Abstract
The Asian citrus psyllid Diaphorina citri (Insecta: Hemiptera: Psylloidea), a serious pest of citrus species worldwide, harbors vertically transmitted intracellular mutualists, Candidatus Profftella armatura (Profftella_DC, Gammaproteobacteria: Burkholderiales) and Candidatus Carsonella ruddii (Carsonella_DC, Gammaproteobacteria: Oceanospirillales). Whereas Carsonella_DC is a typical nutritional symbiont, Profftella_DC is a unique defensive symbiont with organelle-like features, including intracellular localization within the host, perfect infection in host populations, vertical transmission over evolutionary time, and drastic genome reduction down to much less than 1 Mb. Large parts of the 460-kb genome of Profftella_DC are devoted to genes for synthesizing a polyketide toxin; diaphorin. To better understand the evolution of this unusual symbiont, the present study analyzed the genome of Profftella_Dco, a sister lineage to Profftella_DC, using Diaphorina cf. continua, a host psyllid congeneric with D. citri. The genome of coresiding Carsonella (Carsonella_Dco) was also analyzed. The analysis revealed nearly perfect synteny conservation in these genomes with their counterparts from D. citri. The substitution rate analysis further demonstrated genomic stability of Profftella which is comparable to that of Carsonella. Profftella_Dco and Profftella_DC shared all genes for the biosynthesis of diaphorin, hemolysin, riboflavin, biotin, and carotenoids, underlining multiple roles of Profftella, which may contribute to stabilizing symbiotic relationships with the host. However, acyl carrier proteins were extensively amplified in polyketide synthases DipP and DipT for diaphorin synthesis in Profftella_Dco. This level of acyl carrier protein augmentation, unprecedented in modular polyketide synthases of any known organism, is not thought to influence the polyketide structure but may improve the synthesis efficiency.
Keywords: Diaphorina, defensive symbiont, reduced genome, secondary metabolite, diaphorin, hemolysin
Significance
Profftella is a highly unique bacterial symbiont found in the Asian citrus psyllid, a notorious agricultural pest. It has features like organelles but unusually produces toxins to protect the host insect. To better understand the evolution of this enigmatic symbiont, we sequenced the genome of a novel strain of Profftella recently found in another psyllid species. The comparative analysis of the two Profftella strains revealed that they share genes for the biosynthesis of toxins, vitamins, and carotenoids, highlighting multiple roles of Profftella. These collective benefits to the host may contribute to the stabilization of symbiotic relationships, leading to the organelle-like status of Profftella. The analysis also revealed unusual structures of toxin-producing enzymes, which may potentially improve the toxin production efficiency.
Introduction
Animals exhibit diverse symbiotic relationships with microbes, among which the most intimate ones predominantly have a nutritional basis (Moran et al. 2008; McFall-Ngai et al. 2013; Morris et al. 2019). For animals feeding only on diets that are deficient in essential nutrients, supplementation of nutritional deficiencies is essential for their survival. Thus, microbes that are able to supply such nutrients can easily become vital to the host animals, resulting in evolutionarily stable nutritional symbionts. Extreme cases are exemplified by bacteriome-associated symbionts in insects (Douglas 1989; Moran et al. 2008; Moran and Bennett 2014; McCutcheon et al. 2019). Various insect lineages that feed on nutritionally restricted diets, including plant sap and vertebrate blood, have a specialized organ called a bacteriome, and depend on nutritional supply by “primary symbionts” housed therein. The primary symbionts are taxonomically diverse in distinct host lineages (e.g., aphids, weevils, tsetse flies, etc.), indicating that they have evolved repeatedly from diverse free-living microbes (Moran et al. 2008; Moran and Bennett 2014; McCutcheon et al. 2019). They are mostly bacterial and are characterized by organelle-like features, including intracellular localization within the host cell, perfect infection in host populations, host–symbiont cospeciation reflecting strictly vertical transmission over evolutionary time, and drastic genome reduction down to <1 Mb (Moran et al. 2008; Moran and Bennett 2014; McCutcheon et al. 2019). Mutually indispensable associations between the hosts and the primary symbionts have been demonstrated by physiological (Nogge 1981; Douglas 1989; Nakabachi and Ishikawa 1999) and omics analyses (Nakabachi et al. 2005; Moran et al. 2008; Ramsey et al. 2010; Shigenobu et al. 2010; Moran and Bennett 2014; Wilson and Duncan 2015; McCutcheon et al. 2019). In some cases, metabolic complementarity is achieved, at least in part, through horizontal gene transfer between the host and symbionts (Nikoh and Nakabachi 2009; Nikoh et al. 2010; Husnik et al. 2013; Sloan et al. 2014; Nakabachi et al. 2014; Nakabachi 2015; Mao et al. 2018).
Another major category of animal-associated microbes is represented by defensive symbionts (Piel 2009; Flórez et al. 2015; Helfrich and Piel 2016; Rust et al. 2020). They protect hosts from natural enemies, including predators, parasites, parasitoids, and microbial pathogens, using biologically active compounds such as toxins and antibiotics. In contrast to nutritional symbionts, the status of defensive symbionts tends to be unstable, probably because 1) they are only conditionally beneficial and not essential to the host, 2) the status of natural enemies in the environment can vary continuously, and 3) defensive toxins potentially place a burden on the host. Thus, most defensive symbionts, reported from various insects and marine invertebrates thus far, exhibit imperfect infection frequencies in host populations (Moran et al. 2008; Piel 2009; Oliver et al. 2010; Kwan et al. 2012; Flórez et al. 2015; Johnson 2015; Ballinger and Perlman 2019), and their genomes are often larger than 1 Mb (Wu et al. 2004; Degnan et al. 2009; Kwan et al. 2012; Wilson et al. 2014; Lopera et al. 2017; Flórez et al. 2018; Waterworth et al. 2020), even when they can be vertically transmitted. By contrast, an unprecedented type of defensive symbiont was found in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Psylloidea: Liviidae).
Diaphorina citri is an important agricultural pest that transmits Candidatus Liberibacter spp. (Alphaproteobacteria: Rhizobiales), which cause a devastating disease of citrus crops known as huanglongbing or greening disease (Grafton-Cardwell et al. 2013). Diaphorina citri possesses a bacteriome (Nakabachi et al. 2010) that harbors two distinct species of vertically transmitted intracellular symbionts: Candidatus Carsonella ruddii (Gammaproteobacteria: Oceanospirillales) and Candidatus Profftella armatura (Gammaproteobacteria: Burkholderiales) (Nakabachi, Ueoka, et al. 2013; Dan et al. 2017). The primary symbiont Carsonella, which is thought to be present in all psyllid species, is a typical nutritional symbiont, providing its host with essential amino acids that are scarce in the diet of phloem sap (Nakabachi et al. 2006; Sloan and Moran 2012b; Nakabachi, Ueoka, et al. 2013). The secondary symbiont Profftella is found in all D. citri individuals across the world and has a very much reduced genome of 460 kb (Nakabachi, Ueoka, et al. 2013). Although this is generally characteristic of nutritional symbionts associated with bacteriomes (Moran et al. 2008; Moran and Bennett 2014; McCutcheon et al. 2019), the genome of Profftella encodes only a few genes required to complement the psyllid diet. Instead, large parts of the genome are composed of genes for synthesizing a secondary metabolite; diaphorin (Nakabachi, Ueoka, et al. 2013). Diaphorin is a polyketide belonging to the pederin family of cytotoxins that are found in a diverse array of host–symbiont systems, including beetles, lichens, and sponges harboring phylogenetically diverse bacterial producers (Helfrich and Piel 2016; Rust et al. 2020). Physiological studies have shown that diaphorin is significantly toxic to various organisms, including natural enemies of D. citri (Nakabachi, Ueoka, et al. 2013; Nakabachi and Fujikami 2019; Nakabachi and Okamura 2019; Yamada et al. 2019). Therefore, Profftella is considered to be an unprecedented type of defensive symbiont with organelle-like features. Moreover, the Liberibacter lineage, except for the most basal species Liberibacter crescens, is shown to have horizontally acquired a gene for a putative amino acid transporter from the Profftella lineage (Nakabachi, Nikoh, et al. 2013). This indicates ecological and evolutionary interactions between the huanglongbing pathogen and the bacteriome symbiont. Thus, comparative genomics of Profftella lineages aiming to better understand the evolutionary trajectory of this unique symbiont is desired.
Our previous study using Illumina sequencing of 16S rRNA gene amplicons demonstrated that Diaphorina cf. continua, a psyllid species closely related to D. citri, possesses a bacterial lineage that is sister to the Profftella of D. citri (Nakabachi et al. 2020). Diaphorina citri and D. cf. continua are different in their geographical distributions and host plants. Whereas D. citri is native to tropical and subtropical South to East Asia, and absent from Europe (Grafton-Cardwell et al. 2013), D. cf. continua occurs in the Mediterranean region (Nakabachi et al. 2020). Also, whereas D. citri feeds and develops on rutaceous plants including Citrus spp. (Grafton-Cardwell et al. 2013), D. cf. continua is associated with Thymelaea tartonraira (Thymelaeaceae), which is distantly related to the Rutaceae (Nakabachi et al. 2020). In the present study, to obtain insights into the evolution of Profftella, the genome of Profftella from D. cf. continua (Profftella_Dco) was analyzed along with that of Carsonella (Carsonella_Dco).
Materials and Methods
Insects and DNA Preparation
The material of Diaphorina cf. continua was collected in Corsica island (France, Haute-Corse department) near Moltifao village (42°29′12″N, 9°8′22″E, 300 m a.s.l.) on April 9, 2017. Many adults and several nymphs were present on T. tartonraira subsp. thomasii (Thymelaeaceae), suggesting that this plant taxon is the host for the psyllid species. The specimens have been tentatively identified as D. continua Loginova, based on morphology and host plant data (Loginova 1972; Burckhardt 1984; Rapisarda 1991; Conci et al. 1993), but their identity needs to be confirmed by a taxonomic revision (Nakabachi et al. 2020). DNA was extracted from whole bodies of mixed sex (three males and eight females) of adult D. cf. continua using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The quality of extracted DNA was assessed using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and the quantity was assessed using a Qubit 2.0 Fluorometer with a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific).
Sequencing and Assembly
DNA extracted from D. cf. continua was sheared to ∼500 bp using a Covaris M220 Focused-ultrasonicator (Covaris, Woburn, MA). After a paired-end library was generated with 15 cycles of polymerase chain reaction amplification using a KAPA HyperPrep Kit (KAPA Biosystems, Wilmington, MA), 300 bp of each end of the library was sequenced using a MiSeq instrument (Illumina, San Diego, CA) and MiSeq Reagent Kit v3 (600 cycles; Illumina). Subsequently, BlastN searches were performed using the obtained reads as queries and the genome sequences of Carsonella from ten psyllid lineages (AP009180.1, CP003541.1, CP003542.1, CP003543.1, CP003544.1, CP003545.1, CP003467.1, CP012411.1, CP019943.1, CP024798.1) and Profftella from two strains of D. citri (chromosome: CP003468.1 and CP012591.1; plasmid: CP003469.1 and CP012592.1) as subjects. Read pairs with e-value scores lower than 1.0E-5 in either read, and those with GC content below 30% were collected. Adapters of filtered reads were removed using cutadapt (Martin 2011) with -m 10 -e 0.2 options. After sequencing errors were corrected using bfc (Li 2015) with default parameters, the reads were assembled using Newbler v2.9 (Roche Diagnostics, Rotkreuz, Switzerland) with -mi 99 -ml 100 -large options. Obtained scaffolds and contigs were manually combined using GenoFinisher (Ohtsubo et al. 2012). Gaps were closed using polymerase chain reaction and Sanger sequencing.
Annotation and Structural Analysis of the Genomes
Initial gene predictions and annotations were conducted using DFast pipeline (Tanizawa et al. 2018), followed by manual corrections with the aid of RNAmmer 1.2 Server (Lagesen et al. 2007), the National Center for Biotechnology Information (NCBI) ORFfinder (Wheeler et al. 2003), and BLAST searches. Functional annotation of the predicted genes was conducted using EggNOG 4.5.1 (Huerta-Cepas et al. 2017). Protein structures were analyzed using NCBI Conserved Domain Database (CDD) (Lu et al. 2020), Pfam (El-gebali et al. 2019), and PROSITE (Sigrist et al. 2013). Metabolic pathways were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa 2019) and UniProt Knowledgebase (UniProtKB) (UniProt Consortium 2019). Dinucleotide bias and GC skew were analyzed using ArcWithColor 1.47 (Ohtsubo et al. 2008). The codon adaptation index (CAI) was calculated using the CAIcal server (Puigbo et al. 2008). Pairwise comparisons between the symbiont genomes from D. cf continua and D. citri were performed using GenomeMatcher 2.3 (Ohtsubo et al. 2008), in which BlastN of all-against-all bl2seq similarity searches was conducted with the parameter set “-F F -W 21 -e 1.0e-10.”
Substitution Rate Analysis
Amino acid sequences were deduced from the protein-coding sequences (CDSs) shared between the symbiont genomes from D. cf. continua and D. citri and then aligned with MAFFT 7.452 (Katoh and Standley 2013) using the E-INS-i algorithm with default parameters. The resulting protein alignments were converted to nucleotide alignments using PAL2NAL 13.0 (Suyama et al. 2006). Nonsynonymous (dN) and synonymous (dS) substitution rates and dN/dS ratios between orthologous pairs were calculated using the KaKs_Calculator 1.2 package (Zhang et al. 2006) implementing the YN model (Yang and Nielsen 2000). All statistical analyses were performed using the R software version 3.6.3 (R Core Team 2020, https://www.r-project.org; last accessed August 24, 2020).
Analysis of Polyketide Synthase Genes
The domain architecture and function of polyketide synthase (PKS) gene products were derived by analyses with the trans-acyltransferase (AT) PKS annotation and structure prediction tool TransATor (Helfrich et al. 2019) (https://transator.ethz.ch/; last accessed August 24, 2020), protein alignments with the pederin PKS using the MUSCLE algorithm (Edgar 2004) as implemented in Geneious 8 (https://www.geneious.com/; last accessed August 24, 2020), and NCBI BLAST searches.
Phylogenetic Analysis
Protein sequences were aligned with MAFFT 7.452 (Katoh and Standley 2013) using the E-INS-i algorithm with default parameters. Amino acid sites corresponding to alignment gap(s) were omitted from the data set. Phylogenetic trees were inferred by the maximum likelihood method using RAxML 8.2.12 (Stamatakis 2014) with 1,000 replicates using the WAG (Whelan and Goldman) matrix of the amino acid replacements, assuming a proportion of invariant positions and four gamma-distributed rates (WAG+I+gamma model). Taxonomic assignment of bacteria was based on the Genome Taxonomy Database (GTDB), in which the former class Betaproteobacteria is reclassified as Burkholderiales, an order within the class Gammaproteobacteria (Parks et al. 2018).
Results and Discussion
Basic Features of the Profftella_Dco Genome
The analysis using a total of 716 Mb in 1.54 million MiSeq reads followed by Sanger sequencing identified the complete chromosome sequence of Profftella_Dco consisting of 469,264 bp (24.4% GC), and its plasmid sequence consisting of 5,952 bp (23.1% GC) with a single gap (table 1, supplementary fig. S1, Supplementary Material online). The gap left in the plasmid corresponds to an ∼900-bp region encoding two hypothetical proteins in the plasmid of Profftella_DC (Nakabachi, Ueoka, et al. 2013). This nearly complete genome encodes 355 predicted protein-coding sequences (CDSs), 10 putative pseudogenes, a single rRNA operon, and 34 tRNAs (table 1, supplementary table S1, Supplementary Material online). A genome-wide alignment of homologous nucleotide sequences in Profftella_Dco and Profftella_DC revealed that these genomes retain highly conserved synteny (fig. 1), indicating that most genes are shared between Profftella_Dco and Profftella_DC, and essentially no genome rearrangements have occurred since they diverged. Between these genomes, a total of 377 pairs of orthologous genes are 91.7% identical on average at the nucleotide level, and amino acid sequences of 341 pairs of orthologous CDSs are 90.0% identical on average. Profftella_Dco and Profftella_DC share all the genes involved in the biosynthesis of diaphorin, hemolysin, riboflavin, biotin, and carotenoids, which will be further discussed later.
Table 1.
Genomic Features of Profftella and Carsonella from Diaphorina cf. continua and Diaphorina citri
| Profftella_Dco | Profftella_DC | Carsonella_Dco | Carsonella_DC | |
|---|---|---|---|---|
| Chromosome size (bp) | 469,264 | 459,399 | 173,853 | 174,014 |
| Plasmid size (bp) | >5,952 | 5,458 | — | — |
| G+C content [plasmid] (%) | 24.4 [23.1] | 24.2 [23.9] | 17.9 | 17.6 |
| CDS [plasmid] | 355 [4] | 365 [6] | 202 | 207 |
| rRNA | 3 | 3 | 3 | 3 |
| tRNA | 34 | 34 | 28 | 27 |
Fig. 1.
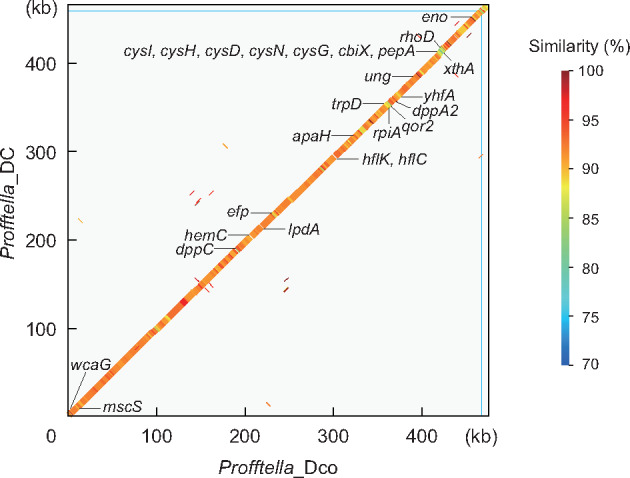
Comparison of the genomic structures of Profftella_Dco and Profftella_DC. The genomes of Profftella_Dco and Profftella_DC are represented by the x and y axes, respectively. The thick line indicates the shared synteny between the two genomes. The color of the line indicates the percentage similarity between the nucleotide sequences. For each genome, the chromosome and the plasmid were concatenated, the borders of which are indicated by thin blue lines. The genes found in Profftella_Dco, but not in Profftella_DC, are presented below the line plot; the genes present in Profftella_DC, but not in Profftella_Dco, are shown above the line plot.
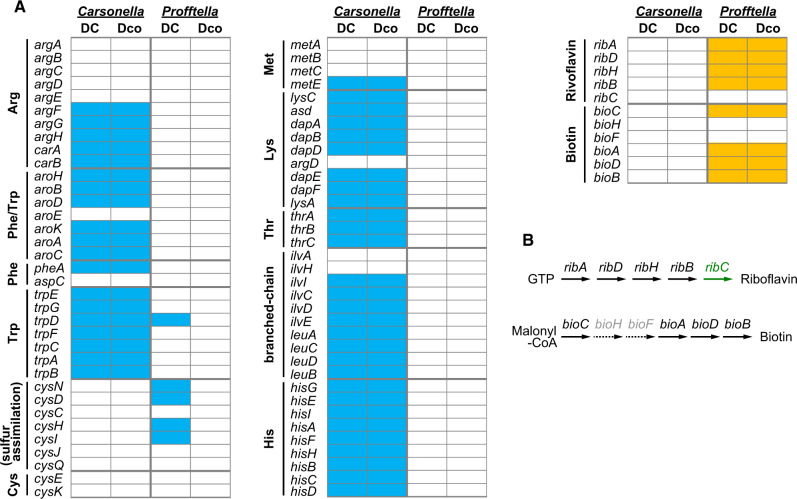
Despite this high level of conservation between the genomes, random gene silencing appears to be still ongoing in Profftella_Dco and Profftella_DC. The genes found in one of the Profftella lineages but not in the other are shown in figure 1 and supplementary table S2, Supplementary Material online. BLAST searches and phylogenetic analyses of these genes revealed no sign of horizontal acquisition following the divergence of these symbionts, indicating that the different gene sets reflect gene silencing on either genome. The functional categories of these genes vary (supplementary table S2, Supplementary Material online), implying that the gene silencing has been randomly occurring. However, it may be notable that the genes for sulfur assimilation (cysDHIN) are retained in Profftella_DC but are lost in Profftella_Dco (figs. 1 and 2A). These genes could potentially contribute to the synthesis of sulfur-containing amino acids, cysteine and methionine, the latter of which is an essential amino acid that the host psyllids are unable to synthesize. However, no other genes related to amino acid synthesis are retained in the Profftella genome. Moreover, Carsonella_Dco and Carsonella_DC lack genes for the biosynthesis of cysteine/methionine other than metE, which converts homocysteine into methionine (fig. 2A). Therefore, the cysteine/methionine synthesis pathway appears to be incomplete, even with the aid of the host psyllids (Sloan et al. 2014), thus making the role of the conservation of cysDHIN genes in Profftella_DC unclear.
Fig. 2.
The repertoire of Carsonella and Profftella genes that are potentially involved in providing nutrition. (A) Amino acid and vitamin biosynthetic genes in the genomes of Carsonella and Profftella from Diaphorina cf. continua and Diaphorina citri. Filled and open boxes indicate the presence and absence of genes, respectively. (B) B vitamin synthetic pathways reconstructed from genes retained in the genomes of Profftella_Dco and Profftella_DC. Genes shown in black type are found in the Profftella genomes. Those in gray type appear to be absent in the genomes. The ribC shown in green type is a host psyllid gene that was horizontally acquired from an unknown bacterium.
Polyketide Biosynthesis in Profftella_Dco
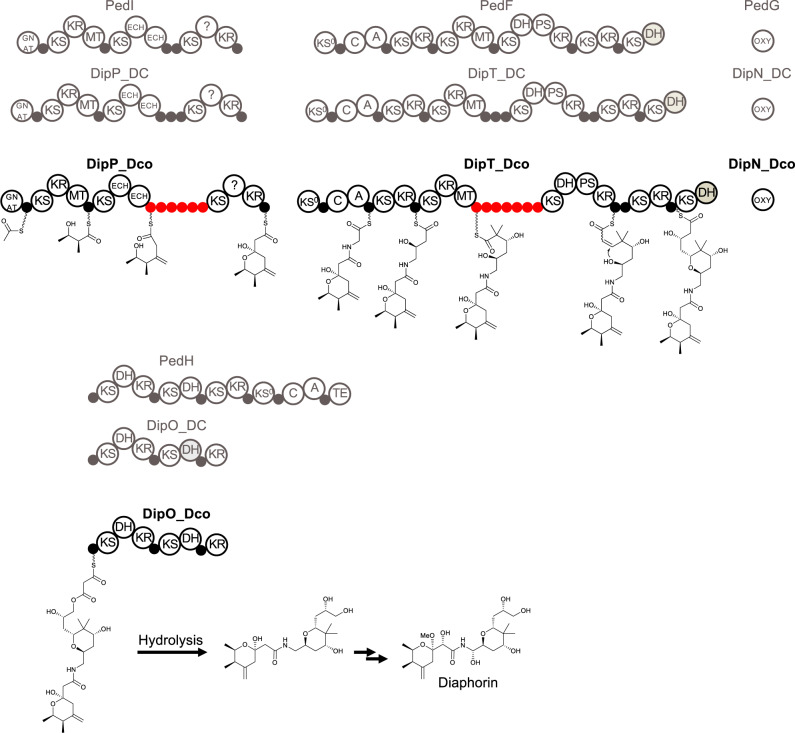
Twenty genes (dipA-T) constituting the PKS biosynthetic gene clusters are perfectly conserved in Profftella_Dco, manifesting that polyketide synthesis is an important function of the Profftella lineages (supplementary fig. S1A and table S1, Supplementary Material online). As in Profftella_DC (Nakabachi, Ueoka, et al. 2013), the PKS system is separated into two loci within the Profftella_Dco chromosome (supplementary fig. S1A, Supplementary Material online). These loci correspond to 15.4% (72,207/469,264 bp) of the chromosome. The genes from Profftella_Dco and Profftella_DC form highly supported clades in the constructed phylogenetic tree (supplementary fig. S2, Supplementary Material online), and domain structures of their encoded proteins are also perfectly conserved, except that acyl carrier proteins (ACPs) are extensively amplified in DipP and DipT of Profftella_Dco (fig. 3, supplementary text S1, Supplementary Material online). DipP, which is orthologous to PedI of the Pseudomonas (Gammaproteobacteria: Pseudomonadales) symbiont of Paederus rove beetles (Coleoptera: Staphylinidae) (Piel et al. 2004), is an enzyme with four modules that catalyzes the initiation of the synthesis, and three steps of the extension, of the polyketide chain (fig. 3). DipT, which is orthologous to PedF of the Paederus symbiont (Piel et al. 2004), is an enzyme with seven modules that assembles the largest part of the diaphorin molecule by catalyzing the extension and modification of the polyketide chain received from DipP (fig. 3). The dip and ped PKS belong to the trans-AT PKS family, in which the recombination of module-encoding gene regions is a common mechanism of metabolic diversification (Nguyen et al. 2008; Ueoka et al. 2015). However, an analysis with the prediction tool TransATor (Helfrich et al. 2019), which proposes polyketide structures based on ketosynthase domains in trans-AT PKSs, demonstrated that both Profftella pathways generate the same compound diaphorin (fig. 3).
Fig. 3.
Architecture of the PKS proteins from Profftella_Dco (Dip_Dco, shown in bold), with the predicted biosynthetic pathway for diaphorin in Profftella_Dco. For comparison, PKS orthologs from Profftella_DC (Dip_DC) and the Paederus symbiont (Ped) are shown in gray, above Dip_Dcos. A, nonribosomal peptide synthetase (NRPS) adenylation domain; C, NRPS condensation domain; DH, dehydratase; GNAT, GCN5-related N-acetyltransferase superfamily (usually serving as acetyltransferase in PKSs); ECH, enoyl-CoA reductase-like domain; KR, ketoreductase; KS, ketosynthase; KS0, nonelongating KS; MT, C-methyltransferase; PS, putative pyrane synthase; OXY, oxygenase; TE, thioesterase; ?, region of unknown function. The domains in gray are predicted to be inactive due to missing active-site residues. The small circles denote carrier protein domains including ACPs and PCPs (NRPS peptidyl carrier proteins). Unusually amplified ACPs in DipP_Dco and DipT_Dco are highlighted in red (see also supplementary text S1, Supplementary Material online).
Whereas PedI has two ACP domains between the enoyl-CoA reductase-like domain and the ketosynthase domain, DipP of Profftella_DC (DipP_DC) has three ACPs at the corresponding site. Furthermore, there are as many as six ACP domains at the corresponding site of DipP from Profftella_Dco (DipP_Dco) (fig. 3, supplementary text S1, Supplementary Material online). Similarly, whereas PedF has only a single ACP domain between the C-methyltransferase and ketosynthase domains, DipT_DC has three, and DipT_Dco has as many as seven ACP domains at the corresponding site (fig. 3, supplementary text S1, Supplementary Material online). Large ACP series also occurs in the PKS-like monomodular polyunsaturated fatty acid synthases (Jiang et al. 2008; Hayashi et al. 2016), and ACP pairs or triplets have been identified in some trans-AT PKS modules (Gu et al. 2011; Helfrich and Piel 2016). However, to our knowledge, extremely amplified ACPs in modular PKSs like these are not known from any other organism. It is shown for polyunsaturated fatty acid synthases as well as trans-AT PKSs that the production titers can correlate with ACP numbers in repeat regions (Jiang et al. 2008; Gu et al. 2011; Hayashi et al. 2016; Helfrich and Piel 2016). The amplification of ACP domains in the Profftella_Dco as compared with Profftella_DC may therefore improve the efficiency of diaphorin synthesis. It would be interesting to assess this possibility in future studies.
Another Gene Potentially Related to Toxicity
Profftella_Dco and Profftela_DC are shown to have tlyC ortholog encoding a protein with the CorC_HlyC domain, the DUF21 domain, and two tandem repeats of the cystathionine beta-synthase (CBS pair) domains (fig. 4). The set of these domains constitutes the characteristic feature of the TlyC superfamily of hemolysin and related proteins (Bateman 1997). Hemolysins are bacterial toxins that cause lysis of red blood cells by destroying their cell membrane, and the TlyC (hemolysin C) of Brachyspira hyodysenteriae (Spirochaetota), a causative agent of swine dysentery, was first found to exhibit hemolytic and cytotoxic activities (ter Huurne et al. 1994). Subsequently, similar activities including disruption of cytoplasmic and intracellular membranes were observed for TlyC orthologs of typhus pathogens, Rickettsia typhi (Alphaproteobacteria) and Rickettsia prowazekii (Radulovic et al. 1999; Whitworth et al. 2005). These findings suggest that TlyCs interact with the host cell membrane and rupture it although its mechanism is not defined (ter Huurne et al. 1994; Radulovic et al. 1999; Whitworth et al. 2005). The TlyCs of Profftella_Dco and Profftella_DC are 34.6% and 32.8% identical, respectively, to that of B. hyodysenteriae (fig. 4). Therefore, they may also have membranolytic activity, which could potentially function as a defensive toxin, like diaphorin, to protect the holobiont (host + symbionts) from natural enemies. Another possibility is that the TlyCs are involved in manipulating the host membrane, for example, at the process of escaping from a large central syncytium of the bacteriome and entering into the oocytes during transovarial transmission (Dan et al. 2017).
Fig. 4.
Amino acid sequences of TlyC orthologs from Profftella_Dco and Profftella_DC, which are aligned with that from Brachyspira hyodysenteriae, whose membranolytic activity has been experimentally confirmed (ter Huurne et al. 1994). Sequences are aligned with MAFFT 7.452 using the E-INS-i algorithm with default parameters. Identical, conservative, and semiconservative residues are marked with asterisks, double dots, and single dots, respectively. DUF21, CBS pair, and CorC_HlyC domains are indicated with blue, green, and orange shades, respectively. Conserved domain structures are based on information of the NCBI Conserved Domain Database implemented with models imported from external databases, including Pfam, SMART, and COG (Lu et al. 2020).
Carotenoid Biosynthesis in Profftella
The plasmid of Profftella_Dco encodes genes for biosynthesis of carotenoids (crtB, crtI, and crtY) (table 2, fig. 5, and supplementary fig. S1B, Supplementary Material online), which are also found in the plasmid of Profftella_DC (Nakabachi, Ueoka, et al. 2013). As shown in figure 5, crtB encodes phytoene synthase (EC:2.5.1.32), which catalyzes the condensation of two molecules of geranylgeranyl diphosphate to give prephytoene diphosphate and the subsequent rearrangement of the cyclopropylcarbinyl intermediate to yield the 15-cis-phytoene isomer (Fraser and Bramley 2004). The crtI gene encodes phytoene desaturase (EC:1.3.99.31), which converts 15-cis-phytoene into all-trans-lycopene via the intermediates phytofluene, zeta-carotene, and neurosporene, through the introduction of four double bonds (Fraser and Bramley 2004). The crtY gene encodes lycopene beta-cyclase (EC:5.5.1.19), which catalyzes the cyclization of both ends of lycopene to form beta-carotene (fig. 5). Carotenoids are yellow, orange, and red organic pigments that are widely distributed in diverse lineages of organisms on Earth (Fraser and Bramley 2004; Mussagy et al. 2019). In animals, these compounds play important roles, including antioxidation and pigmentation for photoprotection, camouflage, or ornamentation (Fraser and Bramley 2004; Mussagy et al. 2019). They are synthesized by various lineages of bacteria, archaea, protists, fungi, and plants, but animals generally lack the ability to produce carotenoids and must obtain them through their diet. Unique exceptions are aphids and mites, which have horizontally acquired carotenoid biosynthesis genes from fungi (Moran and Jarvik 2010; Altincicek et al. 2012). In addition, the chromosome of Candidatus Portiera aleyrodidarum (Gammaproteobacteria: Oceanospirillales), the primary symbiont of whiteflies (Hemiptera: Aleyrodoidea), is shown to encode crtB, crtI, and crtY (Santos-Garcia et al. 2012; Sloan and Moran 2012a, 2013), which are the same set of gene orthologs found in the Profftella lineages. The gene sets encoded in the genomes of Profftella and Portiera appear to be of bacterial origin and independent of each other (table 2, supplementary fig. S3, Supplementary Material online). The plasmids of Profftella may have been horizontally acquired from other bacterial lineages, but neither phylogenetic analysis of CrtI encoded therein (supplementary fig. S3, Supplementary Material online), GC content (23.1% vs. 24.4% of the chromosome), nor CAI (0.769 ± 0.011 vs. 0.774 ± 0.002 of the chromosome) shows clear evidence of recent horizontal acquisition. In any case, conservation of the carotenoid biosynthetic plasmid in both Profftella_Dco and Profftella_DC indicates that carotenoid synthesis is a crucial function for Profftella.
Table 2.
Carotenoid Biosynthesis Genes in the Profftella_Dco Genome
| Gene ID | Product | Length (aa) | Top BLAST Hit | Source Organism | E-value | Identity (aa %) |
|---|---|---|---|---|---|---|
| crtB | Phytoene synthase | 330 | WP_105077524 | Pantoea ananatis (Gammaproteobacteria) | 4E-96 | 45.8 |
| crtI | Phytoene desaturase | 493 | WP_154058492 | Pseudescherichia vulneris (Gammaproteobacteria) | 0 | 57.8 |
| crtY | Lycopene beta-cyclase | 392 | GAL56634 | Pseudescherichia vulneris (Gammaproteobacteria) | 3E-117 | 44.4 |
Fig. 5.
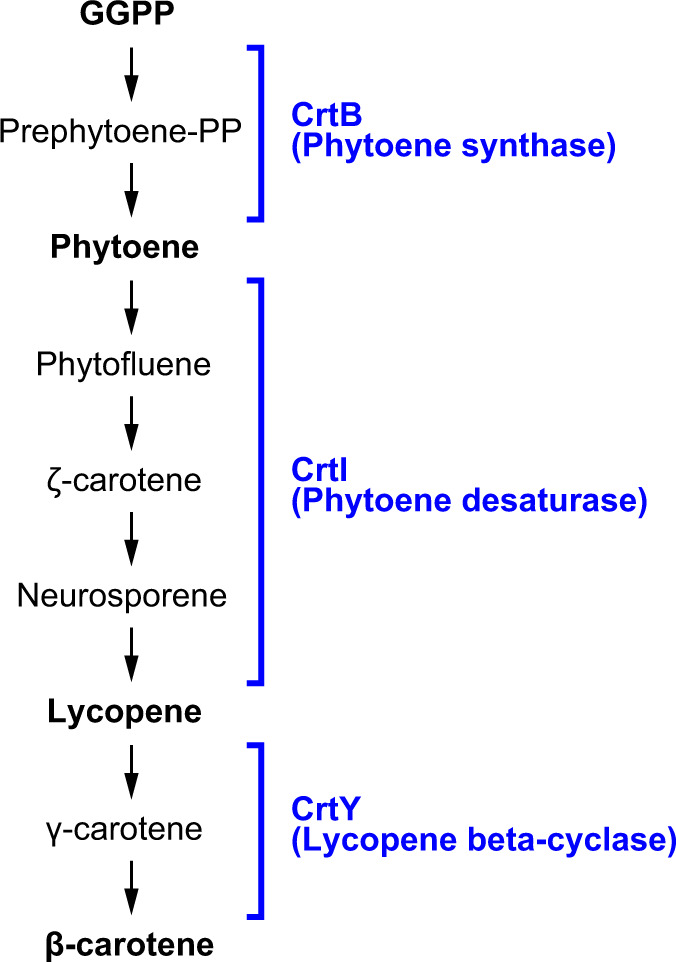
Biosynthetic pathway of carotenoids, the steps of which are catalyzed by CrtB (phytoene synthase), CrtI (phytoene desaturase), and CrtY (lycopene beta-cyclase). GGPP, geranylgeranyl diphosphate.
Previous studies have suggested sister relationships not only between whiteflies and psyllids but also between their primary symbionts Portiera and Carsonella (Spaulding and von Dohlen 1998; Thao and Baumann 2004; Sloan and Moran 2012a). Therefore, the common ancestor of Portiera and Carsonella may have possessed carotenoid biosynthesis genes. However, all Carsonella lineages sequenced to date including Carsonella_Dco (supplementary table S3, Supplementary Material online) and Carsonella_DC lack them (Nakabachi et al. 2006; Sloan and Moran 2012b; Nakabachi, Ueoka, et al. 2013). Thus, the gene set found in the Profftella lineage appears to add another important role to this secondary symbiont. Future studies should focus on the analysis of carotenoid products and their functional roles in Diaphorina psyllids.
Nutritional Contribution of Profftella
Sixteen genes for “coenzyme transport and metabolism (COG category H)” found in Profftella_DC are perfectly conserved in Profftella_Dco (supplementary table S1, Supplementary Material online). The genes in this category include those for the synthesis of riboflavin (ribA, ribB, ribD, and ribH) and biotin (bioA, bioB, bioC, and bioD) (fig. 2). Although both Profftella lineages lack ribC that is required for the final step of riboflavin synthesis, a previous study demonstrated that host psyllids possess ribC that has been horizontally acquired from a bacterium (Sloan et al. 2014), which can potentially complete the biosynthetic pathway of riboflavin (fig. 2B). On the other hand, the pathway for biotin synthesis appears to be incomplete in the psyllid–Profftella–Carsonella symbiotic system (fig. 2B). Still, besides the bio genes, the Profftella lineages possess the birA gene encoding a biotin–[acetyl-CoA-carboxylase] ligase, which is involved in the metabolism of biotin (supplementary table S1, Supplementary Material online). This pattern of gene conservation implies that biosynthesis and metabolism of biotin are important processes in Profftella.
These genes mentioned above are essentially the only contributing factors to Profftella’s role as a nutritional symbiont, which possibly play a crucial role in stabilizing symbiotic relationships between the host and this unique organelle-like defensive symbiont.
Features of the Carsonella_Dco Genome
A total of 0.15 million MiSeq reads yielding 38 Mb were assembled into the complete Carsonella_Dco genome consisting of a single circular chromosome of 173,853 bp with a GC content of 17.9% (table 1, supplementary fig. S4A, Supplementary Material online). It encodes 202 CDSs, a single putative pseudogene, a single rRNA operon, and 28 tRNAs (table 1, supplementary table S3, Supplementary Material online). The whole-genome alignment of Carsonella_Dco and Carsonella_DC (CP003467.1) revealed a strong colinearity between these genomes (supplementary fig. S4B, Supplementary Material online), indicating that there have been no genome rearrangements since the divergence of these symbiont lineages. Similar colinear patterns have also been observed among other Carsonella genomes (Sloan and Moran 2012b; Nakabachi, Ueoka, et al. 2013), as well as in the genomes of other bacteriome-associated primary symbionts (Tamas et al. 2002; Degnan et al. 2005; Rio et al. 2012; Bennett and Moran 2013; Koga and Moran 2014; Husnik and McCutcheon 2016; Mao et al. 2017). All genes with functional annotations are shared between Carsonella_Dco and Carsonella_DC (supplementary table S3, Supplementary Material online). In these genomes, a total of 233 pairs of orthologous genes exhibited 91.9% nucleotide identity on average, and 202 pairs of orthologous CDSs exhibited 89.6% amino acid identity on average.
As previously shown for Carsonella_DC (Nakabachi, Ueoka, et al. 2013), the gene inventory of Carsonella_Dco (fig. 2A) indicates that this symbiont is a typical nutritional symbiont that provides essential amino acids to its host psyllid. Carsonella_Dco and Carsonella_DC share seven genes (trpA–G) constituting the intact tryptophan biosynthesis pathway, and nine genes (hisA–I) encoding the intact histidine biosynthesis pathway, whereas the other Carsonella lineages analyzed to date partially or completely lack these genes (Nakabachi et al. 2006; Sloan and Moran 2012b). In two psyllid species Ctenarytaina eucalypti (Aphalaridae) and Heteropsylla cubana (Psyllidae), missing genes are complemented by secondary symbionts (Gammaproteobacteria: Enterobacterales) that are distantly related to Profftella (Sloan and Moran 2012b). The conservation of the biosynthetic pathways of tryptophan and histidine in Carsonella_Dco and Carsonella_DC indicates the broader metabolic capacities of these primary symbionts and highlights the limited nutritional capacity of Profftella.
Genome-Wide Substitution Rates in Profftella and Carsonella
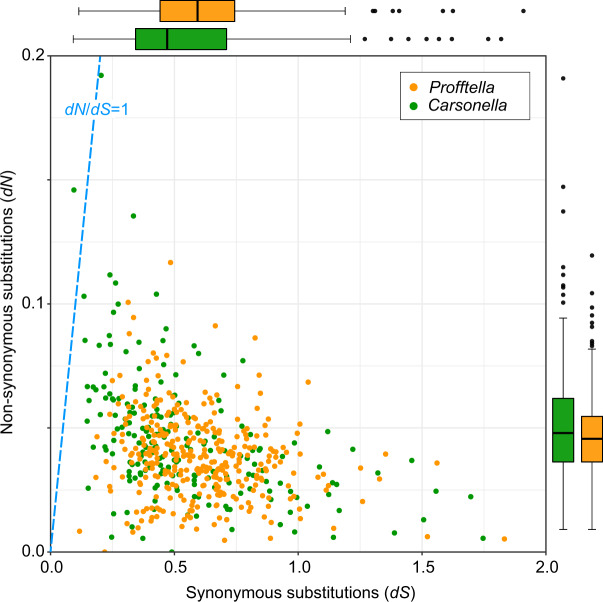
Whereas various lineages of primary symbionts have been repeatedly shown to have highly stable genomic structures (Tamas et al. 2002; Degnan et al. 2005; Rio et al. 2012; Sloan and Moran 2012b; Bennett and Moran 2013; Nakabachi, Ueoka, et al. 2013; Koga and Moran 2014; Husnik and McCutcheon 2016; Mao et al. 2017), this is often not the case for secondary or more recently acquired symbionts (Burke and Moran 2011; Manzano-marin et al. 2012; Bennett et al. 2014; Koga and Moran 2014; Campbell et al. 2015; McCutcheon et al. 2019). However, as mentioned above, the synteny analysis revealed that not only Carsonella but also Profftella retain highly conserved genomic structures (fig. 1 and supplementary fig. S4B, Supplementary Material online). This highly conserved synteny in Profftella may be indicative of genomic stasis in this symbiont and may simply reflect a short divergence time. Thus, to evaluate the stability of these genomes, we further analyzed genome-wide rates of synonymous (dS) and nonsynonymous (dN) substitutions for Profftella and Carsonella, for which the same divergence time can be applied (fig. 6, supplementary tables S1 and S3, Supplementary Material online). The average dS rate was higher (Mann–Whitney U test, P < 0.001) in Profftella (0.605 ± 0.013) than in Carsonella (0.545 ± 0.022). No genes appeared to be at or near saturation for dS (>2.0). On the other hand, the average dN was higher (Mann–Whitney U test, P < 0.05) in Carsonella (0.045 ± 0.002) than in Profftella (0.039 ± 0.001). Accordingly, the average dN/dS ratio was higher (Mann–Whitney U test, P < 0.001) in Carsonella (0.130 ± 0.011) than in Profftella (0.076 ± 0.003). With a single exception of a Carsonella gene encoding a hypothetical protein, all genes were estimated to have dN/dS <1, indicating purifying selection for most genes in these symbionts. Whereas the differences in dS and dN between Profftella and Carsonella were statistically significant, they were much less than double, indicating that Profftella and Carsonella have a similar level of nucleotide substitutions (fig. 6). This is a stark contrast to the cases of Ca. Sulcia muelleri (Bacteroidota) and various lineages of coresident symbionts in which DNA substitution rates are 1–2 orders of magnitude higher in more recently acquired coresidents than in Sulcia (Bennett et al. 2014; Campbell et al. 2015; Silva and Santos-Garcia 2015). The highly conserved synteny and substitution rates that are similar to those of Carsonella imply that Profftella has acquired a relatively stable status, which is comparable to that of obligate primary symbionts.
Fig. 6.
Synonymous (dS) and nonsynonymous (dN) substitution rates inferred from pairwise comparisons of orthologous CDSs in lineages of Profftella (orange dots) and Carsonella (green dots). A total of 341 and 202 orthologous pairs in Profftella and Carsonella, respectively, were used for the analysis. Box plots (Profftella, orange; Carsonella, green) on the x and y axes indicate distributions (median, quartiles, minimum, maximum, and outliers) of dS and dN values, respectively.
Conclusions
The present study revealed that Profftella_Dco and Profftella_DC share all the genes involved in the biosynthesis of diaphorin, hemolysin, riboflavin, biotin, and carotenoids, underlining multiple roles of Profftella, which may contribute to the stabilization of symbiotic relationships with their host psyllids. However, ACPs were extensively amplified in enzymes to synthesize diaphorin in Profftella_Dco. This ACP augmentation, which is unprecedented in modular PKSs of any organism, is not thought to influence the polyketide structure but may improve the efficiency of synthesis. Analyses of synteny and genome-wide substitution rates revealed that the Profftella genome is stable similarly to the Carsonella genome, implying that Profftella has acquired the status that is comparable to that of primary nutritional symbionts.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Naruo Nikoh at the Open University of Japan for his advice on nucleotide substitution analyses. This work was supported by the Japan Society for the Promotion of Science (https://www.jsps.go.jp; last accessed August 24, 2020) KAKENHI (Grant Nos. 26292174 and 20H02998), and research grants from Tatematsu foundation and Nagase Science and Technology Foundation to A.N., and the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (Grant Agreement No. 742739) to J.P. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data deposition: This project has been deposited at DDBJ/EMBL/GenBank databases under the accession numbers AP023215 (Profftella_Dco chromosome), AP023216 (Profftella_Dco plasmid), and AP023214 (Carsonella_Dco).
Literature Cited
- Altincicek B, Kovacs JL, Gerardo NM. 2012. Horizontally transferred fungal carotenoid genes in the two-spotted spider mite Tetranychus urticae. Biol Lett. 8(2):253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger MJ, Perlman SJ. 2019. The defensive Spiroplasma. Curr Opin Insect Sci. 32:36–41. [DOI] [PubMed] [Google Scholar]
- Bateman A. 1997. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 22(1):12–13. [DOI] [PubMed] [Google Scholar]
- Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio 5(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5(9):1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt D. 1984. The Mediterranean species of Diaphorina Loew (Homoptera, Psylloidea). Phytophaga 2:1–30. [Google Scholar]
- Burke GR, Moran NA. 2011. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 3:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, et al. 2015. Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. Proc Natl Acad Sci U S A. 112(33):10192–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conci C, Rapisarda C, Tamanini L. 1993. Annotated catalogue of the Italian Psylloidea. First part. (Insecta Homoptera). Atti Acad Roveretana Agiati, Ser VII. 242(2B):33–135. [Google Scholar]
- Dan H, Ikeda N, Fujikami M, Nakabachi A. 2017. Behavior of bacteriome symbionts during transovarial transmission and development of the Asian citrus psyllid. PLoS One 12(12):e0189779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Lazarus AB, Wernegreen JJ. 2005. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 15(8):1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A. 106(22):9063–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 1989. Mycetocyte symbiosis in insects. Biol Rev. 64(4):409–434. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-gebali S, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórez L, Biedermann PHW, Engl T, Kaltenpoth M. 2015. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 32(7):904–936. [DOI] [PubMed] [Google Scholar]
- Flórez LV, et al. 2018. An antifungal polyketide associated with horizontally acquired genes supports symbiont- mediated defense in Lagria villosa beetles. Nat Commun. 9(1):2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. 2004. The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res. 43(3):228–265. [DOI] [PubMed] [Google Scholar]
- Grafton-Cardwell EE, Stelinski LL, Stansly PA. 2013. Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu Rev Entomol. 58(1):413–432. [DOI] [PubMed] [Google Scholar]
- Gu L, et al. 2011. Tandem acyl carrier proteins in the curacin biosynthetic pathway promote consecutive multienzyme reactions with a synergistic effect. Angew Chem Int Ed Engl. 50(12):2795–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Satoh Y, Ujihara T, Takata Y, Dairi T. 2016. Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci Rep. 6(1):35441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich E, Piel J. 2016. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 33(2):231–316. [DOI] [PubMed] [Google Scholar]
- Helfrich EJN, et al. 2019. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat Chem Biol. 15(8):813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 34(8):2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153(7):1567–1578. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A. 113(37):E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, et al. 2008. The role of tandem acyl carrier protein domains in polyunsaturated fatty acid biosynthesis. J Am Chem Soc. 130(20):6336–6337. [DOI] [PubMed] [Google Scholar]
- Johnson KN. 2015. The impact of Wolbachia on virus infection in mosquitoes. Viruses 7(11):5705–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. 2019. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11):1947–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8(6):1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JC, et al. 2012. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci U S A. 109(50):20655–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2015. BFC: correcting Illumina sequencing errors. Bioinformatics 31(17):2885–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loginova M. 1972. On the fauna of Psylloidea from Morocco (Homoptera). Comment Biol. 47:1–37. [Google Scholar]
- Lopera J, Miller IJ, McPhail KL, Kwan JC. 2017. Increased biosynthetic gene dosage in a genome-reduced defensive bacterial symbiont. mSystems 2(6):e00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, et al. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48(D1):D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-marin A, Lamelas A, Moya A, Latorre A. 2012. Comparative genomics of Serratia spp.: two paths towards endosymbiotic life. PLoS One 7(10):e47274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Yang X, Bennett GM. 2018. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc Natl Acad Sci U S A. 115(50):E11691–E11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Yang X, Poff K, Bennett G. 2017. Comparative genomics of the dual-obligate symbionts from the treehopper, Entylia carinata (Hemiptera: Membracidae), provide insight into the origins and evolution of an ancient symbiosis. Genome Biol Evol. 9(6):1803–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol. 29(11):R485–R495. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 110(9):3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68(1):195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328(5978):624–627. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42(1):165–190. [DOI] [PubMed] [Google Scholar]
- Morris LA, Voolstra CR, Quigley KM, Bourne DG, Bay LK. 2019. Nutrient availability and metabolism affect the stability of coral–Symbiodiniaceae symbioses. Trends Microbiol. 27(8):678–689. [DOI] [PubMed] [Google Scholar]
- Mussagy CU, Winterburn J, Santos-Ebinuma VC, Pereira JFB. 2019. Production and extraction of carotenoids produced by microorganisms. Appl Microbiol Biotechnol. 103(3):1095–1114. [DOI] [PubMed] [Google Scholar]
- Nakabachi A. 2015. Horizontal gene transfers in insects. Curr Opin Insect Sci. 7:24–29. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Fujikami M. 2019. Concentration and distribution of diaphorin, and expression of diaphorin synthesis genes during Asian citrus psyllid development. J Insect Physiol. 118:103931. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima S. 2014. Aphid gene of bacterial origin encodes protein transported to obligate endosymbiont. Curr Biol. 24(14):R640–R641. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. 1999. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 45(1):1–6. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Koshikawa S, Miura T, Miyagishima S. 2010. Genome size of Pachypsylla venusta (Hemiptera: Psyllidae) and the ploidy of its bacteriocyte, the symbiotic host cell that harbors intracellular mutualistic bacteria with the smallest cellular genome. Bull Entomol Res. 100(1):27–33. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Malenovsky I, Gjonov I, Hirose Y. 2020. 16S rRNA sequencing detected Profftella, Liberibacter, Wolbachia, and Diplorickettsia from relatives of the Asian citrus psyllid. Microb Ecol. 80(2):410–422. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Okamura K. 2019. Diaphorin, a polyketide produced by a bacterial symbiont of the Asian citrus psyllid, kills various human cancer cells. PLoS One 14(6):e0218190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci U S A. 102(15):5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314(5797):267. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Nikoh N, et al. 2013. Horizontal gene acquisition of Liberibacter plant pathogens from a bacteriome-confined endosymbiont of their psyllid vector. PLoS One 8(12):e82612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Ueoka R, et al. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 23(15):1478–1484. [DOI] [PubMed] [Google Scholar]
- Nguyen T, et al. 2008. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 26(2):225–233. [DOI] [PubMed] [Google Scholar]
- Nikoh N, Nakabachi A. 2009. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6(2):e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogge G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82:101–104. [Google Scholar]
- Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9(1):376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M. 2012. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol. 194(24):6970–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 55(1):247–266. [DOI] [PubMed] [Google Scholar]
- Parks DH, et al. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 36(10):996–1004. [DOI] [PubMed] [Google Scholar]
- Piel J. 2009. Metabolites from symbiotic bacteria. Nat Prod Rep. 26(3):338–362. [DOI] [PubMed] [Google Scholar]
- Piel J, Wen G, Platzer M, Hui D. 2004. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. Chembiochem 5(1):93–98. [DOI] [PubMed] [Google Scholar]
- Puigbo P, Bravo IG, Garcia-Vallve S. 2008. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct. 3(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic S, Troyer JM, Beier MS, Lau AOT, Azad AF. 1999. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect Immun. 67(11):6104–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JS, et al. 2010. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 19(Suppl 2):241–248. [DOI] [PubMed] [Google Scholar]
- Rapisarda C. 1991. Faunistic and ecological notes on the psyllids of Sardinia. Mem Soc Entomol Ital. 69:7–52. [Google Scholar]
- Rio RV, et al. 2012. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. mBio 3(1):e00240–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M, et al. 2020. A multi-producer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc Natl Acad Sci U S A. 117(17):9508–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Garcia D, et al. 2012. Complete genome sequence of ‘Candidatus Portiera aleyrodidarum’ BT-QVLC, an obligate symbiont that supplies amino acids and carotenoids to Bemisia tabaci. J Bacteriol. 194(23):6654–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, et al. 2010. A full-length cDNA resource for the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. 19:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, et al. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41(D1):D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FJ, Santos-Garcia D. 2015. Slow and fast evolving endosymbiont lineages: positive correlation between the rates of synonymous and non-synonymous substitution. Front Microbiol. 6:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2012. a. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. 8(6):986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2012. b. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 29(12):3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol Evol. 5(5):783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31(4):857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding AW, von Dohlen CD. 1998. Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol Biol Evol. 15(11):1506–1513. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science (80-.) 296(5577):2376–2379. [DOI] [PubMed] [Google Scholar]
- Tanizawa Y, Fujisawa T, Nakamura Y. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34(6):1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Baumann P. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol. 70(6):3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Huurne A, et al. 1994. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb Pathog. 16(4):269–282. [DOI] [PubMed] [Google Scholar]
- Ueoka R, et al. 2015. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat Chem Biol. 11(9):705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth SC, et al. 2020. Horizontal gene transfer to a defensive symbiont with a reduced genome in a multipartite beetle microbiome. mBio 11(1):e02430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, et al. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 31(1):28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth T, Popov VL, Yu X, Walker DH, Bouyer DH. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect Immun. 73(10):6668–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ACC, Duncan RP. 2015. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci U S A. 112(33):10255–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MC, et al. 2014. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506(7486):58–62. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3):E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Hamada M, Floreancig P, Nakabachi A. 2019. Diaphorin, a polyketide synthesized by an intracellular symbiont of the Asian citrus psyllid, is potentially harmful for biological control agents. PLoS One 14(5):e0216319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 17(1):32–43. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4(4):259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.