Abstract
In recent years, it has been revealed that Parkinson’s disease pathology may begin to manifest in the gastrointestinal track at a much earlier time point than in the brain. This paradigm shift has been suggested following evidence in humans that has been reproduced in animal models. Since rodent models cannot recapitulate many of the human disease features, human induced pluripotent stem cells derived from Parkinson’s patients have been used to generate brain organoids, greatly contributing to our understanding of the disease pathophysiology. To understand the multifaced aspects of Parkinson’s disease, it may be desirable to expand the complexity of these models, to include different brain regions, vasculature, immune cells as well as additional diverse organ-specific organoids such as gut and intestine. Furthermore, the contribution of gut microbiota to disease progression cannot be underestimated. Recent biotechnological advances propose that such combinations may be feasible. Here we discuss how this need can be met and propose that additional brain diseases can benefit from this approach.
Subject terms: Neuroscience, Diseases
Parkinson’s disease
The synucleinopathy Parkinson’s disease (PD), is a neurodegenerative disease characterized by abnormal accumulation of the alpha-synuclein (α-Syn) protein in the brain. A key neuropathological hallmark of PD are neuronal inclusions positive for the protein α-synuclein known as Lewy bodies and Lewy neurites. Most patients present a movement disorder that can include tremor, slowness of movement, rigidity, and postural instability [1]. Additional nonmovement symptoms include neuropsychiatric problems, altered smell sense, sleeping difficulties as well as orthostatic hypotension, constipation, and urinary incontinence [2]. The motor symptoms are attributed to dopaminergic cell loss within the substantia nigra (SN) pars compacta, resulting in subsequent dysfunction of the basal ganglia, a cluster of deep nuclei that participate in the initiation and execution of movements [3]. Genetics plays an important role in PD, with disease–susceptibility loci including more than 90 genes, including SNCA (Synuclein Alpha), LRRK2 (Leucine Rich Repeat Kinase 2), GBA (Glucosylceramidase Beta), and MAPT (Microtubule Associated Protein Tau) [4]. However, genetics is not the only contributing factor to the disease; it is likely that the interactions between age, genetics, epigenetics, and environmental factors can trigger the disease [5–7]. It has been recently suggested that COVID-19 may enhance disease progression, yet additional studies are needed to investigate this suggestion in depth [8–10]. The genetic information facilitated modeling of some of the common monogenic Parkinson’s mutations using genetic approaches in cellular and animal systems. For example, it has been demonstrated that GBA mutations in mouse models result in increased levels of α-Syn [11–13]. Forced expression of the GBA enzyme in mouse brains ameliorated histopathological and memory aberrations [13]. Accumulation of the α-Syn protein in the brain occurs rather late in the disease. Interestingly, about 14 years after fetal dopaminergic neurons are implanted in the striatum of PD patients, these neurons exhibit Lewy pathology in the cell bodies and axons [14, 15]. These findings and others resulted in the theory that PD may be a prion-related disorder, yet some of the criteria fall short for the full definition [16, 17]. Postmortem brain sections of these PD patients at different time points post-implantation suggest the inflammation and microglial activation in the grafts are present long before the accumulation of α-Syn (Fig. 1a). These findings contributed to the concept that microglia participate in the propagation and spread of α-Syn pathology [16, 17]. In addition, a number of studies suggest the adaptive immune system is involved in disease progression [18]. In recent years, it has become evident that accumulated α-Syn protein can be observed in the gastrointestinal (GI) tract in the early stages of the disease [19]. Interestingly, neuronal cell loss in the enteric nervous system (ENS) of PD patients and animal models has not been observed, despite the accumulation of α-Syn aggregates in the GI tract [20, 21]. It has been proposed that there could be transfer of the aggregated α-Syn protein from the GI tract to the brain [22].
Fig. 1. Schematic presentation of PD in human patients and mouse models.
a Comparative changes in the gut-brain axis of a PD patient compared to a healthy individual. b α-Synuclein can be transported in a retrograde fashion from the gut to the brain, recapitulating PD pathology in the mouse. c Transplantation of gut microbiome from PD patient into mice leads to several PD phenotypes, including motor deficets and chronic inflamation.
Indeed, animal models have demonstrated that α-Syn protein aggregates can move from the gut to the brain via the vagus nerve, particularly in aged mice [20, 23–25]. This transfer was also observed in a mouse model exposed to an environmental toxin that induced the production and secretion of the α-Syn protein, and that the recession of the autonomic nerve halted this process [26] (see Fig. 1b). Supporting this notion, longitudinal analysis of people that have undergone bilateral vagotomy suggested a decreased risk for the development of PD [27]. Introduced expression of the GBA enzyme using an inducible viral expression system with high affinity for the peripheral nervous system in enteric neurons partially restored the GI phenotype observed in mice overexpressing the α-Syn protein in Thy1-positive projection neurons [25]. However, not only is the ENS relevant to the disease etiology in the GI, rather both the gut endothelium [28], and gut microbiome contribute significantly [29–33]. The human body hosts a rich collection of microorganisms, most of them residing in the gut, where they are involved in food digestion as well as providing the host different by-products [34]. The brain gut-axis is bidirectional. The brain affects the intestinal activity and function [35] and the gut microbiome is involved in the maintenance of the mucus in the gut epithelium, and its metabolites affect the immune system and brain function [34]. Interestingly, PD patients have been found to have a different composition of microbiota [32, 36, 37] and the patients exhibit altered concentrations of short chain fatty acids, as well as altered plasma concentrations of different cytokines, suggesting the involvement of the immune system [31, 36]. The intestinal microbiota has an instructive role in PD, and is required for the motor deficits, microglia activation, and α-Syn pathology [30]. Supporting this notion is the finding that the introduction of specific microbial metabolites is sufficient to induce the pathology [30]. Implantation of PD fecal microbes into mice resulted in a more pronounced phenotype in comparison with those obtained from healthy controls [30] (Fig. 1c). It has been proposed that the exposure to amyloid proteins existing in gut microbes can promote the aggregation of α-Syn [38].
The complex interactions between PD, gut microbes, and the immune system has been demonstrated in PTEN Induced Kinase 1 (Pink1) knockout mice [39].
Mutations in either PINK1, a ubiquitin kinase, or in Parkin RBR E3 Ubiquitin Protein Ligase (PRKN), also known as Parkinson Disease Protein 2, PARK2, are associated with PD. The function of the encoded proteins is related to mitophagy and they were found to contribute to the understanding of the gut-brain axis in PD pathology [40, 41]. In relation to PD, knockout mice for both Pink1 and Park2 demonstrated a pronounced inflammatory response to exhaustive exercise [42]. Another study revealed there is an increase in the presentation of mitochondrial antigens in immune cells in the absence of PINK1 or Parkin, suggesting that autoimmune mechanisms are involved in the development of PD [40]. Furthermore, autoimmune mechanisms evolve when the intestines of Pink1 knockout mice are infected with bacteria, resulting in the establishment of a group of cytotoxic mitochondria-specific T cells in the periphery and in the brain [39]. These specific T cells are able to kill dopaminergic neurons in vitro. The mice develop motor impairment, which can improve following L-DOPA treatment [39]. L-DOPA (l-3,4-dihydroxyphenylalanine), an amino acid precursor that passes the blood-brain barrier (BBB) to be taken up by the dopaminergic neurons and converted into dopamine, is commonly used for treatment of PD patients. The gut microbiota can also be involved in metabolism of L-DOPA, which can interfere with the disease management [43]. Eradicating some bacteria, such as Helicobacter pylori, more commonly found in PD patients, has been shown to improve patients’ symptoms and enhanced the effectiveness of L-DOPA treatment [37]. A systematic study identified microbial species that convert L-DOPA to dopamine by tyrosine decarboxylase enzymes, then determined which species can dehydroxylate dopamine to m-tyramine. This second activity was found to be related to a single nucleotide polymorphism that induced an amino-acid substitution. To facilitate L-DOPA application and reduce its processing, the investigators identified a small molecule inhibitor that can inhibit the dehydroxylation of dopamine [43]. Despite all these studies indicating the role of the microbiome in PD, it should be noted that mice lacking the microbiome do not show any major neuronal dysfunction or PD like symptoms.
Development of organoid models to study PD
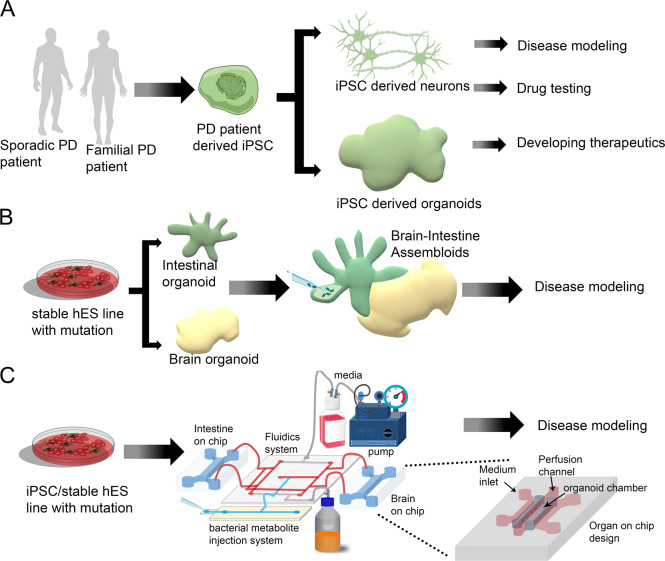
Organoids are 3D structures grown from stem cells, consisting of organ-specific cell types that self-organize through cell sorting and spatially restricted lineage commitment [44–46]. The popularity of human derived organoids for modeling development and disease has been increasing in recent years (Fig. 2a). Prerequisite for the development of organoid models for human diseases was the introduction of cell reprogramming by Yamanaka [47]. This technological breakthrough was used to generate iPSCs from PD patients, which were then differentiated into dopaminergic neurons [48, 49]. Nevertheless, it should be noted that neurons and other tissue-like structures that are derived from iPSCs usually lack maturity, and this is an issue that has not been completely solved yet [50]. Some approaches to solve this caveat has been coculturing of different cell types, for example coculturing of human neuronal progenitors with rodent astrocytes resulted in mutually synergistic maturation [51]. Generation of isogenic lines in which a particular mutation was corrected, or a new mutation was introduced into a control line, markedly reduced the variability due to different genetic backgrounds [52, 53]. Most of these studies investigated a common mutation in LRRK2 [48, 49, 53]. Genetic engineering of human cells was dependent upon the introduction of efficient genome editing tools as zinc-finger nucleases, transcription activator-like effector nucleases, and more recently clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas-based RNA-guided DNA endonucleases [54]. An additional technological breakthrough was the ability to generate different types of human organoids from iPSCs derived from patients, human embryonic stem cells, or organ-restricted stem cells [44]. Early success has been noted with the establishment of organoids from endodermal-derived organs such as the esophagus, gut, stomach, liver, pancreas, and lung [55–57]. Mesodermal-derived organs include kidney, heart, cartilage, bone, reproductive organs, and muscle. Successful renal and endometrium organoids have also been generated [55, 58, 59]. Ectoderm-derived organs include two main tissues; the surface ectoderm that will develop into skin and associated glands and hair, and the neural ectoderm that will develop into the brain, the spinal cord, and the neural crest [60]. Most relevant to current research on PD are brain organoids. Pioneering research from the lab of the late Yoshiki Sasai demonstrated that stem cells can recapitulate several features of organogenesis, including cell differentiation, spatial patterning and morphogenesis, and successfully generated organoids resembling different brain regions and retina [45]. Subsequent research from Lancaster and Knoblich demonstrated that it is possible to obtain a mixed regional identity using a relatively simple media [46, 61]. This field has increased dramatically over the last few years with multiple protocols [62]. Scientists can now generate connections between different brain regions by fusing structures known as “assembloids”, thus mimicking a higher organization level, which may prove critical in modeling diseases [63]. There have also been some advances in regards to generating functional networks [64]. Multiple studies have characterized the cell repertoire and diversity in organoids obtained from different protocols using single-cell (sc) analyses [65]. Overall, the general notion in the field is that although brain organoids are not identical to the developing human brain, these are useful models. Comparisons of sc-RNA-seq data from multiple brain organoids to data derived from the developing human brain indicate that the developmental trajectories and cell types in the organoids resemble those observed in the human embryonic brain [66]. A different study using both their own data combined with published data doubts the fidelity of the model and claims that despite presenting a broad cell classes, brain organoids do not recapitulate distinct cellular subtype identities or the appropriate progenitor maturation [67].
Fig. 2. Present and future in vitro models for PD.
a Existing approaches to model PD using iPSC-derived neurons/brain organoids. b Future prospect can combine several organoids (gut & brain organoids) to make an assembloid system where microbiota derived metabolites could be injected and thus it can mimic the gut brain axis in PD. c Further usage of the organ on chip system (the magnified view depicts the layout of organ on chip system) could mimic the gut brain axis where the sophisticated fluidics system can establish a connection between two on chip organs. In one chip it is possible to create intestine and brain in the other. The microfuidic system would establish the connection between the two and there would be an injection site for bacterial metabolites as depicted in the image. These approaches will improve the PD modeling and can be used to study disease progression or it can help in bulk drug screening.
Midbrain organoids containing dopaminergic neurons are of special interest for understanding PD. Two studies aimed at the generation of midbrain organoids started with 3D aggregates of neuroepithelial stem cells treated with activators of the WNT and Hedgehog pathways, embedding them in Matrigel droplets, followed by inducing the differentiation into human midbrain organoids [68, 69]. In these midbrain organoids they observed a large population of TH-, LMX1A-, and FOXA2-positive neurons, which were also positive for other ventral midbrain identity markers. They further demonstrated the presence of both A9 and A10 subtypes of midbrain dopaminergic neurons (GIRK2 and TH; CALBINDIN and TH, respectively). The presence of astrocytes and oligodendrocytes was also verified, and myelination was observed. Synaptic connectivity and electric activity were also demonstrated [68, 69]. A more recent study has optimized the protocol for midbrain organoid generation which could enable efficient drug testing [70].
The relevance of this model is underscored by studies demonstrating that midbrain-specific organoids derived from PD patients carrying the LRRK2-G2019S mutation can recapitulate disease-relevant phenotypes [71, 72]. In one study, isogenic 3D midbrain organoids with or without the PD LRRK2-G2019S mutation were generated and shown to recapitulate pathological hallmarks [71]. These mutant organoids exhibited increased susceptibility to induced neurotoxic damage, resulting in increased apoptosis. Phosphorylated α-Syn was localized in endosomes, and there was an increase in mitophagy. Reduction in the expression of a specific thiol-oxidoreductase, TXNIP, can significantly decrease aggregated α-Syn [71]. Another study focused on the cause for the decreased number and complexity of midbrain dopaminergic neurons in LRRK2-G2019S mutant organoids compared to controls [72]. The floor plate marker FOXA2, required for midbrain dopaminergic neuron generation, is increased in PD patient-derived midbrain organoids, suggesting a neurodevelopmental defect in midbrain dopaminergic neurons expressing LRRK2-G2019S [72].
Future possibilities
Despite being a movement disorder, it is clear that PD etiology is not restricted to the brain. PD is a multisystem condition that involves several organs and systems. Evidence of early involvement of the intestine and its’ interaction with the microbiome are accumulating. In parallel the understanding of the involvement of the immune system both innate and adaptive components are revealed. The cross-talk between the gut and the brain unfolds through transfer of substances/proteins/metabolites via the ENS and the vagus nerve to the brain, and with a possible contribution of vasculature system. This view dictates a more complex approach to PD modeling. Is it possible to combine all of these systems for in vitro studies? (see Fig. 2b, c for possible schemes).
Intestinal organoids have already been used in personalized medicine approaches, such as predicting the efficacy of Cystic Fibrosis drug treatments [73, 74]. Bioengineering approaches has enabled incorporating human intestinal organoids into small micro-engineered chips [75]. Both intestinal and epithelial organoids can be initiated from single adult stem cells obtained from patient epithelial biopsies, thus skipping the long reprogramming process required when starting from a somatic cell. However, the introduction of the microbiome to this culture is not a trivial task. One possibility may be to introduce specific bacterial-derived metabolites to the media, which would first require in-depth analysis and screening of multiple bioactive metabolites. Bacterial metabolites could be sensed either by G-protein-coupled receptors that are often expressed by intestinal epithelial cells, specific subsets of immune cells, or by tissues and/or cell types that are central to the host metabolism, such as pancreatic islet cells, adipocytes, and enteroendocrine cells of the gut [76]. A large proportion of the endogenous microbiota is anaerobic, and are localized on the apical cell surface, facing the lumen. However in culture, micro-organisms added to the media will only be able to interact with the basal and not the apical side of the cells, and be in an environment with relatively high oxygen concentration. To overcome these challenges, one approach has been to reverse the polarity, creating an “inside out” organization of the organoids [77]. Another approach has been to introduce the microorganisms into the lumen by microinjection [78]. This study managed to achieve a complex and stable microbiome by implementing innovations in bioengineering, using advanced organoid culture, microfabricated culturing devices, computer vision and semiautomated injection devices [78]. Organs-on-chip are microfluidic cell cultures that were generated as possible alternatives for the use of animal models with the promise that they can recapitulate the structure, function, physiology, and pathology of living human organs in vitro and possibly mimic interorgans interactions [79–83]. Most of the organoid systems cannot be precisely controlled and they have a limited capacity to provide for instructive cues that are required for organogenesis [82]. The initial intestinal organoids are limited as they are lacking endothelium-lined blood vessels and immune cells and are not exposed to fluid flow and other mechanical constraints. These deficits may be overcome in gut chip models [79, 84]. It is possible to generate one layer of intestinal epithelial cells on top of a lower layer of microvascular endothelial cells to fabricate a three-dimensional villi structure [84]. Furthermore, it is possible to co-culture these villi structures with living microbes [85, 86]. It should be noted that organs-on-chip and organoids represent two different but complementary approaches and the possibility to integrate these two approaches in a synergistic way is extremely exciting [82]. More recently, the organoid and organ chip approaches have been combined to develop a microfluidic primary human intestine chip model [87]. A possible caveat is that most microfabricated devices rely on the silicone-based polymer polydimethylsiloxane (PDMS), which could absorb small and hydrophobic molecules.
Introduction of the ENS into these complex models may prove to be a challenge [88, 89]. The ENS develops both from the vagal and the sacral neural crest which can be differentiated from human pluripotent stem cells (ES). Following their production, the cells were cocultured in the presence of smooth muscle cells, and were able to form interconnections [90]. These cells were also transplanted into immunodeficient mice and were found to repopulate the host colon. A tissue-engineering approach was used to develop human intestinal organoids incorporating ENS [91]. To incorporate vagal neural crest cells and ENS precursors into the developing intestinal organoids, mid/hindgut spheroids and neural crest cells were mechanically coaggregated. The crest cells were derived from human stem cells by low-speed centrifugation and then the aggregates were transferred to three-dimensional growth conditions for twenty-eight days. To achieve a complete maturation and add vascularization, these cultures were transplanted in mice. This approach yielded a tissue that was highly organized, integrated into the intestinal smooth muscle and drove NO-dependent relaxation. The authors noted that although CHAT-positive neurons were detected in vitro, they were not detected following transplantation, possibly reflecting the fetal nature of the transplanted organoids. Another study improved the maturity of the organoids by using an early in vivo coimplantation of the stem cell derived enteric neural crest cells with the intestinal organoids [92]. Thus, so far there has not been an in vitro solution for combing the ENS with the intestine as there was a need to implant different components to mice.
Advances have also been made in introducing the immune system into organoid cultures [93]. In one study, human intestinal stem cell-derived enteroid monolayers were cocultured with human monocyte-derived macrophages [94]. The addition of macrophages changed the physiology of enteroid monolayers by enhancing their barrier function and maturity. Another study added human polymorphonuclear leukocytes, a cell population comprised mainly of neutrophils, to human intestinal organoids, and then introduced either commensal or pathogenic bacteria [95]. While the commensal bacteria did not harm the development of the intestinal organoids, the pathogenic bacteria induced the loss of epithelial integrity and the production of interleukin 8 (IL-8). IL-8 induced rapid recruitment of neutrophils [95]. Another study used coculturing of intestinal organoids with fetal TNF-α producing CD4+ T cells, demonstrating that when T cells are introduced in low numbers they promote epithelium development, while in high numbers they mediate inflammation [96]. In a different study, human intestinal organoids were cocultured with human T lymphocytes, inducing the in vitro maturation of the organoids [97]. IL-2 was identified as the major factor that induced maturation [97]. A different approach involves transplanting the human organoids under the kidney capsule of immunocompromised mice in order to recruit missing cell types [98].
For PD studies, the main focus has been on midbrain-specific organoids, yet the ability to increase the complexity and the representation of additional cell types as well as defining the conditions that support the growth of other brain regions may be advantageous. Microglia are one of the target cell types to be included into PD models, and is of great importance for studying the involvement of innate immunity in the disease onset and progression [99]. Activation of microglia are thought to promote the disease or alternatively be involved in some neuroprotective aspects [100, 101]. Here we will discuss two potential methods for including microglia (reviewed in [99, 102]). The ability to generate functional microglia is not trivial and requires careful characterization of the cells [99, 102]. The first method is to develop brain organoids that contain endogenous microglia. Minimal modifications to the original Lancaster cerebral organoid protocol [61] resulted in efficient development of microglia from mesodermal progenitors [103]. These cells resemble adult microglia by gene expression and likely reached maturation through the interaction with the other cell types in the culture. However, the organoids represent a relatively early stage of development in a limited portion of the brain [103]. Another protocol used microglia-like cells that were derived from hiPSCs using a simplified protocol with stage-wise growth factor induction [104]. The second method is to exogenously add iPSC-derived microglia to the brain organoids. For generation of iPSC-derived microglia, several protocols have been developed, and there are commercially available cells as well [99]. In one study, the microglia-like cells were cocultured with brain organoids [105]. Adaptive immune system cells are also to be considered in the context of a complex modeling entity. CD4+ T cells have a role in the intestine but T cells that recognize self-antigen that are CNS derived can invade the brain during PD progression [106, 107]. These immune cells invade the brain via a dysfunctional BBB which has been reported in PD patients [108–110]. The BBB is a highly polarized interface between the brain and the vasculature, composed of tightly connected endothelial cells that are strongly associated with astrocytic endfeet processes and pericytes. There have been many advances in the in vitro modeling of this neurovascular unit in recent years [111]. Organ-on-chip technology has been used to generate a human BBB derived from iPSCs [112, 113]. The unit was composed of brain microvascular endothelial-like cells, astrocytes, and neurons maintaining a “brain-side” and a “blood-side.” An alternative configuration used human hippocampal neural stem cells, cortical microvascular endothelial cells, astrocytes, and pericytes of cortical origin in a microfluidic BBB-vasculature-brain chip [114]. The study identified a previously unknown metabolic coupling between the BBB and neurons [114]. Further studies introduced hypoxia during the development of the BBB in vitro, resulting in improved functionality of the barrier [115]. A recent study used the organ-on-chip technology to engineer a human Brain-Chip representative of the SN area of the brain containing dopaminergic neurons, astrocytes, microglia, pericytes, and microvascular brain endothelial cells, cultured under fluid flow. They were capable of reproducing several key aspects of PD, including accumulation of phosphorylated αSyn (pSer129-αSyn), mitochondrial impairment, neuroinflammation, and compromised barrier function [116]. However, the BBB is not the only entry point for immune cells into the brain, as CD4+ T cells can enter the brain via the choroid plexus (CP) [117, 118]. The CP is formed in each of the four ventricles of the brain, consisting of epithelium cells connected by tight junctions, blood vessels, and other cell types. The CP forms a barrier between the blood and the cerebrospinal fluid (CSF). The CP produces CSF and also secretes many other factors and proteins that have the potential to affect the proliferation of adult stem cells [119]. During the aging process, the normal functions and morphology of the CP are compromised. These changes are further intensified in neurodegenerative diseases such as Alzheimer’s disease [120]. Choroid plexus-like structures have been noticed in cerebral organoids in the Lancaster’s protocol [121]. One protocol that was designed towards generation of this dorsal-medial structure from stem cells was developed in Yoshiki Sasai’s lab is based on regulating WNT and BMP (bone morphogenetic protein) signaling, and is applicable for either mouse or human cells [122, 123]. Recently, Lancaster modified the original protocol to enhance the formation of the CP and demonstrated formation of a barrier and CSF [124].
Will it ever be possible to combine the gut and the brain with all the auxiliary cell types and organs that are relevant for the disease onset and progression in one cultured system? Such an achievement requires further understanding of the biological components and advances in bioengineering. Efforts in this direction are seen in recent years with the emergence of “patient-on-chip” model systems [82, 125, 126]. Initial reports describe mimicking single organs, each requires its own specialized conditions, that are than combined in innovative manners to form a more complex model. One approach establishes the initial growth of each component individually before combining the different organs in a Lego-like system [80]. More recently, human organ-chip models of the gut, liver, and kidneys were fluidically coupled by vascular endothelium lined channels [127]. These channels are separated by a porous extracellular-matrix-coated membrane, lined with human organ-specific parenchymal cells. The fluid path has an integrated arteriovenous reservoir that mimicked the systemic circulation. The presence of the vascular endothelium-covered channels enables the use of a blood substitute, whereas the parenchymal-covered channels of each organ uses a different organ-specific optimized media. The system was designed to conduct quantitative measurements of pharmacokinetic responses to drugs [127]. An alternative design involves an automated culture that includes liquid-handling robotics, custom software with an integrated mobile microscope, perfusion, medium addition, fluidic linking, sample collection, and in situ microscopy imaging of eight organ chips inside a standard tissue-culture incubator [128]. The organs, including intestine, liver, kidney, heart, lung, skin, BBB, and brain, are cultured for 3 weeks with their fluids intermittently coupled using a blood substitute. The system is modular and the configurations can be changed. This system has been used to model drug kinetics and metabolism in different organs. Another multiorgan-on-a-chip platform known as MINERVA (MIcrobiota-Gut-BraiN EngineeRed platform to eVAluate intestinal microflora impact on brain functionality) has been designed to model neurodegenerative diseases such as PD and Alzheimer’s disease [129]. Overall, we envision that investigating human brain diseases should be viewed in the context of a similar multi-organ configuration, and could be studied either individually or in various creative combinations.
Acknowledgements
OR is an incumbent of the Bernstein-Mason Chair of Neurochemistry, Head of the M. Judith Ruth Institute for Preclinical Brain Research, TS is incumbent of the Leir Research Fellow Chair in Autism Spectrum Disorder Research. We thank Dr. Samara Brown for editing the first version of this manuscript. Our research is supported by The Jeanne and Joseph Nissim Center for Life Sciences Research at the Weizmann Institute of Science, the Helen and Martin Kimmel Institute for Stem Cell Research, the Nella and Leon Benoziyo Center for Neurological Diseases, the David and Fela Shapell Family Center for Genetic Disorders Research, the Brenden-Mann Women’s Innovation Impact Fund, the Richard F. Goodman Yale/Weizmann Exchange Program, The Irving B. Harris Fund for New Directions in Brain Research, The Irving Bieber, M.D. and Toby Bieber, M.D. Memorial Research Fund, The Leff Family, Barbara & Roberto Kaminitz, Sergio & Sônia Lozinsky, Debbie Koren, Jack and Lenore Lowenthal, and the Dears Foundation. The research has been supported by the Israel Science Foundation (Grant No. 347/15), the Legacy Heritage Biomedical Program of the Israel Science Foundation (Grant No. 2041/16), Israel Science Foundation (ISF)—National Natural Science Foundation of China (NSFC) (grant No. 2449/16), grant No. 2397/18 from the Canadian Institutes of Health Research (CIHR), the International Development Research Centre (IDRC), the Israel Science Foundation (ISF) and the Azrieli Foundation, a grant from the Ministry of Science & Technology, Israel & The Ministry of Science and Technology of the People’s Republic of China, German-Israeli Foundation (GIF; Grant no. I-1476-203.13/2018), and United States-Israel Binational Science Foundation (BSF; Grant No. 2017006).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pang SY, Ho PW, Liu HF, Leung CT, Li L, Chang EES, et al. The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson’s disease. Transl Neurodegener. 2019;8:23. doi: 10.1186/s40035-019-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Prim. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 3.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 4.Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020;19:170–8. doi: 10.1016/S1474-4422(19)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan-Montojo F, Schwarz M, Winkler C, Arnhold M, O’Sullivan GA, Pal A, et al. Environmental toxins trigger PD-like progression via increased alpha-synuclein release from enteric neurons in mice. Sci Rep. 2012;2:898. doi: 10.1038/srep00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lau LM, Schipper CM, Hofman A, Koudstaal PJ, Breteler MM. Prognosis of Parkinson disease: risk of dementia and mortality: the Rotterdam Study. Arch Neurol. 2005;62:1265–9. doi: 10.1001/archneur.62.8.1265. [DOI] [PubMed] [Google Scholar]
- 7.Tanner CM, Kamel F, Ross GW, Hoppin JA, Goldman SM, Korell M, et al. Rotenone, paraquat, and Parkinson’s disease. Environ Health Perspect. 2011;119:866–72. doi: 10.1289/ehp.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hainque E, Grabli D. Rapid worsening in Parkinson’s disease may hide COVID-19 infection. Parkinsonism Relat Disord. 2020;75:126–7. [DOI] [PMC free article] [PubMed]
- 9.Victorino DB, Guimaraes-Marques M, Nejm M, Scorza FA, Scorza CA. COVID-19 and Parkinson’s Disease: Are We Dealing with Short-term Impacts or Something Worse? J Parkinsons Dis. 2020;10:899–902. [DOI] [PMC free article] [PubMed]
- 10.Helmich RC, Bloem BR. The impact of the COVID-19 pandemic on Parkinson’s disease: hidden sorrows and emerging opportunities. J Parkinsons Dis. 2020;10:351–4. doi: 10.3233/JPD-202038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishbein I, Kuo YM, Giasson BI, Nussbaum RL. Augmentation of phenotype in a transgenic Parkinson mouse heterozygous for a Gaucher mutation. Brain. 2014;137:3235–47. doi: 10.1093/brain/awu291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sardi SP, Clarke J, Kinnecom C, Tamsett TJ, Li L, Stanek LM, et al. CNS expression of glucocerebrosidase corrects alpha-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc Natl Acad Sci USA. 2011;108:12101–6. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14:504–6. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 15.Li JY, Englund E, Holton JL, Soulet D, Hagell P, Lees AJ, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 16.Surmeier DJ, Obeso JA, Halliday GM. Parkinson’s disease is not simply a prion disorder. J Neurosci. 2017;37:9799–807. doi: 10.1523/JNEUROSCI.1787-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olanow CW, Prusiner SB. Is Parkinson’s disease a prion disorder? Proc Natl Acad Sci USA. 2009;106:12571–2. doi: 10.1073/pnas.0906759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garretti F, Agalliu D, Lindestam Arlehamn CS, Sette A, Sulzer D. Autoimmunity in Parkinson’s disease: the role of alpha-synuclein-specific T cells. Front Immunol. 2019;10:303. doi: 10.3389/fimmu.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkes CH, Del Tredici K, Braak H. A timeline for Parkinson’s disease. Parkinsonism Relat Disord. 2010;16:79–84. doi: 10.1016/j.parkreldis.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Holmqvist S, Chutna O, Bousset L, Aldrin-Kirk P, Li W, Bjorklund T, et al. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014;128:805–20. doi: 10.1007/s00401-014-1343-6. [DOI] [PubMed] [Google Scholar]
- 21.Corbille AG, Coron E, Neunlist M, Derkinderen P, Lebouvier T. Appraisal of the dopaminergic and noradrenergic innervation of the submucosal plexus in PD. J Parkinsons Dis. 2014;4:571–6. doi: 10.3233/JPD-140422. [DOI] [PubMed] [Google Scholar]
- 22.Braak H, Rub U, Gai WP, Del, Tredici K. Idiopathic Parkinson’s disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm. 2003;110:517–36. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- 23.Uemura N, Yagi H, Uemura MT, Hatanaka Y, Yamakado H, Takahashi R. Inoculation of alpha-synuclein preformed fibrils into the mouse gastrointestinal tract induces Lewy body-like aggregates in the brainstem via the vagus nerve. Mol Neurodegener. 2018;13:21. doi: 10.1186/s13024-018-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Kwon SH, Kam TI, Panicker N, Karuppagounder SS, Lee S, et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron. 2019;103:627–41. doi: 10.1016/j.neuron.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challis C, Hori A, Sampson TR, Yoo BB, Challis RC, Hamilton AM, et al. Gut-seeded alpha-synuclein fibrils promote gut dysfunction and brain pathology specifically in aged mice. Nat Neurosci. 2020;23:327–36. doi: 10.1038/s41593-020-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poewe W, Mahlknecht P. Pharmacologic treatment of motor symptoms associated with Parkinson disease. Neurol Clin. 2020;38:255–67. doi: 10.1016/j.ncl.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Svensson E, Horvath-Puho E, Thomsen RW, Djurhuus JC, Pedersen L, Borghammer P, et al. Vagotomy and subsequent risk of Parkinson’s disease. Ann Neurol. 2015;78:522–9. doi: 10.1002/ana.24448. [DOI] [PubMed] [Google Scholar]
- 28.Chandra R, Hiniker A, Kuo YM, Nussbaum RL, Liddle RA. alpha-Synuclein in gut endocrine cells and its implications for Parkinson’s disease. JCI Insight. 2017;2:e92295. [DOI] [PMC free article] [PubMed]
- 29.Felice VD, Quigley EM, Sullivan AM, O’Keeffe GW, O’Mahony SM. Microbiota-gut-brain signalling in Parkinson’s disease: implications for non-motor symptoms. Parkinsonism Relat Disord. 2016;27:1–8. doi: 10.1016/j.parkreldis.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–80. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unger MM, Spiegel J, Dillmann KU, Grundmann D, Philippeit H, Burmann J, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Bedarf JR, Hildebrand F, Coelho LP, Sunagawa S, Bahram M, Goeser F, et al. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson’s disease patients. Genome Med. 2017;9:39. doi: 10.1186/s13073-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JY, Tuazon JP, Ehrhart J, Sanberg PR, Borlongan CV. Gutting the brain of inflammation: A key role of gut microbiome in human umbilical cord blood plasma therapy in Parkinson’s disease model. J Cell Mol Med. 2019;23:5466–74. doi: 10.1111/jcmm.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elfil M, Kamel S, Kandil M, Koo BB, Schaefer SM. Implications of the Gut Microbiome in Parkinson’s Disease. Mov Disord. 2020;35:921–33. [DOI] [PubMed]
- 36.Lin CH, Chen CC, Chiang HL, Liou JM, Chang CM, Lu TP, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflammation. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrov VA, Saltykova IV, Zhukova IA, Alifirova VM, Zhukova NG, Dorofeeva YB, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med. 2017;162:734–7. doi: 10.1007/s10517-017-3700-7. [DOI] [PubMed] [Google Scholar]
- 38.Sampson TR, Challis C, Jain N, Moiseyenko A, Ladinsky MS, Shastri GG et al. A gut bacterial amyloid promotes alpha-synuclein aggregation and motor impairment in mice. Elife. 2020;9:e53111. [DOI] [PMC free article] [PubMed]
- 39.Matheoud D, Cannon T, Voisin A, Penttinen AM, Ramet L, Fahmy AM, et al. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1(-/-) mice. Nature. 2019;571:565–9. doi: 10.1038/s41586-019-1405-y. [DOI] [PubMed] [Google Scholar]
- 40.Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, et al. Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell. 2016;166:314–27. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–62. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364:eaau6323. [DOI] [PMC free article] [PubMed]
- 44.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 45.Eiraku M, Sasai Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr Opin Neurobiol. 2012;22:768–77. doi: 10.1016/j.conb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–80. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012;4:380–95. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delsing L, Herland A, Falk A, Hicks R, Synnergren J, Zetterberg H. Models of the blood-brain barrier using iPSC-derived cells. Mol Cell Neurosci. 2020;107:103533. doi: 10.1016/j.mcn.2020.103533. [DOI] [PubMed] [Google Scholar]
- 51.Burke EE, Chenoweth JG, Shin JH, Collado-Torres L, Kim SK, Micali N, et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat Commun. 2020;11:462. doi: 10.1038/s41467-019-14266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–67. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Qing X, Walter J, Jarazo J, Arias-Fuenzalida J, Hillje AL, Schwamborn JC. CRISPR/Cas9 and piggyBac-mediated footprint-free LRRK2-G2019S knock-in reveals neuronal complexity phenotypes and alpha-Synuclein modulation in dopaminergic neurons. Stem Cell Res. 2017;24:44–50. doi: 10.1016/j.scr.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lancaster MA, Huch M. Disease modelling in human organoids. Dis Model Mech. 2019;12:dmm039347. [DOI] [PMC free article] [PubMed]
- 56.Kechele DO, Wells JM. Recent advances in deriving human endodermal tissues from pluripotent stem cells. Curr Opin Cell Biol. 2019;61:92–100. doi: 10.1016/j.ceb.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 57.In JG, Foulke-Abel J, Estes MK, Zachos NC, Kovbasnjuk O, Donowitz M. Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nat Rev Gastroenterol Hepatol. 2016;13:633–42. doi: 10.1038/nrgastro.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H. Development and application of human adult stem or progenitor cell organoids. Nat Rev Nephrol. 2015;11:546–54. doi: 10.1038/nrneph.2015.118. [DOI] [PubMed] [Google Scholar]
- 59.Takasato M, Little MH. A strategy for generating kidney organoids: recapitulating the development in human pluripotent stem cells. Dev Biol. 2016;420:210–20. doi: 10.1016/j.ydbio.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nugraha B, Buono MF, von Boehmer L, Hoerstrup SP, Emmert MY. Human cardiac organoids for disease modeling. Clin Pharm Ther. 2019;105:79–85. doi: 10.1002/cpt.1286. [DOI] [PubMed] [Google Scholar]
- 61.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelava I, Lancaster MA. Stem cell models of human brain development. Cell Stem Cell. 2016;18:736–48. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Marton RM, Pasca SP. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2019;30:133–43. [DOI] [PubMed]
- 64.Seto Y, Eiraku M. Toward the formation of neural circuits in human brain organoids. Curr Opin Cell Biol. 2019;61:86–91. doi: 10.1016/j.ceb.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 65.Arlotta P, Pasca SP. Cell diversity in the human cerebral cortex: from the embryo to brain organoids. Curr Opin Neurobiol. 2019;56:194–8. doi: 10.1016/j.conb.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park IH. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–9. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–8. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 2017;8:1144–54. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–57. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwak TH, Kang JH, Hali S, Kim J, Kim KP, Park C, et al. Generation of homogeneous midbrain organoids with in vivo-like cellular composition facilitates neurotoxin-based Parkinson’s disease modeling. Stem Cells. 2020;38:727–40. [DOI] [PubMed]
- 71.Kim H, Park HJ, Choi H, Chang Y, Park H, Shin J, et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Rep. 2019;12:518–31. doi: 10.1016/j.stemcr.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smits LM, Reinhardt L, Reinhardt P, Glatza M, Monzel AS, Stanslowsky N, et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019;5:5. doi: 10.1038/s41531-019-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schutgens F, Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu Rev Pathol. 2020;15:211–34. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- 74.Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8:344ra384. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 75.Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, et al. Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell Mol Gastroenterol Hepatol. 2018;5:669–77. doi: 10.1016/j.jcmgh.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan JK, McKenzie C, Marino E, Macia L, Mackay CR, Metabolite-Sensing G. Protein-coupled receptors-facilitators of diet-related immune regulation. Annu Rev Immunol. 2017;35:371–402. doi: 10.1146/annurev-immunol-051116-052235. [DOI] [PubMed] [Google Scholar]
- 77.Co JY, Margalef-Catala M, Li X, Mah AT, Kuo CJ, Monack DM, et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 2019;26:2509–20. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol. 2018;6:301–19. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol. 2018;5:659–68. doi: 10.1016/j.jcmgh.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loskill P, Marcus SG, Mathur A, Reese WM, Healy KE. muOrgano: a lego(R)-like plug & play system for modular multi-organ-chips. PLoS One. 2015;10:e0139587. doi: 10.1371/journal.pone.0139587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prantil-Baun R, Novak R, Das D, Somayaji MR, Przekwas A, Ingber DE. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. Annu Rev Pharm Toxicol. 2018;58:37–64. doi: 10.1146/annurev-pharmtox-010716-104748. [DOI] [PubMed] [Google Scholar]
- 82.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–5. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu F, Hunziker W, Choudhury D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines. 2019;10:165. [DOI] [PMC free article] [PubMed]
- 84.Shim KY, Lee D, Han J, Nguyen NT, Park S, Sung JH. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices. 2017;19:37. doi: 10.1007/s10544-017-0179-y. [DOI] [PubMed] [Google Scholar]
- 85.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–74. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 86.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113:E7–15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a primary human small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonaz B, Bazin T, Pellissier S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front Neurosci. 2018;12:49. doi: 10.3389/fnins.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Min S, Kim S, Cho SW. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp Mol Med. 2020;52:227–37. doi: 10.1038/s12276-020-0386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fattahi F, Steinbeck JA, Kriks S, Tchieu J, Zimmer B, Kishinevsky S, et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. 2016;531:105–9. doi: 10.1038/nature16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schlieve CR, Fowler KL, Thornton M, Huang S, Hajjali I, Hou X, et al. Neural crest cell implantation restores enteric nervous system function and alters the gastrointestinal transcriptome in human tissue-engineered small intestine. Stem Cell Rep. 2017;9:883–96. doi: 10.1016/j.stemcr.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol. 2019;20:279–93. [DOI] [PubMed]
- 94.Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep. 2017;7:45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karve SS, Pradhan S, Ward DV, Weiss AA. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One. 2017;12:e0178966. doi: 10.1371/journal.pone.0178966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schreurs R, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, et al. Human fetal TNF-alpha-cytokine-producing CD4(+) effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. 2019;50:462–76. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 97.Jung KB, Lee H, Son YS, Lee MO, Kim YD, Oh SJ, et al. Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat Commun. 2018;9:3039. doi: 10.1038/s41467-018-05450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watson CL, Mahe MM, Munera J, Howell JC, Sundaram N, Poling HM, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20:1310–4. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hasselmann J, Blurton-Jones M. Human iPSC-derived microglia: a growing toolset to study the brain’s innate immune cells. Glia. 2020;68:721–39. doi: 10.1002/glia.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yun SP, Kam TI, Panicker N, Kim S, Oh Y, Park JS, et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat Med. 2018;24:931–8. doi: 10.1038/s41591-018-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herrera AJ, Espinosa-Oliva AM, Carrillo-Jimenez A, Oliva-Martin MJ, Garcia-Revilla J, Garcia-Quintanilla A, et al. Relevance of chronic stress and the two faces of microglia in Parkinson’s disease. Front Cell Neurosci. 2015;9:312. doi: 10.3389/fncel.2015.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haenseler W, Rajendran L. Concise review: modeling neurodegenerative diseases with human pluripotent stem cell-derived microglia. Stem Cells. 2019;37:724–30. doi: 10.1002/stem.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, et al. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci Rep. 2019;9:11055. doi: 10.1038/s41598-019-47444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–93. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gonzalez H, Contreras F, Pacheco R. Regulation of the neurodegenerative process associated to Parkinson’s disease by CD4+ T-cells. J Neuroimmune Pharm. 2015;10:561–75. doi: 10.1007/s11481-015-9618-9. [DOI] [PubMed] [Google Scholar]
- 107.Brochard V, Combadiere B, Prigent A, Laouar Y, Perrin A, Beray-Berthat V, et al. Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J Clin Invest. 2009;119:182–92. doi: 10.1172/JCI36470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Desai BS, Monahan AJ, Carvey PM, Hendey B. Blood-brain barrier pathology in Alzheimer’s and Parkinson’s disease: implications for drug therapy. Cell Transpl. 2007;16:285–99. doi: 10.3727/000000007783464731. [DOI] [PubMed] [Google Scholar]
- 109.Gray MT, Woulfe JM. Striatal blood-brain barrier permeability in Parkinson’s disease. J Cereb Blood Flow Metab. 2015;35:747–50. doi: 10.1038/jcbfm.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cabezas R, Avila M, Gonzalez J, El-Bacha RS, Baez E, Garcia-Segura LM, et al. Astrocytic modulation of blood brain barrier: perspectives on Parkinson’s disease. Front Cell Neurosci. 2014;8:211. doi: 10.3389/fncel.2014.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bhalerao A, Sivandzade F, Archie SR, Chowdhury EA, Noorani B, Cucullo L. In vitro modeling of the neurovascular unit: advances in the field. Fluids Barriers CNS. 2020;17:22. doi: 10.1186/s12987-020-00183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sances S, Ho R, Vatine G, West D, Laperle A, Meyer A, et al. Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep. 2018;10:1222–36. doi: 10.1016/j.stemcr.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vatine GD, Barrile R, Workman MJ, Sances S, Barriga BK, Rahnama M, et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019;24:995–1005. doi: 10.1016/j.stem.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 114.Maoz BM, Herland A, FitzGerald EA, Grevesse T, Vidoudez C, Pacheco AR, et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol. 2018;36:865–74. doi: 10.1038/nbt.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park TE, Mustafaoglu N, Herland A, Hasselkus R, Mannix R, FitzGerald EA, et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat Commun. 2019;10:2621. doi: 10.1038/s41467-019-10588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pediaditakis I, Kodella K, Manatakis D, Hinojosa C, Manolakos E, Rubin L et al. Modeling alpha-synuclein pathology in a human brain-chip to assess blood-brain barrier disruption in Parkinson’s disease. bioRxiv2020. [DOI] [PMC free article] [PubMed]
- 117.Meeker RB, Williams K, Killebrew DA, Hudson LC. Cell trafficking through the choroid plexus. Cell Adh Migr. 2012;6:390–6. doi: 10.4161/cam.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kivisakk P, Mahad DJ, Callahan MK, Trebst C, Tucky B, Wei T, et al. Human cerebrospinal fluid central memory CD4+ T cells: evidence for trafficking through choroid plexus and meninges via P-selectin. Proc Natl Acad Sci USA. 2003;100:8389–94. doi: 10.1073/pnas.1433000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell. 2016;19:643–52. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 120.Van Cauwenberghe C, Gorlé N, Vandenbroucke RE. Roles of the Choroid Plexus in Aging. In: Praetorius J, Blazer-Yost B, Damkier H (eds). Role of the Choroid Plexus in Health and Disease. Springer US: New York, NY, 2020, pp 209–32.
- 121.Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–29. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 123.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, Lancaster MA. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020;369:eaaz5626. [DOI] [PMC free article] [PubMed]
- 125.Williamson A, Singh S, Fernekorn U, Schober A. The future of the patient-specific Body-on-a-chip. Lab Chip. 2013;13:3471–80. doi: 10.1039/c3lc50237f. [DOI] [PubMed] [Google Scholar]
- 126.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–92. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 127.Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Novak R et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng. 2020;4:421–36. [DOI] [PMC free article] [PubMed]
- 128.Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng. 2020;4:407–20. [DOI] [PMC free article] [PubMed]
- 129.Raimondi MT, Albani D, Giordano C. An organ-on-a-chip engineered platform to study the microbiota-gut-brain axis in neurodegeneration. Trends Mol Med. 2019;25:737–40. doi: 10.1016/j.molmed.2019.07.006. [DOI] [PubMed] [Google Scholar]