Abstract
Short tandem repeats (STRs) contribute to structural variation in plant mitochondrial genomes, but the mechanisms underlying their formation and expansion are unclear. In this study, we detected high polymorphism in the nad7-1 region of the Pinus tabuliformis mitogenome caused by the rapid accumulation of STRs and rearrangements over a few million years ago. The STRs in nad7-1 have a 7-bp microhomology (TAG7) flanking the repeat array. We then scanned the mitogenomes of 136 seed plants to understand the role of microhomology in the formation of STR and mitogenome evolution. A total of 13,170 STRs were identified, and almost half of them were associated with microhomologies. A substantial amount (1,197) of microhomologies was long enough to mediate structural variation, and the length of microhomology is positively correlated with the length of tandem repeat unit. These results suggest that microhomology may be involved in the formation of tandem repeat via microhomology-mediated pathway, and the formation of longer duplicates required greater length of microhomology. We examined the abundance of these 1,197 microhomologies, and found 75% of them were enriched in the plant mitogenomes. Further analyses of the 400 prevalent microhomologies revealed that 175 of them showed differential enrichment between angiosperms and gymnosperms and 186 differed between angiosperms and conifers, indicating lineage-specific usage and expansion of microhomologies. Our study sheds light on the sources of structural variation in plant mitochondrial genomes and highlights the importance of microhomology in mitochondrial genome evolution.
Keywords: structural variation, microhomology, short tandem repeat, mitochondrial genome, Pinus
Significance
Short tandem repeats are ubiquitous and play important roles in the evolution of plant mitogenomes, but the mechanisms underlying their origin and proliferation remain unclear. In this study, we reveled that tandem repeats were associated with microhomologies in seed plants mitogenomes, and the accumulation of microhomologies was lineage specific showing differential enrichment among angiosperm and gymnosperm. These results suggest that microhomologies may be involved in the formation of tandem repeat via microhomology-mediated pathway, and have undergone lineage-specific usage and expansion. Our results highlight the high prevalence of microhomology and its important role in mitogenome evolution.
Introduction
The plant mitochondrial genome (mitogenome) is characterized by an unusually low silent substitution rate along with extensive structural variation (Palmer et al. 2000). In the vast majority of plant mitogenomes, synonymous substitution rates are up to 20 times lower than those of nuclear genomes (Drouin et al. 2008; Wang and Wang 2014). In contrast, frequent length variation and rearrangements associated with repetitive sequences have been detected throughout mitogenomes (Marechal and Brisson 2010; Jaramillo-Correa et al. 2013; Cole et al. 2018), leading to multiple alternative isoforms (Unseld et al. 1997; Kozik et al. 2019). Short tandem repeats (STRs) account for a substantial proportion of repetitive content in mitogenomes and are lineage- and gene region-specific, leading to substantial structural differences among species and populations (Godbout et al. 2005; Jaramillo-Correa et al. 2013; Potter et al. 2013; Wang and Wang 2014). In conifers, STRs serve as mutagenic hotspots with elevated substitution rates (Jaramillo-Correa et al. 2013). Although STRs are ubiquitous and play important roles in the evolution of plant mitogenomes, the mechanisms underlying their origin and proliferation remain unclear.
Recent studies suggest that sequence duplications and rearrangements in plant mitogenomes are linked to the repair of double-strand breaks (DSBs) (Shedge et al. 2007; Davila et al. 2011; Gualberto and Newton 2017) via either nonhomologous end-joining (NHEJ) or microhomology-mediated end-joining (MMEJ) pathways (McVey and Lee 2008; Lieber 2010). In the NHEJ pathway, broken DNA strands are usually processed by degradation of the 5′-end and subsequent blunt-end ligation, leading to insertions and/or deletions of variable lengths (supplementary fig. S1A, Supplementary Material online) (Lieber 2010). In the MMEJ pathway, microhomologous sequences anneal to each other before the joining of broken ends, resulting in deletions, and/or insertions flanking the breakpoints (supplementary fig. S1B, Supplementary Material online) (McVey and Lee 2008; Garcia-Medel et al. 2019). Although MMEJ relies on substantial microhomology for the repair of DSBs, this is not essential for NHEJ. Other mechanisms, including fork stalling and template switching (FoSTeS), and microhomology-mediated break-induced replication (MMBIR), also generate structural variation and rely on microhomology (Hastings et al. 2009; Ottaviani et al. 2014; Taylor et al. 2015). In the FoSTeS pathway, the lagging strand (which is formed by the stalling of the replication fork) disengages from the stalled fork, invades the other active replication fork by annealing to a microhomologous sequence, and restarts synthesis at the invaded fork (supplementary fig. S1C, Supplementary Material online) (Ottaviani et al. 2014). The process of template switching continues until the lagging strand returns to the original replication fork, resulting in DNA deletion or duplication (Ottaviani et al. 2014). In the MMBIR pathway, a DSB is created by the collapse of the replication fork, and the 3′ overhang at the broken end invades a new microhomologous template and reinitiates replication of the invaded template (supplementary fig. S1D, Supplementary Material online) (Hastings et al. 2009). As in the FoSTeS pathway, the extended end may switch to multiple new templates until annealing back to the original fork, leading to complex structural variation. These mechanisms have been investigated by comparative bioinformatics analyses of population genomic data of plant nuclear genomes (Vaughn and Bennetzen 2014), and examined by in vitro (Garcia-Medel et al. 2019) and in vivo experiments in plant chloroplast (Kwon et al. 2010) and nuclear genomes (Schiml et al. 2016). However, little is known about the process of structural variation in plant mitogenomes although different hypotheses have been proposed (Palmer and Herbon 1988; Davila et al. 2011; Christensen 2013; Gualberto and Newton 2017). Moreover, the relative contributions of various mechanisms underlying the formation of rearrangements have not been evaluated.
Our understanding of the mitogenome structure and variation in gymnosperms is very limited. Only a handful of mitogenomes have been assembled to date, and the size of these genomes differs by more than 17-fold (Chaw et al. 2008; Guo et al. 2016; Kan et al. 2020; Sullivan et al. 2020) (0.35–5.99 Mb; supplementary table S1, Supplementary Material online). This size variation might be related to the abundance of repeats, including STRs, in each genome. In this study, we report a highly variable region in nad7-1 (NADH dehydrogenase subunit 7 intron 1) in the Pinus tabuliformis mitogenome caused by complex rearrangements of STRs (fig. 1). The plant nad7 genecontains five exons and encodes a subunit of respiratory complex I (NADH-ubiquinone oxidoreductase), which transfers electrons from NADH to ubiquinone (Pineau et al. 2005). We detected a large number of haplotypes in 157 individual trees of P. tabuliformis. To understand whether the observed high haplotype diversity was due to the retention of ancestral polymorphisms or the accumulation of new mutations following speciation, we examined the sequences in this region in 550 individuals of two closely related species and in multiple accessions of 21 additional pine species. We modeled the evolution of STRs in the region by close examination of sequence and structural variation among haplotypes. Finally, we scanned the mitogenomes of 136 seed plants to assess the role of microhomology in the formation of STRs and structural variation in plant mitogenomes.
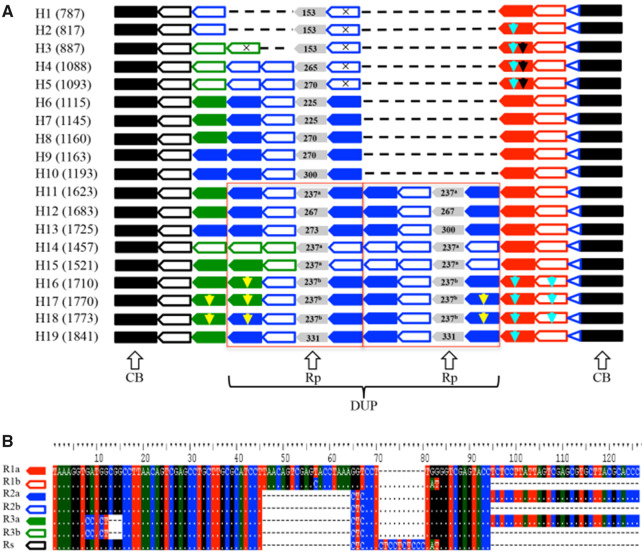
Fig. 1.
Sequence structure of 19 haplotypes identified in nad7-1 of P. tabuliformis. (A) The two conserved blocks are indicated by a black rectangle; seven motifs are indicated by pentagons with colors corresponding to those used in (B); the highly variable motif Rp is indicated by gray pentagons with the length labeled. The superscript latter "a" and "b" indicated two different Rp motifs with same length; and the TAG7 microhomology is indicated by a blue open triangle. Three insertions specific to a subset of motifs are indicated by yellow, light blue, and black arrows. A large sequence block (DUP) duplicated in H11–19 is outlined in red. (B) Alignment of the seven motifs.
Materials and Methods
Sampling and Sequencing
Pinus tabuliformis is a major coniferous forest species in northern and central China, with a range of 2,000 km from east to west and 1,200 km from north to south (Ying et al. 2004; Mao and Wang 2011). Studies of the biogeography of P. tabuliformis based on maternally inherited mitochondrial (mt) markers have provided valuable insights into its migration and colonization history (Chen et al. 2008; Hao et al. 2018; Xia et al. 2018). However, the few conserved mtDNA fragments used in previous studies limit our understanding of mitogenome evolution in this species.
In this study, we characterized diversity in a highly variable region, nad7-1, in 17 populations of P. tabuliformis, as well as in 23 and 11 representative populations of two closely related species, Pinus densata and Pinus yunnanensis, respectively. The distribution of the 17 sampled populations of P. tabuliformis is shown in supplementary figure S2, Supplementary Material online. The name, location, and sample size of all 51 populations are listed in supplementary table S2, Supplementary Material online. In addition, we included 21 species representing all five recognized subsections of the subgenus Pinus for comparison (Gernandt et al. 2005). For nine species, multiple accessions were collected from documented individuals grown by different institutions (supplementary table S3, Supplementary Material online). For other species, nad7-1 sequences were obtained from previous publications (supplementary table S3, Supplementary Material online).
Genomic DNA was extracted from needles or seedlings using a Plant Genomic DNA Kit (Tiangen, Beijing, China). A sequence-specific primer pair was designed on the basis of a conserved region of nad7-1 (Neale et al. 2014) (nad7-1F, 5′-GAGGGACAACCCTGGAATA-3′; nad7-1R, 5′-AAGGCCTCTCCATTTCCAAT-3′). The PCR amplified region of this primer pair was between 176,897 and 176,916 bp of the Pinus taeda mitogenome. The PCR products were examined by agarose gel electrophoresis (1.5% in TAE). The desired bands were cut from the gel, purified using a TIANGEN DNA Purification Kit (Tiangen, Beijing, China), and sequenced using an ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA).
Phylogenetic Analysis
Sequences were aligned using ClustalX (Larkin et al. 2007) and manually adjusted using BioEdit v. 7.2 (https://bioedit.software.informer.com/7.2/). A median joining network of mitotypes was produced using Network 5 (Bandelt et al. 1999), and a neighbor-joining (NJ) tree of mitotypes was constructed using MEGA for macOS (Stecher et al. 2020). Topological robustness of the NJ tree was assessed using 1,000 nonparametric bootstrap replicates. Indels were coded as binary characters using gap-coder (Young and Healy 2003).
Tandem Repeat Analysis
To assess tandem repeats in mitogenomes of seed plants, assemblies of mitogenomes of 128 angiosperms and eight gymnosperms were retrieved from GenBank (supplementary table S1, Supplementary Material online). For species with multiple mitogenome assemblies, the assembly with the highest quality was used in this study. Tandem repeat sequences were identified using Tandem Repeats Finder (Benson 1999) with the following default parameters: maximum period size = 500 bp; detection matching probability (Pm) = 0.8; detection indel probability (Pi) = 0.1; and alignment weights of match, mismatch, and indel = 2, 7, and 7, respectively. We filtered tandem repeat arrays with overall matches <60% or alignment scores <50. Among overlapped repeat arrays, the one with the highest alignment score was retained.
Delimiting a microhomology is not straightforward, and various thresholds are used in previous studies (Ottaviani et al. 2014; Bhargava et al. 2016; Ceccaldi et al. 2016). In this study, we defined a microhomology as a set of short (<70 bp) homologous DNA sequences at breakpoint junctions and flanking regions (Ottaviani et al. 2014). To search for microhomology in tandem repeat regions, the sequence of the last unit of the repeat array was compared with the consensus sequence of repeats. The last unit was considered as a microhomologous sequence if its length was less than 70 bp, and less than half of the consensus repeat. For example, in a tandem repeat array with the last unit of length n (n < 70 bp) and consensus sequence of length m, if n < m/2, the sequence of the last unit was regarded as a microhomology in the repeat array (supplementary fig. S3, Supplementary Material online). Microsatellites (also known as simple sequence repeats) containing repeats of 1–6 bp were not included in microhomology analyses because they have complex mutation models (e.g. stepwise mutation and two-phase models) (Ellegren 2004; Takezaki 2017), which were not the focus of this study.
To quantify the enrichment of microhomologous sequences, we developed a statistic termed as enrichment of microhomology (ECH). First, we counted the observed number of microhomologous sequences (ObsMH) on both DNA strands of the mitogenome (no mismatch allowed). Then, we shuffled (per base) the mitogenome 1,000 times, and randomly sampled the same number of bases (without replacement) at each shuffling. The expected number of microhomologous sequences (ExpMH) on both strands was then counted in each replicate, and the mean and 95% confidence interval (CI) of the ExpMH were calculated across the 1,000 replicates. The ECH was defined as the ratio of ObsMH to the mean of ExpMH (ECH = ObsMH/mean of ExpMH). The significance of ECH was determined by comparing the value of ObsMH with the 95% CI of ExpMH; the ECH was considered significant (P < 0.05) when ObsMH was larger than the upper limit of the 95% CI of ExpMH. For each microhomology, we calculated the value of ECH and tested the significance of enrichment in the mitogenome.
To further test whether microhomologies were differentially accumulated among seed plants, we calculated the ECH values for 400 microhomologies in the mitogenomes of 136 seed plants, and examined the enrichment of these microhomologies among angiosperms, gymnosperms, and conifers (supplementary table S4, Supplementary Material online). Among these 400 microhomologies, 397 were common in plant mitogenomes (length ≥6 bp; ObsMH ≥100; and ECH >1.00), one was detected in the nad7-1 region of P. tabuliformis (“TAAAGGT”; see Results and Discussion section), and two were previously reported in sugar beet (“CCATACT” in the rrn26 gene region) (Nishizawa et al. 2000) and Norway spruce (“GAAGAA” in the mh44 gene region) (Bastien et al. 2003). The significance (P value) of over-representation of microhomology was calculated using Mann–Whitney U test and then corrected for multiple comparisons using Benjamini–Hochberg false discovery rate (FDR) adjustment (Benjamini and Hochberg 1995).
Results and Discussion
Extremely High Variation in nad7-1 of P. tabuliformis
Analysis of the nad7-1 region in 157 individuals of P. tabuliformis identified 19 haplotypes with lengths ranging from 782 to 1,835 bp (fig. 1; supplementary table S2 and data S1, Supplementary Material online). Sixteen of these haplotypes were population-specific, seven of which had frequencies of >50% in the population. The high polymorphism in nad7-1 was in contrast to the low diversity observed in other mtDNA regions in this species, with fewer than five haplotypes detected over three segments (NADH dehydrogenase subunit 1 intron 2, NADH dehydrogenase subunit 4 intron 3, and NADH dehydrogenase subunit 5 intron 1) spanning a total length of 2,800 bp in range-wide samples (Chen et al. 2008; Wang et al. 2011; Hao et al. 2018).
The observed polymorphism could reflect either ancestral polymorphisms or new mutations that occurred after speciation. Although ancestral polymorphisms are often shared by closely related species before complete lineage sorting, new mutations are usually lineage specific. To distinguish between these two alternatives, we further sequenced nad7-1 in 375 and 175 individuals of two closely related species, P. densata and P. yunnanensis, respectively (supplementary table S2, Supplementary Material online). Four mitotypes (H1, H9, H20, and H21) were detected in P. densata, of which two (H1 and H9) were shared with P. tabuliformis (supplementary table S2, Supplementary Material online). In the 175 samples of P. yunnanensis, only two species-specific mitotypes (H21 and H22) were found, and none of these were shared with P. tabuliformis (supplementary table S2, Supplementary Material online). We extended our survey to an additional 21 Pinus species (supplementary table S3, Supplementary Material online) and found that none of the haplotypes in P. tabuliformis were shared with other species. In addition, the P. tabuliformis haplotypes were highly divergent from those of other species, differing by multiple deletions and substitutions. Taken together, these results supported the hypothesis that variations in nad7-1 in P. tabuliformis were formed after the divergence of the species. Alternatively, mitotypes specific to P. tabuliformis could be explained by incomplete lineage sorting of ancestral polymorphisms during species diversification. However, this hypothesis is very unlikely considering the low rate of lineage sorting in the mitogenome of Pinus; mitotypes could be shared between pine species that diverged millions of years ago (Zhou et al. 2010). The extensive population-specific haplotypes in P. tabuliformis are more likely to have originated recently, after the divergence of populations dated at 0.58–3.67 Ma (Xia et al. 2018). High polymorphism has also been reported in the nad7-1 region of three other pine species (supplementary table S3, Supplementary Material online): Pinus banksiana (14 haplotypes; Godbout et al. 2005), Pinus armandii (11 haplotypes; Liu et al. 2014), and Pinus kwangtungensis (nine haplotypes; Tian et al. 2010). Unlike in P. tabuliformis, mitotypes in these species were usually shared with sister species, suggestive of incomplete sorting of ancestral polymorphisms.
Tandem Duplication in nad7-1 of P. tabuliformis
Examination of sequence structure of the nad7-1 region in the P. tabuliformis revealed two conserved blocks surrounding one highly variable block. The upstream conserved block was 223 bp with a single substitution in three haplotypes (H4–H6), and the downstream conserved block was 39 bp without any variation (fig. 1A). The variable block was characterized by a set of perfect or imperfect (differed by 1–4 indels or substitutions) tandem repeats with different copy numbers among haplotypes (fig. 1).
We identified seven motifs (R1a, R2a, R3a, R1b, R2b, R3b, and Rs) in the variable block (fig. 1B). Because this highly variable block is absent from the mitogenomes of other pine species, we cannot infer the ancestral sequence based on homology with outgroups. The Rs motif was basic and shared among haplotypes (fig. 1B); it is thus possible that Rs (or its ancestral sequence) duplicated and one of the copies acquired substitutions. Among the motifs, R2b was most similar to Rs and thus likely evolved from Rs, followed by further changes to generate the other motifs (see our hypothetical evolutionary pathway for these motifs in supplementary fig. S4, Supplementary Material online). Consistent with this hypothesis, H1 containing the Rs-R2b array is the most frequent and probably most ancient haplotype.
All haplotypes ended with the same R1a-R1b array in the variable block (fig. 1). R1a and R1b differed by three substitutions and a 32 bp deletion. One plausible evolutionary pathway is that R1a was duplicated to generate R1b. Alternatively, R1b was derived from Rs, and R1a was derived from R1b. The evolutionary history of these motifs may be more complex than we described (supplementary fig. S4, Supplementary Material online), and multiple pathways might have been involved in the formation of duplicates.
The structural variation in nad7-1 involved not only the stepwise expansion of a single motif and subsequent modification, as discussed above, but also a large duplication including multiple motifs. A sequence block (referred to as the DUP block), composed of one highly variable region (referred to as Rp hereafter) surrounded by three motifs, was duplicated in haplotypes H11–H19 (fig. 1A). The Rp motif varied among haplotypes with lengths ranging from 153 to 331 bp and more than eight substitutions differentiating the most differentiated copies (fig. 1A). In contrast, the two copies of the Rp motif within each haplotype were identical, except for a 27 bp indel in the Rp motif of H13. This observation that two Rp copies within a haplotype were more similar to each other than to copies from different haplotypes suggests that the divergence of the Rp copies predated the duplication of the DUP block.
We found a 7-bp sequence (“TAAAGGT”; hereafter referred to as the TAG7) at the 5′ end of each motif as well as directly downstream of the variable block (fig. 1). The presence of this motif in tandem repeats seems consistent with models of microhomology-mediated sequence duplications, including MMEJ, FoSTeS, and MMBIR (McVey and Lee 2008; Ottaviani et al. 2014). The MMEJ pathway may be important for the repair of DSBs, resulting in tandem duplications (McVey and Lee 2008), whereas FoSTeS and MMBIR can produce duplications and complex rearrangements by microhomology-mediated repair of broken replication forks (Ottaviani et al. 2014). Slippage strand replication caused by the mispairing of existing microhomologous sequences can also account for the formation of tandem duplicates (Darmon and Leach 2014). These mechanisms have different genetic bases but are characterized by a similar signature of microhomology at the junction (Ottaviani et al. 2014). NHEJ can also repair DSBs and generate tandem duplications, either through blunt end-joining or 1–4 bp microhomology (Lieber 2010), inconsistent with the length of the TAG7 observed in the nad7-1 region. In summary, we found complex structural variation due to the expansion and contraction of tandemly arrayed duplicates in the nad7-1 region of P. tabuliformis, and the TAG7 motif most likely served as a microhomologous sequence in the formation of structural variants. Short direct repeats flanking tandem arrays have been reported in other plant mitogenomes, and suggested to be involved in the generation of tandem repeat array (Nishizawa et al. 2000; Bastien et al. 2003). However, microhomology-mediated pathways cannot be reliably distinguished based only on the sequences surrounding breakpoint junctions. Future studies using highly reliable reporters are needed to elucidate the detailed biochemical and genetic underpinnings of sequence duplications as well as universal features associated with each of the underlying mechanisms.
Tandem Duplication Associated with Microhomology in Plant Mitogenomes
To assess the importance of microhomology in tandem duplication and mitogenome evolution, we investigated the abundance of tandem repeats and their associations with microhomologies in 136 seed plants. The in silico search yielded 13,170 STRs, with 3– 872 STRs per genome among angiosperms and 54–2,358 STRs per genome among gymnosperms (supplementary table S1, Supplementary Material online). The median length of an STR unit was 22 bp, and median copy number was 2.2 per STR (supplementary fig. S5A and B, Supplementary Material online). Both the length of the repeat unit and total length of the repeat array of tandem repeats were shorter than those of nontandem repeats reported previously in plant mitogenomes (Chaw et al. 2008; Guo et al. 2016; Wynn and Christensen 2019; Kan et al. 2020). Overall, tandem repeats were more abundant in larger genomes, producing a positive correlation between genome size and STR length (Pearson’s correlation coefficient r = 0.708, P < 2.2e−16; supplementary fig. S5C, Supplementary Material online).
The tandem repeats observed in mitogenomes were generally associated with microhomologies. In angiosperms, 52.6% of the tandem repeats ended in short homologies; this value was marginally higher than that in gymnosperms (42.6%) after controlling the genome size (F1,133 = 3.622, P = 0.0592; fig. 2A). It is worth noting that our estimation of microhomology abundance may be conservative because of two reasons. First, we did not consider sequences with lengths more than half of the repeat unit as a microhomology (see Materials and Methods section). When considering all imperfect end-repeats as microhomologies, 79.4% and 77.8% of tandem repeats were associated with microhomologous sequences in angiosperms and gymnosperms, respectively (supplementary table S1, Supplementary Material online). Second, tandem repeats may have been lost or reduced in size to below the detection threshold during the repairing of DSBs via microhomology-mediated pathway or asymmetrical recombination (Davila et al. 2011; Gualberto and Newton 2017).
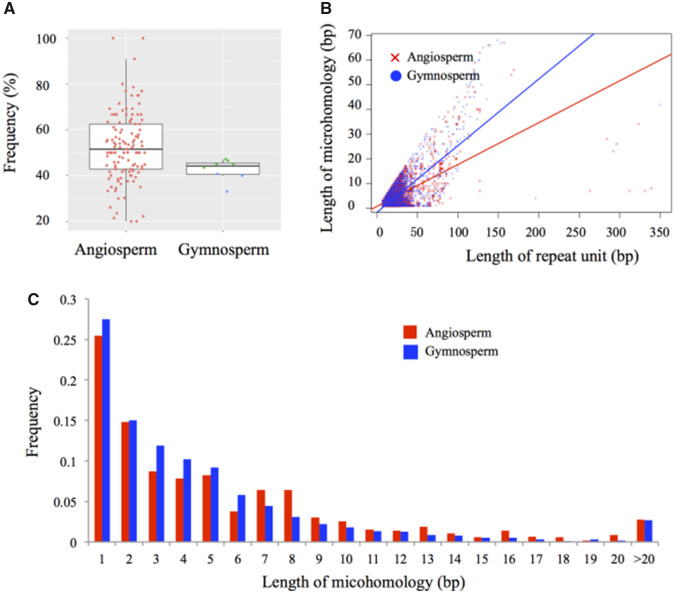
Fig. 2.
Tandem repeats associated with end homology in plant species. (A) Frequency of tandem repeats associated with end homology in angiosperm and gymnosperm species. In each box, horizontal lines from top to bottom refer to the first quartile, median, and third quartile. Each red, green, and blue dot represents an angiosperm, conifer, and gymnosperm (excluding conifers), respectively. (B) Length of end homology against repeat units. Correlations were evaluated by Pearson’s correlation coefficient tests. (C) Distribution of end homology lengths. Values of ≥6 bp suggest microhomology-mediated tandem duplication.
Size distribution of microhomologies showed that 34.9% and 26.3% of all microhomologies in angiosperms and gymnosperms, respectively were longer than 6 bp (fig. 2C). A previous study has suggested that homologous sequences with lengths of ≥6 bp could form stable loops and result in structural variations (Montgomery et al. 2013). Organellar DNA polymerases can effectively repair DSBs using microhomologous sequences as short as 6 bp in Arabidopsis thaliana (Garcia-Medel et al. 2019). An in vitro study has also shown that the minimum primer length for extension is 6 bp for DNA polymerase Klenow fragment (Zhao and Guan 2010). In addition, we found that microhomology length is positively correlated with the length of the repeat unit (Pearson’s correlation coefficient r = 0.634, P < 2.2e−16 in angiosperms; and r = 0.746, P < 2.2e−16 in gymnosperms; fig. 2B and supplementary fig. S6, Supplementary Material online), consistent with the expectation that the longer the duplications, the greater the length of microhomologous sequences is needed to stabilize the annealed end before ligation (Ottaviani et al. 2014; Vaughn and Bennetzen 2014). In summary, these results suggest that microhomology may be involved in the generation of tandem duplications, probably via microhomology-mediated repairing of DSBs (McVey and Lee 2008; Ottaviani et al. 2014) or slippage strand replication (Darmon and Leach 2014). The prevalence of tandem repeats associated with microhomologous sequences in plant mitogenomes differs from the finding in the rice nuclear genome, in which tandem duplications rarely associate with microhomology (Vaughn and Bennetzen 2014). In the case of rice nuclear genome, tandem duplications are suggested to have formed through the repairing of DSBs via NHEJ (Vaughn and Bennetzen 2014). Future studies should evaluate whether the low abundance of microhomologous sequences is a rule in plant nuclear genomes, and whether different models govern tandem duplications in mitochondrial and nuclear genomes. Such studies would require both genome-wide and population-level scans of representative lineages.
Abundance of Microhomologous Sequences in Plant Mitogenomes
If the presence of microhomology can facilitate tandem duplication, it is then necessary to investigate the abundance of microhomologous sequences across plant mitogenome, and the relationship between microhomologies and genome sizes. In this study, we focused on 1,197 microhomologous sequences with length ≥6 bp, because these sequences were more likely to facilitate the formation of tandem duplicates than the shorter sequences (see above). These microhomologies were found in 1,716 STRs, which tend to be more abundant in intergenic regions (1,211 STRs), followed by RNA genes (60 STRs), and lastly exons (41 STRs) and introns (38 STRs) of protein-coding genes (supplementary table S5, Supplementary Material online). These results suggest that STRs and the associated microhomologies are more prone to accumulation in intergenic mtDNA, most likely because structural variations in intergenic regions are less functional constrained whereas such variations in coding regions will likely be removed by selection (Christensen 2013, 2014). In addition, the structural variations in intergenic regions could be due to different processes of replication, recombination and repair that participate in the maintenance of plant mitogenomes stability, considering that plant mitogenomes are very different in isoforms (lineal, branched, and circular) in contrast for example with human mitogenome (completely circular) (Oldenburg and Bendich 2015).
We noticed that most microhomologies were found in lineage- or mtDNA region-specific STRs. Of the 1,197 screened microhomologies, 1,066 (89.1%) were species specific, found only in STR(s) of one species, and 998 occurred in only one STR (supplementary table S5, Supplementary Material online). Eighteen microhomologies showed high abundance (involved in 5–42 STRs) in mitogenomes of four species, including Nymphaea colorata (12 microhomologies), Cucumis sativus (three microhomologies), and Physochlaina orientalis (two microhomologies) (supplementary table S5, Supplementary Material online). Studies suggest that these three species experienced recent expansion of STRs (Alverson et al. 2011; Dong et al. 2018; Gandini et al. 2019). The lineage- and region-specific use of microhomology is probably determined by availability of microhomologies around breakpoints or secondary structure that determine microhomology usage, and supports fast structural evolution of mitogenome after speciation.
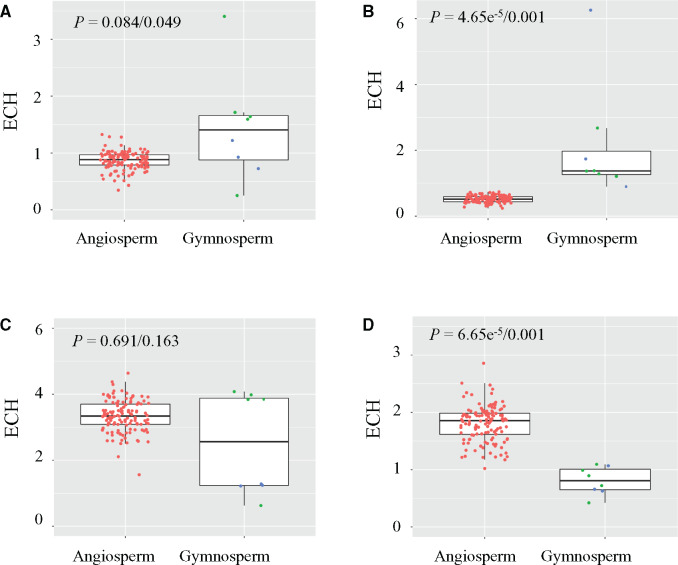
We further examined the enrichment of microhomologies across plant mitogenomes, and found 75% of the 1,197 tested microhomologies were more abundant than expected in at least one mitogenome where the microhomology sequences formed STRs (supplementary table S5, Supplementary Material online). Among the 400 prevalent microhomologies (see Materials and Methods section), 175 showed differential enrichment between angiosperms and gymnosperms and 186 differed between angiosperms and conifers (P < 0.05; Mann–Whitney U test with Benjamini–Hochberg FDR adjustment; fig. 3, supplementary table S4, Supplementary Material online). Moreover, microhomologies identified in angiosperm STRs were more abundant in angiosperm species than in gymnosperm, and vice versa. For example, the TAG7 microhomology identified in the nad7-1 region of P. tabuliformis was more highly enriched in conifers (mean ECH = 1.72) than in angiosperms (mean ECH = 0.867; P = 0.0427, Mann Whitney U test with Benjamini–Hochberg FDR adjustment; fig. 3A, supplementary table S4, Supplementary Material online). The estimated ECH value (3.40) was the highest for the mitogenome of P. taeda, with 595 observed TAG7 copies, which was 3-fold greater than that expected by chance (175 copies). These results suggest that microhomologies have undergone lineage-specific expansion likely caused by microhomology-mediated tandem duplications in the mitogenome. Previous studies also show that expansion of STRs can occur in a group of closely related species (Gandini et al. 2019). Consider all these findings, PCR amplification of mtDNA segments using primers derived from a reference genome can be unstable and unpredictable due to the high rate of structural variation in plants.
Fig. 3.
Differential enrichment of microhomologies in mitogenomes of angiosperms, gymnosperms, and conifers. (A) Microhomology “TAAAGGT” (TAG7); (B) “ATATACG”; (C) “AGCAAGC”; (D) microhomology “AGTCTTC.” Each red, green, and blue dot represents a representative angiosperm, conifer, and nonconifer gymnosperm, respectively. P values of Mann Whitney U tests after Benjamini–Hochberg correction for ECH values of angiosperm vs. gymnosperm and angiosperm vs. conifer are shown in each boxplot. The top, middle, and bottom horizontal lines in each boxplot indicate the first quartile, median, and third quartile, respectively.
Conclusions
This study investigated the structural evolution of mitogenomes in pines and other plant taxa. We show that STRs can result in high polymorphism characterized by complex structural variation. These variants are species- and population-specific and form rapidly following speciation and during subsequent divergence. We illustrate that tandem repeats contribute to mitogenome expansion in plants, and that many repeats are associated with microhomologies. A substantial portion of microhomologies in plant mitogenomes are long enough to result in structural variation, and the length of microhomology is positively correlated with the length of repeat units. These results suggested that microhomology is involved in the formation of tandem duplication. Most of the STR-associated microhomologies are differentially enriched between angiosperms and gymnosperms, indicating the lineage-specific usage and expansion of microhomology. Our results highlight the high prevalence of microhomology and its important role in generating structural variation in mitogenomes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers and Associate Editor for their insightful comments. This work was supported by grants from the Guangdong Natural Science Fund for Distinguished Young Scholar (2018B030306040), Foundation for young talents in Zhongkai University of Agriculture and Engineering, the National Natural Science Foundation of China (NSFC 31800550 and 31971673), and the Swedish Research Council (VR).
Data deposition: This project has been deposited at Genbank under the accession numbers MT792927–MT792949.
Literature Cited
- Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. 2011. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23(7):2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Bastien D, Favre JM, Collignon AM, Sperisen C, Jeandroz S. 2003. Characterization of a mosaic minisatellite locus in the mitochondrial DNA of Norway spruce Picea abies (L.) Karst. Theor Appl Genet. 107(3):574–580. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 57(1):289–300. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, Stark JM. 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32(9):566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26(1):52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, et al. 2008. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol. 25(3):603–615. [DOI] [PubMed] [Google Scholar]
- Chen K, Abbott RJ, Milne RI, Tian XM, Liu JQ. 2008. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol Ecol. 17(19):4276–4288. [DOI] [PubMed] [Google Scholar]
- Christensen AC. 2013. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 5(6):1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. 2014. Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biol Evol. 6(6):1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LW, Guo WH, Mower JP, Palmer JD. 2018. High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Mol Biol Evol. 35:2773–2785. [DOI] [PubMed] [Google Scholar]
- Darmon E, Leach DRF. 2014. Bacterial genome instability. Microbiol Mol Biol Rev. 78(1):1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, et al. 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, et al. 2018. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics 19(1):614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49(3):827–831. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5(6):435–445. [DOI] [PubMed] [Google Scholar]
- Gandini CL, Garcia LE, Abbona CC, Virginia Sanchez-Puerta M. 2019. The complete organelle genomes of Physochlaina orientalis: insights into short sequence repeats across seed plant mitochondrial genomes. Mol Phylogen Evol. 137:274–284. [DOI] [PubMed] [Google Scholar]
- Garcia-Medel PL, et al. 2019. Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining. Nucleic Acids Res. 47:3028–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernandt DS, Lopez GG, Garcia SO, Liston A. 2005. Phylogeny and classification of Pinus. Taxon 54(1):29–42. [Google Scholar]
- Godbout J, Jaramillo-Correa JP, Beaulieu J, Bousquet J. 2005. A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range North American conifer. Mol Ecol. 14(11):3497–3512. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ. 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol. 68(1):225–252. [DOI] [PubMed] [Google Scholar]
- Guo WH, et al. 2016. Ginkgo and welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol. 33(6):1448–1460. [DOI] [PubMed] [Google Scholar]
- Hao Q, et al. 2018. The critical role of local refugia in postglacial colonization of Chinese pine: joint inferences from DNA analyses, pollen records, and species distribution modeling. Ecography 41(4):592–606. [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5(1):e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Correa JP, Aguirre-Planter E, Eguiarte LE, Khasa DP, Bousquet J. 2013. Evolution of an ancient microsatellite hotspot in the conifer mitochondrial genome and comparison with other plants. J Mol Evol. 76(3):146–157. [DOI] [PubMed] [Google Scholar]
- Kan SL, Shen TT, Gong P, Ran JH, Wang XQ. 2020. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): eight protein-coding genes have transferred to the nuclear genome. BMC Evol Biol. 20(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik A, et al. 2019. The alternative reality of plant mitochondrial DNA: one ring does not rule them all. PLoS Genet. 15(8):e1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Huq E, Herrin DL. 2010. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci U S A. 107(31):13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 79(1):181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, et al. 2014. Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines. PLoS One 9(1):e85920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JF, Wang XR. 2011. Distinct niche divergence characterizes the homoploid hybrid speciation of Pinus densata on the Tibetan Plateau. Am Nat. 177(4):424–439. [DOI] [PubMed] [Google Scholar]
- Marechal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186(2):299–317. [DOI] [PubMed] [Google Scholar]
- McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24(11):529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, et al. 2013. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 23(5):749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15(3):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa S, Kubo T, Mikami T. 2000. Variable number of tandem repeat loci in the mitochondrial genomes of beets. Curr Genet. 37(1):34–38. [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. 2015. DNA maintenance in plastids and mitochondria of plants. Front Plant Sci. 6:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D, LeCain M, Sheer D. 2014. The role of microhomology in genomic structural variation. Trends Genet. 30(3):85–94. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. 1988. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol. 28(1–2):87–97. [DOI] [PubMed] [Google Scholar]
- Palmer JD, et al. 2000. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci U S A. 97(13):6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Mathieu C, Gérard-Hirne C, De Paepe R, Chétrit P. 2005. Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J Biol Chem. 280(28):25994–26001. [DOI] [PubMed] [Google Scholar]
- Potter KM, Hipkins VD, Mahalovich MF, Means RE. 2013. Mitochondrial DNA haplotype distribution patterns in Pinus ponderosa (Pinaceae): Range-wide evolutionary history and implications for conservation. Am J Bot. 100(8):1562–1579. [DOI] [PubMed] [Google Scholar]
- Schiml S, Fauser F, Puchta H. 2016. Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proc Natl Acad Sci U S A. 113(26):7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen A, Mackenzie S. 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19(4):1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher G, Tamura K, Kumar S. 2020. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 37(4):1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AR, et al. 2020. The mitogenome of Norway spruce and a reappraisal of mitochondrial recombination in plants. Genome Biol Evol. 12(1):3586–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N. 2017. CNVs and microsatellite DNA polymorphism In: Saitou N, editor. Evolution of the human genome I: the genome and genes. Tokyo, Japan: Springer; p. 143–155. [Google Scholar]
- Taylor ZN, Rice DW, Palmer JD. 2015. The complete moss mitochondrial genome in the Angiosperm Amborella is a chimera derived from two moss whole-genome transfers. PLoS One 10(11):e0137532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, López-Pujol J, Wang H-W, Ge S, Zhang Z-Y. 2010. Molecular evidence for glacial expansion and interglacial retreat during Quaternary climatic changes in a montane temperate pine (Pinus kwangtungensis Chun ex Tsiang) in southern China. Plant Syst Evol. 284(3–4):219–229. [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 15(1):57–61. [DOI] [PubMed] [Google Scholar]
- Vaughn JN, Bennetzen JL. 2014. Natural insertions in rice commonly form tandem duplications indicative of patch-mediated double-strand break induction and repair. Proc Natl Acad Sci U S A. 111(18):6684–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mao JF, Gao J, Zhao W, Wang XR. 2011. Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata. Mol Ecol. 20(18):3796–3811. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang XR. 2014. Mitochondrial DNA capture and divergence in Pinus provide new insights into the evolution of the genus. Mol Phylogen Evol. 80:20–30. [DOI] [PubMed] [Google Scholar]
- Wynn EL, Christensen AC. 2019. Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3 (Bethesda) 9(2):549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, et al. 2018. Combining mitochondrial and nuclear genome analyses to dissect the effects of colonization, environment, and geography on population structure in Pinus tabuliformis. Evol Appl. 11(10):1931–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying TS, Chen ML, Chang HC. 2004. Atlas of the gymnosperms of China. Beijing: China Science and Technology Press. [Google Scholar]
- Young ND, Healy J. 2003. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Guan Y. 2010. Polymerization behavior of Klenow fragment and Taq DNA polymerase in short primer extension reactions. Acta Biochim Biophys Sin. 42(10):722–728. [DOI] [PubMed] [Google Scholar]
- Zhou YF, et al. 2010. Gene flow and species delimitation: a case study of two pine species with overlapping distributions in southeast China. Evolution 64:2342–2352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.