Abstract
Background
Mitochondrial encephalomyopathy caused by bi-allelic deleterious variants in TARS2 is rare. To date, only two pedigrees were reported in the literature and the connection between the gene and disease needs further study.
Case presentation
We report one infant who presented with limb hypertonia, epilepsy, developmental delay, and increased serum lactate from a non-consanguineous Chinese family. Whole-genome sequencing was performed to help to underlie the cause. We identified compound heterozygous variants c.470C > G, p.Thr157Arg and c.2143G > A, p.Glu715Lys in TARS2 and the variants were confirmed by Sanger sequencing. The patient was diagnosed with combined oxidative phosphorylation deficiency 21 according to the Online Mendelian Inheritance in Man (OMIM) database based on the clinical data and the deleterious effect of the two variants in TARS2 predicted by in silico tools.
Conclusions
We presented one case diagnosed with combined oxidative phosphorylation deficiency 21 based on clinical characteristics and genetic analysis. This is the first case in China and the fourth case in the world based on our document retrieval. This study facilitates the understanding of combined oxidative phosphorylation deficiency disease and demonstrates that the next-generation sequencing has a high potential to study inherited disease with high phenotypic heterogeneity and genetic heterogeneity including mitochondrial diseases such as combined oxidative phosphorylation deficiency.
Keywords: Mitochondrial diseases, Encephalomyopathy, TARS2, Whole genome sequencing, Case report
Background
Mitochondrial diseases are a group of genetic disorders with high heterogeneous that are characterized by dysfunctional mitochondria [1]. The minimum prevalence of all mitochondrial diseases is 1:5000 [2] and the clinical manifestations may present at any age [3]. Most of the mitochondrial disease patients harbor a defect in the oxidative phosphorylation system (OXPHS) which is the core and very important step for energy production [4].
TARS2 (*MIM 612,805) gene was first described by Bonnefond and mapped the gene to chromosome 1 in 2005 [5]. The gene encodes a mitochondrial aminoacyl-tRNA synthetase, a group of ubiquitous ‘house-keeping’ enzymes that perform an integral step in the initiation of translation by charging tRNAs with their cognate amino acids [6]. The enzymes are connected to many kinds of specific diseases including cancer, neuronal pathologies, autoimmune disorder, and disrupted metabolic conditions [7]. Many disease-causing mutations have been reported [8–11]. The first and the only report connecting TARS2 with the disease was published in 2014. Diodato et al. identified two compound heterozygous mutations in 2 siblings by whole-exome sequencing (WES) who presented with axial hypertonia, limb hypertonia, psychomotor delay, and increased serum lactate. Both of them died a few months after birth because of a metabolic crisis [12].
Nowadays, next-generation sequencing (NGS) or massively parallel sequencing has emerged as a powerful tool for testing genetically heterogeneous conditions including mitochondrial disorders [13–16]. The Mitochondrial Medicine Society demonstrated that this technology should be the preferred methodology when testing mtDNA and should be performed when mitochondrial disease is suspected [17]. Here we presented a patient with combined oxidative phosphorylation deficiency 21 who was diagnosed through clinical and molecular investigation.
Case presentation
The proband was a 4-month-old boy at the time of admission. He was born at full-term in an uneventful pregnancy. His birth weight, height, and head circumference were 2200 g (3th percentile), 48 cm (15th percentile), 33 cm (5th percentile) respectively. He had no so much cry after birth and was first referred to the local hospital because of afebrile convulsion about four times a day 1 month before he came to our hospital. The seizures often lasted for one minute and were characterized by staring, motion arrest, cyanosis, and rigidity of the extremities. The lactic acid level was elevated above the normal level (8.7 mmol/L, normal range < 2.0 mmol/L) and no obvious abnormalities were found in the routine examination for cerebrospinal fluid (CSF) and blood. The seizure wasn’t controlled with valproate (VPA, 21 mg/kg/d) and his parents stopped the treatment on their own after discharge. He had a fever 20 days ago. The seizures were more frequent than before accompanied by poor mental response and the lactic acid level was still higher than normal level (3.2 mmol/L). After cephalosporin treatment in the local hospital, his temperature returned to normal, but seizures were not relieved and his mental reaction was still poor.
He was then referred to our hospital. The global development delay with the parameters including the head circumference: 38 cm (3rd percentile), body length: 56 cm (3rd percentile), and weight: 5.1 kg (5th percentile) were revealed. His vital signs were still stable, although he had poor eye-eye contact and poor responses, and increased muscle tone of the limbs. The lactic acid level remained high (3.6 mmol/L) at initial admission, while CSF with mild polycytosis (white blood cell count = 30 × 106/L), mild increased protein level (0.66/L), normal glucose and chloride levels were detected. The CSF cultures were negative. Blood routine test, C-reactive protein and organs function were generally normal. Blood procalcitonin was 0.845 ng/ml, and sputum culture was positive for klebsiella pneumoniae. Plasma amino acid analysis showed an increase in alanine and proline content, indicating a metabolic disorder of lactate-pyruvate. Analysis of acyl-carnitine showed a significant increase in C2 and C4-OH, suggesting ketosis. The brain magnetic resonance imaging (MRI) showed an atrophy-like changes in the bilateral hemisphere and subdural effusion in the frontotemporal (Fig. 1). Electroencephalogram showed abnormal background activity with diffuse low and moderate amplitude slow activity of 1.5–2 Hz. The posterior head was dominated by frequent multi-focal sharp wave, spike wave, and slow spike wave, with medium voltage suppression of 1–2 s (Fig. 2). Brainstem auditory evoked potential revealed no obvious differentiation of bilateral I, III and V waves and bilateral auditory response thresholds were not differentiated. Echocardiography showed left to right shunt in the horizontal chamber, with a diameter about 3.5 mm. The initial diagnosis was suspected bacterial meningoencephalitis, epilepsy and psychomotor delay. After antibiotic therapy and anti-epileptic treatment (topiramate), the baby still had frequent seizures. Repeated examination of brain MRI showed abnormal symmetry signals in bilateral basal ganglia, thalamus and pedunculus cerebri. There was no significant changes in bilateral hemispheric atrophy and subdural effusion (Fig. 1). Mitochondrial energy metabolism disorders were considered and cocktail therapy was administrated including intravenous L-carnionc and oral coenzyme Q10, folic acid, vitamin B1, B2 and E. Antiepileptic treatment with levetiracetam was then added. The seizure reduced significantly and mental reaction improved. He was well fed and then discharged. Two months after discharge, he was readmitted to our hospital for poor mental response, during which he developed recurrent apnea with cyanosis and the heart rate below 60 beats/min. Despite adequate treatment of cocktail therapy, the apnea became progressively worse, and eventually the parents gave up the further treatment. The patient was followed up for 6 months after discharge and only levetiracetam was used during this period. At the last follow-up, he still had frequent seizures, and could not eat independently and remained restricted growth and development.
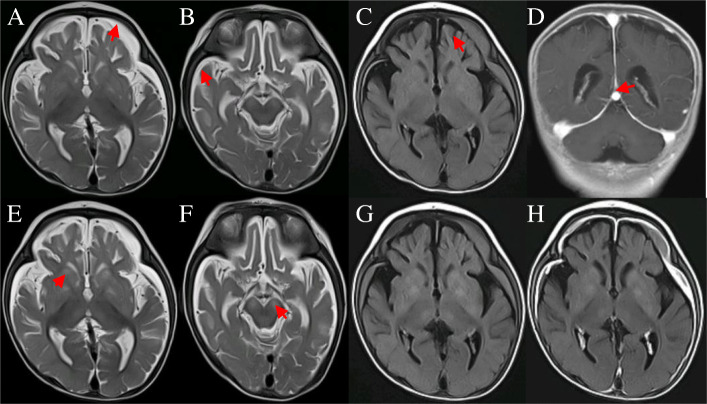
Fig. 1.
3T Brain MRI of the patient: a to d were performed when he was first transfer to our hospital at 4-month-old. a and b: axial T2WI showed arc-shaped subdural empyema was seen in bilateral frontotemporal parietal lobes. c: axial T2 Flair showed arc-shaped subdural empyema and bilateral cerebral hemisphere atrophy. d: Coronal T1WI enhanced scanning showed bilateral frontotemporal parietal dura enhanced which was called Mercedes-Benz sign. e to h was reexamined 7 days after hospitalization. e and f: axial T2WI showed new symmetrical hypersignal lesions in bilateral basal ganglia, thalamus and cerebral peduncle. g: the lesions had a slightly higher signal on T2Flair. h: the lesions had no obvious enhancement on T2Flair
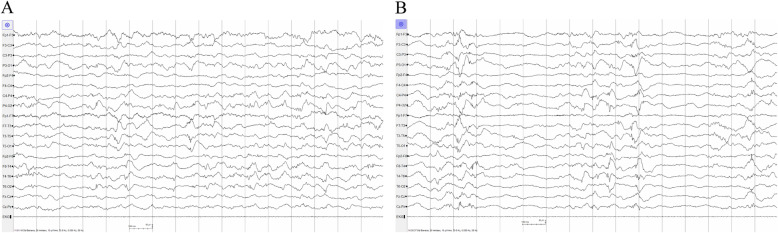
Fig. 2.
Electroencephalogram: a: abnormal background activity with diffuse low and moderate amplitude slow activity of 1.5-2 Hz. b the posterior head was dominated by frequent multi-focal sharp wave, spike wave, and slow spike wave, with medium voltage suppression of 1–2 s
Genomic DNA was extracted from whole blood using QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) and then sequenced on Illumina HiSeq X Ten platform (Illumina, Inc., San Diego, CA, USA). The reads were analyzed and annotated by an in-house pipeline. According to the biological significance of the mutation site, combined with the results of HGMD, ClinVar disease database and various protein function prediction software for analysis, priority is given to candidate variables related to disease phenotype. The putative mutation was further confirmed by Sanger DNA sequencing. All variant pathogenicity assessments are based on the joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology [18]. For copy number variants (CNV) analysis, we focused on the ultra-rare ones which were not been recorded in the Database of Genomic Variants (DGV) and the size which beyond 500 kb.
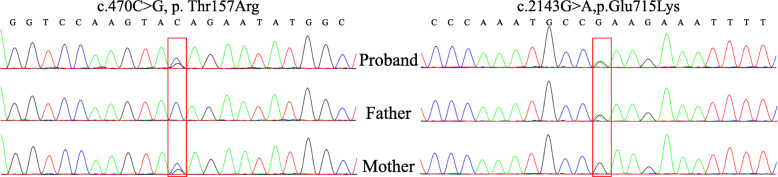
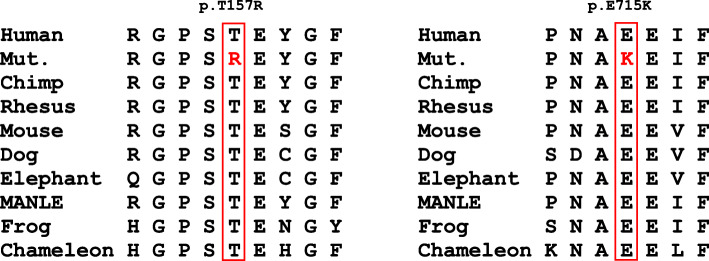
Finally, together with inheritance model analysis, two promising variants in TARS2 (c.470C > G and c.2143G > A) that were inherited from their parents, respectively, draw our attention after strict filtering. Also, the two variants were further validated by Sanger sequencing (Fig. 3). These two variants have not been reported in Pubmed, nor have they been cited by HGMD and ClinVar database. The minor allele frequency (MAF) of these two mutations in the gnomAD, ExAC and 1000 Genomes database is very low or non-existent and multiple lines of computational evidence support a deleterious effect on the gene or gene product (Table 1). The two residues are conserved among species, including humans, Chimp, Rhesus, Mouse, Dog, and Elephant respectively (Fig. 4). These two variants were interpreted as likely pathogenic based on ACMG guidelines (PM1, PM2, PP1 and PP4).
Fig. 3.
The compound heterozygous variants were detected by whole-genome sequencing and confirmed by Sanger sequencing
Table 1.
The analysis of pathogenicity of the variants in TARS2
| Gene | Transcript | Mutation | Variant origin | MAF | CADD | GERP + + | SIFT | PolyPhen | Mutation Taster | PROVEAN | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD (EAS AC) | ExAC (EAS AC) | 1000 Genomes | ||||||||||
| TARS2 | NM_025150.4 | c.470C > G p.Thr157Arg | paternal | 0.000163 | 0.000116 | NE | 22.6 | 5.23 | D | PD | D | D |
| TARS2 | NM_025150.4 | c.2143G > A p.Glu715Lys | maternal | NE | NE | NE | 25.7 | 5.25 | D | PD | D | D |
MAF minor allele frequency, NE not exist, D disease causing or deleterious or damaging, PD possibly damaging, EAS AC East Asian ExAC
Fig. 4.
The variants region is conserved among human, Chimp, Rhesus, Mouse, Dog, Elephant, MANLE, Frog, and Chameleon. Mut., mutant; MANLE, Mandrillus leucophaeus
Discussion and Conclusions
COXPD-21(Phenotype MIM number: 615918) is rare mitochondrial encephalomyopathy caused by mutation of the TARS2 gene on chromosome 1q21.2. The first pedigree was reported in 2014 by Diodato. They identified compound heterozygous variants c.845C > T p. Pro282Leu and c.695 + 3A > G in two siblings with early-onset mitochondrial encephalopathies by whole-exome sequencing. The two variants segregated within the family and the amount of TARS2 proteins and threonyl-tRNA levels was decreased in samples of afflicted patients according to the genetic defect. Wang et al. further analyzed the mechanism of the p.Pro282Leu mutation and clarified the molecular consequence of the mutation [19]. They found that the mutation induced both structural and functional defects by vitro and vivo analyses.
A PubMed search for reported cases of TARS2 and COXPD-21 yield no result but we find one abstract through Google Scholar search. Lucas Bartl et al. (2018) reported one pediatric patient with a putative splice-site mutation and missense mutation in TARS2 who present with hypotonia, cerebellar atrophy, psychomotor delay, and increased blood-lactate underwent whole-exome sequencing [20]. Both the two variants are predicted to cause loss of mtThrRS function and the author demonstrated that the two mutations cause mitochondrial dysfunction and are therefore causal for the disorder. Unfortunately, there were no further details about the two variants in this abstract.
TARS2 gene encodes a 718 length protein that belongs to mitochondrial aminoacyl-tRNA syntheses. This protein family has been associated with diverse clinical presentations such as hypertrophic cardiomyopathy, leukoencephalopathy with brain stem, and spinal cord involvement and lactate elevation(LBSL), Pontocerebellar hypoplasia type 6, progressive sensorineural hearing loss and ovarian dysgenesis and Charcot-Marie-Tooth disease [9, 10, 21–24]. As reviewed by González-Serrano et al., the majority of clinical manifestations are in the central nervous system [25]. Unfortunately, the pathogenic mechanism is not fairly well understood.
Here we described one infant suspected with congenital metabolic disease. By WGS analysis, we identified two putative variants in TARS2 and no other promising variant was identified. These two variants were novel and classified as likely pathogenic according to ACMG guidelines. Certainly, further study, such as measuring the enzyme activity of mitochondrial respiratory chain complex, is required to understand functional and structural changes of these two variants. Including our case, until now, there were 4 patients with TARS2 mutations reported, and they are summarized in Table 2.
Table 2.
Cases with TARS2 mutations including reported and the case in our study
| Pedigree | patient | Clincal phenotype | Pathogenic mutation | Reference |
|---|---|---|---|---|
| Family 1 | Two siblings (Female/Male) | Axial hypotonia, limb hypertonia, psychomotor delay, high levels of blood lactate, both died a few months after birth of a metabolic crisis | NM_025150.4:c.845C> T(p.Pro282Leu), Maternal / NM_025150.4:c.695 + 3A > G, Paternal | [12] |
| Family 2 | Not available | Hypotonia, cerebellar atrophy, psychomotor delay, and increased blood‐lactate | Not available | [20] |
| Family 3 | Male | Limb hypertonia, epilepsy, psychomotor delay, and high levels of blood lactate | NM_025150.4:c.470C > (p. Thr157Arg), Maternal / c.2143G > A p. Glu715Lys, Paternal | [This Study] |
In summary, this is the fourth case with COXPD-21 disease worldwide and the first in China which was diagnosed by a clinical and molecular investigation. We expand the mutation spectrum of combined oxidative phosphorylation deficiency. This contributes to the understanding of the connections between tRNA synthetase function and health. Besides, this study also suggests that WGS is a powerful technology that permits researchers to study the disease with highly phenotypic heterogeneity and will become more widely used in clinical settings.
Acknowledgements
The authors are very grateful to the participating family for their courtesy and humanity.
Abbreviations
- WGS
Whole-genome sequencing
- NGS
Next-generation sequencing
- CSF
Cerebrospinal fluid
- MRI
Magnetic resonance imaging
- ACMG
American College of Medical Genetics and Genomics
- HGMD
The Human Gene Mutation Database
- MAF
Minor allele frequency
- MIM
Mendelian Inheritance in Man
Authors’ contributions
Study Design, Analysis and Revision of the Manuscript, WC; Follow-up of Patient’s Information, XL, BP and CH; Original Draft Preparation, XL, BP and CH; Reanalysis of WGS Data, JL; Collection of Clinical and WGS Data, YZ, YL, WW and YT; Literature Search and Sanger Sequencing, YZ, YL, WW and YT. All authors have read and agreed to the published version of the manuscript.
Funding
Not applicable.
Availability of data and materials
Sanger sequencing data of the variant in TARS2 was deposited in the DDBJ database (Project ID: 5f7de96aa3c8820eeb82a9df). Human reference genome used in this study (UCSC hg19) can be downloaded at the following url: https://hgdownload.soe.ucsc.edu/downloads.html. All other data and materials mentioned in this manuscript, except patient’s private information, can be promptly available to readers by a reasonable request to the corresponding author.
Ethics approval and consent to participate
This research was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center, conforming to the Helsinki Declaration. Informed written consent for clinical and genetic analyses was obtained from the patient's parents.
Consent for publication
Written informed consent was obtained from the parents for publication of this case report and any accompanying tables/images.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaojing Li, Bingwei Peng and Chi Hou are co-first author and contributed equally to this work.
References
- 1.Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, Suomalainen A, Thorburn DR, Zeviani M, Turnbull DM. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders–past, present and future. BiochemBiophysActa. 2004;1659(2–3):115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Adam M, Ardinger H, Pagon R, Wallace S, Bean L, Stephens K, Amemiya A. Mitochondrial disorders overview--GeneReviews®.
- 4.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnefond L, Fender A, Rudinger-Thirion J, Giegé R, Florentz C, Sissler M. Toward the full set of human mitochondrial aminoacyl-tRNAsynthetases: characterization of AspRS and TyrRS. Biochemistry. 2005;44(12):4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- 6.Yao P, Fox PL. Aminoacyl-tRNAsynthetases in medicine and disease. EMBO Mol Med. 2013;5(3):332–343. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SG, Schimmel P, Kim S. AminoacyltRNAsynthetases and their connections to disease. ProcNatlAcadSci USA. 2008;105(32):11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. GlycyltRNAsynthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72(5):1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edvardson S, Shaag A, Kolesnikova O, Gomori JM, Tarassov I, Einbinder T, Saada A, Elpeleg O. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81(4):857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheper GC, van der Klok T, van Andel RJ, van Berkel CG, Sissler M, Smet J, Muravina TI, Serkov SV, Uziel G, Bugiani M, et al. Mitochondrial aspartyl-tRNAsynthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet. 2007;39(4):534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 11.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNAsynthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38(2):197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 12.Diodato D, Melchionda L, Haack TB, Dallabona C, Baruffini E, Donnini C, Granata T, Ragona F, Balestri P, Margollicci M, et al. VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum Mutat. 2014;35(8):983–989. doi: 10.1002/humu.22590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy KW. Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med. 2019;380(2):200–201. doi: 10.1056/NEJMc1814955. [DOI] [PubMed] [Google Scholar]
- 14.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, Yeung A, Peters H, Mordaunt D, Cowie S, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18(11):1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 15.Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet. 2018;19(5):325. doi: 10.1038/nrg.2018.12. [DOI] [PubMed] [Google Scholar]
- 16.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. SciTransl Med. 2012;4(118):118ra110. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikh S, Goldstein A, Koenig MK, Scaglia F, Enns GM, Saneto R, Anselm I, Cohen BH, Falk MJ, Greene C, et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med. 2015;17(9):689–701. doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhou XL, Ruan ZR, Liu RJ, Eriani G, Wang ED. A human disease-causing point mutation in mitochondrial threonyl-tRNAsynthetase induces both structural and functional defects. J BiolChem. 2016;291(12):6507–6520. doi: 10.1074/jbc.M115.700849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartl LDB, Reider J, Vitiello SPJTFJ. The genetic origin of a rare mitochondrial disorder. FASEB J. 2018;32(1_supplement):6. [Google Scholar]
- 21.Götz A, Tyynismaa H, Euro L, Ellonen P, Hyötyläinen T, Ojala T, Hämäläinen RH, Tommiska J, Raivio T, Oresic M, et al. Exome sequencing identifies mitochondrial alanyl-tRNAsynthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet. 2011;88(5):635–642. doi: 10.1016/j.ajhg.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce SB, Chisholm KM, Lynch ED, Lee MK, Walsh T, Opitz JM, Li W, Klevit RE, King MC. Mutations in mitochondrial histidyltRNAsynthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. ProcNatlAcadSci USA. 2011;108(16):6543–6548. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K, Iyer R, Cruz P, Cherukuri PF, Hansen NF, et al. Compound heterozygosity for loss-of-function lysyl-tRNAsynthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87(4):560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonellis A, Oprescu SN, Griffin LB, Heider A, Amalfitano A, Innis JW. Compound heterozygosity for loss-of-function FARSB variants in a patient with classic features of recessive aminoacyl-tRNAsynthetase-related disease. Hum Mutat. 2018;39(6):834–840. doi: 10.1002/humu.23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González-Serrano LE, Chihade JW, Sissler M. When a common biological role does not imply common disease outcomes: disparate pathology linked to human mitochondrial aminoacyl-tRNAsynthetases. J BiolChem. 2019;294(14):5309–5320. doi: 10.1074/jbc.REV118.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sanger sequencing data of the variant in TARS2 was deposited in the DDBJ database (Project ID: 5f7de96aa3c8820eeb82a9df). Human reference genome used in this study (UCSC hg19) can be downloaded at the following url: https://hgdownload.soe.ucsc.edu/downloads.html. All other data and materials mentioned in this manuscript, except patient’s private information, can be promptly available to readers by a reasonable request to the corresponding author.