are important for neurogenesis, as represented in human diseases such as Meier-Gorlin syndrome and Wolf-Hirschhorn syndrome (Kerzendorfer et al. 2013). Animals that live underground have developed unique sensory systems such as the somatosensory vibrissa-like body hairs on the body of the naked mole-rat (Crish et al. 2003), the middle ear structure of airborne and seismic stimuli in the Cape golden mole (Willi et al. 2006), and the “star” of the star-nosed mole (Gould et al. 1993). Among Microchiroptera, a group that uses echolocation, the vampire bat has extremely sensitive neurons that detect not only echolocation signals but also low frequency sounds, presumably for foraging prey in the dark (Schmidt et al. 1991). Our results provide insights into additional possible development of cancer resistance in mammals.
Differences in sites under positive selection in the protein functional domains may reflect species unique adaptation (fig. 5E). As discussed previously, two Megachiroptera species have been identified under positive selection with TCF3, which is also associated with cancer (Patel et al. 2015) and neuronal differentiation (Kuwahara et al. 2014). We observed multiple positively selected sites on the functional motif basic helix-loop-helix (bHLH) of TCF3 in the large flying fox (P. vampyrus), among the largest species of bat with a wingspan of up to 1.5 meter (Kunz and Jones 2000). Potential mechanisms of reducing cancer risk through response to DNA damage has been reported in elephants (Abegglen et al. 2015; Vazquez et al. 2018). Interestingly, we also observed positively selected sites on the bHLH of TCF3 in elephants, implying the resolution to Peto's paradox and the underlying convergent evolution between species that have developed a larger body size compared with their phylogenetic relatives. We identified positively selected sites on the transactivation domain, 9aaTAD in the Rousette bat (R. aegyptiacus), which has been reported to exhibit enhanced infection tolerance (Pavlovich et al. 2018). Whereas the previous study revealed genetic signatures of unique signaling in NK cell receptors (Pavlovich et al. 2018), E protein encoded by TCF3 plays a critical role in B and T lymphocyte development (Engel et al. 2001; Seet et al. 2004), suggesting multiple strategies for antiviral defense in the rousette.
The roles of transposable elements in genomic rearrangement, gene regulation, and epigenetics have been extensively studied to understand primate evolution (Lee et al. 2015). Our findings of the relatively frequent positive selection with epigenetic regulators among Primates may reflect the various impacts of transposable elements on the primate genome. Among Primates, we observed that two epigenetic regulators were under positive selection in the gibbon (Nomascus leucogenys), whose chromatin interactions and epigenetic landscape has been remarkably conserved in spite of extensive genomic shuffling (Lazar et al. 2018). Interestingly, we also observed an event of positive selection at the common ancestor of Primates and the flying lemur with LEFTY2, with which DNA methylation plays a critical role during early embryogenesis in vertebrates (Wang et al. 2017), although the results need to be treated with caution.
Due to the habitat transition from terrestrial to aquatic environment, cetaceans have achieved a remarkable changes in their morphology (Uhen 2010). The minke whale (Balaenoptera acutorostrata) genome has provided support for genetic changes in HOX genes in this species (Yim et al. 2014), which have an important role in the body plan and embryonic development (Pearson et al. 2005). Consequently, we observed that the minke whale has been under positive selection with KAT6A, which regulates the expression of HOX gene (Voss et al. 2009), implying the morphological adaptation of the whale to the aquatic environment. However, our analyses did not indicate significant evidence of positive selection with KAT6A in other cetacean species. Because KAT6A is also associated with senescence and tumor growth (Baell et al. 2018), our findings with the minke whale may imply the adaptation of longevity and resistance to age-related diseases as illustrated with the genome and transcriptomes of the bowhead whale (Keane et al. 2015).
Overall, our data indicated that the PGRN genes positively selected in some species are also involved in their unique adaptations, which may subsequently alter their regulatory function in PSCs. Further efforts, such as in vitro genetic modification and characteristics observation, are necessary to test the functional consequences of the genetic mutations discovered in this study.
Conclusions
Our analyses illustrate the evolutionary patterns in the PGRN, underlying the similarities and variations in characteristics among mammalian PSCs. This study is one of the first to compare the PGRN genes across major taxa. We showed the evolutionary conservation profiles of the mammalian PGRN and uncovered the evolutionary variable PGRN subcircuits. We identified phylogenetic inference of positive selection of genes involved in the PGRN which has enabled insights into development of lineage-specific PGRN and linkage between PGRN genes and species adaptation. These genes and the associated subcircuits will be plausible targets for future investigations exploring mammalian PSCs.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Rob Ogden and Rebecca N. Johnson for constructive feedback on manuscript and English correction; Kristin Havercamp for valuable editorial input and English correction. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI (17H03624, 20H00420 to M.I.-M., 17H02083 to K.K.), and Kyoto University Supporting Program for Interaction-based Initiative Team Studies (SPIRITS) to M.I.-M.
Author Contributions
Y.E. designed the original concept and scientific objectives of the project; performed data collection and data analyses; and wrote the manuscript. M.I.-M. and K.K. coordinated and participated in the design of the study, and helped draft the manuscript. All authors gave final approval for publication.
Contributor Information
Yoshinori Endo, Wildlife Research Center, Kyoto University, Japan.
Ken-ichiro Kamei, Institute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Japan.
Miho Inoue-Murayama, Wildlife Research Center, Kyoto University, Japan; Institute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Japan; Wildlife Genome Collaborative Research Group, National Institute for Environmental Studies, Tsukuba, Ibaraki, Japan.
Literature Cited
- Abegglen LM, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314(17):1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, et al. 2018. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 46(D1):D477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB, et al. 2018. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560(7717):253–257. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57(1):289–300. [Google Scholar]
- Ben-Nun IF, et al. 2011. Induced pluripotent stem cells from highly endangered species. Nat Methods 8(10):829–831. [DOI] [PubMed] [Google Scholar]
- Boroviak T, et al. 2015. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell 35(3):366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A, et al. 2013. Derivation and characterization of induced pluripotent stem cells from equine fibroblasts. Stem Cells Dev. 22(4):611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. 2008. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B 178(4):439–445. [DOI] [PubMed] [Google Scholar]
- Caulin AF, Maley CC. 2011. Peto’s paradox: evolution’s prescription for cancer prevention. Trends Ecol Evol. 26(4):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. TCF3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22(6):746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crish SD, Rice FL, Park TJ, Comer CM. 2003. Somatosensory organization and behavior in naked mole-rats I: vibrissa-like body hairs comprise a sensory array that mediates orientation to tactile stimuli. Brain Behav Evol. 62(3):141–151. [DOI] [PubMed] [Google Scholar]
- Cuella-Martin R, et al. 2016. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol Cell 64(1):51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney MA, Nagy L, Kinsel MJ, Treuting PM. 2013. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): a retrospective survey of lesions in a zoo population. Vet Pathol. 50(4):607–621. [DOI] [PubMed] [Google Scholar]
- Endo Y, Kamei K-I, Inoue-Murayama M. 2018. Genetic signatures of lipid metabolism evolution in Cetacea since the divergence from terrestrial ancestor. J Evol Biol. 31(11):1655–1665. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 194(6):733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 10(2):141–148. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292(5819):154–156. [DOI] [PubMed] [Google Scholar]
- Ezashi T, et al. 2009. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci. 106(27):10993–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Yuan Y, Roberts RM. 2016. Pluripotent stem cells from domesticated mammals. Annu Rev Anim Biosci. 4(1):223–253. [DOI] [PubMed] [Google Scholar]
- Fuet A, Pain B. 2017. Chicken induced pluripotent stem cells: establishment and characterization In: Sheng G, editor. Avian and reptilian developmental biology: methods and protocols. New York, (NY: ): Springer New York; p. 211–228. [DOI] [PubMed] [Google Scholar]
- Gould E, McShea W, Grand T. 1993. Function of the star in the star-nosed mole, Condylura cristata. J Mammal. 74(1):108–116. [Google Scholar]
- Han J, et al. 2010. TBX3 improves the germ-line competency of induced pluripotent stem cells. Nature 463(7284):1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, et al. 2011. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 21(10):1509–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Maruyama T, Fan G. 2014. The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15(4):410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, et al. 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc Natl Acad Sci U S A. 108(38):15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10(1):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C, Colnaghi R, Abramowicz I, Carpenter G, O’Driscoll M. 2013. Meier–Gorlin syndrome and Wolf–Hirschhorn syndrome: two developmental disorders highlighting the importance of efficient DNA replication for normal development and neurogenesis. DNA Repair (Amst.) 12(8):637–644. [DOI] [PubMed] [Google Scholar]
- Kim J, Liu Y, Qiu M, Xu Y. 2016. Pluripotency factor Nanog is tumorigenic by deregulating DNA damage response in somatic cells. Oncogene 35(10):1334–1340. [DOI] [PubMed] [Google Scholar]
- Kumar R, Cheok CF. 2014. RIF1: a novel regulatory factor for DNA replication and DNA damage response signaling. DNA Repair (Amst.) 15:54–59. [DOI] [PubMed] [Google Scholar]
- Kunz TH, Jones DP. 2000. Pteropus vampyrus. Mamm Species 642(1):1–6. [Google Scholar]
- Kuwahara A, et al. 2014. TCF3 represses Wnt-β-catenin signaling and maintains neural stem cell population during neocortical development. PLoS One 9(5):e94408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti DC, et al. 2009. Relaxed selection in the wild. Trends Ecol Evol. 24(9):487–496. [DOI] [PubMed] [Google Scholar]
- Latos PA, et al. 2015. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat Commun. 6(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar NH, et al. 2018. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28(7):983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HE, Ayarpadikannan S, Kim HS. 2015. Role of transposable elements in genomic rearrangement, evolution, gene regulation and epigenetics in primates. Genes Genet Syst. 90(5):245–257. [DOI] [PubMed] [Google Scholar]
- Lee S-G, et al. 2017. Naked mole rat induced pluripotent stem cells and their contribution to interspecific chimera. Stem Cell Rep. 9(5):1706–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE. 2017. What is adaptation by natural selection? Perspectives of an experimental microbiologist. PLoS Genet. 13(4):e1006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Belmonte JCI. 2017. Ground rules of the pluripotency gene regulatory network. Nat Rev Genet. 18(3):180–191. [DOI] [PubMed] [Google Scholar]
- Liao J, et al. 2009. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 4(1):11–15. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. 2012. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology 77(2):338–346. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102(30):10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, et al. 1997. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-β. Nature 388(6644):778–782. [DOI] [PubMed] [Google Scholar]
- Manor YS, Massarwa R, Hanna JH. 2015. Establishing the human naïve pluripotent state. Curr Opin Genet Dev. 34:35–45. [DOI] [PubMed] [Google Scholar]
- Marchetto MCN, et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503(7477):525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, et al. 2012. Esrrb is a pivotal target of the GSK3/TCF3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11(4):491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 78(12):7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Serrano M. 2008. The Arf/p53 pathway in cancer and aging. Cancer Res. 68(15):6031–6034. [DOI] [PubMed] [Google Scholar]
- Menzorov AG, et al. 2015. Comparison of American mink embryonic stem and induced pluripotent stem cell transcriptomes. BMC Genomics 16(S13):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334(6055):521–524. [DOI] [PubMed] [Google Scholar]
- Miyawaki S, et al. 2016. Tumour resistance in induced pluripotent stem cells derived from naked mole-rats. Nat Commun. 7(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Li N, Wu S. 2014. Generation and characterization of bat-induced pluripotent stem cells. Theriogenology 82(2):283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP. 2010. Phenotypic plasticity and diversity in insects. Phil Trans R Soc B 365(1540):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. 2007. Recent and ongoing selection in the human genome. Nat Rev Genet. 8(11):857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12(13):2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteil P, et al. 2013. Induced pluripotent stem cells derived from rabbits exhibit some characteristics of naive pluripotency. Biol Open 2(6):613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Chinaranagari S, Chaudhary J. 2015. Basic helix loop helix (bHLH) transcription factor 3 (TCF3, E2A) is regulated by androgens in prostate cancer cells. Am J Cancer Res. 5(11):3407–3421. [PMC free article] [PubMed] [Google Scholar]
- Paterson YZ, Kafarnik C, Guest DJ. 2018. Characterization of companion animal pluripotent stem cells. Cytometry 93(1):137–148. [DOI] [PubMed] [Google Scholar]
- Pavlovich SS, et al. 2018. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell 173(5):1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. 2005. Modulating HOX gene functions during animal body patterning. Nat Rev Genet. 6(12):893–904. [DOI] [PubMed] [Google Scholar]
- Pereira L, Yi F, Merrill BJ. 2006. Repression of Nanog gene transcription by TCF3 limits embryonic stem cell self-renewal. MCB 26(20):7479–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, et al. 2011. A molecular phylogeny of living primates Brosius, J, editor. PLoS Genet. 7(3):e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R. 2015. Quantitative implications of the approximate irrelevance of mammalian body size and lifespan to lifelong cancer risk. Phil Trans R Soc B 370(1673):20150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R, Roe FJ, Lee PN, Levy L, Clack J. 1975. Cancer and ageing in mice and men. Br J Cancer 32(4):411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Pronobis MI, Rusan NM, Peifer M. 2015. A novel GSK3-regulated APC: Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient βcatenin destruction. eLife 4:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy K, et al. 2015. Derivation of induced pluripotent stem cells from orangutan skin fibroblasts. BMC Res Notes 8(1):577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Schlegel P, Schweizer H, Neuweiler G. 1991. Audition in vampire bats, Desmodus rotundus. J Comp Physiol A 168(1):45–51. [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL. 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 199(12):1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, et al. 2009. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol Reprod Dev. 77(1):2. [DOI] [PubMed] [Google Scholar]
- Takahashi K, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami M, Yamanaka S. 2005. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem Soc Trans. 33(6):1522–1525. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2016. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 17(3):183–193. [DOI] [PubMed] [Google Scholar]
- Thompson D, Regev A, Roy S. 2015. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu Rev Cell Dev Biol. 31(1):399–428. [DOI] [PubMed] [Google Scholar]
- Thomson JA. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282(5391):1145–1147. [DOI] [PubMed] [Google Scholar]
- Tian X, et al. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499(7458):346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka I, et al. 2010. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells 15(9):959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhen MD. 2010. The origin(s) of whales. Annu Rev Earth Planet Sci. 38(1):189–219. [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. 2018. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 24(7):1765–1776. [DOI] [PubMed] [Google Scholar]
- Verma R, et al. 2013. Nanog is an essential factor for induction of pluripotency in somatic cells from endangered felids. BioRes Open Access 2(1):72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Holland MK, Temple-Smith P, Verma PJ. 2012. Inducing pluripotency in somatic cells from the snow leopard (Panthera uncia), an endangered felid. Theriogenology 77(1):220–228. [DOI] [PubMed] [Google Scholar]
- Voss AK, Collin C, Dixon MP, Thomas T. 2009. Moz and retinoic acid coordinately regulate H3K9 acetylation, HOX gene expression, and segment identity. Dev Cell 17(5):674–686. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. 2017. Epigenetic regulation of left–right asymmetry by DNA methylation. EMBO J. 36(20):2987–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, et al. 2006. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 25(19):2697–2707. [DOI] [PubMed] [Google Scholar]
- Weeratunga P, Shahsavari A, Ovchinnikov DA, Wolvetang EJ, Whitworth DJ. 2018. Induced pluripotent stem cells from a marsupial, the Tasmanian devil (Sarcophilus harrisii): insight into the evolution of mammalian pluripotency. Stem Cells Dev. 27(2):112–122. [DOI] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. 2016. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 17(3):155–169. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth DJ, et al. 2019. Platypus induced pluripotent stem cells: the unique pluripotency signature of a monotreme. Stem Cells Dev. 28(3):151–164. [DOI] [PubMed] [Google Scholar]
- Whitworth DJ, Ovchinnikov DA, Sun J, Fortuna PRJ, Wolvetang EJ. 2014. Generation and characterization of leukemia inhibitory factor-dependent equine induced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 23(13):1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi UB, Bronner GN, Narins PM. 2006. Ossicular differentiation of airborne and seismic stimuli in the Cape golden mole (Chrysochloris asiatica). J Comp Physiol A 192(3):267–277. [DOI] [PubMed] [Google Scholar]
- Wunderlich S, et al. 2014. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. 12(3):622–629. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22(4):1107–1118. [DOI] [PubMed] [Google Scholar]
- Yi F, Pereira L, Merrill BJ. 2008. TCF3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells 26(8):1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H-S, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 46(1):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.