Abstract
The extensive sequence data generated from SARS-CoV-2 during the 2020 pandemic has facilitated the study of viral genome evolution over a brief period of time. This has highlighted instances of directional mutation pressures exerted on the SARS-CoV-2 genome from host antiviral defense systems. In this brief review we describe three such human defense mechanisms, the apolipoprotein B mRNA editing catalytic polypeptide-like proteins (APOBEC), adenosine deaminase acting on RNA proteins (ADAR), and reactive oxygen species (ROS), and discuss their potential implications on SARS-CoV-2 evolution.
Keywords: SARS-CoV-2, Virus evolution, Genome editing, ADAR, APOBEC, ROS
1. Introduction
During the 2020 pandemic, SARS-CoV-2 has so far infected more than 33 million people, resulting in over 1 million deaths attributed to COVID-19 [1,2]. SARS-CoV-2 is a positive sense (+) single-stranded (ss) RNA virus belonging to the Coronaviridae family, and it possesses one of the largest genomes (∼30 kb) among RNA viruses [3]. Unusually for RNA viruses, coronaviruses have proofreading machinery, thus possessing only a moderate mutation rate relative to other RNA viral genomes [4].
Genetic variation arises by replication errors, recombination, and shuffling of genomic segments providing the raw material upon which natural selection will act. Yet, certain host antiviral defense systems edit the viral genome in very specific patterns, thus creating genetic variation in a highly directed manner, which, in turn, restricts the sequence space that the virus may occupy. This brief review discusses three host factors that can directly edit the SARS-CoV-2 genome and thereby impact upon the genomic evolution of this pathogen (Fig. 1 ).
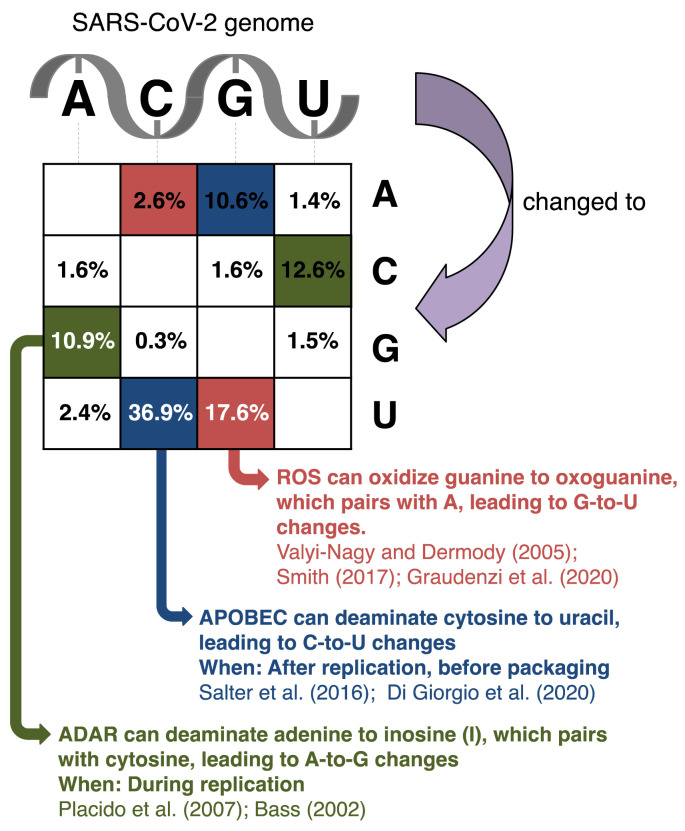
Fig. 1.
Matrix showing the distribution of genomic changes in SARS-CoV-2 sequences deposited at GISAID (https://www.gisaid.org/; [71]) as of October 2nd, 2020. Changes are accumulated across 79,887 samples and mapped onto the reference SARS-CoV-2 genome sequence, and the percentages of changes were recorded as previously described [21]. Changes at individual sites may therefore represent multiple independent events, and the most prominent changes are most likely underestimated [21]. The three types of changes resulting from the activity of ROS, APOBEC, and ADAR (G-to-U, C-to-U, and A-to-G, see main text) are highlighted in red, blue, and green, respectively. The types of changes that would result of the same host factors acting on the complement strand on double-stranded RNA are similarly colored but in a checkered pattern.
2. APOBEC
The human genome encodes eleven APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) proteins, a family of zinc-dependent deaminases [5]. These are APOBEC1, APOBEC2, APOBEC3 (with family members A, B, C, D, F, G, and H), APOBEC4, and AID (activation-induced cytidine deaminase). Most of the APOBEC proteins are shown to catalyze cytosine deamination to uracil (C-to-U) of foreign single-stranded DNA and RNA [6]. This hypermutational effect of APOBEC proteins has been extensively studied in HIV-1 infection [[7], [8], [9], [10]] but is also reported for other retroviruses as well as DNA viruses (reviewed in Harris & Dudley 2015 & Moris et al., 2014) [11,12].
APOBEC proteins show a preference for deaminating cytosines following thymine/uracil, 5’-[T/U]C-3′, for both DNA and RNA substrates [5,[13], [14], [15]]. A notable exception is human APOBEC3G, which preferentially targets a 5′–CC–3′ motif [7].
CpG dinucleotides are prominent targets for host antiviral defenses such as the zinc finger antiviral protein (ZAP) [16]. ZAP targets CpG dinucleotides, inhibiting a range of viruses (reviewed in Chemudupati et al., 2019) [17] as well as endogenous retroelements [18]. Consistently, CpG-creating mutations were found to be evolutionary costly in a range of viruses [19]. Although APOBEC proteins do not specifically target CpG motifs, it is noteworthy that their activity at [T/U]CG sites could result in CpG depletion [20,21].
Interestingly, APOBEC proteins, in particular APOBEC3F and APOBEC3G, have been shown to restrict HIV replication in a non-deaminase dependent matter – hence not resulting in hypermutation [8,10,11]. All proteins of the APOBEC3 family inhibit propagation of the abundant human endogenous retroelement, LINE-1 [22], with deaminase-dependent and -independent mechanisms being deployed by different APOBEC3 proteins [23].
2.1. APOBECs editing the SARS-CoV-2 genome
An excess of C-to-U substitutions was observed early in the SARS-CoV-2 pandemic [24], and the genomic context of these substitutions was enriched for APOBEC target sites [25]. Testing SARS-CoV-2 and six other human coronaviruses, Wei and colleagues found that only viruses regularly infecting tissues with high expression of APOBEC – and other antiviral proteins – exhibited CpG-depletion and U-rich genomes [26].
A prerequisite for APOBEC-driven hypermutation of SARS-CoV-2 is APOBEC gene expression activity during infection. In the alphacoronavirus, HCoV-229E, associated with the common cold, an early up-regulation of APOBEC3B has been observed [27]. Further, Blanco-Melo et al. (2020) found APOBEC3A to be among the most abundant mRNAs in COVID-19 patients [28].
Variation in APOBEC genes exists among different human populations. Seven human APOBEC3H haplotypes have been reported, conferring differences in host resistance to HIV infections [29]. Besides APOBEC3H, polymorphisms in APOBEC3D, APOBEC3F, and APOBEC3G have been reported from a HIV-infected cohort [30]. Seven single-nucleotide polymorphisms affecting the gene expression of APOBEC3D and APOBEC3G have further been hypothesized to affect SARS-CoV-2 susceptibility [31]. Whether these haplotypes impact SARS-CoV-2 infection, disease severity and clinical outcome is unknown and warrants further study in an analogous manner to prior studies on HIV.
The APOBEC1 complementation factor, A1CF, part of the APOBEC1 editing complex [32], was recently experimentally found to interact with SARS-CoV-2 [33] (see below). Although A3C, A3F, and A3H were previously shown to inhibit infection of HCoV-NL63, another alphacoronavirus member inducing bronchiolitis [34], this was not caused by hypermutation through editing [35].
2.2. SARS-CoV-2 genome evolution driven by host APOBEC proteins
Global samples of SARS-CoV-2 genome sequences reveal extensive C-to-U mutations, a pattern that can be followed progressively throughout the pandemic [21]. One study estimated that 52% of the observed non-synonymous mutations in SARS-CoV-2 were the result of C-to-U changes [36], and the progressive loss of genomic cytosines may result in the depletion of alanine, histidine, glutamine, proline, and threonine codons severely restricting the evolutionary trajectory of SARS-CoV-2 [37].
3. ROS and SARS-CoV-2 genome
Reactive oxygen species (ROS), which oxidize proteins, lipids, and nucleic acids can lead to virus inactivation and mutagenesis, represents another strategy utilized by a host cell to combat viral infections [38,39]. ROS have been found to play either beneficial or deleterious roles during different viral infections, enhancing viral replication during the early stages for certain viral species, while helping in the immunomodulation and inactivation of other viruses via oxidative burst [40]. Interestingly, ROS can also cause the oxidation of guanine to 7,8-dihydro-8-oxo-2′-deoxyguanine (oxoguanine) that can readily base pair with adenine, yielding G-to-T transversions [41]. It was recently hypothesized that G-to-U and C-to-A changes – the second most common observed substitutions in SARS-CoV-2 genomes – might be associated with the mutagenic activity of ROS [42].
4. ADAR
Adenosine deamination in dsRNAs is attributed to adenosine deaminase acting on RNA (ADAR) enzymes, which are able to convert A-to-I (inosine), but mainly in Alu sequences (an endogenous retroelement) [43]. There are three ADAR genes encoded in the human genome; the first two, ADAR1 and ADAR2, are interferon-inducible and catalytically active for adenosine deamination, while ADAR3, which is expressed mainly in the brain, has no reported ADAR activity [44,45]. The human ADAR1 gene is expressed in most tissues [46]. It has two isoforms, ADAR1p110, constitutively expressed in the majority of cell types, principally acting in the nucleus, and interferon-stimulated ADAR1p150, which primarily operates in the cytoplasm [47]. ADAR1 is considered a “master regulator” of cytoplasmic innate immunity regulating multiple sensors, such as Mda5, RIG-I, OAS and PKR, which detect intracellular dsRNA (which can arise during the replication-transcription process of (+)ssRNA viruses including SARS-CoV-2) and these sensors are essential in fighting viral infections [45].
Mutations at the ADAR1 locus have been linked to human genetic diseases, including the Aicardi–Goutières syndrome, an inflammatory disorder that phenocopies congenital viral infection [48], and the pigmentation disorder, dyschromatosis symmetrica hereditaria [49].
4.1. ADAR-mediated editing of the viral genome
Di Giorgio et al. (2020) analyzed publicly available RNA-seq data of bronchoalveolar lavage fluids from COVID-19 patients [25]. They found that A-to-G changes were distributed equally across the viral genome, and hypothesized that ADAR could be active against the SARS-CoV-2 genome. Similarly, Picardi et al. (2020) used RNA-seq data from infected human cell lines, Vero cells, and clinical samples from time-series experiments [50]. By analyzing the extent of editing at Alu sequences (known to be targets of ADAR1), the authors estimated ADAR1 activity and found evidence for this on both viral and human transcripts. Low levels of editing were observed at early timepoints (4 h post-infection), where both the activity of ADAR and interferon activation is low. However, after 24 h post-infection, higher levels of A-to-I editing were recorded, although accounting for <1% of sites. Clearly, nucleotide variation due to sequencing or polymerase errors might also contribute to the observed substitutions [50].
Besides sequence analysis approaches to show ADAR1 activity, direct interaction between viral RNA and proteins has been established. Using RNA antisense purification and mass spectrometry (RAP-MS), Schmidt and colleagues identified RNA-protein interactions in SARS-CoV-2-infected human cells [33]. From this, notably both ADAR and APOBEC were found as frequent interactors with SARS-CoV-2 RNA.
In contrast to the above findings, a few studies have found no evidence of ADAR1 activity acting on SARS-CoV-2. DNA nanoball sequencing from Vero cells infected with SARS-CoV-2 suggested no ADAR-mediated editing [51]. Further, the same study performed an independent analysis of the previous dataset presented by Kim et al. (2020) [51] and did not detect any A-to-G editing [50]. Using predicted secondary structures of target sites, Klimczak et al. (2020) found statistically significant ADAR editing in rubella virus genomes, another (+)ssRNA virus), but not in SARS-CoV-2 genomes [52].
4.2. Is there a link between ADAR1 and autoimmune diseases after COVID recovery?
Double-stranded RNAs are the pathogen-associated molecular pattern associated with the strong induction of cellular stress and interferon responses. ADAR1 is one of the major regulators of self-tolerance and innate immune activation that involves recognition of dsRNA and its further processing by downstream antiviral pathways [45,53]. ADAR1 can exert either antiviral or pro-viral effects dependent upon the infecting virus [54]. For example, hyperediting of HCV and LCMV viral genomes lead to antiviral effects, while ADAR1 editing of influenza A RNA enhances viral protein expression [54].
It was recently shown that COVID-19 leads to suboptimal interferon responses in comparison to other respiratory viruses [28], and one might speculate that this may arise if ADAR1 activity contributes to the low type I interferon response. ADAR marks the dsRNA with the A-to-I deamination, allowing the edited RNA duplexes to escape other molecular sensors of dsRNA [45]. There is a threshold for the tolerable number of dsRNAs present within the cytosol, which, when exceeded leads to autoimmunity but favors viral infection [55,56]. ADAR-mediated editing of viral RNAs might result in levels of edited RNAs above this threshold. It is not uncommon that some viral infections can lead to autoimmune complications after recovery [57]. The high incidence of different autoimmune conditions has been recorded after the resolution of COVID-19 in adult and, also, pediatric patients are in line with this observation [[58], [59], [60], [61], [62]]. Besides a potential contribution of ADAR in the evasion of type I interferon responses, the virus also has various proteins that inhibit type I interferon induction and signaling [63]. Previously, knockdown of ADAR1 was shown to lead to viral inhibition, which enhanced interferon stimulation in primary macrophages [64]. ADAR1 inhibitors might thus be another strategy to boost antiviral response in viruses that trigger suboptimal interferon responses as seen during SARS-CoV-2 infection.
5. Strandedness of SARS-CoV-2
Upon entry into the host cell, SARS-CoV-2 exists as a single-stranded, positive-sense RNA. This strand is then replicated – during which the virus exists as a double-stranded RNA – and the resulting negative-strand then acts as a template for both replication and transcription (both types of products, thus being positive-sense) [65,66].
From short-read sequences mapped onto the SARS-CoV-2 reference genome, Graudenzi et al. (2020) identified C-to-U changes [42]. C-to-U change occurring on the negative strand will result in observed G-to-A changes using this approach. The authors observed a ratio of 17:1 between C-to-U and G-to-A changes, consistent with APOBEC predominantly working on the positive strand RNA. Remarkably, a similar ratio of 17:1 was observed between C-to-A and G-to-U changes, which would be the result of ROS-induced mutagenesis on the positive and negative strands, respectively [42]. This consistency prompted the speculation that the 17:1 ratio reflects the molar ratio between the two viral strands.
The percentages presented in Fig. 1 are derived from comparing approximately 80,000 assembled consensus genomes to the SARS-CoV-2 reference genome (MN908947.1) and registering all detected changes [21]. This means that a change needs to be present in the majority of viral transcripts in order to be included in the consensus genome, and that it will only be counted once regardless if it is present in one or all of the sample genomes [21]. Therefore, it is not expected that these percentages will reflect the ratios reported by Graudenzi et al. (2020) [42]. It is noteworthy, however, that the percentages in Fig. 1 are similar for A-to-G and U-to-C changes, potential hallmarks of ADAR activity (that works on double-stranded RNA), whereas this is not the case for C-to-U and G-to-A, neither for C-to-A and G-to-U, consistent with APOBEC and ROS, respectively, predominantly acting on the positive stranded viral RNA.
6. Summary
Global travel, societal interactions and the interconnected modern world provide abundant opportunities for the rapid spreading of viral infections that are becoming severe health and an economic burden for humanity [67]. A better understanding of viral genome changes can help design better diagnostics, therapeutics and prophylactic vaccines [68,69]. Herein, we have discussed how host innate immune defenses might drive nucleotide substitutions and genomic evolution in SARS-CoV-2 in a directional manner. These described hypermutation patterns despite the relatively moderate mutation rate is evidence of the adaptation process of a virus of recent zoonotic origin [70].
Di Giorgio et al. [25] have identified a third class of changes comprising A-to-T/T-to-A transversions in SARS-CoV-2 genomes, but the mechanistic basis for this is currently unknown and requires further study.
APOBEC, ROS and ADAR are effective sources of nucleotide changes and from Fig. 1, it is evident that these editing agents may potentially account for the vast number of observed changes, certainly if assuming that they may also act on dsRNAs. The SARS-CoV-2 pandemic offers a challenge of global dimensions and successfully controlling the spread of the virus will heavily depend on insight into the biology of the virus. In this respect, host-directed genome editing is likely to play a substantial role and may hypothetically confer susceptibility and potentially a degree of innate resistance for individuals harboring certain haplotypes. As SARS-CoV-2 will inevitably become the most closely monitored virus in terms of real-time sequence data, this concerted effort of the global scientific community is essential to ameliorate the profound burden of disease elicited by this zoonotic pathogen.
Declaration of competing interest
None declared.
Acknowledgments
We thank all laboratories which have contributed sequences to the GISAID database.
This work was supported by funding from King Abdullah University of Science and Technology (KAUST), Office of Sponsored Research (OSR). Work in AP’s laboratory is supported by the KAUST faculty baseline fund (BAS/1/1020-01-01) and the R3T initiative of KAUST.
References
- 1.WHO Coronavirus disease (COVID-2019) situation reports. World Heal Organ. 2020 Retrieved from October 5, 2020. [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr A.R., Perlman S. Coronaviruses Methods Protoc.; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salter J.D., Smith H.C. Modeling the embrace of a mutator: APOBEC selection of nucleic acid ligands. Trends Biochem. Sci. 2018;43:606–622. doi: 10.1016/j.tibs.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter J.D., Bennett R.P., Smith H.C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 8.Holmes R.K., Koning F.A., Bishop K.N., Malim M.H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 9.Kim E.Y., Lorenzo-Redondo R., Little S.J., Chung Y.S., Phalora P.K., Maljkovic Berry I., et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman E.N.C., Holmes R.K., Craig H.M., Klein K.C., Lingappa J.R., Malim M.H., et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 11.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;(479–480):131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moris A., Murray S., Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thielen B.K., McNevin J.P., McElrath M.J., Hunt B.V.S., Klein K.C., Lingappa J.R. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S., Patnaik S.K., Thomas Taggart R., Kannisto E.D., Enriquez S.M., Gollnick P., et al. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015;6 doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S., Wang J., Alqassim E., Portwood S., Cortes Gomez E., Maguire O., et al. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takata M.A., Gonçalves-Carneiro D., Zang T.M., Soll S.J., York A., Blanco-Melo D., et al. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chemudupati M., Kenney A.D., Bonifati S., Zani A., McMichael T.M., Wu L., et al. From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:382–394. doi: 10.1016/j.bbamcr.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moldovan J.B., Moran J.V. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caudill V.R., Qin S., Winstead R., Kaur J., Tisthammer K., Pineda E.G., et al. CpG-creating mutations are costly in many human viruses. Evol. Ecol. 2020;34:339–359. doi: 10.1007/s10682-020-10039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020;37:2699–2705. doi: 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadykov M., Mourier T., Guan Q., Pain A. Short sequence motif dynamics in the SARS-CoV-2 genome suggest a role for cytosine deamination in CpG reduction. BioRxiv. 2020 doi: 10.1101/2020.06.19.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinomoto M., Kanno T., Shimura M., Ishizaka Y., Kojima A., Kurata T., et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y., Goubran M.H., Follack T.B., Chelico L. Deamination-independent restriction of LINE-1 retrotransposition by APOBEC3H. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RATG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts. Genes. 2020;11:1–13. doi: 10.3390/genes11070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgio S Di, Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv. 2020;6 doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Y., Silke J.R., Aris P., Xia X. Coronavirus genomes carry the signatures of their habitats. BioRxiv. 2020 doi: 10.1101/2020.06.13.149591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman N., Jacob-Hirsch J., Drori Y., Eran E., Kol N., Nayshool O., et al. Transcriptomic profiling of human corona virus (HCoV)-229E -infected human cells and genomic mutational analysis of HCoV-229E and SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.08.17.253682. 2020.08.17.253682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Refsland E.W., Hultquist J.F., Luengas E.M., Ikeda T., Shaban N.M., Law E.K., et al. Natural polymorphisms in human APOBEC3H and HIV-1 vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matume N.D., Tebit D.M., Gray L.R., Turner S.D., Rekosh D., Bessong P.O., et al. Characterization of APOBEC3 variation in a population of HIV-1 infected individuals in northern South Africa. BMC Med. Genet. 2019;20 doi: 10.1186/s12881-018-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotroneo C.E., Mangano N., Dragani T.A., Colombo F. Lung expression of genes encoding SARS-CoV-2 cell entry molecules and antiviral restriction factors: interindividual differences are associated with age and germline variants. BioRxiv. 2020 doi: 10.1101/2020.06.24.168534. [DOI] [Google Scholar]
- 32.Dance G.S.C., Sowden M.P., Cartegni L., Cooper E., Krainer A.R., Smith H.C. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J. Biol. Chem. 2002;277:12703–12709. doi: 10.1074/jbc.M111337200. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt N., Lareau C.A., Keshishian H., Melanson R., Zimmer M., Kirschner L., et al. A direct RNA-protein interaction atlas of the SARS-CoV-2 RNA in infected human cells. BioRxiv. 2020 doi: 10.1101/2020.07.15.204404. 2020.07.15.204404. [DOI] [Google Scholar]
- 34.Abdul-Rasool S., Fielding B.C. Understanding human coronavirus HCoV-NL63. Open Virol. J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milewska A., Kindler E., Vkovski P., Zeglen S., Ochman M., Thiel V., et al. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-24448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmonds P. Rampant C->U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses - causes and consequences for their short and long evolutionary trajectories. mSphere. 2020;5 doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danchin A., Marlière P. Cytosine drives evolution of SARS-CoV-2. Environ. Microbiol. 2020;22:1977–1985. doi: 10.1111/1462-2920.15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valyi-Nagy T., Dermody T.S. Role of oxidative damage in the pathogenesis of viral infections of the nervous system. Histol. Histopathol. 2005;20:957–967. doi: 10.14670/HH-20.957. [DOI] [PubMed] [Google Scholar]
- 39.Smith E.C. The not-so-infinite malleability of RNA viruses: viral and cellular determinants of RNA virus mutation rates. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reshi M.L., Su Y.C., Hong J.R. RNA viruses: ROS-mediated cell death. Int J Cell Biol. 2014 doi: 10.1155/2014/467452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.David S.S., O’Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graudenzi A., Maspero D., Angaroni F., Piazza R., Ramazzotti D. Mutational signatures and heterogeneous host response revealed via large-scale characterization OF SARS-COV-2 genomic diversity. BioRxiv. 2020 doi: 10.1101/2020.07.06.189944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D.D.Y., Kim T.T.Y., Walsh T., Kobayashi Y., Matise T.C., Buyske S., et al. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C.X., Cho D.S.C., Wang Q., Lai F., Carter K.C., Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/S1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamers M.M., van den Hoogen B.G., Haagmans B.L. ADAR1: “editor-in-chief” of cytoplasmic innate immunity. Front. Immunol. 2019;10:1763. doi: 10.3389/fimmu.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picardi E., Manzari C., Mastropasqua F., Aiello I., D’Erchia A.M., Pesole G. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci. Rep. 2015;5 doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savva Y.A., Rieder L.E., Reenan R.A. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallo A., Vukic D., Michalík D., O’Connell M.A., Keegan L.P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017;136:1265–1278. doi: 10.1007/s00439-017-1837-0. [DOI] [PubMed] [Google Scholar]
- 49.Miyamura Y., Suzuki T., Kono M., Inagaki K., Ito S., Suzuki N., et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am. J. Hum. Genet. 2003;73:693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picardi E., Mansi L., Pesole G. A-to-I RNA editing in SARS-COV-2: real or artifact? BioRxiv. 2020 doi: 10.1101/2020.07.27.223172. 2020.07.27.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimczak L.J., Randall T.A., Saini N., Li J.-L., Gordenin D.A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. BioRxiv. 2020 doi: 10.1101/2020.08.03.234005. 2020.08.03.234005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakahama T., Kato Y., Kim J.I., Vongpipatana T., Suzuki Y., Walkley C.R., et al. ADAR 1-mediated RNA editing is required for thymic self-tolerance and inhibition of autoimmunity. EMBO Rep. 2018;19 doi: 10.15252/embr.201846303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomaselli S., Galeano F., Locatelli F., Gallo A. Adars and the balance game between virus infection and innate immune cell response. Curr. Issues Mol. Biol. 2015;17:37–52. [PubMed] [Google Scholar]
- 55.Pfaller C.K., Donohue R.C., Nersisyan S., Brodsky L., Cattaneo R. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1 p150. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad S., Mu X., Yang F., Greenwald E., Park J.W., Jacob E., et al. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell. 2018;172:797–810. doi: 10.1016/j.cell.2017.12.016. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smatti M.K., Cyprian F.S., Nasrallah G.K., Al Thani A.A., Almishal R.O., Yassine H.M. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11 doi: 10.3390/v11080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talarico R., Stagnaro C., Ferro F., Carli L., Mosca M. Symmetric peripheral polyarthritis developed during SARS-CoV-2 infection. Lancet Rheumatol. 2020;2 doi: 10.1016/S2665-9913(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mateu-Salat M., Urgell E., Chico A. SARS-COV-2 as a trigger for autoimmune disease: report of two cases of Graves’ disease after COVID-19. J. Endocrinol. Invest. 2020;43:1527–1528. doi: 10.1007/s40618-020-01366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Restivo D.A., Centonze D., Alesina A., Marchese-Ragona R. Myasthenia gravis associated with SARS-CoV-2 infection. Ann. Intern. Med. 2020 doi: 10.7326/l20-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalakas M.C. Guillain-Barré syndrome: the first documented COVID-19-triggered autoimmune neurologic disease: more to come with myositis in the offing. Neurol Neuroimmunol Neuroinflammation. 2020;7 doi: 10.1212/NXI.0000000000000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ribero M.S., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pujantell M., Riveira-Muñoz E., Badia R., Castellví M., Garcia-Vidal E., Sirera G., et al. RNA editing by ADAR1 regulates innate and antiviral immune functions in primary macrophages. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-13580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) BT - coronavirus disease 2019 (COVID-19): epidemiology, pathogenesis, diagnosis, and therapeutics. Coronavirus Dis. 2019;2020:23–31. doi: 10.1007/978-981-15-4814-7_3. [DOI] [Google Scholar]
- 67.Mas-Coma S., Jones M.K., Marty A.M. COVID-19 and globalization. One Heal. 2020;9 doi: 10.1016/j.onehlt.2020.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rice A.M., Morales A.C., Ho A.T., Mordstein C., Mühlhausen S., Watson S., et al. Evidence for strong mutation bias towards, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 2020 doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-COV-2 infection of animal hosts. Pathogens. 2020;9:1–27. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Challenges. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]