Abstract
Transposable elements (TEs) constitute a large fraction of plant genomes and are mostly present in a transcriptionally silent state through repressive epigenetic modifications, such as DNA methylation. TE silencing is believed to influence the regulation of adjacent genes, possibly as DNA methylation spreads away from the TE. Whether this is a general principle or a context-dependent phenomenon is still under debate, pressing for studying the relationship between TEs, DNA methylation, and nearby gene expression in additional plant species. Here, we used the grass Brachypodium distachyon as a model and produced DNA methylation and transcriptome profiles for 11 natural accessions. In contrast to what is observed in Arabidopsis thaliana, we found that TEs have a limited impact on methylation spreading and that only few TE families are associated with a low expression of their adjacent genes. Interestingly, we found that a subset of TE insertion polymorphisms is associated with differential gene expression across accessions. Thus, although not having a global impact on gene expression, distinct TE insertions may contribute to specific gene expression patterns in B. distachyon.
Keywords: transposable elements, DNA methylation, gene expression, Brachypodium distachyon
Significance
Transposable elements (TEs) are a major component of plant genomes and a source of genetic and epigenetic innovations underlying adaptation to changing environmental conditions. Yet, molecular evidence linking TE silencing and nearby gene expression is lacking for many plant species. We show that in the model grass Brachypodium DNA methylation spreads over very short distances around TEs, with an influence on gene expression for a small subset of TE families.
Introduction
Since their discovery by McClintock (1947) in the late 1940s, transposable elements (TEs) have been shown to populate a large fraction of eukaryotic genomes and to be an important source of genetic and phenotypic diversity. This is explained by the observation that TEs have the ability to modulate nearby gene expression and can remodel genomes when activated by stress (Grandbastien 1998, 2005; Thieme et al. 2017), thereby providing raw diversity necessary for adaptation to changing environments (Bonchev and Parisod 2013; Rey et al. 2016). Remarkable examples of single-trait modulation by a TE insertion providing a selective advantage have been reported in both plants and animals (Guio et al. 2014; Van’t Hof et al. 2016; Horváth et al. 2017).
Although TEs can provide selective advantages, their mutagenic properties may at the same time represent a threat for the integrity of the host genome (McClintock 1950; Shang et al. 2017; Thind et al. 2018). In response, genomes have evolved several layers of defense to contain TE activity. In plants, transcriptional repression of TEs is mediated by the RNA-directed DNA methylation pathway (for review, Sigman and Slotkin 2016; Zhang et al. 2018). Initiated by the plant-specific RNA polymerase IV (Pol IV), TE transcription leads to the formation of 24-nt siRNA targeting DNA methylation at the TE source locus. Acting as a double-edged sword, DNA (and histone) methylation may spread to neighboring regions and possibly contribute the alteration of adjacent gene expression (Hollister et al. 2011; Wang et al. 2013).
Recent studies reported on the impact of TEs on DNA methylation and expression of nearby genes in different plant species (Eichten et al. 2012, 2016; Makarevitch et al. 2015; Choi and Purugganan 2018; Wang et al. 2018). Yet, most of our knowledge on the regulatory mechanisms largely rely on the eudicot model Arabidopsis thaliana (for review, Sigman and Slotkin 2016; Zhang et al. 2018). In A. thaliana, the presence of TEs tends to be associated with a reduced expression of adjacent genes (Wang et al. 2013), but whether this also occurs in other species remains to be demonstrated (Choi and Purugganan 2018). In this context, we contributed an analysis of DNA methylation and gene expression in the model grass Brachypodium distachyon.
Established as an important model for functional genomics, the small genome of B. distachyon (272 Mb; 30% of TEs) is fully sequenced and assembled into five chromosomes (International Brachypodium Consortium 2010). Comparative genomics and cytogenetic analyses (Hasterok et al. 2019) showed that it underwent multiple and independent nested chromosome fusions which resulted in a higher gene density at the former distal regions of the inserted chromosomes. Furthermore, TEs are much more pervasive than in A. thaliana, and although their dynamics is well characterized in B. distachyon (Stritt et al. 2018, 2020), little is known about their functional impact. We, therefore, asked to what extent TEs influence DNA methylation and gene expression in B. distachyon, and how these patterns differ from what is observed in A. thaliana. In A. thaliana, the impact of TEs on adjacent gene expression is extremely consistent across genetically distinct ecotypes originating from the United States (Col-0), Portugal (C24), and Ireland (Bur0; Wang et al. 2013). We thus chose Col-0 as a representative of the species. For B. distachyon, less information is available at such a broad geographical scale. We, therefore, included in our analysis the reference Bd21, which originates from Iraq, as well as ten additional accessions from Spain, France, Iraq, and Turkey to further investigate potential within-species variation.
Results and Discussion
TEs Largely Shape Genome-Wide DNA Methylation Patterns in Brachypodium
We produced bisulfite- and RNA-sequencing data for 11 B. distachyon accessions. Single nucleotide polymorphisms (Gordon et al. 2017; Stritt et al. 2018) and methylation profiling support their split into three distinct genetic clusters (supplementary fig. S1, Supplementary Material online): Western (ABR2, Luc1, Bd30-1, and Sig2; Spain and France), Eastern (the reference accession Bd21, Bd21-3, Bd3-1, BdTR12C, and Koz3; Iraq and Turkey), and accessions from a third cluster (BdTR7a and Bd1-1; Turkey) showing a delay in flowering time (Gordon et al. 2017; Stritt et al. 2018). In agreement with the genetic distances calculated with single nucleotide polymorphisms (Gordon et al. 2017; Stritt et al. 2018), the methylation-based PCA confirms that accessions from the Eastern cluster are more closely related to accessions from the Western cluster than to the two other Turkish accessions BdTR7a and Bd1-1 (supplementary fig. S1B, Supplementary Material online). We also used publically available bisulfite- and RNA-sequencing data for the reference accession Col-0 of A. thaliana (supplementary table S1, Supplementary Material online).
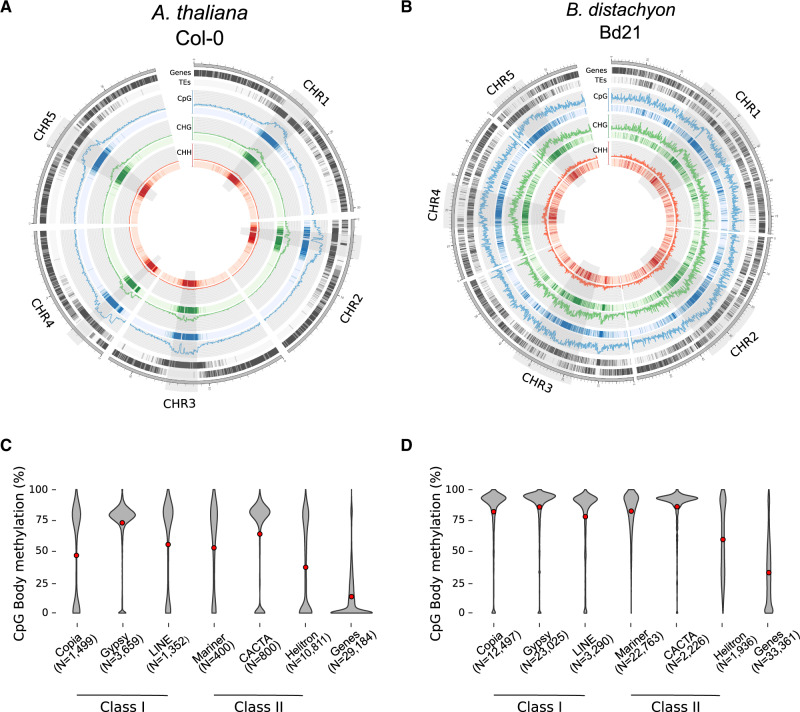
We first estimated methylation levels (expressed as the percentage of methylated cytosines), gene, and TE density in 100-kb windows for the reference genomes of A. thaliana (Col-0) and B. distachyon (Bd21). The circular visualization of these three genomic features reveals two distinct architectures: Genes and TEs are clearly compartmentalized in A. thaliana (fig. 1A) whereas largely interspersed in B. distachyon (fig. 1B). Yet, we found that DNA methylation is positively correlated with TE density and negatively correlated with gene density in both species (supplementary fig. S2, Supplementary Material online). We then estimated the methylation level of genes and TEs, aggregating the latter at the superfamily level. TE body methylation is significantly higher than gene body methylation in the two species, although variability is observed among TE superfamilies (fig. 1C and D, supplementary tables S2–S4, Supplementary Material online, for P-values). Genome-wide DNA methylation patterns are thus largely shaped by TEs in B. distachyon, like in other flowering plant species (Zhang et al. 2006; Regulski et al. 2013; Takuno and Gaut 2013; Seymour et al. 2014). We also found that most TE superfamilies and genes show a higher level of body methylation in B. distachyon than in A. thaliana (fig. 1C and D, supplementary table S5, Supplementary Material online, for P-values), confirming a low genome-wide level of body methylation in A. thaliana compared with other species (Seymour et al. 2014; Eichten et al. 2016).
Fig. 1.
Genome organization and methylation distribution of Col-0 and Bd21. Circos plot displaying methylation levels as well as gene and TE density in 100-kb windows in (A) A. thaliana and (B) B. distachyon. Centromeric and pericentromeric regions are highlighted in gray. Panels (C) and (D) display CpG methylation levels (expressed as % of methylated cytosines) for the most common TE superfamilies and for genes in A. thaliana and B. distachyon, respectively.
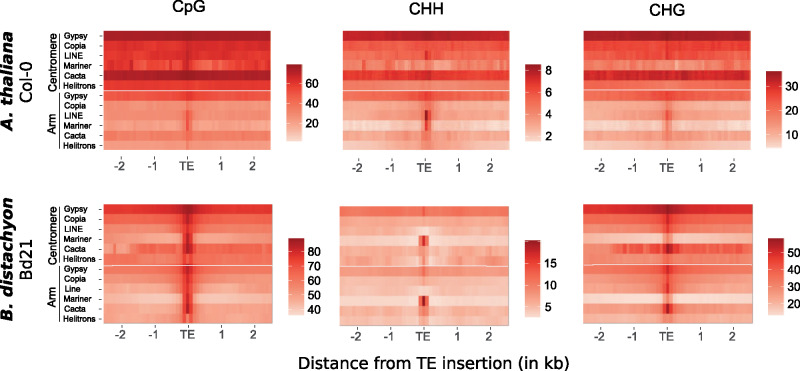
We finally assessed how far methylation spreads around TEs. To do so, we estimated methylation levels in 100-bp windows spanning 2.5 kb around each TE insertion and averaged the values per TE superfamily. We also categorized TE insertions depending on their location on the chromosome (arm vs. centromere) and visualized the results by producing heatmaps. For the three contexts, we observe that DNA methylation around TEs spreads over longer distances in gene-poor centromeric regions than in distal regions of the chromosome arm in A. thaliana (fig. 2). In B. distachyon, spreading mostly reaches a few 100 bp, except around centromeric Gypsy elements where methylation tends to spread beyond 2 kb (fig. 2). Thus, in contrast to our expectations, high levels of TE body methylation in B. distachyon are generally not accompanied with a long spreading of methylation, neither in the centromeres nor on the chromosome arms. This is surprising given that extended methylation spreading around TE is associated with gene-poor pericentromeric regions in rice (Choi and Purugganan 2018), a species closely related to B. distachyon. Because patterns are consistent across CpG, CHH, and CHG contexts (fig. 2), we concentrated the rest of the analysis on CpG methylation.
Fig. 2.
Methylation spreading around transposable elements. Heatmaps representing methylation (CpG) spreading around TEs located in the pericentromere/centromere regions or on the arm of the chromosomes for A. thaliana (N = 18,521 classified TEs) and B. distachyon (N = 62,708 classified TEs). Scales were adapted for the six panels to account for the lower genome-wide methylation level in A. thaliana and the different methylation levels for the three contexts.
Impact of TEs on Nearby Gene Expression
In A. thaliana, genes flanked by a TE within 2 kb show reduced expression compared with genes devoid of TEs in their vicinity (Wang et al. 2013). It was proposed that this may result from a combined effect of DNA methylation spreading and DNA structure variation (Wang et al. 2013). For both the reference accessions Col-0 and Bd21, we estimated the average level of expression for genes having a TE (regardless of the type of TE) in their sequence or within 200-bp windows ranging from near vicinity (0–200 bp) to up to 2 kb (supplementary fig. S3, Supplementary Material online). Although we reproduced the pattern described above with publically available data in A. thaliana, our analysis shows that TEs do not have such an effect on nearby gene expression in B. distachyon. To rule out potential technical artifacts, we extended the analysis to the ten additional B. distachyon accessions we sampled but did not observe any overall effect of TEs on nearby gene expression either (supplementary fig. S3, Supplementary Material online). This is also surprising as the distance between a given gene and its closest TE is significantly smaller (Wilcoxon test, P-value <0.0001; supplementary fig. S3, Supplementary Material online) in Bd21 (1,074 bp) than in Col-0 (5,457bp).
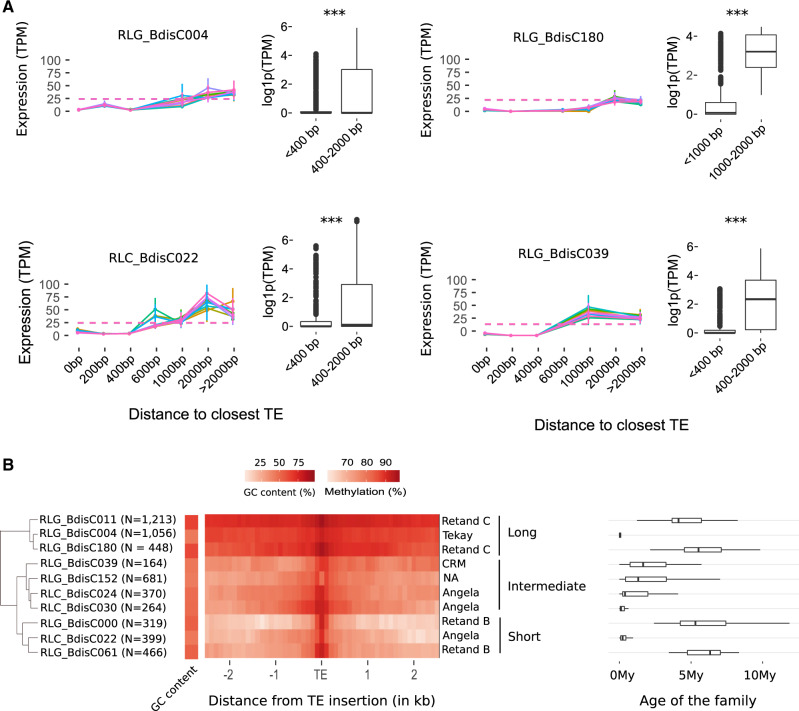
The effect of TEs on methylation and gene expression is known to be family dependent (Eichten et al. 2012; Choi and Purugganan 2018; Noshay et al. 2019), which might explain why no effect is observed when all TEs are pooled. We, therefore, repeated this analysis separately for the ten most active TE families (Stritt et al. 2018) in all 11 B. distachyon accessions to explore potential genetic background effects. We also produced heatmaps as described above to assess a potential link between methylation spreading around these ten TE families and nearby gene expression. Methylation spreading was in a second step used to statistically cluster the ten families. Eventually, as young TEs might be more efficiently targeted by silencing mechanisms, we extracted the age of each copy from Stritt et al. (2020). For four of the ten families, this analysis allowed to uncover clear effects on gene expression following two distinct patterns (fig. 3A): a short-distance effect (<400 bp; all P-values <0.001) conveyed by two Gypsy families (RLG_Bdis0039 and RLG_Bdis004) and one Copia (RLC_Bdis022) family; and a long-distance effect (1 kb; P-values <0.001) for the Gypsy element RLG_Bdis180. However, methylation spreading is unlikely to be involved as our hierarchical clustering shows that spreading varies from long (RLG_Bdis004, RLG_Bdis180) to intermediate (RLG_Bdis0039) and short (RLG_Bdis022) around these four families (fig. 3B). In the same manner, there is no clear association neither with the age of the family (fig. 3B) nor with the genetic background as all accessions show comparable gene expression patterns (fig. 3A). It is nonetheless worth noting that RLG_Bdis004 and RLC_Bdis022 are two of the most recently active families in the genome of B. distachyon (Stritt et al. 2020), suggesting a possible scenario where de novo silencing of these two elements may influence nearby-gene regulation. Altogether, not more than 131 genes are influenced by one of these four families, confirming that the impact of TEs on gene expression is limited in B. distachyon.
Fig. 3.
Impact of transposable elements on gene expression. (A) Average gene expression as a function of the distance to the nearest TE belonging to the four LTR retrotransposons, RLG_BdisC039, RLG_BdisC004, RLG_BdisC180, and RLC_BdisC022, in the natural accessions of B. distachyon. Each colored line represents a different accession. The dashed line represents the genome-wide level of expression. Boxplots display the expression of genes harboring a TE in their vicinity for two distance classes only, all accessions pooled together. Stars indicate Wilcoxon test levels of significance: ***<0.001. (B) Heatmap representing methylation (CpG) spreading around the ten most active LTR retrotransposon families in Bd21. Families were clustered based on the level of methylation spreading (long, intermediate, and short) around each insertion. Names on the right of the heatmap describe the TE lineage to which each family belongs. Boxplots display the age of each family estimated by Stritt et al. (2020).
We eventually investigated the impact of TE insertion polymorphisms (TIPs) on gene expression. Overall, we identified 1,833 high-confidence TIPs, that is, insertions present in at least one of the ten natural accessions but absent from the reference genome Bd21 (supplementary table S6, Supplementary Material online). Due to the relative small number of TIPs located nearby genes, we lacked the power to investigate their impact on gene expression at the family level. To circumvent this difficulty, we measured the log-fold change in gene expression for loci harboring a TIP in their vicinity (<1 kb) compared with the same loci without a TIP in a different accession. We found that the majority of the TIPs have no effect on adjacent gene expression (supplementary fig. S4, Supplementary Material online), consistent with previous findings that TE polymorphisms have little impact on methylation spreading in B. distachyon (Eichten et al. 2016). These results contrast again with what is found in A. thaliana, where polymorphic TEs are associated with differential expression of nearby genes among natural accessions (Stuart et al. 2016). Despite the lack of global effect, a subset of 32 TIPs show a significant effect on nearby gene expression at high confidence (supplementary table S7, Supplementary Material online), providing candidates for further functional validation.
Genome Organization and Functional Impact of TEs
The differences observed between the two species may result from the nature of their TEs. Notably, the genome of B. distachyon displays a much higher ratio of solo-LTRs: intact copies for both conserved elements and TIPs (up to 14:1 for some families; Stritt et al. 2020) compared to A. thaliana, where the number of intact copies approximate the number of solo-LTRs (Devos et al. 2002). This may reflect a much broader aspect of the TE dynamics in the two species and one could assume that B. distachyon harbors overall a much larger proportion of nonfunctional TEs than A. thaliana. As nonfunctional elements might be less efficiently targeted by silencing, they might have in return a limited influence on their neighboring genes. This hypothesis, however, does not hold for rice. There, the ratio solo-LTR:intact elements is 1.6:1 (Tian et al. 2009) and therefore does not differ much from the one found in A. thaliana. Yet, TEs also have little impact on gene expression in rice (Choi and Purugganan 2018).
As an alternative hypothesis, methylation spreading has been suggested to be under purifying selection (Choi and Purugganan 2018) and such a mechanism may largely constrain the impact of TEs on adjacent genes. Compared with A. thaliana, TEs and genes are largely interspersed in both B. distachyon and rice (Garg et al. 2015). Selection against the deleterious effect of methylation spreading around TEs on genes might be stronger in such systems and explain why limited spreading is observed in these two species. We, therefore, suggest that the differences we observed between A. thaliana and B. distachyon with regard to the functional impact of TEs are largely inherent to the contrasting architecture of their genomes. In the light of the current results and recent studies (Seymour et al. 2014; Choi and Purugganan 2018; Quesneville 2020), it appears that genome organization varies greatly between Brassicacea and grasses. With more than 12,000 species (Christenhusz and Bying 2016), grasses cover about 40% of the planet (Gibson 2009). Generalizing the patterns reported in A. thaliana to all plants, as commonly seen in the literature, is therefore arguable. Our study is calling for the analysis of a larger set of plant genomes to reflect the tremendous diversity present in this kingdom.
Materials and Methods
Plant Material
We used 11 accessions of B. distachyon originating from Iraq (Bd21, Bd21-3, Bd3-1), Turkey (BdTR12c, BdTR7a, Koz3, Bd1-1), and Spain (ABR2, Sig2, Luc1, Bd30-1). Seeds were sown on the soil–sand mixture in 50-ml pots. For bisulfite and RNA sequencing, three biological replicates per accession were grown. The second and third leaves of each replicate were sampled after 24 days of 16/8 h of day/night conditions.
Whole-Genome Bisulfite Sequencing and RNA-Sequencing Analysis
For B. distachyon, WGBS (whole-genome bisulfite sequencing) libraries were prepared as described in the supplementary method S1, Supplementary Material online. For A. thaliana, we used publicly available data (supplementary table S1, Supplementary Material online, for references). Raw data were processed as described in supplementary method S1, Supplementary Material online, and mapped onto the reference genomes Bd21 for B. distachyon (Phytozome 12, https://phytozome.jgi.doe.gov) and Col-0 for A. thaliana (TAIR10, www.arabidopsis.org). Methylation levels were estimated following the method described in Schultz et al. (2012) (supplementary method S1, Supplementary Material online) and by averaging the percentage of methylated cytosines over the three replicates per genotype within each focal unit (e.g., 100 kb/100 bp genomic windows, TE and gene body). We drew Circos plots (Connors et al. 2009) to visualize the architecture of the two reference genomes. The PCA based on methylation data was performed using ggplot2 in R (R Development Core Team 2013). We eventually used GLM models with logit transformation to characterize the relationship between gene or TE density and methylation levels. P-values were obtained with the R package multcomp (Hothorn et al. 2008).
For each of the 11 B. distachyon accessions, mRNA was extracted using the RNAeasy Plant Mini Kit from Qiagen. Libraries were prepared by the D-BSSE Basel, and paired-end sequencing was performed on an Illumina HiSeq 2500. For A. thaliana, we again used publicly available data (supplementary table S1, Supplementary Material online). Reads were mapped to the reference genomes with Salmon quant v.0.12.0 (Patro et al. 2017). The abundance of gene expression was quantified averaging transcripts per million per accession for each single genotype over the three replicates.
Methylation Spreading
We estimated methylation spreading by calculating methylation levels in 100-bp windows spanning 2.5 kb upstream and downstream of each TE insertion. TE insertions were categorized with regard to their position in centromeric/pericentromeric regions or on chromosome arms and aggregated at the family- (for the ten most active TE families) or superfamily levels. Heatmaps depicting methylation levels around TEs were drawn in R using ggplot2 (Wickham 2009). We also used the levels of methylation spreading around TEs to calculate Euclidean distances between the ten most active TE families and to perform a hierarchical clustering using hclust in R (R Development Core Team 2013).
Impact of TEs on Gene Expression
We assessed the potential link between the presence of a TE in the vicinity of a gene and expression by assigning genes to different categories depending on whether they harbor a TE in their sequence or within windows ranging from near vicinity (0–200 bp) to up to 2 kb. Average gene transcripts per million and standard errors were calculated per distance class with all TEs pooled together as well as for the ten most active families separately and plotted using the R package plotrix (Lemon 2006). For a subset of candidate families, we also compared the expression level (log1p transformed) of genes displaying a TE in their vicinity (0–400 bp for RLG_Bdis0039, RLG_Bdis004, and RLC_Bdis022; 0–1 kb for RLG_Bdis180) with the expression level of genes displaying a TE further away (between 400 bp and 2 kb and 1 and 2 kb, respectively) with a Wilcoxon test.
TIP Analysis
Detection of TIPs was performed with detettore (Stritt and Roulin forthcoming) using sequencing data from Gordon et al. (2017). Detettore was specifically developed on B. distachyon. It allows to output different features, including the presence of target site duplication, which can be used to filter out false positive. Filtering criteria were adjusted so that no TIP is found when the reads of Bd21 are mapped against the reference genome. The expression of genes harboring a TIP in their vicinity (<1 kb) in at least one of the accessions was compared with the corresponding genes in the other accessions. For each single gene, the difference in expression between groups was tested using a t-test with a confidence interval of 0.95. Change in gene expression was considered significant when the P-value was <0.05 and the log2 change was >1.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the PSC-Syngenta fellowship, the University Research Priority Program Evolution in Action of the University of Zurich and the Swiss National Science Foundation (PZ00P3_154724). The authors would like to thank the D-BSSE for the sequencing effort, the Genetic Diversity Center at ETH Zurich for providing access to their computing resources, and Michael Thieme for his comments on the manuscript.
Data Availability
All the data produced for the current study are available under the ENA project number PRJEB39651. For Arabidopsis thaliana, references are provided in the Supplementary Material online. Scripts are available at https://github.com/mwylerCH.
Literature Cited
- Bonchev G, Parisod C. 2013. Transposable elements and microevolutionary changes in natural populations. Mol Ecol Resour. 13(5):765–775. [DOI] [PubMed] [Google Scholar]
- Choi JY, Purugganan MD. 2018. Evolutionary epigenomics of retrotransposon-mediated methylation spreading in rice. Mol Evol Biol. 35(2):365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz MJM, Bying JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa 261(3):201–217. [Google Scholar]
- Connors J, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. 2002. Genome size reduction though illegitimate recombination counteracts expansion in Arabidopsis. Genome Res. 12(7):1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Stuart T, Srivastava A, Lister R, Borevitz JO. 2016. DNA methylation profiles of diverse Brachypodium distachyon aligns with underlying genetic diversity. Genome Res. 26(11):1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, et al. 2012. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8(12):e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Chevala N, Shankar R, Jain M. 2015. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci Rep. 5(1):14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DJ. 2009. Grasses and grassland ecology Oxford/New York: Oxford University Press. [Google Scholar]
- Gordon SP, et al. 2017. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun. 8(1):2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M-A. 1998. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3(5):181–187. [Google Scholar]
- Grandbastien M-A, et al. 2005. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res. 110(1–4):229–241. [DOI] [PubMed] [Google Scholar]
- Guio L, Barron MG, Gonzalez J. 2014. The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol. 23(8):2020–2030. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Wang K, Jenkins G. Forthcoming 2019. Progressive refinement of the karyotyping of Brachypodium genomes. New Phytol. [DOI] [PubMed] [Google Scholar]
- Hollister JD, et al. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 108(6):2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth V, Merenciano M, González J. 2017. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 33(11):832–841. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom J. 50(3):346–363. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Consortium. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. [DOI] [PubMed] [Google Scholar]
- Lemon J. 2006. Plotrix: a package in the red light district of R. R News. 6:8–12. [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News. 2:18–22. [Google Scholar]
- Makarevitch I, et al. 2015. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 11(1):e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1947. Cytogenetic studies of maize and neurospora. Carnegie Inst Wash Yrbk. 46:146–152. [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 36(6):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshay JM, et al. 2019. Monitoring the interplay between transposable element families and DNA methylation in maize. PLoS Genet. 15(9):e1008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14(4):417–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H. 2020. Twenty years of transposable element analysis in the Arabidopsis thaliana genome. Mobile DNA 11(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: A language and environment for statistical computing. Vienna: [Austria: ]: R Foundation for Statistical Computing; Available from: http://www.R-project.org. [Google Scholar]
- Regulski M, et al. 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23(10):1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey O, Danchin E, Mirouze M, Loot C, Blanchet S. 2016. Adaptation to global change: a transposable adaptation to global change: a transposable element – epigenetics perspective. Trends Ecol Evol (Amst). 31(7):514–526. [DOI] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR. 2012. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28(12):583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. 2014. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10(11):e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, et al. 2017. Gene deletion in barley mediated by LTR-retrotransposon BARE. Sci Rep. 7:43766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman MJ, Slotkin RK. 2016. The first rule of plant transposable element silencing: location, location, location. Plant Cell 28(2):304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Gordon SP, Wicker T, Vogel JP, Roulin AC. 2018. Recent activity in expanding populations and purifying selection have shaped transposable element landscapes. Genome Biol Evol. 10(1):304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Roulin AC. Forthcoming. Detecting signatures of TE polymorphisms in short-read sequencing data. Methods in Molecular Biology Springer Nature Switzerland AG. [DOI] [PubMed] [Google Scholar]
- Stritt C, Wyler M, Gimmi EL, Pippel M, Roulin AC. 2020. Diversity, dynamics and effects of long terminal repeat retrotransposons in the model grass Brachypodium distachyon. New Phytol. 227(6):1736–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, et al. 2016. Population scale mapping of transposable element diversity uncovers novel genetic diversity linked to gene regulation and epigenomic variation. Elife 5:e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2013. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 110(5):1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme M, et al. 2017. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 18(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thind AK, et al. 2018. Chromosome-scale comparative sequence analysis unravels molecular mechanisms of genome dynamics between two wheat cultivars. Genome Biol. 19(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, et al. 2009. Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19(12):2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van’t Hof AE, et al. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534(7605):102–105. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM. 2013. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 9(2):e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liang W, Tang T. 2018. Constant conflict between Gypsy LTR retrotransposons and CHH methylation within a stress-adapted mangrove genome. New Phytol. 220(3):922–935. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis New York: Springer. [Google Scholar]
- Zhang H, Lang Z, Zhu J. 2018. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol. 19(8):489–506. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126(6):1189–1201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data produced for the current study are available under the ENA project number PRJEB39651. For Arabidopsis thaliana, references are provided in the Supplementary Material online. Scripts are available at https://github.com/mwylerCH.