Abstract
Ivermectin (IVM) is a broad-spectrum antiparasitic agent, having inhibitory potential against wide range of viral infections. It has also been found to hamper SARS-CoV-2 replication in vitro, and its precise mechanism of action against SARS-CoV-2 is yet to be understood. IVM is known to interact with host importin (IMP)α directly and averts interaction with IMPβ1, leading to the prevention of nuclear localization signal (NLS) recognition. Therefore, the current study seeks to employ molecular docking, molecular mechanics generalized Born surface area (MM-GBSA) analysis and molecular dynamics simulation studies for decrypting the binding mode, key interacting residues as well as mechanistic insights on IVM interaction with 15 potential drug targets associated with COVID-19 as well as IMPα. Among all COVID-19 targets, the non-structural protein 9 (Nsp9) exhibited the strongest affinity to IVM showing −5.30 kcal/mol and −84.85 kcal/mol binding energies estimated by AutoDock Vina and MM-GBSA, respectively. However, moderate affinity was accounted for IMPα amounting −6.9 kcal/mol and −66.04 kcal/mol. Stability of the protein-ligand complexes of Nsp9-IVM and IMPα-IVM was ascertained by 100 ns trajectory of all-atom molecular dynamics simulation. Structural conformation of protein in complex with docked IVM exhibited stable root mean square deviation while root mean square fluctuations were also found to be consistent. In silico exploration of the potential targets and their interaction profile with IVM can assist experimental studies as well as designing of COVID-19 drugs.
Communicated by Ramaswamy H. Sarma
Keywords: Antiviral agents, SARS-CoV-2, ivermectin, docking, molecular dynamics
Introduction
The coronavirus disease (COVID-19) outbreak has been declared as a global public health emergency by the World Health Organization (WHO). The novel virus, COVID-19, was first reported during the late December 2019, in Wuhan, Hubei Province, People's Republic of China. International Committee on Taxonomy of Viruses (ICTV) named it as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Globally, the data of infected people is alarming and as of September 13, 2020, approximately 28 637 952 confirmed cases have been registered with surpassing 917,417 deaths spanning over 216 countries, areas or territories (WHO, 2020). The pandemic caused by SARS-CoV-2 fueled considerable research efforts towards repurposing or repositioning of existing drugs as possible therapeutic agents (Borkotoky & Banerjee, 2020; de Oliveira et al., 2020; Khan et al., 2020; Mahanta et al., 2020; Sk et al., 2020).
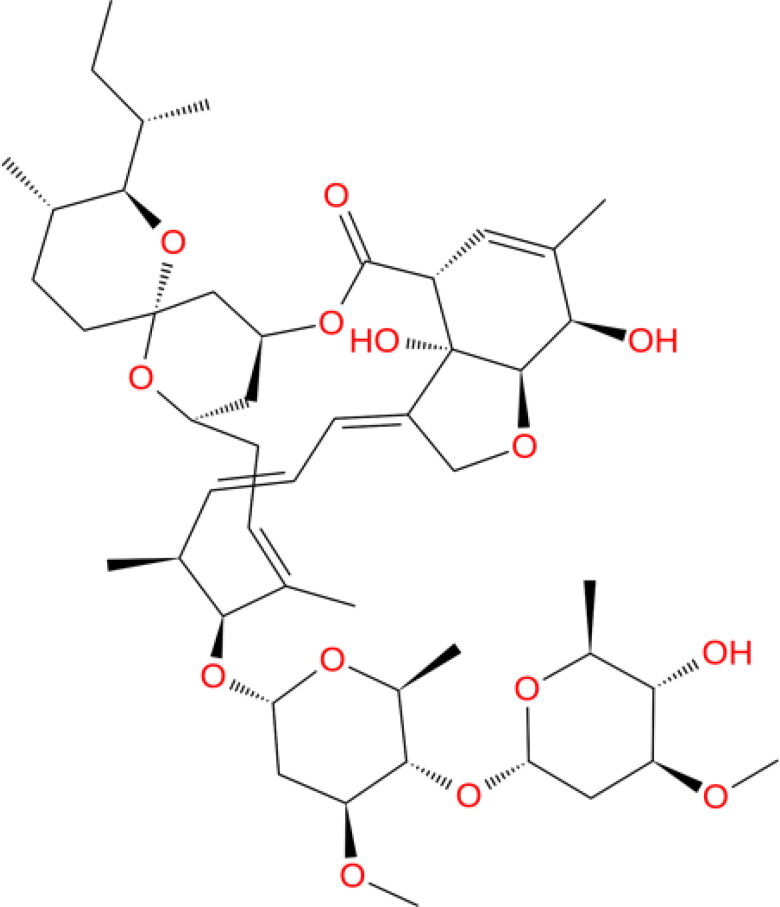
Ivermectin (IVM; Figure 1) is an US Food and Drug Administration (FDA)-approved broad-spectrum anti-parasitic medicine which has been used in humans for the treatment of onchocerciasis, and is also effective against other worm infestations such as strongyloidiasis, ascariasis, trichuriasis and enterobiasis (Luvira et al., 2014). Recently, Caly and co-workers emphasized that a single dose of IVM could control the clinical isolates of SARS-CoV-2 replication in vitro, within 24 to 48 h, making it a suitable candidate for drug repurposing against COVID-19 (Caly et al., 2020). In line with this, a double-blind, randomized controlled trial with two parallel groups that evaluated the efficacy of IVM in reducing nasal viral carriage in seven days after treatment of SARS-CoV-2 infected patients and it is currently planned at a single center in Navarra (ClinicalTrials.gov Identifier: NCT04390022). Earlier, in an effort to treat dengue viral infection, IVM was subjected to phase III clinical trial in Thailand during 2014–2017, wherein it was found safe as a single-daily dose administration of which resulted in a significant diminution of serum levels of viral NS1 protein without any alteration in viremia or clinical benefit (Yamasmith, 2018).
Figure 1.
Chemical structures of the ivermectin used in present study.
Antiviral potential of IVM is well established against several classes of viruses including avian influenza A viruses (Götz et al., 2016), Venezuelan equine encephalitis virus (Lundberg et al., 2013) and 1-4 serotypes of dengue viruses (Tay et al., 2013). In general, antiviral agents which target host-specific mechanisms possess broad-spectrum activity, affecting growth of disparate viruses. Hence, a wide-range of antiviral property associated with IVM is expected to be due to the reliance by several diverse ribonucleic acid (RNA) viruses on importin (IMP) α/β1 during infection (Jans et al., 2019). Also, IVM treatment has been found to decrease HIV-1 and dengue viral replication in cell cultures by inhibiting IMPα/β1 heterodimer, responsible for inhibition of nuclear accumulation of HIV-1 integrase and non-structural protein 5 (NS5) polymerase proteins (Fraser et al., 2014; Wagstaff et al., 2012). Therefore, nuclear targeting of NS5 seems to play a vital role in limiting the host antiviral response; specific inhibitors or mutations curbing NS5 nuclear import significantly restrict viral proliferation. A recent report suggested that the broad-spectrum antiviral activity of IVM could be linked to its ability to target the host IMPα/β1 nuclear transport proteins, intended for nuclear entry of integrase and NS5 cargoes. It was also asserted that IVM could bind to the IMPα armadillo (ARM) repeat domain resulting in dissociation of the preformed IMPα/β1 heterodimer, as well as prevention of its formation (Yang, Atkinson, et al., 2020). The mechanism of action of IVM in reducing the SARS-CoV-2 load in vitro is yet to be elucidated, but an interaction with IMPα/β1 has been proposed by the authors owing to very close resemblance of SARS-CoV-2 with SARS-CoV (Caly et al., 2020).
Studies involving mechanism of action exploration and target-identification of potential drugs are known to have vital impact in the drug design and discovery processes (Schenone et al., 2013). Several putative drug targets of SARS-CoV-2 include both non-structural (Nsp) and structural proteins such as main protease (Mpro), also called as chymotrypsin-like protease (3CLpro), papain-like protease, RNA-dependent RNA polymerase (RdRp), helicase (Nsp13), Nsp14 (N-terminal exoribonuclease and C-terminal guanine-N7 methyl transferase), receptor binding domain of spike protein, spike monomer, spike trimer, post fusion state of spike protein S2, nucleocapsid (N) protein, and ssRNA-binding protein or Nsp9 (Gordon et al., 2020; Kong et al., 2020).
Computer-aided drug design techniques are projected to accelerate the research and development processes in an economical manner (Azam et al., 2018, 2019). In particular, in recent times, molecular docking coupled with molecular dynamics simulation studies have played vital roles in interpreting the mechanism of binding interactions of potential molecules with the target proteins for lead optimization as well as design and the discovery of novel molecules (Eid et al., 2019; Shushni et al., 2013). Therefore, in this study, molecular docking, molecular mechanics generalized Born surface area (MM-GBSA) and molecular dynamics protocols have been exploited to investigate the binding interactions between IVM and 15 potential drug targets associated with COVID-19 as well as IMPα co-crystallized with NS5 fragment. The study is intended to explore potential targets and their interaction profile with IVM which could assist experimental studies as well as the designing of COVID-19 drugs.
Materials and methods
Protein and ligand preparation
Fifteen SARS-CoV-2 targets comprising both the structural and non-structural proteins used in this study include: main protease, papain-like protease, RdRp (RTP site), RdRp (RNA site), helicase (Nsp13; ADP site), helicase (Nsp13; NCB site), Nsp14 (ExoN), Nsp14 (N7-MTase), spike receptor binding domain, spike monomer (close), spike trimer (open), S2 post fusion state protein, N protein (C domain), N protein (N domain), and Nsp9 (Gordon et al., 2020; Kong et al., 2020). The 3D coordinates of the crystal structures were retrieved from the RCSB PDB database as listed in Table 1. In addition, 3D X-ray crystal structure of importin alpha (IMPα) in complex with non-structural protein 5 (NS5; PDB ID: 5FC8) having resolution of 2.1 Å was retrieved from Protein Data Bank (Tay et al., 2016). Chain E consisting of 10 armadillo (ARM) motifs of IMPα was used as protein target. The protein structures were processed in PyMOL 1.7.4 and Biovia Discovery Studio Visualizer 2020 to remove the solvents and the bound molecules. The macromolecules were further processed in MGLTools 1.5.6 for their conversion into pdbqt format after adding Gasteiger charges. Two-dimensional structure of the IVM was obtained from PubChem database in sdf format and converted to its three-dimensional coordinate by using Open Babel (O’Boyle et al., 2011). All non-polar hydrogens were merged, rotatable bonds and torsion tree were defined in MGLTools 1.5.6. The prepared ligand file was then converted into pdbqt format with Gasteiger charges added by using ligand_prepare.py module.
Table 1.
Results obtained after docking of IVM with 15 potential COVID-19 drug targets and host importin α.
| S.N. | Target | PDB ID | ΔGBind |
||
|---|---|---|---|---|---|
| AutoDock Vina 1.1 | AutoDock 4.2 | MM/GBSA | |||
| 1 | Main Protease | 6LU7 | −6.80 | −8.35 | −79.35 |
| 2 | Papain-like protease | 6WUU | −6.70 | −8.17 | −60.61 |
| 3 | RdRp (RTP site) | 7BV2 | −8.10 | −10.17 | −42.57 |
| 4 | RdRp (RNA site) | 7BV2 | −8.00 | −6.75 | −70.06 |
| 5 | Helicase (Nsp13; ADP site) | 6JYT | −8.10 | −5.06 | −58.36 |
| 6 | Helicase (Nsp13; NCB site) | 6JYT | −10.20 | −8.09 | −71.63 |
| 7 | Nsp14 (ExoN) | 5C8S | −7.90 | −7.85 | −63.96 |
| 8 | Nsp14 (N7-MTase) | 5C8S | −9.80 | −8.06 | −78.91 |
| 9 | Spike RBD | 6M0J | −6.70 | −7.32 | −69.13 |
| 10 | Spike monomer (close) | 6VXX | −5.50 | −7.44 | −55.86 |
| 11 | Spike trimer (open) | 6VYB | −7.00 | −8.11 | −72.40 |
| 12 | S2 (post fusion state) | 6LXT | −5.30 | −7.12 | −59.55 |
| 13 | N protein (C domain) | 6YUN | −6.00 | −8.71 | −75.33 |
| 14 | N protein (N domain) | 6YI3 | −7.30 | −7.47 | −69.03 |
| 15 | Nsp9 | 6WXD | −5.30 | −7.52 | −84.85 |
| 16 | Importin α | 5FC8 | −6.90 | −16.02 | −66.04 |
Molecular docking simulation
Molecular docking study was performed by using AutoDock Vina 1.1 (Trott & Olson, 2010) and AutoDock 4.2 (Morris et al., 1998). For AutoDock Vina 1.1, a grid box with a spacing of 1 Å and a size of 30 × 30 × 30 were built around the center of the binding site of all 15 targets of COVID-19 (Kong et al., 2020). Grid box of 60 × 78 × 64 points was used for AutoDock with 0.375 Å grid spacing. For IMPα docking, the grid box of similar dimensions was defined at the center of nuclear localization signal (NLS)-binding domain, known as the major binding pocket of IMPα and constituted armadillo repeats 2–4 (Kobe, 1999; Tay et al., 2016). Other parameters of docking were set to default while exhaustiveness value was adjusted to 12 (Trott & Olson, 2010). At the end of docking, 10 best poses obtained from each target were analyzed individually for binding energy, intermolecular interactions and number in clusters using Biovia Discovery Studio Visualizer 2020, and PyMol 1.7.4 programs (Ahmed et al., 2012; Azam et al., 2012; Fahmy et al., 2020). Ten best poses of IVM obtained from AutoDock Vina 1.1 in complex with each target were further subjected to Molecular mechanics/generalized Born surface area (MM-GBSA) computations.
Prime MM-GBSA calculations
MM-GBSA technique was exploited as a post-docking validation protocol. The binding energy computed by Prime MM-GBSA (Schrödinger, LLC) demonstrates an adequate estimation of binding affinity. The MM-GBSA protocol implemented in Prime combines OPLS molecular mechanics energies, a VSGB solvation model for polar solvation (GSGB), and a nonpolar solvation expression (GNP) involving nonpolar solvent-accessible surface area (SASA) and van der Waals interactions (Vijayakumar et al., 2014). For each docked complex, Prime MM-GBSA estimated the binding free energy (ΔGbind) of each ligand according to the equation (Lyne et al., 2006)
where, ΔEMM is the difference in energy between the complex structure and the sum of the energies of the protein with and without ligand, ΔGsolv is the difference in the GBSA solvation energy of the ligand-protein complex and the sum of the solvation energies for the drug-bound and unbound protein, and ΔGSA is the difference in the energy of surface area for the ligand-protein complex and the sum of the surface area energies for the ligand and un-complexed protein.
Molecular dynamics simulation
The best ranked conformation of IVM obtained from docking and MM-GBSA computations in complex with Nsp9 and IMPα was further examined for assessing their thermodynamic behavior and the stability of binding mode in the target active site using molecular dynamics (MD) simulation studies employing Desmond 6.1 (Bowers et al., 2006; Desmond Molecular Dynamics System, 2020). During system setup, the ligand-protein complexes were placed into an orthorhombic box filled with water molecules simulated by simple point charge (SPC) model and OPLS3 force field was adopted for the MD computations (Harder et al., 2016). The system was neutralized using appropriate numbers of Na+ and Cl− counter ions with fixed salt concentration of 0.15 M that represented the physiological concentration of monovalent ions. Isothermal-isobaric (NPT) ensemble was exercised with temperature and pressure adjusted to 300 K and 1.01325 bar, respectively. A simulation time of 100 ns was adjusted whereas trajectories were saved at every 100 ps. A cut-off radius of 9.0 Å was employed for short-range van der Waals and Coulomb interactions. The temperature and pressure of the system was maintained by Nose–Hoover thermostat (Hoover, 1985) and Martyna–Tobias–Klein (Martyna et al., 1994) methods, respectively. In order to integrate the equations of motion, RESPA integrator was used with an inner time step of 2.0 fs for bonded as well as non-bonded interactions within the short-range cut-off (Humphreys et al., 1994). The system was minimized and equilibrated with the default protocols of the Desmond.
Results and discussion
Validation of docking protocol
The co-crystallized NS5 fragment composed of Asp881-Glu893 of chain C was redocked within the major binding groove of IMPα in order to validate the docking algorithm used in this study because a successful experiment always relies upon validation of the implemented protocol (Azam et al., 2014). The root mean square deviation (RMSD) exhibited by the best docked conformation of NS5 and X-ray crystal structure was within 2 Å, which confirmed the reliability of the scoring algorithms employed in these programs (data not shown). However, the standard RMSD for successful docking according to the reported protocols is ≤2.0 Å (Azam et al., 2015; Hussain et al., 2016). Molecular docking protocols adopted in this study, therefore, could be exploited to predict the molecular interaction mode of IVM with potential COVID-19 drug targets as well as IMPα.
Docking study of IVM with potential COVID-19 drug targets
Computational determination of the binding modes of ligands with their targets by molecular docking is routinely employed in various drug design and discovery programs because of its swiftness and robustness (Ahmed et al., 2016). SARS-CoV-2 is known to possess various conserved non-structural and structural proteins having potential to be exploited in the drug design and discovery field (Gordon et al., 2020; Kong et al., 2020). Estimated binding energies obtained after docking of IVM with 15 potential COVID-19 drug targets and IMPα are presented in Table 1. According to the AutoDock 4.2 results, IMPα was observed as best putative target for IVM exhibiting −16.02 kcal/mol binding energy whereas helicase (Nsp13; ADP site) was rendered as least favourable with −5.06 kcal/mol. However, AutoDock Vina 1.1 predicted helicase (Nsp13; NCB site) as most promising while Nsp9 has been found to be the worst performer having binding energy of −10.2 kcal/mol and −5.3 kcal/mol, respectively. Although numerous docking protocols are routinely exercised to underscore the binding mode and the affinity of a ligand relative to a protein, lack of accurate scoring function cannot be underestimated in these algorithms. Therefore, it is imperative to employ post-docking analyses to avoid false negatives and false positives in order to incorporate the in-silico findings more accurately in rational design of drug candidates (Genheden & Ryde, 2015; Sgobba et al., 2012). Protein-ligand docked complexes computed by AutoDock Vina 1.1 were selected for further optimizations because of improved accuracy of the binding mode predictions when compared with AutoDock 4.2 (Gaillard, 2018; Trott & Olson, 2010). Detailed intermolecular interaction profile of docked IVM with all the targets is listed in Table S1 of supplementary information.
Docking study of IVM with IMPα protein
It has been recently reported that IVM exerts its antiviral effect by interfering with the nuclear transport after binding with IMPα (Yang, Atkinson, et al., 2020). In fact, targeting IMPα emerged as a powerful strategy in the design and development of potential antiviral agents (Jans et al., 2019; Yang et al., 2019). In this study, the IMPα monomer comprising of chain E in the X-ray crystal structure, co-crystallized with non-structural protein 5 (NS5) was used for the docking calculations. ΔG = −6.9 kcal/mol was predicted for IVM interaction with IMPα (Table 1). Results of comprehensive ligand–protein interactions are presented in Table S1 of supplementary information.
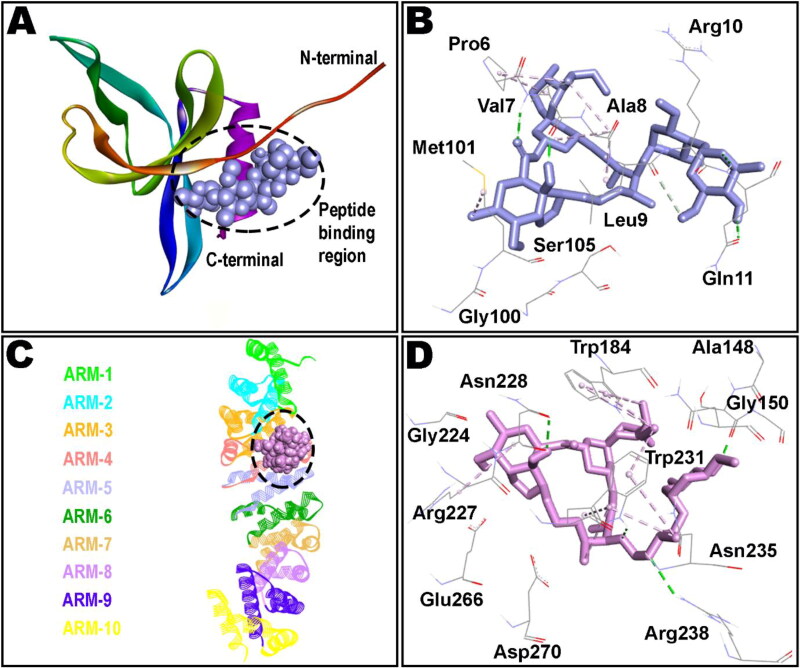
IMPα is a protein involved in the recognition of the import substrates through binding of their NLSs and it is comprised of two functional domains; an importin β binding (IBB) domain, and an NLS-binding domain which is made up of 10 armadillo (ARM) repeats (Görlich et al., 1996; Kobe, 1999). The NLS-binding domain constitutes major binding site, spanning ARM repeats 2–4, while the minor site covers ARM repeats 6–8 (Pumroy & Cingolani, 2015). Interestingly, as shown in Figure 2, docked IVM had ample opportunity within ARMs 2-4 of IMPα to interact through both hydrophobic and hydrophilic interactions. However, minor connections were also noted with ARM 5 of the IMPα. IVM accomodated well within major groove of the binding cavity in a similar manner to the native co-crystallized NS5. Therefore, it seems plausible to predict that after IVM occupation, NS5 interaction with IMPα might be interrupted, jeopardizing vital processes required for viral replication. It is interesting to note that IVM binding to the ARM repeat domains of IMPα not only resulted in the inhibition of IMPα recognition of NLSs through the ARM domain but also curbs tying to IMPβ1 through distinct N-terminal IBB domain of IMPα (Yang, Atkinson, et al., 2020).
Figure 2.
Docked ivermectin in complex of non-structural protein 9, Nsp9 (A and B), and importin α (C and D). ARM repeats 1-10 of importin α are shown in different colors (C). Broken lines define non-bond interactions in the binding pockets; hydrogen bonds in green whereas hydrophobic interactions in purple color.
Detailed analysis of the binding pocket residues highlights the importance of Trp184, Arg227 and Trp231 in hydrophobic connections whereas contribution of Ser149, Asn228, Trp231 and Arg238 in hydrogen bonding seems to be critical in holding the docked IVM within major groove. Residues Trp184, Asn228 and Trp231 are already proven to be vital for affording several intermolecular interactions, buttressing NS5 in the ARM repeats of IMPα (Tay et al., 2016). Molecular interaction of IVM with IMPα is reported to restrict the inherent flexibility of the ARM repeat domain by influencing thermostability as well as α-helicity of this protein, averting binding to IMPβ1/NS5 etc. (Vihinen, 1987). Binding pattern of IVM revealed by molecular docking in this study is intriguing and clearly translates its antiviral properties reported elsewhere (Kosyna et al., 2015; Tay et al., 2013; Wagstaff et al., 2012).
Prime MM/GBSA calculations
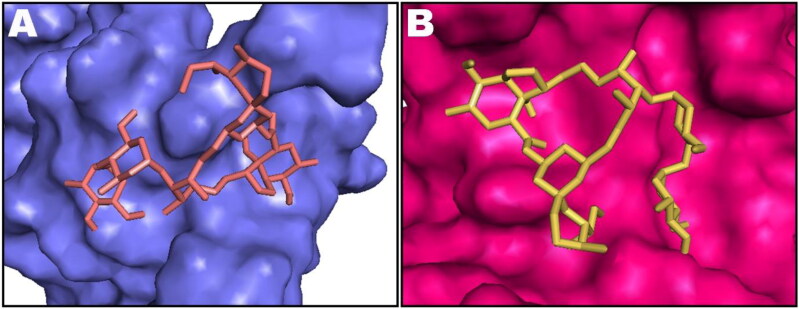
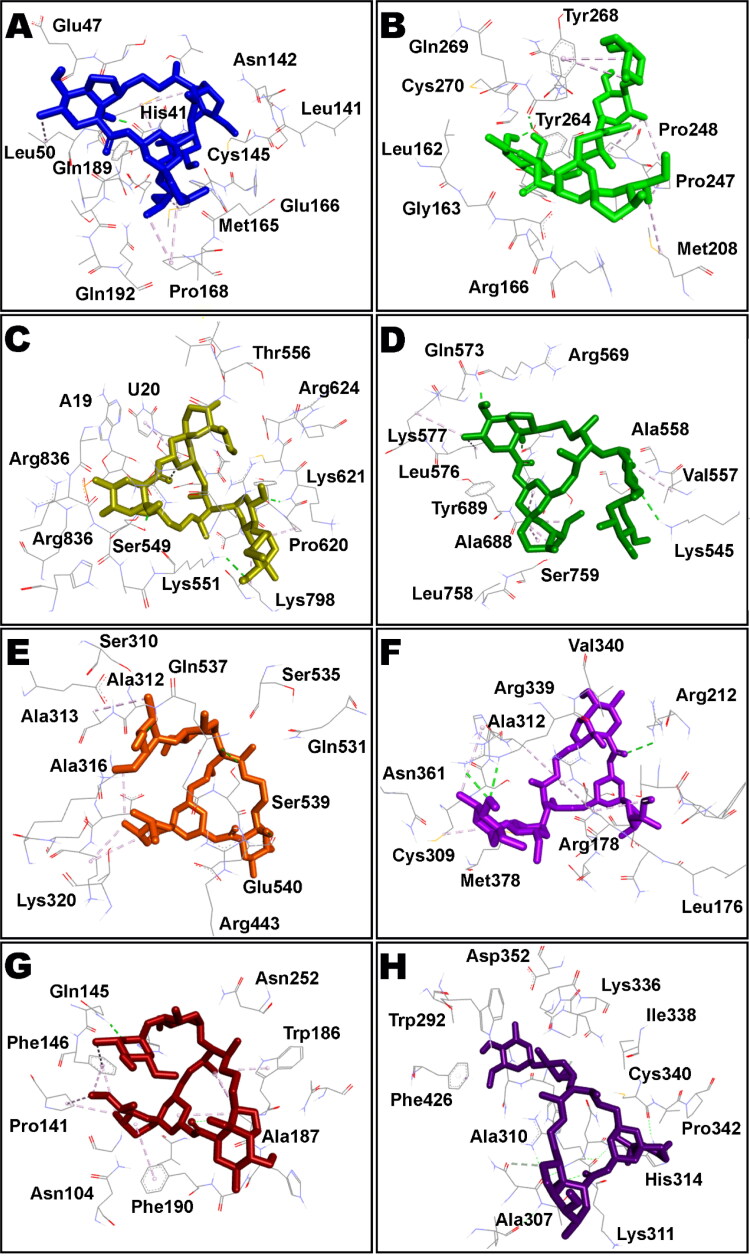
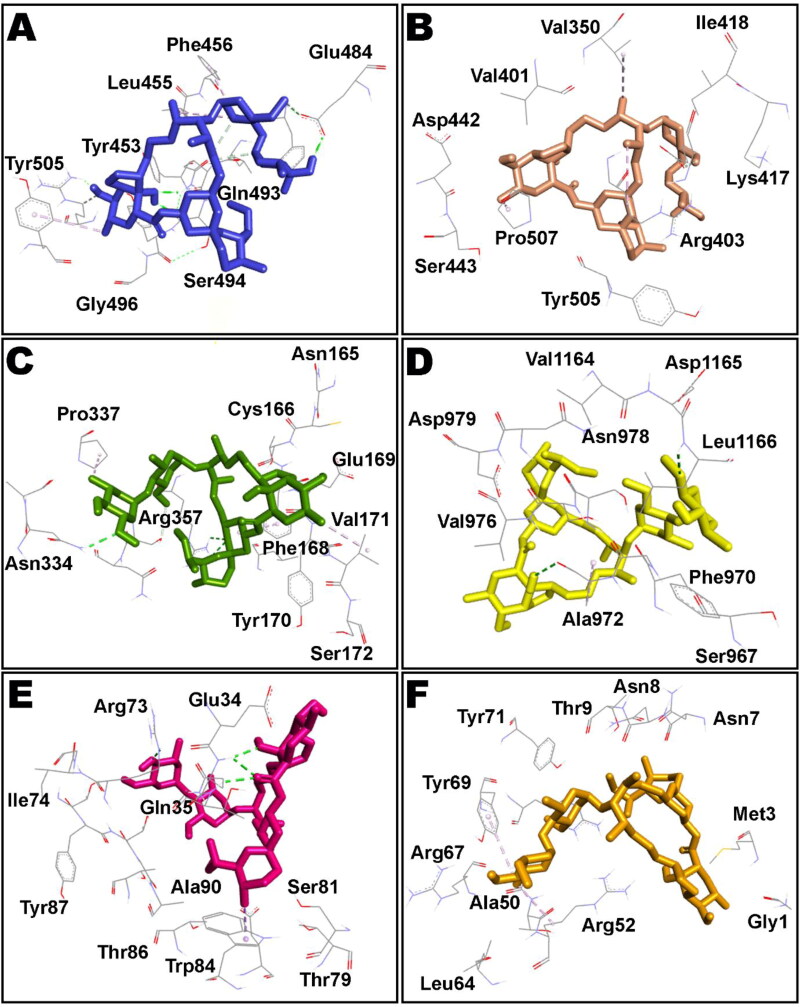
The top ten poses of protein–IVM docked complexes of each target were further analysed by Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) analysis for prediction of more accurate free binding energies. The best poses from each target were selected according to their MM/GBSA binding energy and results are tabulated in Table 2. However, detailed MM/GBSA computation of all poses have been presented as Table S2 in supplementary information. According to the obtained ΔGbind values, Nsp9 target ranked top among all studied proteins showing ΔGbind = −84.85 kcal/mol whereas RdRp (RTP site) has been observed to exhibit minimum affinity with IVM having binding energy of −42.57 kcal/mol. Intermolecular interactions observed after MM/GBSA computation of Nsp9 and IMPα targets are presented in Figure 2 and Figure 3 displays surface view of the binding pockets of these targets. IVM in complex with main protease, papain-like protease, RdRp (RTP site), RdRp (RNA site), helicase (Nsp13; ADP site), helicase (Nsp13; NCB site), Nsp14 (ExoN) and Nsp14 (N7-methyltransferase) are presented in Figure 4. Non-bond contacts of IVM with spike receptor binding domain (RBD), spike monomer, spike trimer, S2 protein (post fusion state), N protein (C domain) and N protein (N domain) are demonstrated in Figure 5. In addition to ΔGBind, MM/GBSA also computes ΔGCoul, ΔGHBond, ΔGLipo, SolvGB, ΔGvdw, and Lig SE representing energies of Coulomb, hydrogen-bonding correction, lipophilic, generalized Born electrostatic solvation energy, Van der Waals, and ligand strain, respectively (Tables 1 and 2).
Table 2.
Results of Prime MM/GBSA calculations.
| S.N. | Target | ΔGCoul | ΔGHBond | ΔGLipo | SolvGB | ΔGvdw | Lig SE |
|---|---|---|---|---|---|---|---|
| 1 | Main Protease | −10.16 | −0.44 | −53.80 | 28.52 | −59.49 | 34.37 |
| 2 | Papain-like protease | −7.97 | −0.44 | −52.07 | 25.94 | −42.95 | 28.76 |
| 3 | RdRp (RTP site) | −28.13 | −1.09 | −23.93 | 79.00 | −69.29 | 13.18 |
| 4 | RdRp (RNA site) | −22.56 | −0.89 | −37.87 | 30.74 | −47.33 | 12.87 |
| 5 | Helicase (Nsp13; ADP site) | −14.00 | −1.08 | −44.85 | 40.20 | −47.14 | 20.30 |
| 6 | Helicase (Nsp13; NCB site) | −10.34 | −0.27 | −83.87 | 49.51 | −52.32 | 34.01 |
| 7 | Nsp14 (ExoN) | −5.31 | −0.14 | −38.29 | 24.52 | −44.44 | 8.19 |
| 8 | Nsp14 (N7-MTase) | −10.49 | −0.27 | −54.97 | 36.66 | −61.37 | 22.25 |
| 9 | Spike RBD | −11.49 | −1.71 | −39.45 | 24.60 | −45.26 | 20.97 |
| 10 | Spike monomer (close) | −4.40 | −0.05 | −39.85 | 17.87 | −33.46 | 7.64 |
| 11 | Spike trimer (open) | −0.33 | −0.81 | −38.38 | 14.86 | −49.10 | 8.60 |
| 12 | S2 (post fusion state) | −8.91 | −0.22 | −30.96 | 27.39 | −45.20 | 10.16 |
| 13 | N protein (C domain) | −9.61 | −0.78 | −50.50 | 27.96 | −50.23 | 10.83 |
| 14 | N protein (N domain) | −3.60 | −0.11 | −54.29 | 31.69 | −58.15 | 5.48 |
| 15 | Nsp9 | −12.53 | −0.79 | −50.48 | 14.06 | −42.88 | 20.54 |
| 16 | Importin α | −24.26 | −1.45 | −35.74 | 37.83 | −45.41 | 6.34 |
Figure 3.
Most stable conformation of IVM shown as stick rendering in the binding pockets of non-structural protein 9, Nsp9 (A), and importin α (B). Binding pockets are shown as surface rendering and docked ivermectin as stick style.
Figure 4.
Intermolecular interactions of IVM (shown as stick style) with different COVID-19 proteins after MM/GBSA analysis. A: main protease; B: papain-like protease; C: RdRp (RTP site); D: RdRp (RNA site); E: helicase (Nsp13; ADP site); F: helicase (Nsp13; NCB site); G: Nsp14 (ExoN); H: Nsp14 (N7-methyltransferase).
Figure 5.
IVM (shown as stick rendering) in complex with several COVID-19 proteins such as spike receptor binding domain (RBD; A), spike monomer (B), spike trimer (C), S2 protein-post fusion state (D), N protein-C domain (E) and N protein-N domain (F) after MM/GBSA computation.
Nsp9 is a member of oligosaccharide/oligonucleotide binding protein superfamily which plays an indispensable role in viral replication. Interestingly, nps9/10 prevents mitochondrial protein synthesis and thus oxidative phosphorylation as a result of its imitation with host ribosomal assembly and also due to methylation of mitochondrial rRNA. The symptoms of SARS-CoV-2 such as reduced blood pressure following coma, induction of platelet aggregation and increased blood coagulability has been observed which was found to be associated with the ATP lowering effect of Nsp9 (Romano et al., 2020). Recently, conivaptan, an arginine vasopressin antagonist has been reported to exhibit high binding energy with Nsp9 and has shown antiviral activity against Human coronavirus OC43 (Chandel et al., 2020; Yang, Peng, et al., 2020). Hence, targeting Nsp9 seems to be a potential strategy for therapeutic armamentarium of the COVID-19 infection (Littler et al., 2020).
Molecular dynamics simulation of ivermectin in complex with Nsp9 and IMPα proteins
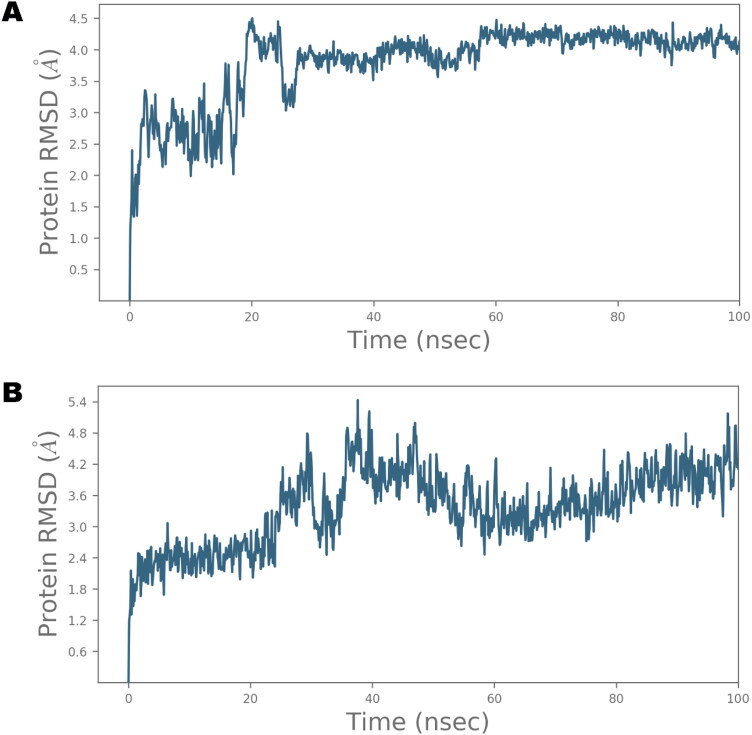
The application of MD simulation in computer-aided drug design field has gained substantial importance for the estimation of dynamic and thermodynamics parameters of living systems under specific situations of physiological milieus (Azam et al., 2018; Hospital et al., 2015). Owing to the maximum affinity of IVM with Nsp9 in MM/GBSA computation, it was selected for further analysis by MD simulation. In addition, IMPα was also chosen for further study because of the established affinity of IVM with this target. The best docked pose of IVM in complex with Nsp9 and IMPα was subjected to MD simulation study in order to investigate the stability of the ligand-protein complex as well as main intermolecular interactions during the period of simulation. Desmond software was employed for the MD simulation of 100 ns for each protein-ligand complex in explicit solvent system. The resulting trajectories of the simulated complex was inspected for different standard simulation parameters such as backbone root mean square deviations (RMSDs) for all Cα carbon atoms of protein, the root mean square fluctuations (RMSFs) of individual amino acid residues, intermolecular interactions involved, and radius of gyration (rGyr). The RMSD plot of simulated complex is presented in Figure 6.
Figure 6.
The Root Mean Square Deviations (RMSD) of Cα relative to the starting frame during 100 ns MD simulation of IVM in complex with Nsp9 (A) and IMPα (B).
The analysis of Nsp9 RMSD indicated that the simulated system equilibrated very well and the fluctuations in the Cα atoms were consistently within 4.5 Å during the entire simulated trajectory whereas up to 4.8 Å oscillations were noted in case of IMPα protein. However, minor fluctuation is obvious during initial period in both proteins which acquired stability throughout the rest of the simulation period. A system showing steady fluctuations is usually considered stable and deemed to be properly equilibrated. However, higher undulations of Cα atoms are regarded as an indication of large conformational changes in protein structure over the course of simulation.
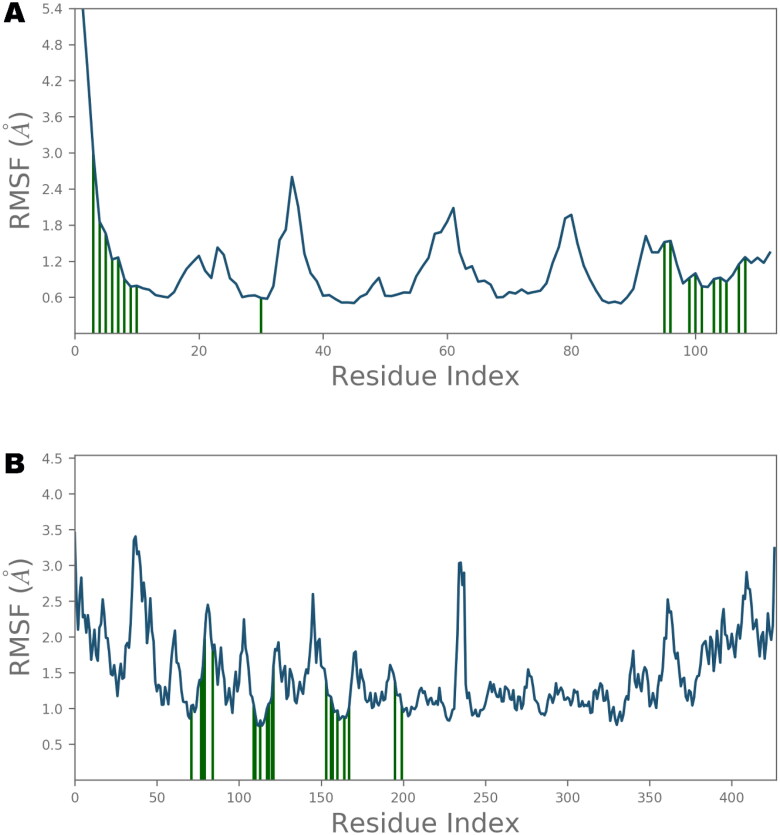
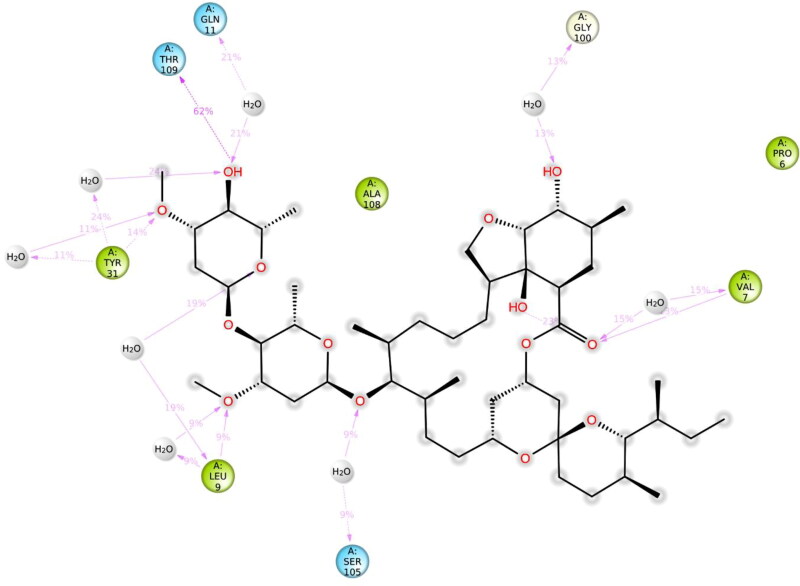
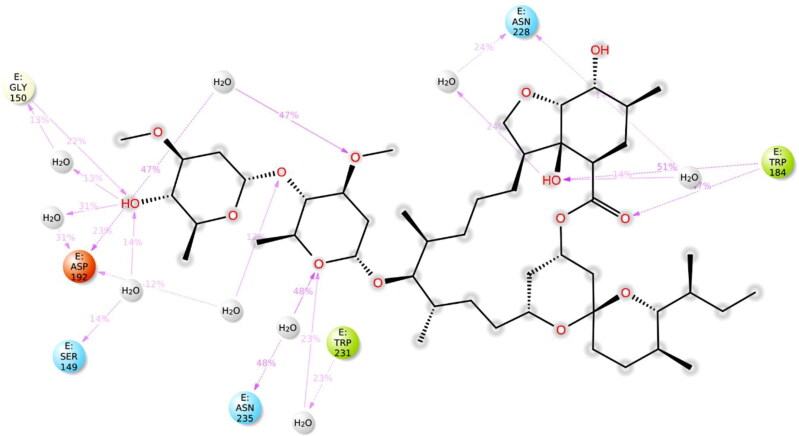
The local conformational alterations along both Nsp9 and IMPα proteins chain were probed by analyzing the root mean square fluctuation (RMSF) during simulation time. As indicated in Figure 7, stable RMSF plots were obtained during entire simulation period. The vertical green lines on the X-axis of the plot illustrated the participation of interacting residues between each protein chain and IVM. Residues of Nsp9 affording polar interactions such as Val7, Leu9, Gln11, Tyr31, Gly100, Ser105 and Thr109 had RMSF values of 1.22, 0.9, 0.79, 0.59, 0.92, 0.93 and 1.27 Å respectively. Chief contributors of non-polar connections with Nsp9 binding pocket such as Leu9, Tyr31 and Leu106 also had less than 1 Å RMSF figures. However, other residues providing van der Waal’s contacts like Pro6, Ala8, Leu97 and Ala108 exhibited RMSF values of approximately 1.5 Å or less. In case of IMPα, key residues for H-bond interactions such as Ala148, Ser149, Gly150, Trp184, Asp192, Asn228, and Arg238 had maximum RMSFs of 1.39, 1.57, 1.97, 0.83, 1.60, 1.05 and 1.01 Å, respectively, whereas main residues participating in hydrophobic interactions like Trp142, Trp184 and Trp231 also exhibited impressive RMSFs of 1.04, 0.83 and 0.97 Å, respectively. Furthermore, residues contributing in ionic bond contacts such as Gly150, Thr155 and Asp192 demonstrated RMSFs of 1.97, 1.81 and 1.60 Å, respectively. All of these values were estimated around the flexible loop regions of target protein. As disclosed in Figure 6, the highest RMSF values of 3.45 and 3.24 Å which correspond to the terminal residues Gln71 and Ser497, respectively, are far from the ligand binding site, and hence, least important.
Figure 7.
Root Mean Square Fluctuations (RMSF) of Nsp9 (A) and importin α (B) residues. The point of contact of ivermectin with protein residues is shown by vertical green lines on X-axis.
Structural compactness of the target protein was ascertained by evaluating the radius of gyration (rGyr) during MD simulation. Time-dependency plot of the radius of gyration for the simulated complex of IVM-Nsp9 and IVM-IMPα protein is presented as Figures S1 and S2 respectively, in supplementary information. As can be seen, no significant deviation was noticed in the values of rGyr and the compactness was maintained throughout the trajectory of 100 ns. In addition, the existence of a stable ligand-protein complexes was also implied by observing the changes in various surface areas such as molecular surface area (MolSA), polar surface area (PSA), and solvent accessible surface area (SASA) of studied complex as a function of simulation time (Figures S1 and S2 of supplementary information).
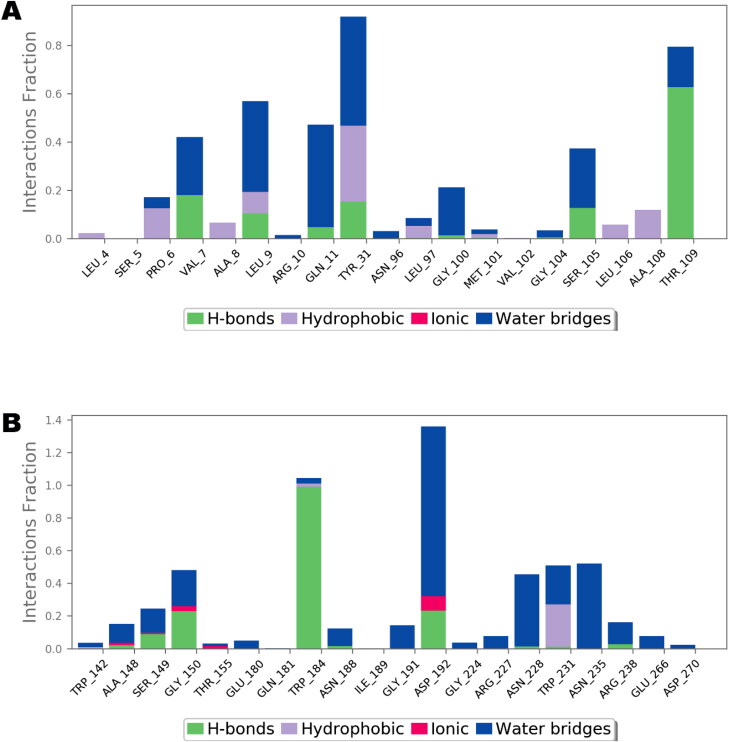
A comprehensive intermolecular interaction of IVM with Nsp9 and IMPα was also studied using simulation interactions diagrams during entire simulation time. The pattern of interaction of IVM clearly showed that the docking predicted main contacts were nearly preserved (Figure 8). Non-bond interactions involving hydrogen bonds, hydrophobic, ionic and water bridges which are considered important in stability of a protein-ligand complex, were involved in supporting IVM in the Nsp9 as well as in NLS binding site of the IMPα.
Figure 8.
Nsp9 (A) and importin α (B) interactions with ivermectin, monitored throughout the simulated trajectory of 100 ns. These interactions are clustered by type and summarized in bar diagram including H-bonds, hydrophobic, ionic and water bridges.
As shown in Figures 8–10, the intermolecular interactions spotted in the docking model were mostly reproduced after MD simulations in both Nsp9 and IMPα targets. Moreover, these proteins also revealed many interesting contacts during simulation of the docked ligand-protein complexes. In Nsp9 site, Leu9, Tyr31, Gly100, Ser105 and Thr109 were identified as additional hydrogen bond providers whereas many water bridges were also noticed with Val7, Leu9, Gln11, Tyr31, Ser105 and Thr109 (Figure 9). Nsp9 is reported to form a homodimer which in turn binds with single stranded viral RNA (Snijder et al., 2016). Moreover, the dimerization of this protein increases its binding affinity with nucleic acids and is considered as important step for viral replication and growth (Zeng et al., 2018). In the peptide binding region, Pro6, Val7, Leu9 and Ser105 are important for hydrogen bond and Van der Waals interactions whereas Tyr31, Met101, Leu106 offer additional hydrophobic contacts (Littler et al., 2020). Analysis of the hydrogen bond lengths between IVM and the key amino acid residues of Nsp9 and IMPα were estimated during 50 ns to 100 ns MD simulations and are presented in Table 3. LigPlot diagrams showing key residues of Nsp9 and IMPα are presented as Figures S3 and S4 respectively, in supplementary information. In Nsp9 site, Ser105 exhibited average bond length of 3.31 Å whereas 3.07 Å bond length was measured for Thr109. However, hydrogen bonds offered by Leu9 and Tyr31 were inconsistent during simulated trajectory (Figure S3, supplementary information).
Figure 9.
2D representation of the atomic interactions between ivermectin and Nsp9 residues during 100 ns molecular dynamics simulation.
Figure 10.
Two-dimensional representation of the atomic interactions between ivermectin and importin α protein residues during 100 ns molecular dynamics simulation.
Table 3.
Hydrogen bond length of key residues during molecular dynamics simulation of ivermectin in complex with Nsp9 and importin α.
| S.No. | Target | Amino acids | H-bond distance (Å) at different time intervals |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 50 ns | 60 ns | 70 ns | 80 ns | 90 ns | 100 ns | Average | |||
| 1. | Nsp9 | Ser105 | 2.89 | 3.07 | 3.22 | 2.93 | 3.74 | 3.98 | 3.31 |
| Thr109 | 2.86 | 2.69 | 2.80 | 3.38 | 3.44 | 3.27 | 3.07 | ||
| 2. | IMPα | Gly150 | 2.97 | 3.61 | 3.07 | 3.16 | 3.02 | 2.75 | 3.09 |
| Trp184 | 2.71 | 3.07 | 2.81 | 2.88 | 3.07 | 2.85 | 2.89 | ||
In case of IMPα, residues such as Ala148, Ser149, Gly150, Trp184, Gly191, Asp192, Asn228, Asn235, Arg238, and Glu266 were important for affording water bridges, and hence, supporting IVM in the binding cavity (Figure 10). For hydrogen bond interactions of IVM with IMPα, Ser149, Gly150, Asp192 and Arg238 were exploited. Gly150 and Trp184 demonstrated average hydrogen bond lengths of 3.09 and 2.89 Å, respectively whereas polar contacts provided by Ser149 and Asp192 were not reliable. A timeline representation of the interactions and contacts in terms of hydrogen bonds, hydrophobic, ionic, and water bridges between ivermectin and residues of Nsp9 and IMPα in each trajectory frame is presented as Figures S5 and S6, respectively, in supplementary material.
Conclusion
Ivermectin, a broad-spectrum antiparasitic agent, has been found to inhibit SARS-CoV-2 replication in vitro. In this study, 15 potential COVID-19 targets were used to underscore IVM affinity by computational techniques employing molecular docking, MM/GBSA computation and molecular dynamics simulation. IMPα protein was also included in this study owing to the reported interaction of IVM with host importin α, resulting in the prevention of nuclear localization signal recognition. Estimation of MM/GBSA based binding energy revealed that Nsp9 possesses highest affinity among COVID-19 targets. Strengths of IVM complex with Nsp9 and IMPα was ascertained by the evaluation of RMSD and RMSF plots obtained after molecular dynamics simulation of 100 ns. Both hydrophobic as well as hydrophilic interactions were identified in anchoring the IVM inside the binding site of Nsp9 as well as major binding groove of the IMPα. Intermolecular interaction profile of IVM disclosed in the current study is expected to assist experimental studies and designing of COVID-19 drugs.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Ahmed, M. A., Azam, F., Rghigh, A. M., Gbaj, A., & Zetrini, A. E. (2012). Structure-based design, synthesis, molecular docking, and biological activities of 2-(3-benzoylphenyl) propanoic acid derivatives as dual mechanism drugs. Journal of Pharmacy & Bioallied Sciences, 4(1), 43. 10.4103/0975-7406.92728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, M., Azam, F., Gbaj, A., Zetrini, A. E., Abodlal, A. S., Rghigh, A., Elmahdi, E., Hamza, A., Salama, M., & Bensaber, S. M. (2016). Ester prodrugs of ketoprofen: Synthesis, in vitro stability, in vivo biological evaluation and in silico comparative docking studies against COX-1 and COX-2. Current Drug Discovery Technologies, 13(1), 41–57. [DOI] [PubMed] [Google Scholar]
- Azam, F., Abodabos, H. S., Taban, I. M., Rfieda, A. R., Mahmood, D., Anwar, M. J., Khan, S., Sizochenko, N., Poli, G., Tuccinardi, T., & Ali, H. I. (2019). Rutin as promising drug for the treatment of Parkinson’s disease: An assessment of MAO-B inhibitory potential by docking, molecular dynamics and DFT studies. Molecular Simulation, 45(18), 1563–1571. 10.1080/08927022.2019.1662003 [DOI] [Google Scholar]
- Azam, F., Alabdullah, N. H., Ehmedat, H. M., Abulifa, A. R., Taban, I., & Upadhyayula, S. (2018). NSAIDs as potential treatment option for preventing amyloid β toxicity in Alzheimer’s disease: An investigation by docking, molecular dynamics, and DFT studies. Journal of Biomolecular Structure and Dynamics, 36(8), 2099–2117. 10.1080/07391102.2017.1338164 [DOI] [PubMed] [Google Scholar]
- Azam, F., Amer, A. M., Rabulifa, A., & Elzwawi, M. M. (2014). Ginger components as new leads for the design and development of novel multi-targeted anti-Alzheimer’s drugs: A computational investigation. Drug Design, Development and Therapy, 8, 2045–2059. 10.2147/DDDT.S67778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam, F., Mohamed, N., & Alhussen, F. (2015). Molecular interaction studies of green tea catechins as multitarget drug candidates for the treatment of Parkinson's disease: Computational and structural insights. Network (Bristol, England), 26(3–4), 97–115. 10.3109/0954898X.2016.1146416 [DOI] [PubMed] [Google Scholar]
- Azam, F., Vijaya Vara Prasad, M., Thangavel, N., Kumar Shrivastava, A., & Mohan, G. (2012). Structure-based design, synthesis and molecular modeling studies of thiazolyl urea derivatives as novel anti-Parkinsonian agents. Medicinal Chemistry (Shariqah (United Arab Emirates)), 8(6), 1057–1068. 10.2174/1573406411208061057 [DOI] [PubMed] [Google Scholar]
- Borkotoky, S., & Banerjee, M. (2020). A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1774419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters. SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. [Google Scholar]
- Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., & Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel, V., Sharma, P. P., Raj, S., Choudhari, R., Rathi, B., & Kumar, D. (2020). Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1811773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira, O. V., Rocha, G. B., Paluch, A. S., & Costa, L. T. (2020). Repurposing approved drugs as inhibitors of SARS-CoV-2S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1772885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond Molecular Dynamics System . (2020). Schrödinger LLC.
- Eid, E., Alanazi, A., Koosha, S., Alrasheedy, A., Azam, F., Taban, I., Khalilullah, H., Sadiq Al-Qubaisi, M., Alshawsh, M., Eid, E., Alanazi, A., Koosha, S., Alrasheedy, A., Azam, F., Taban, I., Khalilullah, H., Sadiq Al-Qubaisi, M., & Alshawsh, M. (2019). Zerumbone induces apoptosis in breast cancer cells by targeting αvβ3 integrin upon co-administration with TP5-iRGD peptide. Molecules, 24(14), 2554. 10.3390/molecules24142554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy, N. M., Al-Sayed, E., Moghannem, S., Azam, F., El-Shazly, M., & Singab, A. N. (2020). Breaking down the barriers to a natural antiviral agent: Antiviral activity and molecular docking of erythrina speciosa extract, fractions, and the major compound. Chemistry & Biodiversity, 17(2), e1900511. 10.1002/cbdv.201900511 [DOI] [PubMed] [Google Scholar]
- Fraser, J. E., Watanabe, S., Wang, C., Chan, W. K. K., Maher, B., Lopez-Denman, A., Hick, C., Wagstaff, K. M., Mackenzie, J. M., Sexton, P. M., Vasudevan, S. G., & Jans, D. A. (2014). A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. The Journal of Infectious Diseases, 210(11), 1780–1791. 10.1093/infdis/jiu319 [DOI] [PubMed] [Google Scholar]
- Gaillard, T. (2018). Evaluation of AutoDock and AutoDock Vina on the CASF-2013 benchmark. Journal of Chemical Information and Modeling, 58(8), 1697–1706. 10.1021/acs.jcim.8b00312 [DOI] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O'Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., … Krogan, N. J. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., Henklein, P., Laskey, R. A., & Hartmann, E. (1996). A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. The EMBO Journal, 15(8), 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Götz, V., Magar, L., Dornfeld, D., Giese, S., Pohlmann, A., Höper, D., Kong, B.-W., Jans, D. A., Beer, M., Haller, O., & Schwemmle, M. (2016). Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Scientific Reports, 6(1), 23138. 10.1038/srep23138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, E., Damm, W., Maple, J., Wu, C., Reboul, M., Xiang, J. Y., Wang, L., Lupyan, D., Dahlgren, M. K., Knight, J. L., Kaus, J. W., Cerutti, D. S., Krilov, G., Jorgensen, W. L., Abel, R., & Friesner, R. A. (2016). OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- Hoover, W. G. (1985). Canonical dynamics: Equilibrium phase-space distributions. Physical Review. A, General Physics, 31(3), 1695–1697. 10.1103/physreva.31.1695 [DOI] [PubMed] [Google Scholar]
- Hospital, A., Goñi, J. R., Orozco, M., & Gelpí, J. L. (2015). Molecular dynamics simulations: Advances and applications. Advances and Applications in Bioinformatics and Chemistry: AABC, 8, 37–47. 10.2147/AABC.S70333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, D. D., Friesner, R. A., & Berne, B. J. (1994). A multiple-time-step molecular dynamics algorithm for macromolecules. The Journal of Physical Chemistry, 98(27), 6885–6892. 10.1021/j100078a035 [DOI] [Google Scholar]
- Hussain, M. S., Azam, F., Ahamed, K. F. H. N., Ravichandiran, V., & Alkskas, I. (2016). Anti-endotoxin effects of terpenoids fraction from Hygrophila auriculata in lipopolysaccharide-induced septic shock in rats. Pharmaceutical Biology, 54(4), 628–636. 10.3109/13880209.2015.1070877 [DOI] [PubMed] [Google Scholar]
- Jans, D. A., Martin, A. J., & Wagstaff, K. M. (2019). Inhibitors of nuclear transport. Current Opinion in Cell Biology, 58, 50–60. 10.1016/j.ceb.2019.01.001 [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y.-J., Chinnasamy, S., & Wei, D.-Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1769733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B. (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nature Structural Biology, 6(4), 388–397. 10.1038/7625 [DOI] [PubMed] [Google Scholar]
- Kong, R., Yang, G., Xue, R., Liu, M., Wang, F., Hu, J., Guo, X., & Chang, S. (2020). COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics, 1–3. 10.1093/bioinformatics/btaa645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosyna, F. K., Nagel, M., Kluxen, L., Kraushaar, K., & Depping, R. (2015). The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways. Biological Chemistry, 396(12), 1357–1367. 10.1515/hsz-2015-0171 [DOI] [PubMed] [Google Scholar]
- Littler, D. R., Gully, B. S., Colson, R. N., & Rossjohn, J. (2020). Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience, 23(7), 101258. 10.1016/j.isci.2020.101258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, L., Pinkham, C., Baer, A., Amaya, M., Narayanan, A., Wagstaff, K. M., Jans, D. A., & Kehn-Hall, K. (2013). Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Research, 100(3), 662–672. 10.1016/j.antiviral.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Luvira, V., Watthanakulpanich, D., & Pittisuttithum, P. (2014). Management of Strongyloides stercoralis: A puzzling parasite. International Health, 6(4), 273–281. 10.1093/inthealth/ihu058 [DOI] [PubMed] [Google Scholar]
- Lyne, P. D., Lamb, M. L., & Saeh, J. C. (2006). Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. Journal of Medicinal Chemistry, 49(16), 4805–4808. 10.1021/jm060522a [DOI] [PubMed] [Google Scholar]
- Mahanta, S., Chowdhury, P., Gogoi, N., Goswami, N., Borah, D., Kumar, R., Chetia, D., Borah, P., Buragohain, A. K., & Gogoi, B. (2020). Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: An in silico based approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1768902 [DOI] [PubMed] [Google Scholar]
- Martyna, G. J., Tobias, D. J., & Klein, M. L. (1994). Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics, 101(5), 4177–4189. 10.1063/1.467468 [DOI] [Google Scholar]
- Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., & Olson, A. J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry, 19(14), 1639–1662. [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy, R. A., & Cingolani, G. (2015). Diversification of importin-α isoforms in cellular trafficking and disease states. The Biochemical Journal, 466(1), 13–28. 10.1042/BJ20141186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells, 9(5), 1267. 10.3390/cells9051267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenone, M., Dančík, V., Wagner, B. K., & Clemons, P. A. (2013). Target identification and mechanism of action in chemical biology and drug discovery. Nature Chemical Biology, 9(4), 232–240. 10.1038/nchembio.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgobba, M., Caporuscio, F., Anighoro, A., Portioli, C., & Rastelli, G. (2012). Application of a post-docking procedure based on MM-PBSA and MM-GBSA on single and multiple protein conformations. European Journal of Medicinal Chemistry, 58, 431–440. 10.1016/j.ejmech.2012.10.024 [DOI] [PubMed] [Google Scholar]
- Shushni, M. A. M., Azam, F., & Lindequist, U. (2013). Oxasetin from Lophiostoma sp. of the Baltic Sea: Identification, in silico binding mode prediction and antibacterial evaluation against fish pathogenic bacteria. Natural Product Communications, 8(9), 1934578X1300800. [PubMed] [Google Scholar]
- Sk, M. F., Roy, R., Jonniya, N. A., Poddar, S., & Kar, P. (2020). Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1768149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., Decroly, E., & Ziebuhr, J. (2016). Chapter Three - The nonstructural proteins directing Coronavirus RNA synthesis and processing. In Ziebuhr V. R. (Ed.), Coronaviruses (Vol. 96, pp. 59–126). Academic Press. 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M. Y. F., Fraser, J. E., Chan, W. K. K., Moreland, N. J., Rathore, A. P., Wang, C., Vasudevan, S. G., & Jans, D. A. (2013). Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Research, 99(3), 301–306. 10.1016/j.antiviral.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Tay, M. Y. F., Smith, K., Ng, I. H. W., Chan, K. W. K., Zhao, Y., Ooi, E. E., Lescar, J., Luo, D., Jans, D. A., Forwood, J. K., & Vasudevan, S. G. (2016). The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathogens, 12(9), e1005886. 10.1371/journal.ppat.1005886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen, M. (1987). Relationship of protein flexibility to thermostability. Protein Engineering, 1(6), 477–480. 10.1093/protein/1.6.477 [DOI] [PubMed] [Google Scholar]
- Vijayakumar, B., Parasuraman, S., Raveendran, R., & Velmurugan, D. (2014). Identification of natural inhibitors against angiotensin I converting enzyme for cardiac safety using induced fit docking and MM-GBSA studies. Pharmacognosy Magazine, 10(Suppl 3), S639–S644. 10.4103/0973-1296.139809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., & Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. The Biochemical Journal, 443(3), 851–856. 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2020). Coronavirus disease 2019 (COVID-19) Situation report. World Health Organization. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200914-weekly-epi-update-5.pdf?sfvrsn=cf929d04_2
- Yamasmith, E. (2018). Efficacy and safety of ivermectin against dengue infection: A phase III, randomized, double-blind, placebo-controlled trial. In He 34th Annual Meeting the Royal College of Physicians of Thailand. Internal Medicine and One Health. [Google Scholar]
- Yang, C.-W., Peng, T.-T., Hsu, H.-Y., Lee, Y.-Z., Wu, S.-H., Lin, W.-H., Ke, Y.-Y., Hsu, T.-A., Yeh, T.-K., Huang, W.-Z., Lin, J.-H., Sytwu, H.-K., Chen, C.-T., & Lee, S.-J. (2020). Repurposing old drugs as antiviral agents for coronaviruses. Biomedical Journal, 43(4), 368–374. 10.1016/j.bj.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. N. Y., Atkinson, S. C., Fraser, J. E., Wang, C., Maher, B., Roman, N., Forwood, J. K., Wagstaff, K. M., Borg, N. A., & Jans, D. A. (2019). Novel flavivirus antiviral that targets the host nuclear transport importin α/β1 heterodimer. Cells, 8(3), 281. 10.3390/cells8030281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. N. Y., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A., & Jans, D. A. (2020). The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Research, 177, 104760. 10.1016/j.antiviral.2020.104760 [DOI] [PubMed] [Google Scholar]
- Zeng, Z., Deng, F., Shi, K., Ye, G., Wang, G., Fang, L., Xiao, S., Fu, Z., & Peng, G. (2018). Dimerization of Coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. Journal of Virology, 92(17), e00692-18. 10.1128/JVI.00692-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.