Abstract
Background
Hepatocellular carcinoma (HCC) remains one of the most common cancers worldwide and tends to be detected at an advanced stage. More effective biomarkers for HCC screening and prognosis assessment are needed and the mechanisms of HCC require further exploration. The role of MAOA in HCC has not been intensively investigated.
Methods
In‐house tissue microarrays, genechips, and RNAsequencing datasets were integrated to explore the expression status and the clinical value of MAOA in HCC. Immunohistochemical staining was utilized to determine MAOA protein expression. Intersection genes of MAOA related co‐expressed genes and differentially expressed genes were obtained to perform functional enrichment analyses. In vivo experiment was conducted to study the impact of traditional Chinese medicine nitidine chloride (NC) on MAOA in HCC.
Results
MAOA was downregulated and possessed an excellent discriminatory capability in HCC patients. Decreased MAOA correlated with poor prognosis in HCC patients. Downregulated MAOA protein was relevant to an advanced TNM stage in HCC patients. Co‐expressed genes that positively related to MAOA were clustered in chemical carcinogenesis, where CYP2E1 was identified as the hub gene. In vivo experiment showed that nitidine chloride significantly upregulated MAOA in a nude mouse HCC model.
Conclusions
A decreased MAOA level is not only correlated with aggressive behaviors in males but also serves as a promising biomarker for the diagnosis and prognosis of HCC patients. Moreover, MAOA may play a role in AFB1 toxic transformation through its synergistic action with co‐expressed genes, especially CYP3A4. MAOA also serves as a potential therapy target of NC in HCC patients.
Keywords: gene chip, HCC, immunohistochemical staining, MAOA, RNA‐seq, tissue microarray
A decreased MAOA level is not only correlated with aggressive behaviors in males but also serves as a promising biomarker for the diagnosis and prognosis of HCC patients. Moreover, MAOA may play a role in AFB1 toxic transformation through its synergistic action with co‐expressed genes, especially CYP3A4. MAOA also serves as a potential therapy target of traditional Chinese medicine nitidine chloride in hepatocellular carcinoma patients.
1. INTRODUCTION
Hepatocellular carcinoma (HCC) threatens the lives of humans worldwide, and it is regarded as one of the most dominant causes of cancer‐related deaths. 1 Surgery, namely liver transplantation and hepatectomy, is currently the most effective approach to treat HCC patients. 2 Despite the major progress made in treating HCC, the prognostic status of HCC patients remains poor because of the lack of significant symptoms until advanced cancer in most HCC patients, thus causing a poor prognosis and a heavy burden on these patients. 3 , 4 To date, many protein markers, such as AFP and PIVKA‐II, have been found to screen for HCC patients. 5 There are also a lot of promising biomarkers for HCC therapy or prognosis evaluation. 6 , 7 Nonetheless, according to the 2018 global cancer statistics, 841 080 new cases and 781 631 deaths of liver carcinoma were reported. 8 Moreover, the incidence of liver cancer is increasing rapidly compared with other forms of cancer based on American cancer statistics in 2020. 9 Therefore, the situation confronting HCC patients remains grim. Substantial efforts have been made to analyze the carcinogenesis and deterioration mechanisms of HCC. 10 , 11 , 12 Accumulating research supports that HCC risk is associated with inter‐individual genetic variation. 13 Moreover, HCC correlates with chronic hepatitis B or C virus infection, aflatoxin, and alcohol abuse. 8 , 14 Despite the expended time and efforts, the precise molecular mechanisms of HCC remain obscure. Deeper insight into the complex molecular mechanisms of HCC is urgently needed. Furthermore, it is important to discover more novel target genes relevant to the pathogenesis and progression of HCC and to provide potential approaches for early diagnosis and effective treatment regimens of HCC. 15
Monoamine oxidase‐A (MAOA), an enzyme distributed in the central nervous system and peripheral tissues and organs, has a profound function of breaking down monoamine substances, namely dopamine, epinephrine, and norepinephrine. 16 Previous studies have documented that the MAOA gene, also known as the warrior gene, is relevant to abnormal behaviors (ie antisocial, aggressive, criminal, etc) in males. 17 Conversely, lower MAOA expression is associated with greater happiness of females after adjusting for possible impact of other factors, namely ages, incomes, and interpersonal relationship. 18 The multiple effects of the MAOA gene have attracted the interest of researchers.
Recently, the overexpression of MAOA has been reported in various cancers, such as prostate cancer, 19 , 20 renal cell carcinoma, 21 classical Hodgkin lymphomas, 22 and glioma. 23 Conversely, other studies have found a downregulation trend of MAOA in several cancers. 24 , 25 MAOA was also found to participate in the formation and progression of tumors. According to Jason et al, MAOA induced the distal metastasis of prostate cancer. 26 MAOA was also proved to be correlated with the worse prognosis of HER‐2 subtype breast cancer. 27 Further studies supported that upregulated MAOA promoted angiogenesis in breast cancer 28 and contributed to the transition from epithelial to mesenchymal in lung cancer. 29 Li et al discovered that MAOA was downregulated in cholangiocarcinoma (ICC) specimens through hypermethylation orinterleukin‐6 (IL‐6) signaling and that MAOA could serve as a prognostic biomarker for ICC patients. 30 However, only one study has mentioned the expression significance of MAOA in HCC to date. 31 They found that MAOA hampers HCC metastasis by inhibiting the epinephrine system, which mediates the metastasis of tumor cells by binding to a specific receptor and transactivating the EGFR signal pathway. 31 The role of MAOA in the carcinogenesis and progression of HCC has not been intensively studied, and the mechanisms underlying MAOA in HCC remain obscure.
In the present study, we evaluated the MAOA expression levels in HCC patients compared with non‐cancer groups by integrating in‐house tissue microarray data and datasets downloaded from Gene Expression Omnibus (GEO), Array Express, Sequence Read Archive (SRA), Oncomine, the Cancer Genome Atlas (TCGA), and the Genotype‐Tissue Expression project (GTEx). According to the MAOA expression value, a diagnostic trial was performed to estimate the clinical value of MAOA in HCC screening. The prognostic capacity of MAOA in HCC patients was also investigated. We also shed light on the clinical significance of the MAOA protein and determined the potential molecular mechanisms of MAOA in HCC. Finally, we addressed the effect of traditional Chinese medicine (TCM) nitidine chloride (NC) on the MAOA expression to provide potential therapy for HCC patients.
2. MATERIALS AND METHODS
2.1. Datasets downloaded from GEO, array express, SRA, oncomine, TCGA, and GTEx databases
HCC datasets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), SRA (https://www.ncbi.nlm.nih.gov/sra), Oncomine (https://www.oncomine.org/resource/main.html), and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), and RNA sequencing (RNA‐seq) data were downloaded from TCGA (https://cancergenome.nih.gov/) and GTEx (https://gtexportal.org/home/) using the following MESH terms: (malignancy OR cancer OR tumor OR neoplasm OR neoplasia OR carcinoma OR carcinomatosis) AND (hepatocellular OR liver OR hepatic OR HCC). HCC datasets meeting the following criteria were included in the analysis: (a) the specimens were collected from humans, (b) both HCC sample tissues and non‐cancer liver tissues were provided, (c) the expression profile could be downloaded, and (d) the expression level of MAOA is available. If the datasets were duplicated, only the latest version would be preserved. Several studies were excluded because of the following reasons: (a) datasets of cell lines, (b) sample size <3, and (c) lack of MAOA.
2.2. Statistical analyses and the clinical significance of MAOA in HCC patients
Independent‐samples t‐test was performed to compare the expression status of MAOA between the HCC and non‐cancer groups using GraphPad Prism v8.0 software. To assess the capacity of MAOA to distinguish HCC patients from people without liver cancer, the receiver operating characteristic (ROC) curve was plotted using GraphPad v8.0 software. The area under the curve (AUC)> 0.70 was recognized to have moderate diagnostic capability, and AUC >0.90 indicated great diagnostic capability. Subsequently, the standard mean difference (SMD) was computed to assess the general expression level of MAOA in HCC patients using STATA v12.0 software. To evaluate the heterogeneity of the included datasets, we performed a heterogeneity test in STATA v12.0, where I 2 >0.50 indicated significant heterogeneity, and a random effect analysis was required. A Begg's funnel plot was drawn to determine whether publication bias occurred, and a P‐value ≥.05 indicated no publication bias. A summary of the receiver operating characteristic (sROC) curve was presented, and the specificity and sensitivity of MAOA were computed in STATA v12.0. A Kaplan‐Meier curve was plotted to obtain further insight into the association between the MAOA expression level and the overall survival condition of HCC patients.
2.3. Genetic alteration and mutation types of MAOA in HCC
The genetic alteration status of MAOA in HCC was explored in the cBioPortal for Cancer Genomics (https://www.cbioportal.org/) database, 32 which provides an interactive interface to visualize the results. The study on liver hepatocellular carcinoma (TCGA, Provisional) consisting of 440 samples was queried for our analysis. The mutation types of MAOA in HCC were interpreted in the Catalogue of Somatic Mutations in Cancer (https://cancer.sanger.ac.uk/cosmic). 33
2.4. Identification of co‐expressed genes and differentially expressed genes of MAOA
The Pearson's correlation coefficient of MAOA and other detected genes in each expression matrices was computed using R software v3.6.1. The genes meeting the following criteria were identified as significantly co‐expressed genes (CEGs) of MAOA: (a) the absolute value of Pearson's correlation coefficient >0.3 and (b) a P‐value <.05. The reshape2 and ggplot2 packages were utilized to plot the heatmap of the CEGs of MAOA in the datasets using R software v3.6.1. Moreover, a relationship network diagram was plotted with Cytoscape v3.6.1. The Limma Voom package was used to identify differentially expressed genes (DEGs). The standards of DEGs identification were as follows: (a) the absolute value of log2FoldChange >1 and (b) an adjusted P‐value <.05. The intersection genes of the MAOA positively related CEGs and downregulated DEGs and those of the MAOA negatively related CEGs and upregulated DEGs were obtained for further analyses.
2.5. Disease Ontology, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes enrichment analyses and the protein‐to‐protein interaction network
The two types of intersection genes acquired were used to undertake the Disease Ontology (DO) (http://disease‐ontology.org/), Gene Ontology (GO) (http://geneontology.org/), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) enrichment analyses respectively. The results were visualized with the aid of DOSE, GO.db, topGO, GSEABase, GOplot, and stringr packages in R v3.6.1 software. The protein‐to‐protein interaction (PPI) network was constructed in STRING (https://string‐db.org/), and the hub genes were detected using Cytoscape v3.6.1 to explore the potential mechanism of MAOA underlying HCC.
2.6. Evaluation of the clinical value of the MAOA protein expression in HCC patients by immunohistochemical (IHC) staining
A total of 315 HCC and 192 control tissue specimens (ie adjacent normal tissues or non‐HCC tissues) were collected and fixed with formalin from patients in First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, P.R.CHINA. The clinicopathological parameters of the patients were recorded. All patients signed informed consent forms. This study complied with all the relevant national regulations and institutional policies and was approved by the ethics committee of The First Affiliated Hospital of Guangxi Medical University. Four tissue microarrays (ie,LVC1505, LVC1531, LVC1601, and LVC1602) were prepared, and IHC staining was performed using the MAOA antibody. All operations complied with the manufacturer's requirements.
2.7. Exploration of the effect of NC on MAOA expression
Our team has been studying the biological function NC has on HCC for a period. Previous in vitro and in vivo studies have verified that TCM NC exhibits a strong anti‐HCC capability by mediating complicated mechanisms, namely TF‐miRNA‐mRNA axis, circRNA‐miRNA‐mRNA network, andJAK1/STAT3 cascade signaling inhibition. 34 , 35 , 36 , 37 , 38 Given the role that MAOA plays in HCC, we supposed that NC could exert its anti‐tumor effect partially by influencing the expression of MAOA. To validate our hypothesis, an in vivo experiment was conducted to study the effect of TCM NC on MAOA expression in a nude mouse HCC model. A total of 48 healthy nude mice were inoculated with SMMC‐7721 HCC cells (1 × 107 cells/L) subcutaneously. The nude mice were divided randomly into three groups (ie NC‐treated group with intraperitoneal injection of NC [7 mg·kg−1], negative control group with intraperitoneal injection of saline, and the blank control group) when the tumor diameter was 15‐30 mm. After administration for 15 days, the nude mice were anesthetized and the tumor tissues were excised. Subsequently, we used next‐generation sequencing methods to determine the mRNA expression profiles in HCC tissues dissected from NC‐treated and control nude mice. The DEGs were identified using the Limma Voom package in R software v3.6.1; the criteria used were described previously. The research related to animals use complied with all the relevant national regulations and institutional policies for the care and use of animals (Ethics number: 2018‐KY‐NSFC‐102). All operations strictly complied with the manufacturer's requirements.
3. RESULTS
3.1. Included datasets and quality assessment
As shown in the flow chart (Figure S1), we retrieved datasets from the GEO, Array Express, SRA, Oncomine, TCGA, and GTEx databases. A total of 109 datasets were obtained after 48 duplicates were removed. Among these datasets, 26 were excluded, the data of which were not available. After verifying the matrices, seven datasets were removed without MAOA expression data. Eventually, 76 datasets were included for the analysis. To solidify the reliability of our study, the data were verified by LJD, GL, and YX independently, and a consensus was reached.
3.2. Dataset information
Table S1 shows the basic information of the 76 included datasets and one in‐house tissue microarray, covering 4348 HCC patients and 3624 non‐cancerous controls from 2007 to 2020. The datasets came from various ethnicities: 42 Asian datasets and 34 non‐Asian datasets. The expression levels of MAOA in the HCC patients and control groups were analyzed either by genechip technology (N = 58) or the RNA sequencing method (N = 18). The expression levels of MAOA in the HCC patients and non‐cancer groups were compared using the mean (M) expression levels and standard deviation (SD). All the specimens were hepatic tissues collected from HCC patients and non‐cancer individuals.
3.3. Decreased expression level of MAOA mRNA in HCC patients
The expression statuses of MAOA in various cancers are shown in Figure S2A. Among the RNA sequencing datasets, seven datasets showed a significant downregulation trend of MAOA in the HCC patients compared with the noncancer groups: GSE63018, GSE56545, GSE63863, GSE77509, GSE77314, TCGA‐GTEx, and GSE69164 (all with P‐values <.05; Figure S2B‐Q). As for the genechip datasets, the scatter plots in Figure S3A‐P show significant lower expression levels of MAOA in the HCC patients compared with the non‐cancer groups in GSE6764, GSE14323, GSE14520‐GPL571, GSE14520‐GPL3921, GSE29721, GSE45436, GSE55092, GSE76297, GSE102079, GSE112790, GSE121248, GSE54236, GSE67764, and GSE59259 (all with P‐values <.05). Subsequently, the heterogeneity test of the included datasets was conducted, and a result of I 2 = 84.9% (P‐value < .0001) indicated a significant heterogeneity. Thus, a random effects analysis was employed to compute the SMD. The outcome exhibited in the MAOA expression levels in patients with HCC was significantly lower than that in the non‐cancer groups (Figure 1, SMD = −0.32, 95% CI: −0.45 to −0.18). Moreover, the outcome of the sensitive analysis showed no significant difference among the included studies (Figure S4A). The Begg's funnel plot indicated no publication bias, with a P‐value = .657 (Figure S4B). Two subgroup analyses were performed to assess the expression status of MAOA from two perspectives (Figures S5 and S6).The results revealed that neither ethnicity nor the methods used to determine the MAOA mRNA expression levels of HCC patients was the source of heterogeneity in the present study, as MAOA was downregulated in all subgroups. To validate the MAOA expression status in HCC, we also downloaded the RNA sequencing data from the Broad Institute Cancer Cell Line Encyclopedia (CCLE). Surprisingly, we found that MAOA was not expressed in the cell lines of C3A_LIVER, ALEXANDERCELLS_LIVER, and HLE_LIVER. Therefore, the evidence confirmed the downregulated MAOA level in HCC.
FIGURE 1.
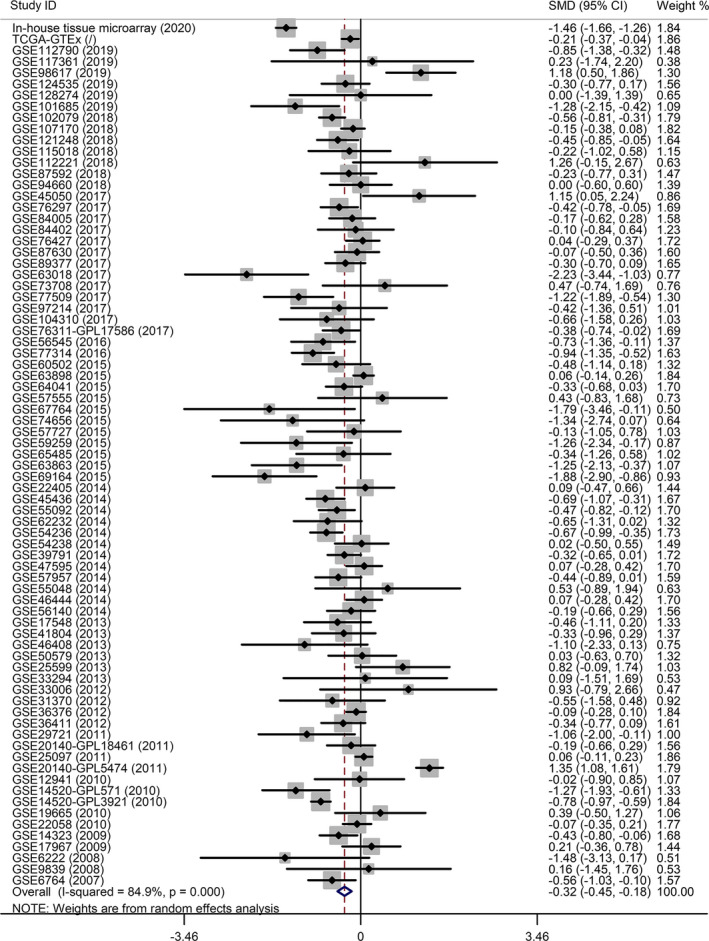
A forest plot based on the included datasets and in‐house tissue microarray. Decreased MAOAmRNA expression levels were found in hepatocellular carcinoma patients compared to non‐cancerous groups
3.4. Promising diagnostic and prognostic value of downregulated MAOA in HCC
The ROC curves of the following six RNA sequencing datasets indicated the discriminatory capacity of MAOA in the HCC patients and non‐HCC groups: GSE63018, GSE56545, GSE63863, GSE77509, GSE77314, and GSE69164 (all with AUC >0.70 and P‐values <.05; Figure S7A‐P). Regarding the gene chip dataset, the remarkable distinguishing ability of MAOA in the HCC patients and non‐cancer individuals was exhibited in GSE14520‐GPL571, GSE14520‐GPL3921, GSE29721, GSE45436, GSE112790, and GSE59259 (all with AUC >0.70 and P‐values <.05; Figure S8A‐I). The AUC of the sROC curve, sensitivity, and specificity were 0.77 (95% CI: 0.73‐0.80), 0.56 (95% CI: 0.51‐0.61), and 0.85 (95% CI: 0.81‐0.88), respectively (Figure 2A). In the Fagan plot, with a pre‐test probability of 20%, the post‐test probability of HCC using MAOA for a positive test result was 48%, and the probability of a negative test result was 11%, suggesting that MAOA could serve as a remarkable marker for HCC screening (Figure 2B). The diagnostic likelihood ratio positive (DLR P), diagnostic likelihood ratio negative (DLR N), diagnostic score, and diagnostic odd ratio were 3.74 (95% CI: 3.01‐4.64), 0.51 (95% CI: 0.46‐0.57), 1.98 (95% CI: 1.73‐2.23), and 7.26 (95% CI: 5.64‐9.34) respectively (Figure 2C and D).The aforementioned outcomes indicated that MAOA had an excellent discriminatory ability for HCC. Furthermore, the downregulated expression level of MAOA was relevant to age (≥ 60), race (Asian), tumor metastasis status (M0), neoplasm histologic grade (G1>G2>G3>G4), and AFP concentration (>20 ng/mL) (all with P‐values <0.05; Table S2; Figure S9A‐E). Interestingly, the Kaplan‐Meier curve indicated that the high MAOA mRNA expression group had a better survival outcome, with a hazard ratio of 0.61 (95% CI: 0.41‐0.93) (Logrank P = .019; Figure 3), indicating that the downregulated MAOAmRNA correlated with a poor prognosis of HCC patients.
FIGURE 2.
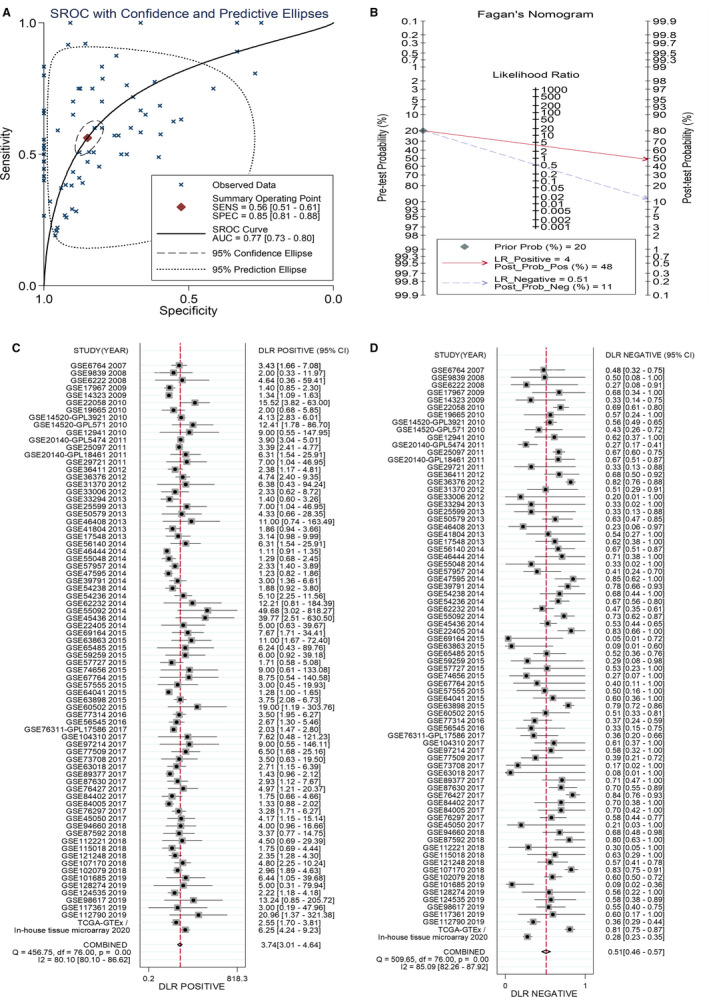
Hepatocellular carcinoma (HCC) patient diagnostic trial by using MAOA as a marker. A, The summary receiver operating characteristic (sROC) curve exhibited a marvelous discriminatory capacity of MAOA in HCC and non‐HCC patients. B, Fagan's nomogram, (C) diagnostic likelihood ratio negative (DLR N), and (D) diagnostic likelihood ratio positive (DLR N) also addressed the diagnostic value of MAOA for HCC patients
FIGURE 3.
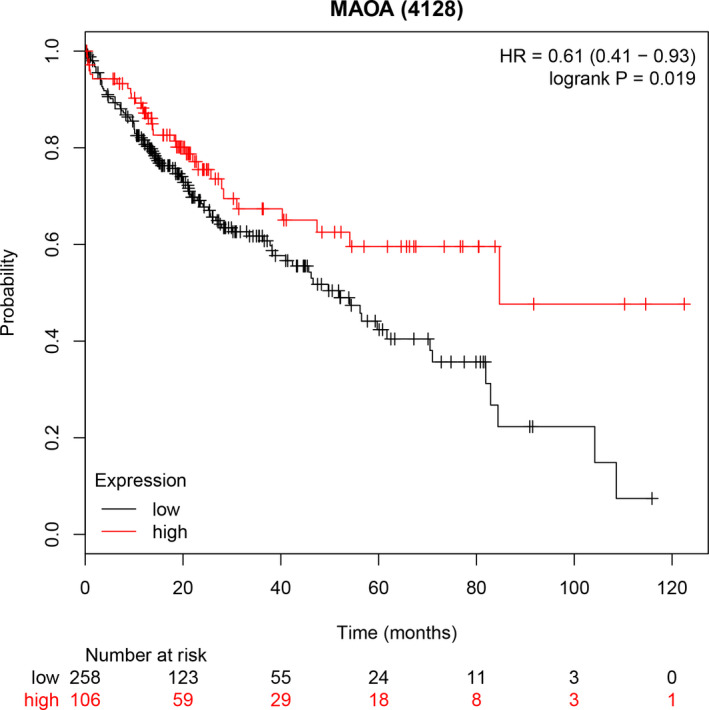
A Kaplan‐Meier curve of the hepatocellular carcinoma patients. The result indicated that the low MAOA group possessed worse survival outcome
3.5. Genetic alterations and mutation types of MAOA in HCC
MAOA was altered in 30 (8%) of the 440 patients in the study on liver hepatocellular carcinoma (TCGA, Provisional) (Figure 4A). Notably, not all genetic alteration information of the patients was available. The genetic alterations of MAOA in HCC were focused on missense mutation, amplification, deep deletion, and the mRNA overexpression level. Further study on the mutation type of MAOA in HCC showed that missense was the predominant mutation type (Table S3).
FIGURE 4.
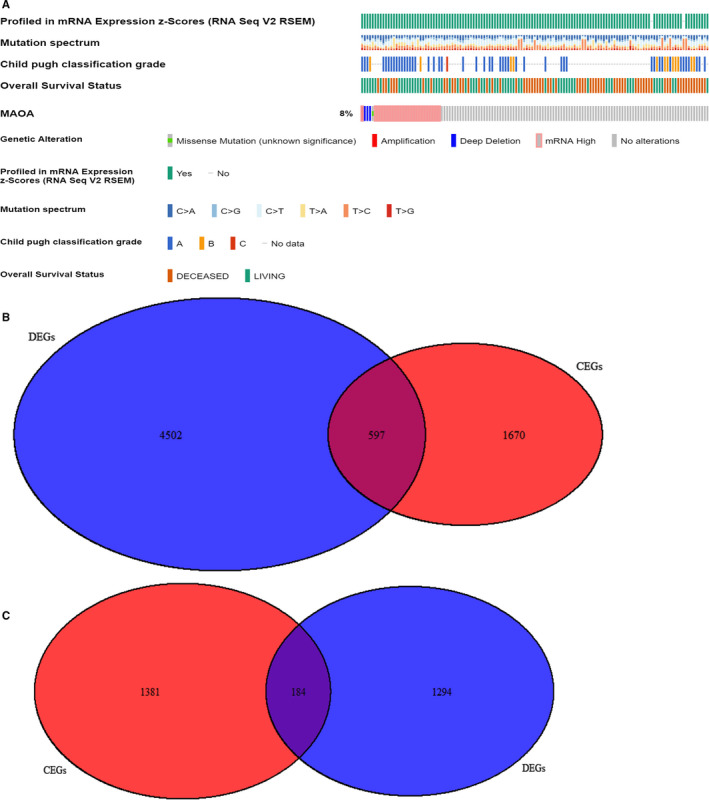
MAOA genetic alteration status based on cBioPortal. A, MAOA is altered in 30 (8%) of the queried hepatocellular patients. B, Venn diagram for the intersection genes of 5099 MAOA positively related co‐expressed genes (CEGs) and 2267 downregulated differentially expressed genes (DEGs). C, Venn diagram for the intersection genes of 1565 MAOA‐negatively‐related CEGs and 1478 up‐regulated DEGs
3.6. Identification of CEGs of MAOA and DEGs
The Pearson's correlation coefficient of MAOA and other genes in each dataset was computed. Two correlation heatmaps were plotted based on the calculated Pearson's correlation coefficients of GSE6222 and GSE19665 (Figure S10A and B). Subsequently, the co‐expressed genes meeting the standards of the absolute value of Pearson's correlation coefficient >0.3 and a P‐value <.05 were obtained for further analysis. A total of 5099 MAOA positively related CEGs and 1565 MAOA negatively related CEGs were identified, all of which appeared in no fewer than nine datasets. Herein, PCOLCE and TBC1D8 were detected to be the most significant CEGs positively related to MAOA, and PKM was detected to be the most significant CEGs positively related to MAOA (Figure S11A and B). In addition, 2267 downregulated genes and 1478 upregulated genes appearing in no fewer than eight datasets were identified. In total, 597 intersection genes of the MAOA positively related CEGs and downregulated DEGs and 184 intersection genes of the MAOA negatively related CEGs and upregulated DEGs were obtained (Figure 4B and C).
3.7. GO, DO, and KEGG enrichment analyses and PPI network construction
According to the 597 intersection genes of the MAOA positively related CEGs and downregulated DEGs, GO analysis showed the importance of the organic acid catabolic process, blood microparticle, and monooxygenase activity in terms of biological process (BP), cellular component (CC), and molecular function (MF) respectively (Figure 5A; Table S4). For the 184 intersection genes of the MAOA negatively related CEGs and upregulated DEGs, T cell activation, MHC class II protein complex, and MHC class II receptor activity were emphasized in terms of BP, CC, and MF respectively (Figure 5B; Table S5). Based on the 597 intersection genes, DO analysis and KEGG enrichment analysis indicated that coronary artery disease (Figure 6A) was the predominantly associated disease and that retinol metabolism and chemical carcinogenesis were the top two clustered pathways (Figure 6B). We wanted to focus on the chemical carcinogenesis pathway. Thus, we analyzed the specific process of the chemical carcinogenesis pathway (Figure S12) and found that MAOA could participate in the pathogenesis and progression of HCC and other cancers through multiple genes, namely CYP1A1, CYP1A2, CYP2A, CYP2E1, CYP3A4, GST, HSD11B1, NAT, NAT2, and UGT. Among these genes, CYP3A4 played an important role in the carcinogenesis process of HCC by promoting the transformation from aflatoxin B1 to AFB1‐exo‐8,9‐epoxide. The 184 intersection genes of the MAOA negatively related CEGs and upregulated DEGs were significantly related to lymphocytic choriomeningitis (Figure 6C) and were primarily enriched in the KEGG pathway of rheumatoid arthritis (Figure 6D). Moreover, PPI networks were constructed based on the genes in the chemical carcinogenesis pathway to show the interplay of proteins (Figure S13). CYP2E1 was selected as the hub gene in the network. We also explored the prognostic value of CYP2E1, and, consistent with MAOA, the results indicating low CYP2E1 correlated with the reduced overall survival in HCC patients (Figure S14A and B).
FIGURE 5.
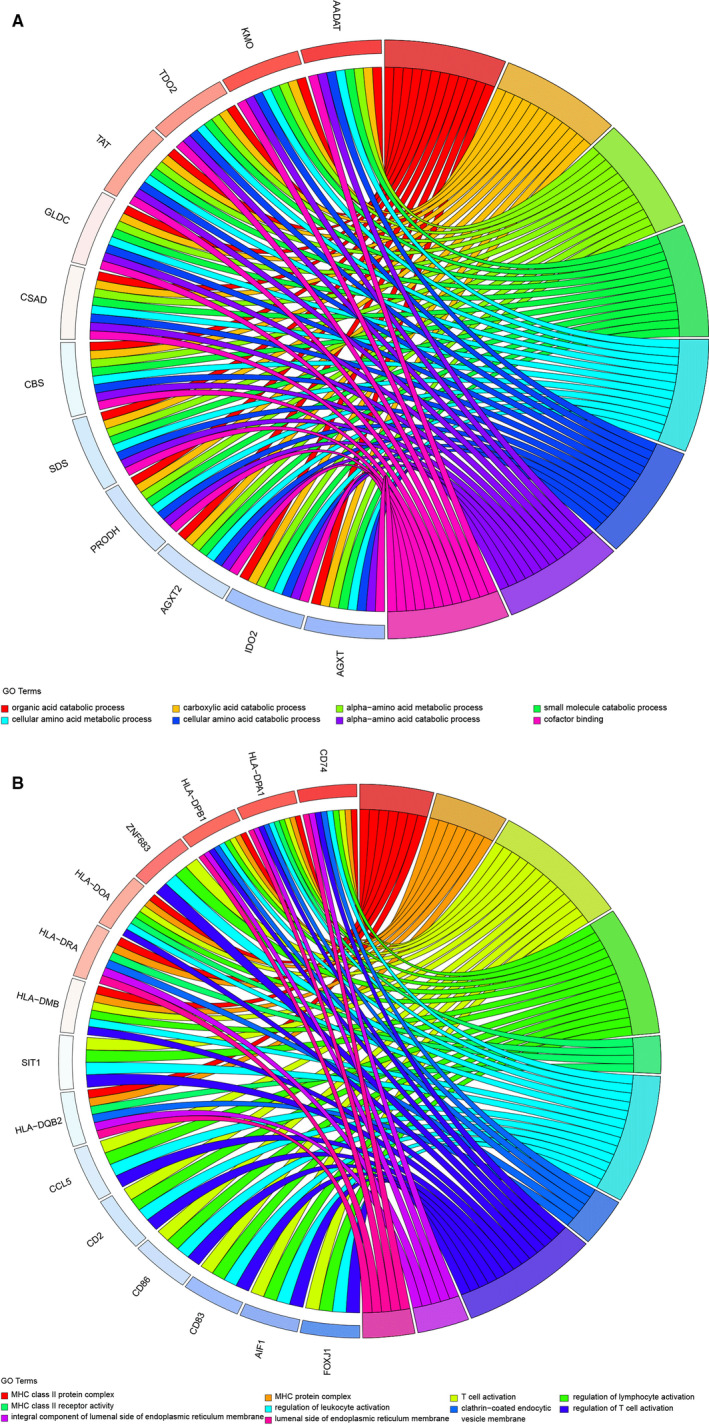
GO analysis based on the intersection genes. A, Chord plot of the 597 intersection genes from MAOA positively related co‐expressed genes (CEGs) and downregulated differentially expressed genes (DEGs). B, Chord plot of the 184 intersection genes of MAOA negatively related CEGs and upregulated DEGs. GO, Gene Ontology
FIGURE 6.
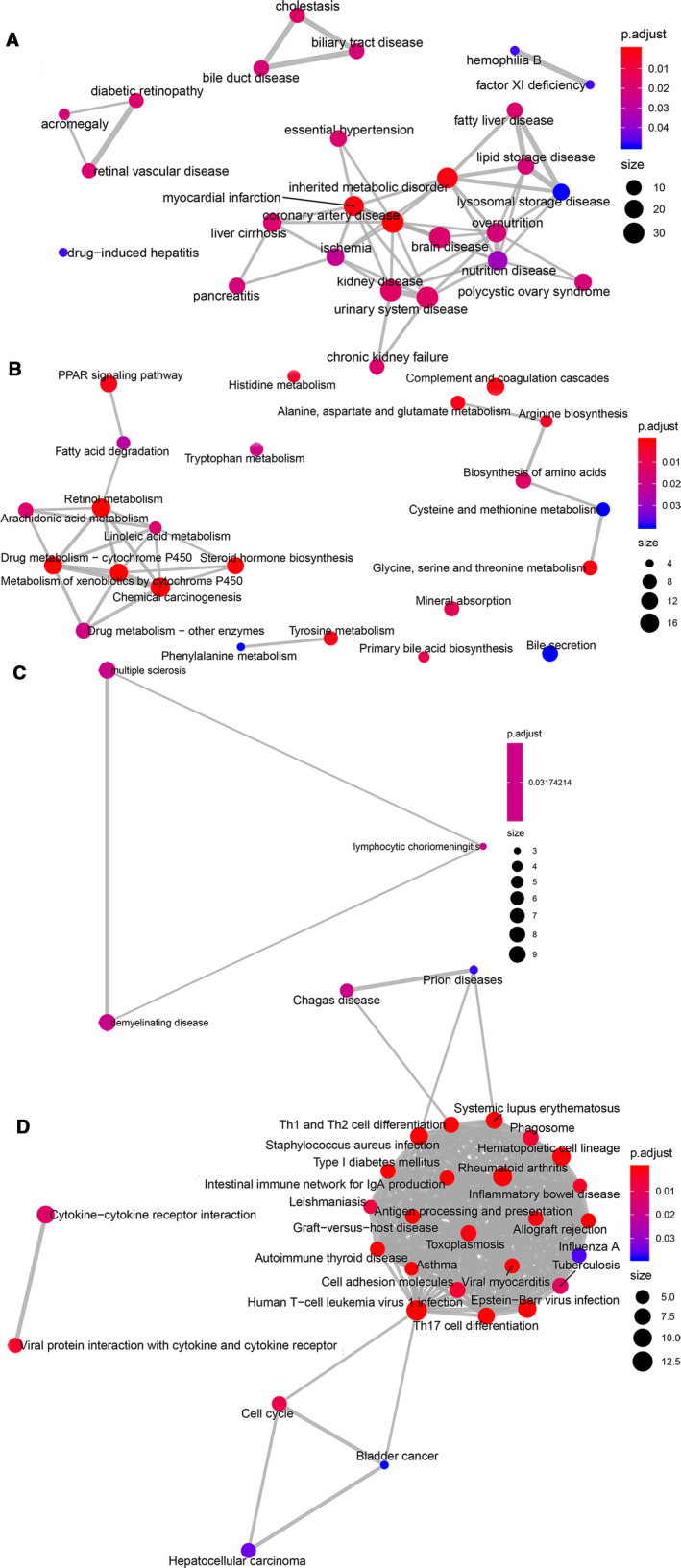
DO enrichment analysis and KEGG pathway. A, DO and (B) KEGG emap plot of the 597 intersection genes from MAOA positively related co‐expressed genes (CEGs) and downregulated differentially expressed genes (DEGs). (C) DO and (D) KEGG emap plot of the 184 intersection genes of MAOA negatively related CEGs and upregulated DEGs. DO, Disease Ontology. KEGG, the Kyoto Encyclopedia of Genes and Genomes
3.8. Protein expression level and clinical significance of MAOA in HCC
The results of the tissue microarrays and IHC staining indicated that the MAOA protein expression level was significantly decreased in HCC compared with non‐HCC specimens (Figure 7A), consistent with the expression status of MAOA from the mRNA level. A Kaplan‐Meier curve of the hepatocellular carcinoma patients based on the MAOA protein expression level was drawn, and no significant difference was identified between the low and high MAOA protein expression groups in HCC patients (Figure 7B). The HCC (Figure 7C, E, and G) and non‐HCC (Figure 7D, F, and H) immunohistochemical IHC staining images under a microscope (×400) indicated a lower MAOA protein expression in HCC compared with non‐HCC tissues, as confirmed by the IHC staining results from Human Protein Atlas
FIGURE 7.
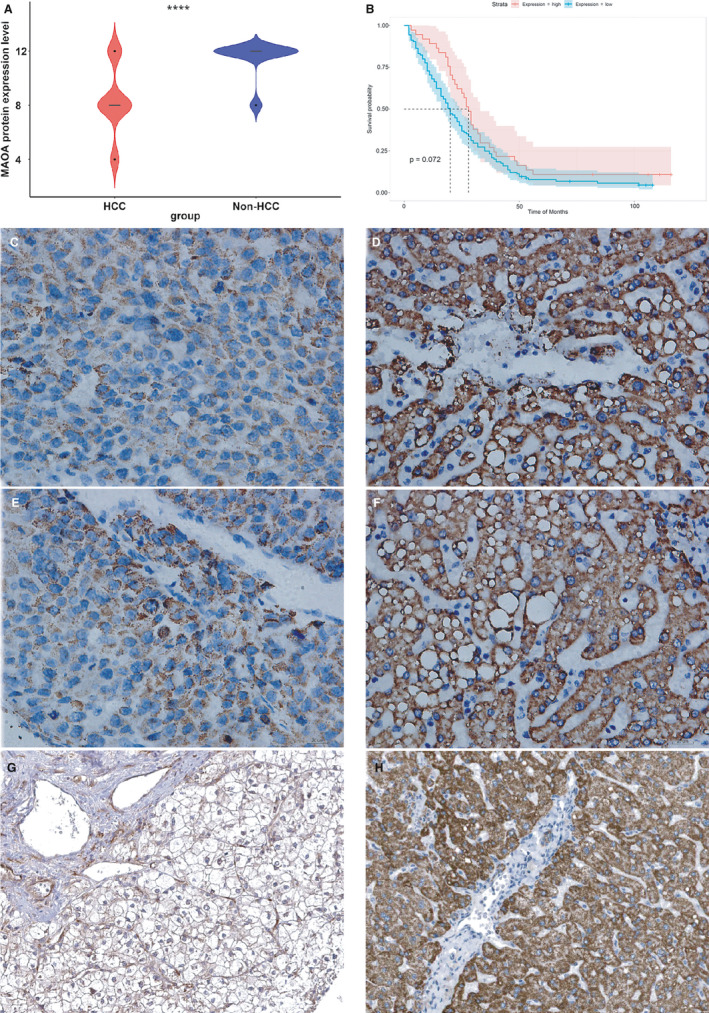
Expression level and clinical significance of MAOA protein in hepatocellular carcinoma (HCC). A, MAOA protein expression was significantly decreased in HCC compared to non‐HCC tissues. B, A Kaplan‐Meier curve of the hepatocellular carcinoma patients based on the MAOA protein expression level. However, no significant difference was identified between low and high MAOA protein expression group in HCC patients. HCC (C, E, and G)and non‐HCC (D, F, and H) immunohistochemical (IHC) staining images under a microscope (x400) signified lower MAOA protein expression in HCC compared to non‐HCC tissues. G and H were provided by HumanProteinAtlas (antibody: CAB009437)
(Figure 7G and H). More importantly, we found that a lower MAOA protein expression was significantly correlated with the advanced TNM stage of HCC patients (Table S6).
3.9. Effect of NC on the MAOA expression in the nude mouse HCC model
The tumor volumes of the NC‐treated nude mice were significantly reduced when compared with control nude mice (P‐value <.05). 39 A total of two NC‐treated and three control HCC tissues were eligible for expression analysis. The mRNA expression profiles in HCC tissues dissected from NC‐treated and control nude mice were determined by next‐generation sequencing. The DEGs were identified, and a volcano plot was drawn (Figure S15). As expected, the MAOA expression level was significantly elevated in the NC‐treated group compared with the control group.
4. DISCUSSION
In this study, we validated the downregulation of MAOA and evaluated its clinical values. We revealed the impact NC has on MAOA for the first time and explored its potential mechanism.
When compared with the previous studies of MAOA concerning HCC, our research appraised the clinical value of MAOA in a more comprehensive manner. As a multiple‐centered study, we have integrated in‐house tissue chip with large‐scale of microarrays and sequencing data sets of HCC tissues. We confirmed that MAOA expression exhibited a downregulated trend after analyzing 4033 HCC and 3432 controls, and the decreased level of MAOA in HCC displayed no significant association with ethnicity or expression profile methods (namely gene chip and high throughout sequencing).
According to the study of Du et al, MAOA mediates a signaling pathway contributing to reactive oxygen species generation, through which cancer‐associated fibroblasts induce the epithelial‐mesenchymal transition, metastasis, and infiltration of prostate cancer. 40 Liao etal demonstrated that MAOA promoted prostate tumor cell proliferation by mediating AKT phosphorylation and that MAOA absence hampered the progression of prostate cancer. 41 In the present study, we found that decreased MAOA in HCC correlated with poor survival outcome. Therefore, MAOA could be a potential prognosis predictor of HCC patients.
We identified the CEGs and DEGs of MAOA in HCC for further analysis of the mechanisms. PCOLCE and TBC1D8 were selected as the most significant CEGs positively related to MAOA, and PKM was detected as the most significant CEGs negatively related to MAOA. According to Li et al, PCOLCE was significantly downregulated in liver cancer and was positively correlated with DAPK1, which is a proven prognostic factor of liver cancer. 42 Although research on TBC1D8 in HCC is lacking, it has been found to be an independent prognosis marker in ovarian cancer. It was reported that circMAT2Bcould accelerate the progression of HCC by miR‐338‐3p/PKM2 axis. 43 A GO enrichment analysis based on the 597 intersection genes of the MAOA positively related CEGs and downregulated DEGs emphasized the importance of the organic acid catabolic process, blood microparticle, and cofactor binding in the GO terms of BP, CC, and MF respectively. These genes were related to coronary artery disease and were prominently clustered in the chemical carcinogenesis pathway. We found downregulated DEGs that positively correlated with MAOA, especially CYP1A1, CYP1A2, CYP2A, CYP2E1, CYP3A4, GST, HSD11B1, NAT, NAT2, and UGT, were significantly aggregated in the chemical carcinogenesis pathway. Interestingly, CYP1A1, CYP1A2, CYP2A, CYP2E1, and CYP3A4 all belong to genes that encode the essential enzymes of the cytochrome P450 system and participate in the metabolism and biological transformation of chemicals. 44 , 45 , 46 , 47 Among these genes, CYP3A4, which is highlighted in the KEGG pathway, played an important role in the carcinogenesis process of HCC by promoting the transformation from aflatoxin B1 to AFB1‐exo‐8,9‐epoxide, consistent with Bonomo et al. 48 There is a consensus that AFB1, 49 the most powerful carcinogen to date, works as a trigger in the carcinogenesis of the liver. In this study, co‐expression analysis revealed the positive relation between MAOA and CYP3A4. Recently, it has been confirmed that MAOA and CYP3A4 were two major enzymes that participated in the formation of metabolite A and D in hepatocytes. 50 Therefore, it can be inferred that MAOA may synergistically participate in the toxic transformation of AFB1 through its positively related CEGs, especially CYP3A4, thus causing pathogenesis and progression in HCC patients. Our findings enrich the understanding of pathogenesis and progression of HCC. More studies are needed to further confirm the interaction between MAOA and CYP3A4 in the future.
Regarding the 184 intersection genes of the MAOA negatively related CEGs and upregulated DEGs, T cell activation, MHC class II protein complex, and MHC class II receptor activity were important in the GO terms of BP, CC, and MF, respectively. These genes were significantly associated with lymphocytic choriomeningitis in DO analysis and were enriched in the KEGG pathway of rheumatoid arthritis. To the best of our knowledge, cytotoxic T lymphocyte (CTL) is considered the most pivotal effector cell in anti‐tumor immunity, 51 and it is capable of specifically killing tumor cells through the Fas‐FasL and TNF‐TNFR pathway and the perforin/granzyme pathway. The activation of CTL relies on two signals: tumor antigen presented by the antigen presentation cell (APC) and co‐stimulatory molecules, especially CD28, located on the surface of the T cell. 52 MHC class II protein complex, a molecule distributed to APC and activated T cell, plays an important role in the indirect activation of CD8+ T cells by presenting the tumor antigen to CD4+ T cells. With the help of CD4+ T cells, CD8+ T cells successfully differentiate into CTL and exert its anti‐tumor effects. Contrary to the carcinogenesis effect of MAOA positively related CEGs, the MAOA negatively related CEGs exhibit an anti‐tumor biological function by promoting T cell activation while participating in coding the MHC class II protein complex. Therefore, we suppose that the imbalance between the biological functions of MAOA positively related CEGs and those of MAOA negatively related CEGs may partially account for the onset of HCC.
Interestingly, we found that NC could exert its partial anti‐tumor effect by influencing the expression of MAOA. As demonstrated previously, a decreased MAOA was detected in HCC patients and correlated with poor prognosis. Our previous studies have confirmed the anti‐HCC effect of NC. 34 , 35 , 37 In this study, in vivo experiment showed that the tumor volumes were reduced and the MAOA expression level was significantly elevated in the NC‐treated group compared with the control group. Taken together, NC increases MAOA expression and plays a protective role in HCC nude mouse model. To date, some mechanisms of the suppressor role of NC in HCC have been proved. Based on in vitro and in vivo studies, TF‐miRNA‐mRNA axis and circRNA‐miRNA‐mRNA network were established to provide clues on the role of NC in HCC. 34 , 35 Another in vitro and in vivo experiment confirmed that the therapeutic effect of NC on HCC correlated with the upregulation of p53 and Bcl‐2. 36 According to an in vivo study of Jun Liao etal, the tumor suppression effect of NC was also attributed to the JAK1/STAT3 cascade signaling inhibition. 38 In light of the present study, it is reasonable to suppose that NC may attenuate tumor cell growth and other malignant behaviors in HCC by increasing the MAOA expression.
The unique advantages of the present study are as follows. First, the clinical value of MAOA was comprehensively estimated by integrating in‐house tissue microarray, genechip, and RNA sequencing data. We not only discovered the moderate capability of MAOA in distinguishing HCC from non‐HCC patients but also showed the relevance between a downregulated MAOA and the poor prognosis of HCC patients. Second, a decreased MAOA expression was verified by immunohistochemical staining, and a downregulated MAOA protein expression was correlated with an advanced TNM stage in HCC patients. Third, we found that TCM NC showed a potential in anti‐HCC by upregulating MAOA.
Nevertheless, this study has its limitations. First, the included datasets had high heterogeneity, and the influence analysis indicated no significant difference. The reasons for this could be due to the different methods and platforms used in the datasets and the patients coming from various countries, such as Singapore, China, the United States, Switzerland, Japan, Turkey, Japan, Canada, South Korea, Spain, Germany, and Italy. To cope with the high heterogeneity, a randomized effect model was employed in our analysis. Second, MAOA had a sensitivity of 0.56 (95% CI: 0.51‐0.61), which is slightly lower than that of AFP (63.3%). Therefore, MAOA is more effective in confirming diagnosis than in screening in HCC. Third, our result was based on clinical data and in vivo mouse model. Therefore, large‐scale clinical experiments are needed to further confirm the diagnostic and prognostic significance of MAOA in HCC in the future, and the potential therapeutic effect of NC by targeting MAOA should be further validated.
In conclusion, a decreased MAOA level is not only correlated with aggressive behaviors in males but also serves as a promising biomarker for the diagnosis and prognosis of HCC patients. Moreover, MAOA may play a role in AFB1 toxic transformation through its synergistic action with co‐expressed genes, especially CYP3A4. MAOA also serves as a potential therapy target of NC in HCC patients.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Design of project study, experiment operation directing, statistics process directing, and modification of paper: Yu‐Yan Pang, Yi‐Wu Dang, Ze‐Feng Lai, Li‐Min Liu, Jie Yang, Hua‐Yu Wu, Rong‐Quan He, Zhi‐Guang Huang, Li‐Hua Yang, Lin Shi, Wei‐Jia Mo, Deng Tang, Hui‐Ping Lu, and Gang Chen; experiment operation of tissue microarray immunohistochemistry, data collection, data analysis, and manuscript drafting: Jian‐Di Li, Li Gao, Xia Yang, and Dan‐Dan Xiong; final approval of manuscript: all the authors.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Figure S9
Figure S10
Figure S11
Figure S12
Figure S13
Figure S14
Figure S15
Table S1‐S6
ACKNOWLEDGMENTS
We are grateful to people who contribute to public biomedical databases.
Pang Y‐Y, Li J‐D, Gao L, et al. The clinical value and potential molecular mechanism of the downregulation of MAOA in hepatocellular carcinoma tissues. Cancer Med. 2020;9:8004–8019. 10.1002/cam4.3434
Yu‐Yan Pang and Jian‐Di Li are contributed equally as first authors.
Funding information
The present study was funded by: Natural Science Foundation of Guangxi, China (2018GXNSFAA294025, 2019GXNSFAA245087), the Promoting Project of Basic Capacity for Young and Middle‐aged University Teachers in Guangxi (2018KY0123), Innovation Project of Guangxi Graduate Education (YCBZ2019041), Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (JGY2019050), Guangxi Medical University Education and Teaching Reform Project (2019XJGZ04), Guangxi Medical University Training Program for Distinguished Young Scholars, Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University.
Contributor Information
Hui‐Ping Lu, Email: luhuiping@gxmu.edu.cn.
Gang Chen, Email: chengang@gxmu.edu.cn.
DATA AVAILABILITY STATEMENT
The original data are available from the corresponding author upon request.
REFERENCES
- 1. Ali ES, Rychkov GY, Barritt GJ. Deranged hepatocyte intracellular Ca(2+) homeostasis and the progression of non‐alcoholic fatty liver disease to hepatocellular carcinoma. Cell Calcium. 2019;82:102057. [DOI] [PubMed] [Google Scholar]
- 2. Zhu Q, Sun Y, Zhou Q, He Q, Qian H. Identification of key genes and pathways by bioinformatics analysis with TCGA RNA sequencing data in hepatocellular carcinoma. Mol Clin Oncol. 2018;9:597‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gougelet A, Desbois‐Mouthon C. Non‐coding RNAs open a new chapter in liver cancer treatment. Clin Res Hepatol Gastroenterol. 2019;43:630‐637. [DOI] [PubMed] [Google Scholar]
- 4. Wang D, Zheng X, Fu B, et al. Hepatectomy promotes recurrence of liver cancer by enhancing IL‐11‐STAT3 signaling. EBioMedicine. 2019;46:119‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoon YJ, Han KH, Kim DY. Role of serum prothrombin induced by vitamin K absence or antagonist‐II in the early detection of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Scand J Gastroenterol. 2009;44:861‐866. [DOI] [PubMed] [Google Scholar]
- 6. Zhou F, Shang W, Yu X, Tian J. Glypican‐3: a promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev. 2018;38:741‐767. [DOI] [PubMed] [Google Scholar]
- 7. Ma X‐L, Tang W‐G, Yang M‐J, et al. Serum STIP1, a novel indicator for microvascular invasion, predicts outcomes and treatment response in hepatocellular carcinoma. Front Oncol. 2020;10:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. . 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 9. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. Cancer J Clin. 2020;70:7‐30. [DOI] [PubMed] [Google Scholar]
- 10. Yuan L, Bing Z, Han J, et al. Study on the anti‐tumor mechanism related to immune microenvironment of Bombyx Batryticatus on viral and non‐viral infections of hepatocellular carcinoma. Biomed Pharmacother. 2020;124:109838. [DOI] [PubMed] [Google Scholar]
- 11. Zhang L, Chen Y, Bao C, Zhang X, Li H. Eukaryotic initiation Factor 4AIII facilitates hepatocellular carcinoma cell proliferation, migration, and epithelial‐mesenchymal transition process via antagonistically binding to WD repeat domain 66 with miRNA‐2113. J Cell Physiol. 2020:1‐11. 10.1002/jcp.29475. [DOI] [PubMed] [Google Scholar]
- 12. Yu Y, Xu P, Cui G, et al. UBQLN4 promotes progression of HCC via activating wnt‐β‐catenin pathway and is regulated by miR‐370. Cancer Cell Int. 2020;20:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stickel F, Lutz P, Buch S, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology. 2019;72(1):88‐102. 10.1002/hep.30996 [DOI] [PubMed] [Google Scholar]
- 14. Cheng K, Shi J, Liu Z, et al. A panel of five plasma proteins for the early diagnosis of hepatitis B virus‐related hepatocellular carcinoma in individuals at risk. EBioMedicine. 2020;52:102638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR‐149‐5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51:2262‐2274. [DOI] [PubMed] [Google Scholar]
- 16. Steuer AE, Boxler MI, Stock L, Kraemer T. Inhibition potential of 3,4‐methylenedioxymethamphetamine (MDMA) and its metabolites on the in vitro monoamine oxidase (MAO)‐catalyzed deamination of the neurotransmitters serotonin and dopamine. Toxicol Lett. 2016;243:48‐55. [DOI] [PubMed] [Google Scholar]
- 17. González‐Tapia MI, Obsuth I. "Bad genes" & criminal responsibility. Int J Law Psychiatry. 2015;39:60‐71. [DOI] [PubMed] [Google Scholar]
- 18. Chen H, Pine DS, Ernst M, et al. The MAOA gene predicts happiness in women. Prog Neuropsychopharmacol Biol Psychiatry. 2013;40:122‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaur S, Gross ME, Liao CP, Qian B, Shih JC. Effect of Monoamine oxidase A (MAOA) inhibitors on androgen‐sensitive and castration‐resistant prostate cancer cells. Prostate. 2019;79:667‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin YC, Chang YT, Campbell M, et al. MAOA‐a novel decision maker of apoptosis and autophagy in hormone refractory neuroendocrine prostate cancer cells. Sci Rep. 2017;7:46338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hodorova I, Rybarova S, Solar P, et al. Monoamine oxidase B in renal cell carcinoma. Med Sci Monitor. 2018;24:5422‐5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li PC, Siddiqi IN, Mottok A, et al. Monoamine oxidase A is highly expressed in classical Hodgkin lymphoma. J Pathol. 2017;243:220‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kushal S, Wang W, Vaikari VP, et al. Monoamine oxidase A (MAO A) inhibitors decrease glioma progression. Oncotarget. 2016;7:13842‐13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang B, Jiang J, Du H, et al. Decreased monoamine oxidase (MAO) activity and MAO‐A expression as diagnostic indicators of human esophageal cancers. Biomarkers. 2009;14:624‐629. [DOI] [PubMed] [Google Scholar]
- 25. Rybaczyk LA, Bashaw MJ, Pathak DR, Huang K. An indicator of cancer: downregulation of monoamine oxidase‐A in multiple organs and species. BMC Genom. 2008;9:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu JB, Yin L, Shi C, et al. MAOA‐dependent activation of Shh‐IL6‐RANKL signaling network promotes prostate cancer metastasis by engaging tumor‐stromal cell interactions. Cancer Cell. 2017;31:368‐382. [DOI] [PubMed] [Google Scholar]
- 27. Sun WY, Choi J, Cha YJ, Koo JS. Evaluation of the expression of amine oxidase proteins in breast cancer. Int J Mol Sci. 2017;18:2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bharti R, Dey G, Das AK, Mandal M. Differential expression of IL‐6/IL‐6R and MAO‐A regulates invasion/angiogenesis in breast cancer. Br J Cancer. 2018;118:1442‐1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu F, Hu L, Ma Y, et al. Increased expression of monoamine oxidase A is associated with epithelial to mesenchymal transition and clinicopathological features in non‐small cell lung cancer. Oncol Lett. 2018;15:3245‐3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang L, Frampton G, Rao A, et al. Monoamine oxidase A expression is suppressed in human cholangiocarcinoma via coordinated epigenetic and IL‐6‐driven events. Lab Invest. 2012;92:1451‐1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J, Yang XM, Wang YH, et al. Monoamine oxidase A suppresses hepatocellular carcinoma metastasis by inhibiting the adrenergic system and its transactivation of EGFR signaling. J Hepatol. 2014;60:1225‐1234. [DOI] [PubMed] [Google Scholar]
- 32. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941‐D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao L, Xiong D‐D, He R‐Q, et al. Identifying TF‐miRNA‐mRNA regulatory modules in nitidine chloride treated HCC xenograft of nude mice. Am J Transl Res. 2019;11:7503‐7522. [PMC free article] [PubMed] [Google Scholar]
- 35. Xiong D‐D, Feng Z‐B, Lai Z‐F, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 2019;10:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ou X, Lu Y, Liao L, et al. Nitidine chloride induces apoptosis in human hepatocellular carcinoma cells through a pathway involving p53, p21, Bax and Bcl‐2. Oncol Rep. 2015;33:1264‐1274. [DOI] [PubMed] [Google Scholar]
- 37. Gao L, Xiong D‐D, He R‐Q, et al. MIR22HG as a tumor suppressive lncRNA In HCC: a comprehensive analysis integrating RT‐qPCR, mRNA‐Seq and microarrays. Onco Targets Ther. 2019;12:9827‐9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao J, Xu T, Zheng J‐X, et al. Nitidine chloride inhibits hepatocellular carcinoma cell growth in vivo through the suppression of the JAK1/STAT3 signaling pathway. Int J Mol Med. 2013;32:79‐84. [DOI] [PubMed] [Google Scholar]
- 39. Fu XH. Exploration the anti‐angiogenesis effect of nitidine chloride based on mTOR signal pathway. Guangxi Med Univ. 2017;1:1‐86. [Google Scholar]
- 40. Du Y, Long Q, Zhang L, et al. Curcumin inhibits cancer‐associated fibroblast‐driven prostate cancer invasion through MAOA/mTOR/HIF‐1α signaling. Int J Oncol. 2015;47:2064‐2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao C‐P, Lin T‐P, Li P‐C, et al. Loss of MAOA in epithelia inhibits adenocarcinoma development, cell proliferation and cancer stem cells in prostate. Oncogene. 2018;37:5175‐5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L, Guo L, Wang Q, et al. DAPK1 as an independent prognostic marker in liver cancer. PeerJ. 2017;5:e3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70:1298‐1316. [DOI] [PubMed] [Google Scholar]
- 44. Huang B, Bao J, Cao YR, Gao HF, Jin Y. Cytochrome P450 1A1 (CYP1A1) catalyzes lipid peroxidation of oleic acid‐induced HepG2 cells. Biochemistry. 2018;83:595‐602. [DOI] [PubMed] [Google Scholar]
- 45. Davydova NY, Dangi B, Maldonado MA, Vavilov NE, Zgoda VG, Davydov DR. Toward a systems approach to cytochrome P450 ensemble: interactions of CYP2E1 with other P450 species and their impact on CYP1A2. Biochem J. 2019;476:3661‐3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimada T, Murayama N, Kakimoto K, et al. Oxidation of 1‐chloropyrene by human CYP1 family and CYP2A subfamily cytochrome P450 enzymes: catalytic roles of two CYP1B1 and five CYP2A13 allelic variants. Xenobiotica. 2018;48:565‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Natsui K, Mizuno Y, Tani N, Yabuki M, Komuro S. Retraction Note to: identification of CYP3A4 as the primary cytochrome P450 responsible for the metabolism of tandospirone by human liver microsomes. Eur J Drug Metab Pharmacokinet. 2019;44:853. [DOI] [PubMed] [Google Scholar]
- 48. Bonomo S, Jorgensen FS, Olsen L. Dissecting the cytochrome P450 1A2‐ and 3A4‐mediated metabolism of aflatoxin B1 in ligand and protein contributions. Chemistry. 2017;23:2884‐2893. [DOI] [PubMed] [Google Scholar]
- 49. Zhou R, Liu M, Liang X, Su M, Li R. Clinical features of aflatoxin B1‐exposed patients with liver cancer and the molecular mechanism of aflatoxin B1 on liver cancer cells. Environ Toxicol Pharmacol. 2019;71:103225. [DOI] [PubMed] [Google Scholar]
- 50. Nirogi R, Bhyrapuneni G, Muddana NR, et al. Absorption, distribution, metabolism, excretion (ADME), drug‐drug interaction potential and prediction of human pharmacokinetics of SUVN‐G3031, a novel histamine 3 receptor (HR) inverse agonist in clinical development for the treatment of narcolepsy. Eur J Pharm Sci. 2020;152:105425. [DOI] [PubMed] [Google Scholar]
- 51. Kim D‐H, Choi J‐M. Chitinase 3‐like‐1, a novel regulator of Th1/CTL responses, as a therapeutic target for increasing anti‐tumor immunity. BMB Rep. 2018;51:207‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hombach A, Sent D, Schneider C, et al. T‐cell activation by recombinant receptors: CD28 costimulation is required for interleukin 2 secretion and receptor‐mediated T‐cell proliferation but does not affect receptor‐mediated target cell lysis. Cancer Res. 2001;61:1976‐1982. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Figure S9
Figure S10
Figure S11
Figure S12
Figure S13
Figure S14
Figure S15
Table S1‐S6
Data Availability Statement
The original data are available from the corresponding author upon request.


