Abstract
Dysregulated neural mechanisms in reward and somatosensory circuits result in an increased appetitive drive for and reduced inhibitory control of eating, which in turn causes obesity. Despite many studies investigating the brain mechanisms of obesity, the role of macroscale whole‐brain functional connectivity remains poorly understood. Here, we identified a neuroimaging‐based functional connectivity pattern associated with obesity phenotypes by using functional connectivity analysis combined with machine learning in a large‐scale (n ~ 2,400) dataset spanning four independent cohorts. We found that brain regions containing the reward circuit positively associated with obesity phenotypes, while brain regions for sensory processing showed negative associations. Our study introduces a novel perspective for understanding how the whole‐brain functional connectivity correlates with obesity phenotypes. Furthermore, we demonstrated the generalizability of our findings by correlating the functional connectivity pattern with obesity phenotypes in three independent datasets containing subjects of multiple ages and ethnicities. Our findings suggest that obesity phenotypes can be understood in terms of macroscale whole‐brain functional connectivity and have important implications for the obesity neuroimaging community.
Keywords: functional connectivity, machine learning, obesity, UK Biobank, whole‐brain connectome
1. INTRODUCTION
Obesity is thought to be caused by uncontrolled eating, which is highly associated with the imbalance in reward and inhibitory control processing of the brain (Martin et al., 2010; Moore, Sabino, Koob, & Cottone, 2017; Moreno‐Lopez, Contreras‐Rodriguez, Soriano‐Mas, Stamatakis, & Verdejo‐Garcia, 2016; Murray, Tulloch, Gold, & Avena, 2014; Val‐Laillet et al., 2015; Van Opstal, Wijngaarden, Grond, & Pijl, 2019; Verdejo‐Román, Vilar‐López, Navas, Soriano‐Mas, & Verdejo‐García, 2017; Ziauddeen, Alonso‐Alonso, Hill, Kelley, & Khan, 2015). Neurobiological studies have found that dysregulation of reward and inhibitory circuits gives rise to an increased threshold for satiation, ultimately leading to overeating (Martin et al., 2010; Murray et al., 2014; Steward, Miranda‐Olivos, Soriano‐Mas, & Fernández‐Aranda, 2019; Val‐Laillet et al., 2015; Van Opstal et al., 2018; Verdejo‐Román et al., 2017; Ziauddeen et al., 2015). Recent neuroimaging studies have increasingly shown associations between obesity and alterations in cortical and subcortical morphology (Herrmann, Tesar, Beier, Berg, & Warrings, 2019; Marqués‐Iturria et al., 2013; Shott et al., 2015), brain activity (Brooks, Cedernaes, & Schiöth, 2013; Goldstone et al., 2009; Gupta et al., 2018; Opel et al., 2015; Park, Hong, & Park, 2017; Steward, Juaneda‐Seguí, et al., 2019; Stoeckel et al., 2008; Van Meer et al., 2019), functional connectivity (García‐García et al., 2013; García‐García et al., 2015; Lips et al., 2014; Park, Seo, & Park, 2016; Park, Seo, Yi, & Park, 2015), and diffusivity (Gupta et al., 2017; Olivo et al., 2017; Steward, Picó‐Pérez, et al., 2019). These studies collectively show that the pathophysiology of obesity is associated with various types of brain measurements.
Despite extensive research on identifying underlying brain mechanisms of obesity, few studies have investigated how macroscopic alterations in functional brain connectivity are related to obesity phenotypes (Ding et al., 2020; Park et al., 2016). Some of the primary challenges that obesity neuroimaging studies face are conflicting reports concerning obesity‐associated brain regions and the magnitude and direction of the observed effects. These inconsistencies may be due to the limited reproducibility of existing studies because of the small sample size (n < 100) from a single site, highlighting the necessity of using large‐scale datasets from multiple centers.
A large amount of data could be analyzed effectively by utilizing recent advances in neuroimaging processing algorithms. With recent advances in neuroimaging acquisition and analytics, it is now possible to assess the whole‐brain functional connectivity (FC) patterns, which may serve as novel biomarkers for neurobiological diseases (Damaraju et al., 2014; Hong et al., 2019; Lee et al., 2019; Park et al., 2017). Indeed, FC derived from functional magnetic resonance imaging (fMRI) approximates the statistical association of brain activity between different regions, which can be used to estimate the importance of each brain region based on the degree distributions (Bullmore & Sporns, 2009; Griffa & Van Den Heuvel, 2018; Rubinov & Sporns, 2010; Van Den Heuvel, Kahn, Goñi, & Sporns, 2012). Machine learning is a powerful tool capable of handling large‐scale datasets for constructing analytical models (Kale, Hamde, & Holambe, 2019; Park, Took, & Seong, 2018). Specifically, a regularized regression framework identifies predictor variables (such as regional FC) that correlate with response variables (such as obesity phenotypes) in a data‐driven way. Thus, consolidating whole‐brain FC information and advanced machine learning techniques may help elucidate how subtle and complex brain organizations associate with obesity phenotypes.
In this study, we identified an obesity phenotype‐associated whole‐brain FC pattern using a large‐scale (n ~ 1,500 for training and n ~ 900 for validation) multi‐center (spanning four independent cohorts) neuroimaging dataset that was processed using state‐of‐the‐art methods. Graph‐theoretical approach and machine learning techniques were used to estimate how inter‐regional brain connectivity correlated with obesity phenotypes. Our results may help us understand how the whole‐brain functional organizations are linked to obesity phenotypes.
2. MATERIAL AND METHODS
2.1. Imaging data and participants
Resting‐state fMRI (rs‐fMRI) data from 2,390 participants in four different cohorts were obtained. The data from cohort 1 were used to derive the obesity phenotype‐associated FC pattern, and data from the other cohorts were used for validation. Cohort 1 consisted of 1,497 participants (54% female) from the UK Biobank database of 13,711 participants (Miller et al., 2016). Participants who lacked rs‐fMRI data or full phenotypic data, or those classified as having neuroticism, depression, or anxiety were excluded. Data from the UK Biobank database were obtained through application number 34613 entitled “Neuroimaging correlates of obesity.” A detailed demographic summary of the participants is presented in Table 1, and the imaging acquisition parameters of the rs‐fMRI data are reported in Table S1. The imaging data were scanned using Siemens 3T Skyra scanner. Rs‐fMRI data from 893 participants were then obtained from three different cohorts for validation. Cohort 2 contained 587 participants (53% female) from the Human Connectome Project database, which contains a total of 1,206 participants (Van Essen et al., 2013). Participants without full rs‐fMRI or phenotypic data or those with a history of drug use or a family history of mental illness were excluded. The imaging data were scanned using Siemens 3T Skyra scanner. The data from cohort 3 were obtained from the enhanced Nathan Kline Institute‐Rockland Sample database of 650 participants using Siemens 3T Magnetom scanner (Nooner et al., 2012). Participants without full rs‐fMRI or phenotypic data were excluded. A total of 276 participants (62% female) from cohort 3 were enrolled in this study. Data from cohort 4 (50% female, n = 30) were obtained locally from St. Vincent's Hospital. Detailed demographic information of the participants is reported in Table 1, and the imaging acquisition parameters of the rs‐fMRI data are reported in Table S1. The imaging data were acquired with Siemens 3T Magnetom scanner. The Institutional Review Board (IRB) of Sungkyunkwan University approved this retrospective study using the UK Biobank, Human Connectome Project, and enhanced Nathan Kline Institute‐Rockland Sample datasets. This study was performed in full accordance with local IRB guidelines. All participants provided written informed consent. The use of the data from St. Vincent's Hospital was approved by the IRB of the Catholic University of Korea, and written consent was obtained from all participants.
TABLE 1.
Demographic summary of the study participants
| Information | UKB (n = 1,497) | HCP (n = 587) | eNKI‐RS (n = 276) | SVH (n = 30) | ||||
|---|---|---|---|---|---|---|---|---|
| NHW (n = 1,007) | HW (n = 490) | NHW (n = 291) | HW (n = 296) | NHW (n = 276) | HW (n = 0) | NHW (n = 27) | HW (n = 3) | |
| Age | 56.78 (8.14) | 55.37 (8.40) | 28.74 (3.56) | 28.49 (3.79) | 48.33 (19.26) | N/A | 38.89 (10.60) | 41.33 (3.79) |
| Sex (male:Female) | 534:473 | 156:334 | 164:127 | 109:187 | 106:170 | N/A | 14:13 | 1:2 |
| Body mass index (kg/m2) | 29.61 (3.97) | 22.82 (1.65) | 28.45 (2.85) | 22.31 (1.78) | 30.65 (4.83) | N/A | 28.19 (2.66) | 23.77 (0.74) |
| Waist circumference (cm) | 96.23 (11.38) | 78.17 (8.08) | N/A | N/A | 96.93 (11.99) | N/A | 93.51 (7.17) | 87.85 (3.11) |
| Waist‐to‐hip ratio | 0.90 (0.09) | 0.81 (0.07) | N/A | N/A | 0.87 (0.09) | N/A | 0.92 (0.05) | 0.86 (0.04) |
| Purpose | FC pattern development | Validation | ||||||
Note: Mean (SD) are reported.
Abbreviations: eNKI‐RS, enhanced Nathan Kline Institute‐Rockland Sample; HCP, Human Connectome Project; HW, healthy weight (18.5 ≤ body mass index <25); N/A, not available; NHW, non‐healthy weight (body mass index ≥25); SVH, St. Vincent's Hospital; UKB, UK Biobank.
2.2. Data preprocessing
The UK Biobank database provided rs‐fMRI data that had been preprocessed using FSL software (Alfaro‐Almagro et al., 2018; Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012). The preprocessing pipeline can be described as follows: distortions caused by gradient nonlinearity and head motion were corrected; intensity normalization of the 4D volumes and high‐pass filtering with 50 s (~0.009 Hz) were applied; and nuisance variables attributable to cerebrospinal fluid, white matter, head motion, and cardiac‐ and large‐vein‐related artifacts were removed using the FIX software (Salimi‐Khorshidi et al., 2014). The nuisance variable‐free rs‐fMRI data were registered onto the T1‐weighted structural data and then subsequently registered onto the Montreal Neurological Institute (MNI152) standard space.
The rs‐fMRI data obtained from the Human Connectome Project had been minimally preprocessed using FSL, FreeSurfer, and Workbench (Fischl, 2012; Glasser et al., 2013; Jenkinson et al., 2012). The gradient distortions and head motion‐corrected data were registered onto the T1‐weighted structural data and then onto the MNI standard space. Magnetic field bias was corrected, and the skull was removed. Intensity normalization of the 4D volumes was applied. Nuisance variables were removed via FIX (Salimi‐Khorshidi et al., 2014). Additional steps of high‐pass filtering with a frequency of 0.01 Hz and spatial smoothing with a full width at half maximum of 3 mm were applied manually.
The rs‐fMRI data from the enhanced Nathan Kline Institute‐Rockland Sample database and St. Vincent's Hospital were preprocessed using a fusion of the neuroimaging preprocessing (FuNP) pipeline integrating AFNI, FSL, and ANTs software (Avants et al., 2011; Cox, 1996; Jenkinson et al., 2012; Park, Byeon, & Park, 2019). Volumes during the first 10 s were discarded to allow for magnetic field saturation. A slice‐timing correction was performed on the data from St. Vincent's Hospital. However, slice‐timing correction was not performed on data from the enhanced Nathan Kline Institute‐Rockland Sample database due to their short (< 1 s) repetition times, as recommended by a previous study (Bijsterbosch, Smith, & Beckmann, 2017). Volumes with large head motion (frame‐wise displacement >0.5 mm) were removed (Power, Barnes, Snyder, Schlaggar, & Petersen, 2012), and head motion correction was performed on the remaining data. Skull was removed and the intensity normalization of the 4D volumes was applied. The rs‐fMRI data were co‐registered to the T1‐weighted structural data and subsequently mapped to MNI152 standard space. The nuisance variables were removed using FIX (Salimi‐Khorshidi et al., 2014). High‐pass filtering with a frequency of 0.01 Hz and spatial smoothing with a full width at half maximum of 3 mm were applied.
2.3. Functional connectivity analysis
FC analysis based on graph theory was used to quantify the strength of interconnection between different brain regions (Bullmore & Sporns, 2009; Rubinov & Sporns, 2010; Watts & Strogatz, 1998). The graph nodes were brain regions defined by the Brainnetome atlas, which divides the whole brain into 246 regions involving frontal, temporal, parietal, insular, limbic, and occipital lobes, as well as subcortical nuclei including amygdala, hippocampus, basal ganglia, and thalamus (Fan et al., 2016). The graph edges were the connections between different brain regions as defined by the partial correlation coefficients of the time‐series between two nodes (Smith et al., 2011; Smith et al., 2013). Partial correlation coefficients were computed using FSLNets (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLNets) with a ridge (i.e., L2) regularization of rho = 0.5 as previously used in the UK Biobank study (Alfaro‐Almagro et al., 2018). The correlation coefficients were entered into a matrix referred to as the connectivity matrix. The correlation coefficients were then soft‐thresholded using the formula {(r + 1)/2}β to satisfy the scale‐free topology, where r was the correlation coefficient and β (set to six) was the scale‐free index (Mumford et al., 2010; Schwarz & McGonigle, 2011). The coefficients were further processed with Fisher's r‐to‐z transformation to ensure the correlation values were normally distributed (Thompson & Fransson, 2016). Degree centrality values representing the strength of FC of a given node were calculated by determining the sum of edge weights connected to a given node (Bullmore & Sporns, 2009; Rubinov & Sporns, 2010). We chose degree centrality as the nodal connectivity measure among many other graph‐theoretical measures including betweenness and eigenvector centrality, as well as local efficiency, because our prior works found its usefulness for associating brain function and body mass index, as well as eating behaviors in individuals with obesity (Park et al., 2015; Park et al., 2016; Park, Lee, Kim, Kim, & Park, 2018).
2.4. Quantification of FC
A schematic for quantifying FC patterns associated with obesity phenotype is shown in Figure 1. We calculated degree centrality values from the preprocessed fMRI data (Figure 1a‐1, a‐2) and fed these values into the elastic net regularization framework to associate nodal degree centrality with an obesity phenotype (i.e., waist circumference) (Figure 1a‐3) (Zou & Hastie, 2005). We used waist circumference as the dependent variable as it is a better measure to assess obesity than body mass index regarding evaluating metabolically unhealthy obesity with obesity‐related complications (Bujalska, Kumar, Bujalska, Kumar, & Stewart, 1997; Després et al., 2008; Després & Lemieux, 2006; Folsom et al., 1993; Folsom et al., 2000). Here, the waist circumference was controlled for age and sex. Elastic net regression is a representative machine learning approach for feature selection, which estimates regression coefficients of brain regions at a given sparsity that maximally correlate with the dependent variable. This approach has shown good performance for feature selection at a given sparsity level compared to L1 (least absolute shrinkage and selection operator)‐ and L2 (ridge)‐norm regularization methods (Zou & Hastie, 2005). The two tuning parameters of the elastic net, corresponding to L1 and L2 penalty terms, were determined with five‐fold cross‐validation. Using the optimized tuning parameters, we applied the elastic net regression to associate degree centrality and obesity phenotypes with five‐fold cross‐validation using strictly separated training and test data to avoid overfitting. For each cross‐validation, we estimated regression coefficients from the training data only. A linear combination of the estimated regression coefficients and regional degree centrality values was computed and further correlated with waist circumference of the test data to assess model performance (Figure 1a‐4). We averaged the regression coefficients across cross‐validations, and the mean coefficient values were considered to be the FC pattern associated with obesity phenotypes (Figure 1a‐5). To assess whether this FC pattern could predict obesity phenotypes in all participants, we first computed FC pattern score by calculating a linear combination of the FC pattern and regional degree centrality values for each participant. Then, we correlated the FC pattern score with obesity phenotypes, including body mass index, waist circumference, and waist‐to‐hip ratio, for all participants in the UK Biobank database. Age and sex were added as covariates to control for their possible effects on obesity for the correlation analysis (Table S2).
FIGURE 1.
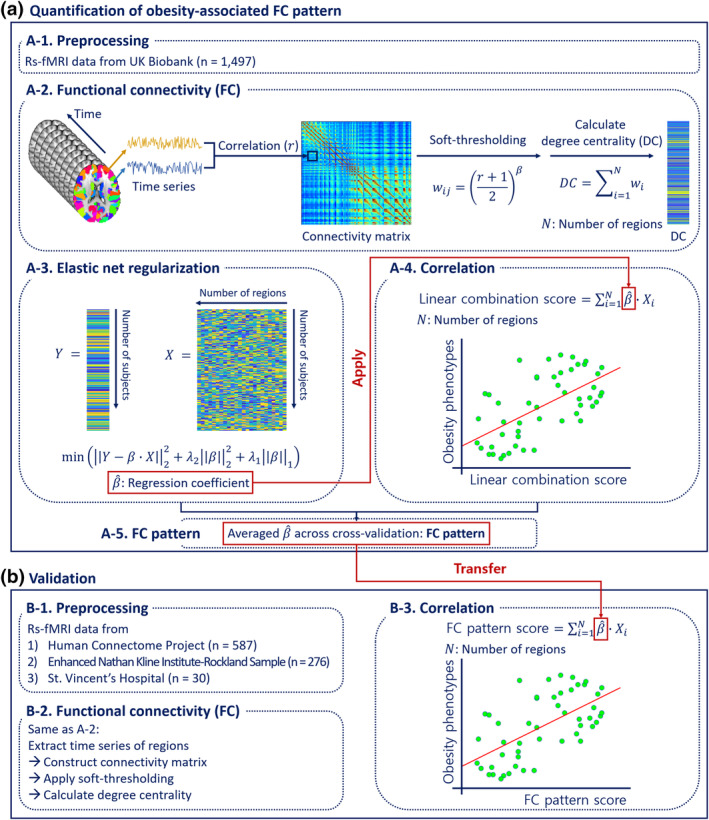
Flowchart of this study. (a) Quantification of FC pattern using data from the UK Biobank database. FC analysis was applied to the preprocessed rs‐fMRI data, and degree centrality values were calculated. Elastic net regularization was used to estimate regression coefficients. A single scalar score was computed using a linear combination of the estimated coefficients and degree centrality values, and it was correlated with obesity phenotypes. (b) Validation procedures. The same FC analysis was performed in independent datasets. The FC pattern developed using the UK Biobank dataset was transferred to the independent dataset to calculate the FC pattern score via a linear combination of the developed FC pattern and regional degree centrality values for all participants, which was correlated with their obesity phenotypes
2.5. Validation of FC pattern
The reliability and reproducibility of the FC pattern (i.e., averaged regression coefficients across cross‐validations) were assessed by applying the same FC analysis used on the UK Biobank dataset to the independent Human Connectome Project, enhanced Nathan Kline Institute‐Rockland Sample, and St. Vincent's Hospital datasets (Figure 1b‐1, b‐2). The FC pattern score was calculated and correlated with obesity phenotypes (Figure 1b‐3). The Human Connectome Project dataset provided only body mass index values, while the enhanced Nathan Kline Institute‐Rockland Sample and St. Vincent's Hospital datasets provided body mass index, waist circumference, and waist‐to‐hip ratio values. Multiple comparisons of correlations were corrected using the false discovery rate (FDR) (Benjamini & Hochberg, 1995). To assess how changing the number of brain regions correlate differently to obesity phenotypes, we tested how well the FC pattern score performed in different sparsity levels between 10 and 106 in steps of 10. The maximum number (i.e., 106) was determined by the number of brain regions with nonzero FC pattern. Changes in sparsity levels lead to choosing how many brain regions to include for computing the FC pattern score. For example, if we set a sparsity level at ten, then ten brain regions with the highest FC pattern were included to calculate the FC pattern score. The FC pattern scores calculated with different sparsity levels were correlated with the obesity phenotypes.
3. RESULTS
3.1. Study participants
The rs‐fMRI data used in this study were from a total of 2,390 participants across four different cohorts. Data from 1,497 participants were obtained from the UK Biobank database (Miller et al., 2016), data from 587 participants were obtained from the Human Connectome Project database (Van Essen et al., 2013), data from 276 participants were obtained from the enhanced Nathan Kline Institute‐Rockland Sample database (Nooner et al., 2012), and data from 30 participants were obtained from a local site of St. Vincent's Hospital, Suwon, Korea. Data from UK Biobank were used to develop the FC pattern, and other datasets were used for validation. The participants' ages spanned from adolescence to late adulthood, and their ethnicities differed across cohorts. Detailed demographic information is reported in Table 1.
3.2. Quantification of the MRI‐based FC pattern of obesity
The obesity‐associated FC pattern was developed using rs‐fMRI data from 1,497 participants from the UK Biobank database after five cross‐validations. For each cross‐validation, a combination score was calculated using a linear combination of the selected regional degree centrality values and the elastic net coefficients. The linear combination score was then correlated with waist circumference. The mean correlation value between the score and waist circumference across cross‐validations was 0.275 (standard deviation [SD] 0.074) for the training set and 0.102 (SD 0.086) for the test set (Table S3). Although the effect size (i.e., correlation value) was reduced for the test data compared to training data, it reached statistical significance (p < .05). Overfitting was minimized as we separated training and test data. The estimated coefficients for the selected brain regions were averaged across cross‐validations, and the averaged value was considered to be the obesity phenotype‐associated FC pattern (Data S1). The FC pattern (i.e., averaged regression coefficients across cross‐validations) was mapped onto the whole brain (Figure 2a). The top 15 brain regions with the highest magnitudes of the FC pattern involved reward and sensory networks, where the ventromedial and ventrolateral prefrontal cortex, superior temporal gyrus, superior parietal lobule, cingulate gyrus, and globus pallidus showed strong positive associations with waist circumference, while regions associated with sensorimotor skills and perception such as the precentral gyrus, fusiform gyrus, and superior temporal sulcus, as well as thalamus, showed negative associations with obesity phenotype (Figure 2b). The associations of all brain regions with waist circumference (i.e., FC pattern) are shown in Figure 2c.
FIGURE 2.
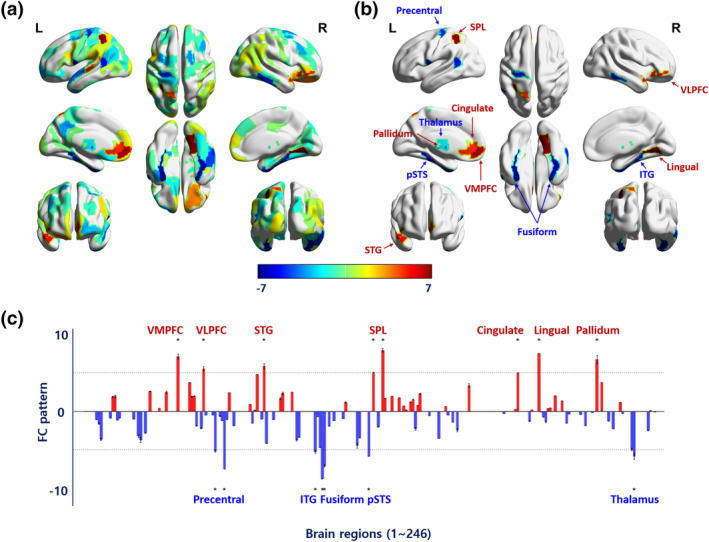
An obesity phenotype‐associated FC pattern derived from the UK Biobank data. (a) The obesity phenotype‐associated FC pattern mapped onto the whole brain. The red/blue colors represent positive/negative FC pattern to associate brain regions and an obesity phenotype (i.e., waist circumference). (b) FC pattern of the regions with the 15 highest magnitudes. (c) FC pattern for all brain regions. Error bars represent ±1 standard error of the mean. The dotted line indicates the threshold for the 15 highest magnitudes of the FC pattern. The brain regions over the threshold are marked with an asterisk. VMPFC, ventromedial prefrontal cortex; VLPFC, ventrolateral prefrontal cortex; STG, superior temporal gyrus; SPL, superior parietal lobule; ITG, inferior temporal gyrus; pSTS, posterior superior temporal sulcus
3.3. Validity of FC pattern
The quality of the obesity phenotype‐associated FC pattern was assessed by calculating the correlation between the FC pattern score from the UK Biobank dataset (n = 1,497) and the obesity phenotypes, which consisted of body mass index, waist circumference, and waist‐to‐hip ratio. The FC pattern score was significantly correlated with all three obesity phenotypes (all p < .05, FDR corrected) (Figure 3a). To further validate this finding, we performed correlation analyses using different sample sizes (200, 700, and 900 samples). Firstly, we randomly selected a subsample of 200 subjects from the 1,497 subjects in the UK Biobank database 1,000 times with different sets of subsamples. We performed the same correlation analysis described above for each 200‐subject subsample. The correlation results from the subsamples were largely similar to the results from the whole dataset (Figure S1). The mean correlation for subsamples between FC pattern score and body mass index was 0.262 (0.265 for the whole dataset), waist circumference was 0.307 (0.308 for the whole dataset), and waist‐to‐hip ratio was 0.236 (0.236 for the whole dataset). All correlation analyses showed significant results on average for subsamples (mean p < .05, FDR corrected). We repeated the procedure using subsamples of 700 and 900 subjects and found that the r‐ and p‐values were similar to those of the whole dataset (Table S4). These results indicate that the significance of the correlation was not driven by the large sample size.
FIGURE 3.
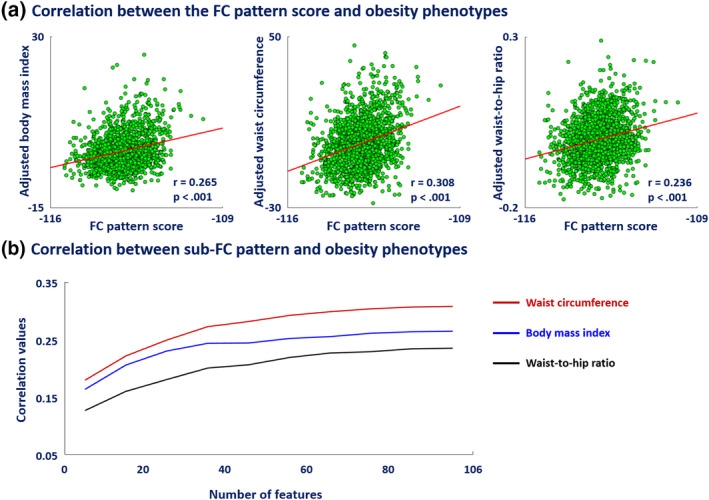
Correlation between FC pattern score and obesity phenotypes controlled for age and sex. (a) Correlation coefficients and FDR‐corrected p‐values. (b) Correlations with obesity phenotypes at various sparsity levels of the FC pattern. The maximum number (=106) was computed from the number of brain regions with nonzero regression coefficients by using the original FC pattern without changing the sparsity
We also tested how well our FC pattern performed against different sparsity levels. Regression coefficients were sorted based on their magnitude from large to small, and we changed the sparsity level by selecting various numbers of coefficients starting from the top. The correlation results showed a monotonically increasing pattern, with saturation occurring at a low sparsity of approximately 30 (Figure 3b). Saturation was defined as the point at which the correlation value exceeded 80% of the maximum value. Brain regions involved in the reward network were consistently observed until saturation at 30 (Table S5). These results indicate that regions involved in the reward network had strong coefficient magnitudes, indicating that they were the primary regions associated with obesity phenotypes.
3.4. Generalization of the FC pattern
3.4.1. Validation to independent dataset
To confirm the generalizability of the obesity‐associated FC pattern, we tested the pattern against independent datasets from the (a) Human Connectome Project database (n = 587), (b) enhanced Nathan Kline Institute‐Rockland Sample database (n = 276), and (c) St. Vincent's Hospital (n = 30). Correlations between the FC pattern score and the obesity phenotypes were significant (all p < .05, FDR corrected) except for the waist‐to‐hip ratio in the enhanced Nathan Kline Institute‐Rockland Sample dataset (p > .4, FDR corrected; Table 2 and Figure S2). The waist circumference and waist‐to‐hip ratio in the St. Vincent's Hospital showed marginal, but strong correlations (p < .1, FDR corrected). The four cohorts used in this study included participants of varying ages. The FC pattern was constructed using data from the UK Biobank database, which consisted of middle‐aged adults (mean age = 56.32 y, range [40–70 y]). The FC pattern was generalizable across cohorts of different ages spanning from adolescents to the elderly (Human Connectome Project: mean 28.61 y; range [22–36 y]; enhanced Nathan Kline Institute‐Rockland Sample: mean 48.33 y; range [13–83 y]; St. Vincent's Hospital: mean 39.13 y; range [23–62 y]).
TABLE 2.
Correlation between the FC pattern score and obesity phenotypes for all datasets
| Purpose | Database | Obesity phenotypes | r | p‐value, FDR corrected |
|---|---|---|---|---|
| FC pattern development | UKB (n = 1,497) | Body mass index | 0.265 | <.001 |
| Waist circumference | 0.308 | <.001 | ||
| Waist‐to‐hip ratio | 0.236 | <.001 | ||
| Validation | HCP (n = 587) | Body mass index | 0.146 | <.001 |
| eNKI‐RS (n = 276) | Body mass index | 0.150 | .020 | |
| Waist circumference | 0.169 | .015 | ||
| Waist‐to‐hip ratio | 0.049 | .450 | ||
| SVH (n = 30) | Body mass index | 0.463 | .020 | |
| Waist circumference | 0.556 | .060 | ||
| Waist‐to‐hip ratio | 0.371 | .068 |
Abbreviations: eNKI‐RS, enhanced Nathan Kline Institute‐Rockland Sample; HCP, Human Connectome Project; SVH, St. Vincent's Hospital; UKB, UK Biobank.
3.4.2. FC pattern with different ethnicity
The participants' ethnicities differed across the cohorts (Table S6 and Figure S3). Participants from the UK Biobank database were primarily Caucasian (93.7% Caucasian, 2.0% African‐American, 2.3% Asian, 0.3% mixed, and 1.6% unknown). The FC pattern was generalizable to the Human Connectome Project and enhanced Nathan Kline Institute‐Rockland Sample cohorts, which had similar ethnic distributions (Human Connectome Project: 79.2% Caucasian, 9.2% African‐American, 7.8% Asian, 2.2% mixed, and 1.5% unknown; enhanced Nathan Kline Institute‐Rockland Sample: 72.5% Caucasian, 19.6% African‐American, 6.5% Asian, and 1.5% mixed) as well as the St. Vincent's Hospital cohort, which had a different ethnicity distribution (100% Asian). These results indicate that the FC pattern is observable across varied populations and may be associated with obesity phenotypes in participants with varying ethnicities.
However, obesity‐related characteristics might vary across different ethnicities. To determine whether the overall results could be replicated in a subsample with a homogeneous ethnicity (i.e., Caucasian), we performed the same analysis only using data from Caucasians in the UK Biobank database. The derived FC pattern had a root mean squared error of 1.231 compared to the previous FC pattern (all ethnicities), indicating that these FC patterns were similar. The brain regions with large FC pattern magnitudes (top 15) were as follows: the ventromedial and ventrolateral prefrontal cortex, superior temporal gyrus, superior parietal lobule, insular gyrus, and nucleus accumbens were positively correlated with obesity, while the superior frontal gyrus, precentral gyrus, inferior temporal gyrus, fusiform gyrus, parahippocampal gyrus, and thalamus were negatively correlated (Figure S4a). These findings were consistent with our overall results, especially those for the ventromedial and ventrolateral prefrontal cortex, superior temporal gyrus, superior parietal gyrus, precentral gyrus, inferior temporal gyrus, fusiform gyrus, superior temporal sulcus, and thalamus. We then calculated the correlation between the FC pattern score and obesity phenotypes (body mass index, waist circumference, and waist‐to‐hip ratio) in Caucasian participants (n = 1,403) from the UK Biobank database. The correlation results in the Caucasian subsample were 0.270 for body mass index (0.265 for all ethnicities), 0.321 for waist circumference (0.308 for all ethnicities), and 0.271 for waist‐to‐hip ratio (0.236 for all ethnicities). All FDR‐corrected p‐values were lower than .05 (Figure S4b). These results indicate that the FC pattern observed in Caucasians was largely similar to the FC pattern observed in all ethnicities. Therefore, the FC pattern observed in Caucasians might effectively validate ethnically diverse datasets.
3.4.3. FC pattern associated with body mass index
We also estimated the FC pattern associated with body mass index (not waist circumference) to compare the whole‐brain connectivity patterns between different obesity phenotypes. A root mean squared error of FC patterns between those derived from waist circumference and body mass index was 1.523. The brain regions with strong FC pattern mostly overlapped (ventrolateral prefrontal cortex, superior parietal lobule, cingulate gyrus, globus pallidus, precentral gyrus, fusiform gyrus, and thalamus), indicating consistency of FC pattern across different obesity phenotypes (Figure S5a). Although the FC pattern score derived from body mass index exhibited significant correlation with obesity phenotypes, the effect sizes (i.e., correlation value) for correlating with obesity phenotypes (i.e., waist circumference and waist‐to‐hip ratio) were slightly reduced compared to the FC pattern score based on waist circumference (Figure S5b).
3.4.4. FC pattern of a non‐healthy weight population
Lastly, we computed the FC pattern using the data from enhanced Nathan Kline Institute‐Rockland Sample database, which contains only non‐healthy weight individuals (body mass index ≥ 25), to compare the pattern with that derived from the UK Biobank dataset. Comparison of FC patterns between the enhanced Nathan Kline Institute‐Rockland Sample and UK Biobank databases yielded quite a large root mean squared error of 3.916. In addition, we found different connectivity patterns, especially in the precentral gyrus, fusiform gyrus, posterior superior temporal sulcus, and cingulate gyrus (Figure S6). The FC pattern developed from enhanced Nathan Kline Institute‐Rockland Sample showed low generalizability when we applied it to the independent datasets to correlate obesity phenotypes (all p > .1; UK Biobank: r = 0.034 for body mass index, r = 0.062 for waist circumference, r = 0.054 for waist‐to‐hip ratio; Human Connectome Project: r = 0.003 for body mass index; St. Vincent's Hospital: r = 0.067 for body mass index, r = 0.043 for waist circumference, r = 0.350 for waist‐to‐hip ratio). This inconsistency may be due to differences in participant demographics between the databases. The UK Biobank contains both healthy and non‐healthy weight populations, while the enhanced Nathan Kline Institute‐Rockland Sample only contains non‐healthy weight subjects. Thus, the FC pattern developed using enhanced Nathan Kline Institute‐Rockland Sample data is biased to the non‐healthy weight subjects, which is not suitable to be applied to the general population.
4. DISCUSSION
Understanding the relation between whole‐brain functional organization and clinical or behavioral phenotypes is a challenge for neuroscience, as reproducible findings are scarce (Masouleh, Eickhoff, Hoffstaedter, & Genon, 2019). Our study identified a robust obesity phenotype‐associated FC pattern that correlated with body mass index, waist circumference, and waist‐to‐hip ratio in a large‐scale rs‐fMRI dataset (n ~ 1,500). Notably, reproducibility was assessed in demographically diverse three independent datasets (n ~ 900).
We found that the FC pattern showed a positive association with obesity phenotypes in brain regions regulating reward processing, while the brain regions that process sensory information showed a negative association. Specifically, reward brain regions including the ventromedial and ventrolateral prefrontal cortex, superior parietal lobule, superior temporal gyrus, and cingulate gyrus, as well as a subcortical structure of globus pallidus showed strong positive FC pattern. The results indicate an increase in functional connectivity in these brain areas as a function of adiposity, supporting prior studies (Chao et al., 2018; García‐García et al., 2013; García‐García et al., 2015; Park et al., 2016; Park, Moon, & Park, 2018). In particular, the ventromedial and orbitofrontal cortex track food‐related reward values (Goldstone et al., 2009; Hare, Camerer, & Rangel, 2009; Holland & Gallagher, 2004; Kable & Glimcher, 2007; O'Doherty, Deichmann, Critchley, & Dolan, 2002; Rolls, 2011). In line with these results, our findings support the fact that individuals with obesity show altered reward system (García‐García et al., 2014; Volkow, Wang, Fowler, & Telang, 2008) by showing higher susceptibility to gustatory and visual cues in a delay discounting paradigm (Morys, Bode, & Horstmann, 2018) and increased responsivity to passive food picture viewing tasks (Pursey et al., 2014; van den Akker, Stewart, Antoniou, Palmberg, & Jansen, 2014). Further, increased macroscale connectivity, particularly in the prefrontal, parietal, and cingulate cortices is known to be related to eating behaviors in people with obesity (Park et al., 2016; Park, Lee, et al., 2018; Park, Moon, et al., 2018). Indeed, imbalance in inhibitory control and food‐related reward system in the prefrontal cortex and paralimbic areas is associated with increased feelings of hunger (Tataranni et al., 1999; Tataranni & DelParigi, 2003) and may lead to overeating and weight gain (Brooks et al., 2013; Ding et al., 2020; Olivo et al., 2017; Steward, Juaneda‐Seguí, et al., 2019; Steward, Picó‐Pérez, et al., 2019; Vainik, Dagher, Dubé, & Fellows, 2013; Van Meer et al., 2019; Van Opstal et al., 2018; Van Opstal et al., 2019; Verdejo‐Román et al., 2017; Volkow, Wang, Telang, et al., 2008; Ziauddeen et al., 2015). In contrast, we found negative associations with obesity phenotypes in brain regions involved in multisensory processing, particularly precentral gyrus, inferior temporal gyrus, fusiform gyrus, superior temporal sulcus, and thalamus. The results indicate that greater adiposity is related to lower functional connectivity within sensory‐related brain regions, which is supported by prior work that found altered sensory processing in people with obesity (Park et al., 2015; Scarpina et al., 2016; Van der Laan, de Ridder, Viergever, & Smeets, 2011; Wang et al., 2002). Indeed, previous studies have suggested the presence of atypical multisensory integration in obesity (Olde Dubbelink et al., 2008; Scarpina et al., 2016; Stice, Spoor, Bohon, Veldhuizen, & Small, 2008). It should be noted that the underlying mechanisms of atypical multisensory processing in obese individuals are still matters of debate, which may need validation based on microcircuit or transcriptomic analysis. Overall, our findings are consistent with a large body of literature that relates obesity to alterations in reward and sensory processing, where increased sensitivity to rewarding food cues via the reward system coupled with altered inhibitory control might be related to overeating and hence lead to obesity. How the neural mechanisms differentially affect macroscale connectome between reward circuits (i.e., positively associated) and sensory processing (i.e., negatively associated) needs to be investigated further.
Our study has made several contributions. First, we used a large‐scale (n ~ 2,400) fMRI dataset spanning four independent sites. Many previous studies had used small sample sizes and single cohorts, perhaps resulting in conflicting reports concerning obesity phenotype‐associated brain regions (Ding et al., 2020; Gupta et al., 2017; Herrmann et al., 2019; Marqués‐Iturria et al., 2013; Van Meer et al., 2019; Opel et al., 2015; Van Opstal et al., 2019; Park et al., 2015; Park et al., 2016; Park, Lee, et al., 2018; Park, Moon, et al., 2018; Shott et al., 2015; Steward, Picó‐Pérez, et al., 2019; Stoeckel et al., 2008). Our study attempted to circumvent these shortcomings by leveraging a large‐scale dataset from different cohorts, which included a wide range of ages and ethnicities. Second, the confounding effects caused by the different acquisition parameters of the four centers in this study were controlled for using standardized data processing pipelines (Table S7). The pipelines adopted state‐of‐the‐art methods and were largely consistent in the major preprocessing components of the preprocessing steps, namely: head motion correction, intensity normalization, spatial registration, nuisance variable removal, and temporal filtering. Despite some differences across the pipelines such as correction for gradient distortions, slice‐timing, and volume scrubbing, our results showed that the FC pattern was well‐validated across demographic characteristics. Third, our findings were relatively robust across a broader age span (from 13 to 83 years) and different ethnicities (Caucasian, African‐American, Asian, and mixed). This finding is even more interesting because both age and ethnicity are known to differentially affect obesity (El‐Hazmi & Warsy, 2002; Menigoz, Nathan, & Turrell, 2016; Rai, Sandell, Cheverud, & Brophy, 2013). We believe the relationship between the FC pattern score and obesity phenotypes could be better described if the effects of age and ethnicity are considered. This issue should also be explored in future studies. Fourth, the FC pattern score is easy to calculate, which means it is feasible to replicate our findings using independent datasets. The full pipeline is available at https://gitlab.com/by9433/fcobesity. Last, our study extended prior obesity‐related neuroimaging studies in terms of identifying whole‐brain FC pattern correlates with obesity phenotypes, not focusing on a few local brain regions. The succinct summary of the whole‐brain FC pattern provides novel insights for the understanding of functional connectivity across a range of obesity phenotypes. This was accomplished by combining graph‐theoretical connectivity analysis and machine learning. Prior obesity‐related neuroimaging studies focused on identifying brain regions that activate for specific tasks (e.g., food‐cue task) (Brooks et al., 2013; Goldstone et al., 2009; Opel et al., 2015; Stoeckel et al., 2008; Van Meer et al., 2019) and comparing intrinsic connectivity of local brain regions between individuals with healthy and non‐healthy weights (García‐García et al., 2013; García‐García et al., 2015; Lips et al., 2014), lacking the probing of FC at the whole‐brain level. Leveraging advanced machine learning techniques with strict cross‐validation, we developed a reliable and reproducible whole‐brain FC pattern that maximally correlates with obesity phenotypes, which can be generally applied to an independent dataset.
In this study, we used the Brainnetome atlas (Fan et al., 2016) rather than other parcellation schemes such as a pre‐defined atlas and data‐driven approaches such as clustering algorithms or independent component analysis (Beckmann, DeLuca, Devlin, & Smith, 2005; Craddock, James, Holtzheimer, Hu, & Mayberg, 2012; Fan et al., 2016; Glasser et al., 2016; Li, Song, Fan, Liu, & Jiang, 2015; Park, Tark, Shim, & Park, 2018). Data‐driven approaches are advantageous in that the defined networks are tailored to the given data. However, they are not easily transferable to other data with different characteristics. The pre‐defined atlas reduces this problem because it divides brain regions based on clear boundaries, which can be reliably registered and identified.
Our study has several limitations. First, we only included neurologically healthy subjects in this study. Further validation is required to generalize the model to people with obesity‐related medical complications. Second, we did not prove a causal relationship between the FC pattern and obesity because our study is cross‐sectional. Future longitudinal work may help identify and characterize such causal associations. Last, we used degree centrality to quantify the connection strength of the given nodes. We selected degree centrality to represent the strength of nodal connectivity because previous studies used it to associate body mass index and eating behaviors in people with obesity (Park et al., 2015; Park et al., 2016; Park, Lee, et al., 2018). Different nodal centrality measures based on graph theory such as betweenness and eigenvector centrality, as well as local efficiency, could be used to characterize the brain networks (Rubinov & Sporns, 2010). By comparing the test–retest reproducibility of these measures, we can find the most stable measure to associate whole‐brain function and obesity phenotypes, from which future studies could benefit. In addition, comparing the segregation and integration patterns of modular architectures according to obesity phenotypes may provide complementary insights for understanding functional connectome organization associated with obesity phenotypes.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Supporting information
Data S1: Supporting Information.
Data S2: Supporting Information.
ACKNOWLEDGMENTS
This work was supported by the Molson Neuro‐Engineering Fellowship from the Montreal Neurological Institute and Hospital (MNI), the Fonds de la Recherche du Québec – Santé (FRQ‐S), the National Research Foundation of Korea (NRF‐2020M3E5D2A01084892), the Institute for Basic Science (grant number IBS‐R015‐D1), the MIST (Ministry of Science and ICT) of Korea under the ITRC (Information Technology Research Center) support program (grant number IITP‐2020‐2018‐0‐01798) supervised by the IITP (Institute for Information & communication Technology Promotion), and the IITP grant funded by the Korean government under the AI Graduate School Support Program (grant number 2019‐0‐00421).
Park B, Byeon K, Lee MJ, et al. Whole‐brain functional connectivity correlates of obesity phenotypes. Hum Brain Mapp. 2020;41:4912–4924. 10.1002/hbm.25167
Funding information Fonds de la Recherche du Québec ‐ Santé (FRQ‐S), Grant/Award Number: Postdoctoral Training; Institute for Basic Science, Grant/Award Number: IBS‐R015‐D1; Korean government under the AI Graduate School Support Program, Grant/Award Number: 2019‐0‐00421; MIST (Ministry of Science and ICT) of Korea under the ITRC (Information Technology Research Center), Grant/Award Number: IITP‐2020‐2018‐0‐01798 National Research Foundation of Korea (NRF‐2020M3E5D2A01084892); Montreal Neurological Institute and Hospital (MNI), Grant/Award Number: Molson Neuro‐Engineering Fellowship
DATA AVAILABILITY STATEMENT
The full imaging and phenotypic data from the UK Biobank, Human Connectome Project, and enhanced Nathan Kline Institute‐Rockland Sample databases are available from each data repository after approval (UK Biobank: https://www.ukbiobank.ac.uk/, Human Connectome Project: https://www.humanconnectome.org/, enhanced Nathan Kline Institute‐Rockland Sample: http://fcon_1000.projects.nitrc.org/indi/enhanced/). Data from St. Vincent's Hospital are not publicly available due to IRB restrictions. The codes for data preprocessing, time‐series extraction, calculation of degree centrality values, and construction of the FC pattern are available at https://gitlab.com/by9433/fcobesity.
REFERENCES
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. R. , Griffanti, L. , Douaud, G. , … Smith, S. M. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UKbiobank. NeuroImage, 166, 400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants, B. B. , Tustison, N. J. , Song, G. , Cook, P. A. , Klein, A. , & Gee, J. C. (2011). A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 54, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann, C. F. , DeLuca, M. , Devlin, J. T. , & Smith, S. M. (2005). Investigations into resting‐state connectivity using independent component analysis. Philosophical Transactions of the Royal Society B, 360, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, 57, 289–300. [Google Scholar]
- Bijsterbosch, J. , Smith, S. M. , & Beckmann, C. F. (2017). Introduction to resting state fMRI functional connectivity. Oxford: Oxford University Press. [Google Scholar]
- Brooks, S. J. , Cedernaes, J. , & Schiöth, H. B. (2013). Increased prefrontal and parahippocampal activation with reduced dorsolateral prefrontal and insular cortex activation to food images in obesity: a meta‐analysis of fMRI studies. PLoS One, 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalska, I. L. , Kumar, S. S. P. , Bujalska, I. J. , Kumar, S. , & Stewart, P. M. (1997). Does central obesity reflect “Cushing's disease of the omentum”? Lancet, 349, 1210–1213. [DOI] [PubMed] [Google Scholar]
- Bullmore, E. , & Sporns, O. (2009). Complex brain networks: Graph theoretical analysis of structural and functional systems. Nature Neuroscience, 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Chao, S. H. , Liao, Y. T. , Chen, V. C. H. , Li, C. J. , McIntyre, R. S. , Lee, Y. , & Weng, J. C. (2018). Correlation between brain circuit segregation and obesity. Behavioural Brain Research, 337, 218–227. [DOI] [PubMed] [Google Scholar]
- Cox, R. W. (1996). AFNI: Software for analysis and visualization of functional magnetic resonance Neuroimages. Computers and Biomedical Research, 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Craddock, R. C. , James, G. A. , Holtzheimer, P. E. , Hu, X. P. , & Mayberg, H. S. (2012). A whole brain fMRI atlas generated via spatially constrained spectral clustering. Human Brain Mapping, 33, 1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju, E. , Allen, E. A. , Belger, A. , Ford, J. M. , McEwen, S. , Mathalon, D. H. , … Calhoun, V. D. (2014). Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage: Clinical, 5, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, J.‐P. , & Lemieux, I. (2006). Abdominal obesity and metabolic syndrome. Nature, 444, 881–887. [DOI] [PubMed] [Google Scholar]
- Després, J. P. , Lemieux, I. , Bergeron, J. , Pibarot, P. , Mathieu, P. , Larose, E. , … Poirier, P. (2008). Abdominal obesity and the metabolic syndrome: Contribution to global cardiometabolic risk. Arteriosclerosis, Thrombosis, and Vascular Biology, 28, 1039–1049. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Ji, G. , Li, G. , Zhang, W. , Hu, Y. , Liu, L. , … Zhang, Y. (2020). Altered interactions among resting‐state networks in individuals with obesity. Obesity, 28, 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Hazmi, M. A. F. , & Warsy, A. S. (2002). Relationship between age and the prevalence of obesity and overweight in Saudi population. Bahrain Medical Bulletin, 24, 49–53. [Google Scholar]
- Fan, L. , Li, H. , Zhuo, J. , Zhang, Y. , Wang, J. , Chen, L. , … Jiang, T. (2016). The human Brainnetome atlas: A new Brain atlas based on connectional architecture. Cerebral Cortex, 26, 3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom, A. R. , Kaye, S. A. , Sellers, T. A. , Hong, C. P. , Cerhan, J. R. , Potter, J. D. , & Prineas, R. J. (1993). Body fat distribution and 5‐year risk of death in older women. JAMA, 269, 483–487. [PubMed] [Google Scholar]
- Folsom, A. R. , Kushi, L. H. , Anderson, K. E. , Mink, P. J. , Olson, J. E. , Hong, C.‐P. , … Prineas, R. J. (2000). Associations of general and abdominal obesity with multiple health outcomes in older women. Archives of Internal Medicine, 160, 2117–2128. [DOI] [PubMed] [Google Scholar]
- García‐García, I. , Horstmann, A. , Jurado, M. A. , Garolera, M. , Chaudhry, S. J. , Margulies, D. S. , … Neumann, J. (2014). Reward processing in obesity, substance addiction and non‐substance addiction. Obesity Reviews, 15, 853–869. [DOI] [PubMed] [Google Scholar]
- García‐García, I. , Jurado, M. Á. , Garolera, M. , Marqués‐Iturria, I. , Horstmann, A. , Segura, B. , … Neumann, J. (2015). Functional network centrality in obesity: A resting‐state and task fMRI study. Psychiatry Research: Neuroimaging, 233, 331–338. [DOI] [PubMed] [Google Scholar]
- García‐García, I. , Jurado, M. Á. , Garolera, M. , Segura, B. , Sala‐Llonch, R. , Marqués‐Iturria, I. , … Junqué, C. (2013). Alterations of the salience network in obesity: A resting‐state fMRI study. Human Brain Mapping, 34, 2786–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Coalson, T. S. , Robinson, E. C. , Hacker, C. D. , Harwell, J. , Yacoub, E. , … Van Essen, D. C. (2016). A multi‐modal parcellation of human cerebral cortex. Nature, 536, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Sotiropoulos, S. N. , Wilson, J. A. , Coalson, T. S. , Fischl, B. , Andersson, J. L. , … Jenkinson, M. (2013). The minimal preprocessing pipelines for the human Connectome project. NeuroImage, 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone, A. P. , Prechtl De Hernandez, C. G. , Beaver, J. D. , Muhammed, K. , Croese, C. , Bell, G. , … Bell, J. D. (2009). Fasting biases brain reward systems towards high‐calorie foods. The European Journal of Neuroscience, 30, 1625–1635. [DOI] [PubMed] [Google Scholar]
- Griffa, A. , & Van Den Heuvel, M. P. (2018). Rich‐club neurocircuitry: Function, evolution, and vulnerability. Dialogues in Clinical Neuroscience, 20, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A. , Mayer, E. A. , Hamadani, K. , Bhatt, R. , Fling, C. , Alaverdyan, M. , … Labus, J. S. (2017). Sex differences in the influence of body mass index on anatomical architecture of brain networks. International Journal of Obesity, 41, 1185–1195. 10.1038/ijo.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A. , Mayer, E. A. , Labus, J. S. , Bhatt, R. R. , Ju, T. , Love, A. , … Kilpatrick, L. A. (2018). Sex commonalities and differences in obesity‐related alterations in intrinsic Brain activity and connectivity. Obesity, 26, 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, T. A. , Camerer, C. F. , & Rangel, A. (2009). Valuation system. Science, 324, 646–649. [DOI] [PubMed] [Google Scholar]
- Herrmann, M. J. , Tesar, A. K. , Beier, J. , Berg, M. , & Warrings, B. (2019). Grey matter alterations in obesity: A meta‐analysis of whole‐brain studies. Obesity Reviews, 20, 464–471. [DOI] [PubMed] [Google Scholar]
- Holland, P. C. , & Gallagher, M. (2004). Amygdala‐frontal interactions and reward expectancy. Current Opinion in Neurobiology, 14, 148–155. [DOI] [PubMed] [Google Scholar]
- Hong, S.‐J. , Vos De Wael, R. , Bethlehem, R. A. I. , Lariviere, S. , Paquola, C. , Valk, S. L. , … Bernhardt, B. C. (2019). Atypical functional connectome hierarchy in autism. Nature Communications, 10, 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. J. , Woolrich, M. W. , & Smith, S. M. (2012). Fsl. NeuroImage, 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Kable, J. W. , & Glimcher, P. W. (2007). The neural correlates of subjective value during intertemporal choice. Nature Neuroscience, 10, 1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale, V. V. , Hamde, S. T. , & Holambe, R. S. (2019). Multi class disorder detection of magnetic resonance brain images using composite features and neural network. Biomedical Engineering Letters, 9, 221–231. 10.1007/s13534-019-00103-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. J. , Park, B. , Cho, S. , Park, H. , Kim, S.‐T. , & Chung, C.‐S. (2019). Dynamic functional connectivity of migraine brain: a resting‐state functional magnetic resonance imaging study. Pain, 160, 2776–2786. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Song M., Fan L., Liu Y., Jiang T. (2015). Parcellation of the primary cerebral cortices based on local connectivity profiles. Frontiers in Neuroanatomy, 9 10.3389/fnana.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips, M. a. , Wijngaarden, M. a. , Van Der Grond, J. , Van Buchem, M. a. , De Groot, G. H. , Rombouts, S. a. R. B. , … Veer, I. M. (2014). Resting‐state functional connectivity of brain regions involved in cognitive control, motivation, and reward is enhanced in obese females. The American Journal of Clinical Nutrition, 100, 524–531. [DOI] [PubMed] [Google Scholar]
- Marqués‐Iturria, I. , Pueyo, R. , Garolera, M. , Segura, B. , Junqué, C. , García‐García, I. , … Jurado, M. Á. (2013). Frontal cortical thinning and subcortical volume reductions in early adulthood obesity. Psychiatry Research: Neuroimaging, 214, 109–115. [DOI] [PubMed] [Google Scholar]
- Martin, L. E. , Holsen, L. M. , Chambers, R. J. , Bruce, A. S. , Brooks, W. M. , Zarcone, J. R. , … Savage, C. R. (2010). Neural mechanisms associated with food motivation in obese and healthy weight adults. Obesity, 18, 254–260. [DOI] [PubMed] [Google Scholar]
- Masouleh, S. K. , Eickhoff, S. B. , Hoffstaedter, F. , & Genon, S. (2019). Empirical examination of the replicability of associations between brain structure and psychological variables. eLife, 8, e43464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menigoz, K. , Nathan, A. , & Turrell, G. (2016). Ethnic differences in overweight and obesity and the influence of acculturation on immigrant bodyweight: Evidence from a national sample of Australian adults. BMC Public Health, 16, 1–13. 10.1186/s12889-016-3608-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. L. , Alfaro‐Almagro, F. , Bangerter, N. K. , Thomas, D. L. , Yacoub, E. , Xu, J. , … Smith, S. M. (2016). Multimodal population brain imaging in the UKbiobank prospective epidemiological study. Nature Neuroscience, 19, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C. F. , Sabino, V. , Koob, G. F. , & Cottone, P. (2017). Neuroscience of compulsive eating behavior. Frontiers in Neuroscience, 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Lopez, L. , Contreras‐Rodriguez, O. , Soriano‐Mas, C. , Stamatakis, E. A. , & Verdejo‐Garcia, A. (2016). Disrupted functional connectivity in adolescent obesity. NeuroImage: Clinical, 12, 262–268. 10.1016/j.nicl.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morys, F. , Bode, S. , & Horstmann, A. (2018). Dorsolateral and medial prefrontal cortex mediate the influence of incidental priming on economic decision making in obesity. Scientific Reports, 8, 17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford, J. a. , Horvath, S. , Oldham, M. C. , Langfelder, P. , Geschwind, D. H. , & Poldrack, R. a. (2010). Detecting network modules in fMRI time series: a weighted network analysis approach. NeuroImage, 52, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, S. , Tulloch, A. , Gold, M. S. , & Avena, N. M. (2014). Hormonal and neural mechanisms of food reward, eating behaviour and obesity. Nature Reviews. Endocrinology, 10, 540–552. [DOI] [PubMed] [Google Scholar]
- Nooner, K. B. , Colcombe, S. J. , Tobe, R. H. , Mennes, M. , Benedict, M. M. , Moreno, A. L. , … Milham, M. P. (2012). The NKI‐Rockland sample: A model for accelerating the pace of discovery science in psychiatry. Frontiers in Neuroscience, 6, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty, J. P. , Deichmann, R. , Critchley, H. D. , & Dolan, R. J. (2002). Neural responses during anticipation of a primary taste reward. Neuron, 33, 815–826. [DOI] [PubMed] [Google Scholar]
- Olde Dubbelink, K. T. E. , Felius, A. , Verbunt, J. P. A. , van Dijk, B. W. , Berendse, H. W. , Stam, C. J. , & Delemarre‐van de Waal, H. A. (2008). Increased resting‐state functional connectivity in obese adolescents; a magnetoencephalographic pilot study. PLoS One, 3, e2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo, G. , Wiemerslage, L. , Swenne, I. , Zhukowsky, C. , Salonen‐Ros, H. , Larsson, E. M. , … Schiöth, H. B. (2017). Limbic‐thalamo‐cortical projections and reward‐related circuitry integrity affects eating behavior: A longitudinal DTI study in adolescents with restrictive eating disorders. PLoS One, 12, e0172129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel, N. , Redlich, R. , Grotegerd, D. , Dohm, K. , Haupenthal, C. , Heindel, W. , … Dannlowski, U. (2015). Enhanced neural responsiveness to reward associated with obesity in the absence of food‐related stimuli. Human Brain Mapping, 36, 2330–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Byeon K., Park H. (2019). FuNP (Fusion of Neuroimaging Preprocessing) Pipelines: A Fully Automated Preprocessing Software for Functional Magnetic Resonance Imaging. Frontiers in Neuroinformatics, 13 10.3389/fninf.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Hong, J. , & Park, H. (2017). Neuroimaging biomarkers to associate obesity and negative emotions. Scientific Reports, 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Lee, M. J. , Kim, M. , Kim, S.‐H. , & Park, H. (2018). Structural and functional Brain connectivity changes between people with abdominal and non‐abdominal obesity and their association with behaviors of eating disorders. Frontiers in Neuroscience, 12, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Moon, T. , & Park, H. (2018). Dynamic functional connectivity analysis reveals improved association between brain networks and eating behaviors compared to static analysis. Behavioural Brain Research, 337, 114–121. [DOI] [PubMed] [Google Scholar]
- Park, B. , Seo, J. , & Park, H. (2016). Functional brain networks associated with eating behaviors in obesity. Scientific Reports, 6, 23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Seo, J. , Yi, J. , & Park, H. (2015). Structural and functional brain connectivity of people with obesity and prediction of body mass index using connectivity. PLoS One, 10, e0141376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. , Tark, K.‐J. , Shim, W. M. , & Park, H. (2018). Functional connectivity based parcellation of early visual cortices. Human Brain Mapping, 39, 1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. , Took, C. C. , & Seong, J. K. (2018). Machine learning in biomedical engineering. Biomedical Engineering Letters, 8, 1–3. 10.1007/s13534-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Barnes, K. A. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursey, K. M. , Stanwell, P. , Callister, R. J. , Brain, K. , Collins, C. E. , & Burrows, T. L. (2014). Neural responses to visual food cues according to weight status: A systematic review of functional magnetic resonance imaging studies. Frontiers in Nutrition, 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, M. F. , Sandell, L. J. , Cheverud, J. M. , & Brophy, R. H. (2013). Relationship of age and body mass index to the expression of obesity and osteoarthritis‐related genes in human meniscus. International Journal of Obesity, 37, 1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, E. T. (2011). Taste, olfactory and food texture reward processing in the brain and obesity. International Journal of Obesity, 35, 550–561. 10.1038/ijo.2010.155 [DOI] [PubMed] [Google Scholar]
- Rubinov, M. , & Sporns, O. (2010). Complex network measures of brain connectivity: Uses and interpretations. NeuroImage, 52, 1059–1069. [DOI] [PubMed] [Google Scholar]
- Salimi‐Khorshidi, G. , Douaud, G. , Beckmann, C. F. , Glasser, M. F. , Griffanti, L. , & Smith, S. M. (2014). Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage, 90, 449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpina, F. , Migliorati, D. , Marzullo, P. , Mauro, A. , Scacchi, M. , & Costantini, M. (2016). Altered multisensory temporal integration in obesity. Scientific Reports, 6, 28382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, A. J. , & McGonigle, J. (2011). Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. NeuroImage, 55, 1132–1146. [DOI] [PubMed] [Google Scholar]
- Shott, M. E. , Cornier, M. , Mittal, V. a. , Pryor, T. L. , Orr, J. M. , Brown, M. S. , & Frank, G. K. W. (2015). Orbitofrontal cortex volume and brain reward response in obesity. International Journal of Obesity, 39, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Miller, K. L. , Salimi‐Khorshidi, G. , Webster, M. , Beckmann, C. F. , Nichols, T. E. , … Woolrich, M. W. (2011). Network modelling methods for FMRI. NeuroImage, 54, 875–891. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , Vidaurre, D. , Beckmann, C. F. , Glasser, M. F. , Jenkinson, M. , Miller, K. L. , … Van Essen, D. C. (2013). Functional connectomics from resting‐state fMRI. Trends in Cognitive Sciences, 17, 666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward, T. , Juaneda‐Seguí, A. , Mestre‐Bach, G. , Martínez‐Zalacaín, I. , Vilarrasa, N. , Jiménez‐Murcia, S. , … Fernandez‐Aranda, F. (2019). What difference does it make? Risk‐taking behavior in obesity after a loss is associated with decreased ventromedial prefrontal cortex activity. Journal of Clinical Medicine, 8, 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward, T. , Miranda‐Olivos, R. , Soriano‐Mas, C. , & Fernández‐Aranda, F. (2019). Neuroendocrinological mechanisms underlying impulsive and compulsive behaviors in obesity: a narrative review of fMRI studies. Reviews in Endocrine & Metabolic Disorders, 20, 263–272. [DOI] [PubMed] [Google Scholar]
- Steward, T. , Picó‐Pérez, M. , Mestre‐Bach, G. , Martínez‐Zalacaín, I. , Suñol, M. , Jiménez‐Murcia, S. , … Fernandez‐Aranda, F. (2019). A multimodal MRI study of the neural mechanisms of emotion regulation impairment in women with obesity. Translational Psychiatry, 9, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice, E. , Spoor, S. , Bohon, C. , Veldhuizen, M. , & Small, D. (2008). Relation of reward from food intake and anticipated food intake to obesity: A functional magnetic resonance imaging study. Journal of Abnormal Psychology, 117, 924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel, L. E. , Weller, R. E. , Cook, E. W. , Twieg, D. B. , Knowlton, R. C. , & Cox, J. E. (2008). Widespread reward‐system activation in obese women in response to pictures of high‐calorie foods. NeuroImage, 41, 636–647. [DOI] [PubMed] [Google Scholar]
- Tataranni, P. A. , & DelParigi, A. (2003). Functional neuroimaging: a new generation of human brain studies in obesity research. Obesity Reviews, 4, 229–238. 10.1046/j.1467-789X.2003.00111.x [DOI] [PubMed] [Google Scholar]
- Tataranni, P. A. , Gautier, J.‐F. , Chen, K. , Uecker, A. , Bandy, D. , Salbe, A. D. , … Ravussin, E. (1999). Neuroanatomical correlates of hunger and satiation in humans using positron emission tomography. Proceedings of the National Academy of Sciences of the United States of America, 96, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, W. H. , & Fransson, P. (2016). On stabilizing the variance of dynamic functional Brain connectivity time series. Brain Connectivity, 6, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainik, U. , Dagher, A. , Dubé, L. , & Fellows, L. K. (2013). Neurobehavioural correlates of body mass index and eating behaviours in adults: A systematic review. Neuroscience and Biobehavioral Reviews, 37, 279–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val‐Laillet, D. , Aarts, E. , Weber, B. , Ferrari, M. , Quaresima, V. , Stoeckel, L. E. , … Stice, E. (2015). Neuroimaging and neuromodulation approaches to study eating behavior and prevent and treat eating disorders and obesity. NeuroImage: Clinical, 8, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Akker, K. , Stewart, K. , Antoniou, E. E. , Palmberg, A. , & Jansen, A. (2014). Food Cue reactivity, obesity, and impulsivity: Are they associated? Current Addiction Reports, 1, 301–308. [Google Scholar]
- Van Den Heuvel, M. P. , Kahn, R. S. , Goñi, J. , & Sporns, O. (2012). High‐cost, high‐capacity backbone for global brain communication. Proceedings of the National Academy of Sciences of the United States of America, 109, 11372–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Laan, L. N. , de Ridder, D. T. D. , Viergever, M. A. , & Smeets, P. A. M. (2011). The first taste is always with the eyes: A meta‐analysis on the neural correlates of processing visual food cues. NeuroImage, 55, 296–303. [DOI] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. J. , Yacoub, E. , & Ugurbil, K. (2013). The WU‐Minn human Connectome project: An overview. NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer, F. , Van der Laan, L. N. , Eiben, G. , Lissner, L. , Wolters, M. , Rach, S. , … Smeets, P. A. M. (2019). Development and body mass inversely affect children's brain activation in dorsolateral prefrontal cortex during food choice. NeuroImage, 201, 116016. [DOI] [PubMed] [Google Scholar]
- Van Opstal, A. M. , Van Den Berg‐Huysmans, A. A. , Hoeksma, M. , Blonk, C. , Pijl, H. , Rombouts, S. A. R. B. , & van Der Grond, J. (2018). The effect of consumption temperature on the homeostatic and hedonic responses to glucose ingestion in the hypothalamus and the reward system. American Journal of Clinical Nutrition, 107, 20–25. [DOI] [PubMed] [Google Scholar]
- Van Opstal, A. M. , Wijngaarden, M. A. , Grond, J. , & Pijl, H. (2019). Changes in brain activity after weight loss. Obesity Science and Practice, 5, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdejo‐Román, J. , Vilar‐López, R. , Navas, J. F. , Soriano‐Mas, C. , & Verdejo‐García, A. (2017). Brain reward system's alterations in response to food and monetary stimuli in overweight and obese individuals. Human Brain Mapping, 38, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow, N. D. , Wang, G. J. , Fowler, J. S. , & Telang, F. (2008). Overlapping neuronal circuits in addiction and obesity: Evidence of systems pathology. Philosophical Transactions of the Royal Society B: Biological Sciences, 363, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow, N. D. , Wang, G.‐J. , Telang, F. , Fowler, J. S. , Thanos, P. K. , Logan, J. , … Pradhan, K. (2008). Low dopamine striatal D2 receptors are associated with prefrontal metabolism in obese subjects: Possible contributing factors. NeuroImage, 42, 1537–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.‐J. , Volkow, N. D. , Felder, C. , Fowler, J. S. , Levy, A. V. , Pappas, N. R. , … Netusil, N. (2002). Enhanced resting activity of the oral somatosensory cortex in obese subjects. Neuroreport, 13, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Watts, D. , & Strogatz, S. (1998). Collective dynamics of “small‐world” networks. Nature, 393, 440–442. [DOI] [PubMed] [Google Scholar]
- Ziauddeen, H. , Alonso‐Alonso, M. , Hill, J. O. , Kelley, M. , & Khan, N. A. (2015). Obesity and the neurocognitive basis of food reward and the control of intake. Advances in Nutrition, 6, 474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H. , & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society, Series B, 67, 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supporting Information.
Data S2: Supporting Information.
Data Availability Statement
The full imaging and phenotypic data from the UK Biobank, Human Connectome Project, and enhanced Nathan Kline Institute‐Rockland Sample databases are available from each data repository after approval (UK Biobank: https://www.ukbiobank.ac.uk/, Human Connectome Project: https://www.humanconnectome.org/, enhanced Nathan Kline Institute‐Rockland Sample: http://fcon_1000.projects.nitrc.org/indi/enhanced/). Data from St. Vincent's Hospital are not publicly available due to IRB restrictions. The codes for data preprocessing, time‐series extraction, calculation of degree centrality values, and construction of the FC pattern are available at https://gitlab.com/by9433/fcobesity.


