Abstract
In MRI practice, it is inevitable to appropriately balance between image resolution, signal-to-noise ratio (SNR), and scan time. It has been shown that super-resolution reconstruction (SRR) is effective to achieve such a balance, and has obtained better results than direct high-resolution (HR) acquisition, for certain contrasts and sequences. The focus of this work was on constructing images with spatial resolution higher than can be practically obtained by direct Fourier encoding. A novel learning approach was developed, which was able to provide an estimate of the spatial gradient prior from the low-resolution (LR) inputs for the HR reconstruction. By incorporating the anisotropic acquisition schemes, the learning model was trained over the LR images themselves only. The learned gradients were integrated as prior knowledge into a gradient-guided SRR model. A closed-form solution to the SRR model was developed to obtain the HR reconstruction. Our approach was assessed on the simulated data as well as the data acquired on a Siemens 3T MRI scanner containing 45 MRI scans from 15 subjects. The experimental results demonstrated that our approach led to superior SRR over state-of-the-art methods, and obtained better images at lower or the same cost in scan time than direct HR acquisition.
Keywords: MRI, Super-resolution, Deep neural networks
1. Introduction
It is inevitable to deal with the trade-off between image resolution, signal-to-noise ratio (SNR), and scan time, in magnetic resonance imaging (MRI) practice [1]. Images of higher resolution provide more details, and correspondingly increase the scan time. Higher SNR renders the signals of interest better from noise contamination. According to MRI physics, SNR is proportional to slice thickness and scan time. In pursuit of higher SNR, however, thick slices lower image resolution, while long scans discomfort subjects and potentially lead to subject motion during the acquisition, which adversely affects image quality. Literature has shown many effective methods to acquire images of high resolution, high SNR, and low scan time, such as parallel imaging [15] and robust K-space sampling [12]. Among these methods, super-resolution reconstruction (SRR) has recently been demonstrated to be capable of obtaining better images than direct high-resolution (HR) acquisition, for certain image contrasts and sequences, such as T2 weighted images [13]. As a result, SRR has become one of the most widely used methods that acquire MR images of high quality.
The idea of SRR originated in [19] for natural images where multiple low-resolution (LR) images were combined into an HR reconstruction. Extensive methods expanded this idea and achieved various SRR schemes in MRI [14,16]. Recently, deep learning-based methods have achieved impressive results for 2D natural images [5]. These 2D learning models were directly applied to 3D MRI volumes by means of slice-by-slice processing [8,23]. However, by the nature that MRI volumes reveal anatomical structures in 3D space, it is straightforward to learn 3D models for capturing richer knowledge. It has been shown that 3D models outperformed their 2D counterparts in MRI [3,11]. To this end, extensive 3D learning models were developed [2–4,11,20]. Unfortunately, these 3D models required large-scale training datasets, that contain HR labels that are practically difficult to obtain at excellent SNR due to subject motion, to learn the patterns mapping from LR to HR images. Although few datasets are publicly available, there is no known theory that indicates a satisfactory amount of training data has been incorporated in the training process. Furthermore, since the LR inputs were manually generated as blurred images, the learned function may be brittle when faced with data from a different scanner or with different intensity properties.
In this work, we aimed at developing the learning model that is not subject to the above limitations. Instead of learning end-to-end mappings from LR to HR images, we turned to learn the inherent structure of the HR reconstruction, and then as prior knowledge the learned structure was incorporated in a forward model-based SRR framework. To this end, we targeted the spatial image gradients in the learning, since they can be equivalently decomposed from 3D space onto 2D space, and correspondingly the required amount of training data was substantially reduced. More importantly, by incorporating the anisotropic acquisition schemes, our learning model was trained over the LR images themselves only, as the training data consisted of the pairs of an in-plane HR slice of an LR image and a through-plane LR slice of another LR image. A gradient-guided SRR framework [17] was leveraged to incorporate the learned gradients for the HR reconstruction.
The SRR method [17] used the gradients of LR images as HR gradient estimates. An LR image containing ns slices can provide ns accurate HR gradients for the HR reconstruction containing Ns slices, as the ns slices are in-plane HR. Since the upscale factor Ns/ns is often 4 or 5 in practice, 75–80% gradients in [17] had to be estimated from interpolated slices. The interpolation led to blurry or displaced image edges, and thus resulted in less accurate gradient localization. In this work, we aimed at improving the localization accuracy of those 75–80% gradients. We used a CNN to learn over the ns in-plane HR slices the patterns mapping from LR slices to HR gradients, enabled by a strategy that is able to decompose a 3D gradient onto 2D space. The learned patterns were used to infer the HR gradients over the rest Ns −ns slices. As high frequencies were gained in the inference, improved gradient localization was achieved, compared to [17], and thus increased the quality of deblurring done in the SRR. In addition, a closed-form solution to the SRR model was developed, instead of an iterative optimization algorithm used in [17] that may be stuck in local optima.
2. Methods
2.1. Gradient-Guided Framework
Given n LR images , acquired from n scans with arbitrary orientations and displacements, the forward model over the HR image , which describes the MRI acquisition process, is formulated as
| (1) |
where x and yk are the column vectors of their volumetric forms and in a lexicographical order of voxels; Tk denotes a rigid transform that characterizes subject motion between scans; Hk denotes a point spread function (PSF); Dk denotes a downsampling operator; μk denotes the noise in the acquisition. According to [7], μk can be assumed to be additive and Gaussian when SNR > 3. As the problem defined by the forward model is ill-posed, prior knowledge is commonly used, known as the regularization, to restrict the HR estimate. By incorporating the gradient guidance regularization [17], the SRR is achieved by
| (2) |
where g denotes the gradient guidance for the HR estimate x, ∇ computes the spatial gradient, σ (·) penalizes the difference between the actual gradient and the guidance, and λ > 0 balances between the data fidelity and the regularization.
2.2. Gradient Guidance Learning
As shown in Eq. (2), the accuracy of the gradient guidance estimate g is critically important to the quality of the HR reconstruction x. Thanks to the fact that a 3D gradient can be decomposed onto 2D space, we are able to compute the gradient of a volume slice by slice. Specifically, the 3D gradient at a scale s in direction d ∈ {x, y, z,−x,−y,−z} is computed from
| (3) |
where I denotes an identity matrix, shifts a volume circularly by s voxels in direction d, and shifts the k-th slice circularly by s pixels in direction d.
Architecture of the Learning Model.
By the above decomposition, the gradient guidance is learned over slices by a convolutional neural network (CNN). The overview of our proposed learning model is shown in Fig. 1. The CNN takes as input 2D LR image patches, and outputs the 2D gradients of HR image patches. ℓ2-loss is used to minimize the difference between the gradient of k-th HR patch and the CNN’s output for the k-th inputted LR patch :
| (4) |
where fθ (·) is the function defined by the CNN, and θ denotes the hyperparameters. The CNN has 3 layers: a layer with 64 filters of size 9 × 9 × 1; a layer with 32 filters of size 1 × 1 × 64; and a with 2 filters of size 5 × 5 × 32. A ReLU is imposed on the outputs of the first 2 layers. Since a slice contains 2D information, the CNN outputs two gradients in two in-plane perpendicular directions.
Fig. 1.
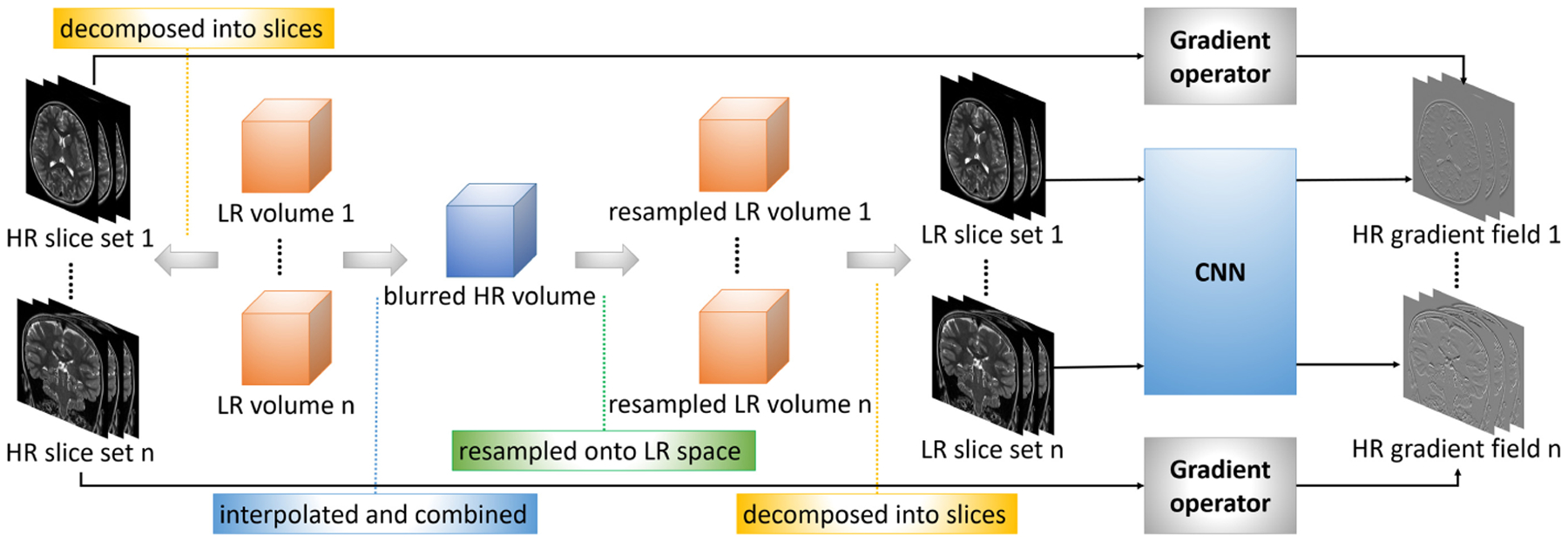
Overview of our proposed approach to learning the gradient guidance.
Pairing LR Inputs and HR Labels for Training Data.
The LR images are of in-plane HR, through-plane LR, and different orientations. Therefore, the gradients of the in-plane HR slices are directly used as the HR labels. The corresponding LR inputs are found in the following steps. First, all the LR images are aligned and interpolated by a third-order B-spline method to the size and resolution the same as the HR reconstruction. These interpolated LR images are combined into a mean image by averaging them out over their voxels. This mean image is then resampled according to the geometric properties of each original LR image, respectively. The slices of the resampled images are HR (small voxel size) but blurry due to the averaging, and are used as the LR inputs. The training dataset is thus constructed from the pairs of the LR inputs and the HR labels.
Training.
The LR-HR pairs in the training are of size 33 × 33 pixels. By adjusting the strides, about 20,000 data pairs are sampled uniformly with overlap from the slices to form the training dataset. The CNN is trained for 2000 epochs on an NVIDIA Quadro P5000 GPU with TensorFlow. Stochastic gradient descent with a learning rate of 1e−4 is used to optimize the hyperparameters θ in Eq. (4).
2.3. Super-Resolution Reconstruction
In the SRR, the CNN is first trained over the LR-HR pairs. The mean image is calculated following the steps mentioned above. The slices from each plane of the mean image are inputted to the trained CNN. The outputted gradients that are in the same direction are averaged into a single gradient. All the gradients form a gradient guidance g for the SRR.
As the gradient guidance is highly accurate due to the learning, a squared error is incorporated in the regularization. Therefore, the SRR is achieved by
| (5) |
where all the gradients at different scales in different directions are organized in a set , and ∇g computes the gradient at the same scale and in the same direction as g. Hk is designed as a sinc function in the slice selection direction of yk (corresponding a boxcar in Fourier domain) convolved with a Gaussian kernel. In this work, the gradient guidance is computed at a scale of 1 in 3 directions of x, y, and z. λ is fixed at 0.5 according to the empirical results.
A closed-form solution to Eq. (5) is developed to obtain the HR reconstruction x from
| (6) |
where denotes the interpolated and aligned form of the LR image yk, denotes 3D discrete Fourier transform, ·* computes complex conjugate, ○ denotes Hardamard product, and the fraction denotes element-wise division.
2.4. Experimental Setup
Simulated Data.
We simulated a dataset by using the MPRAGE data from the Dryad package [10]. We downsampled the original image of isotropic 0.25 mm to the image of isotropic 0.5 mm, and used it as the ground truth. We then downsampled the ground truth by factors of {2, 3, 4, 5, 6, 8} in the directions of x, y, and z, respectively, to form an axial, an coronal, and a sagittal LR image at each factor. Simulated motion with maximum magnitudes of 10 mm for translation and 10% for rotation was randomly generated and applied to each LR image. Gaussian noise was also randomly generated with a power of 10% of mean voxel intensities and added to each LR image. We assessed the quality of the HR reconstruction on this dataset in terms of peak signal-to-noise ratio (PSNR), structural similarity (SSIM) [21], and root square mean error (RMSE).
Real Data.
We acquired 45 MRI scans from 15 subjects with various acquisition schemes on a Siemens 3T MRI scanner. All scans were performed in accordance with the local institutional review board protocol.
Origin-Shifted Axial LR Acquisitions (OSA).
We acquired two axial LR T2 weighted images with their origins shifted by a distance of half slice thickness. The in-plane resolution is 1 mm × 1 mm and the slice thickness is 2 mm. We also acquired an HR image of isotropic 1 mm as the ground truth. The HR reconstruction was evaluated in terms of PSNR, SSIM, and sharpness [6].
Axial and Coronal LR Acquisitions (AC).
We acquired 42 images from 14 subjects. With each subject, an axial and a coronal LR T2-TSE image were acquired with in-plane resolution of 0.4 mm × 0.4 mm and slice thickness of 2 mm. It took about 2 min in acquiring a T2-TSE image. A 3D T2-SPACE image of isotropic 0.9 mm was acquired as the reference. The HR reconstruction was of isotropic 0.4 mm. Since the T2-TSE has different contrast from the 3D T2-SPACE image, we evaluated the HR reconstruction in terms of the blind assessment metrics that focus on image contrast and sharpness, including average edge strength (AES) [22], gradient entropy (GRENT) [9], and diagonal XSML (LAPD) [18]. As the HR reconstructions were assessed blindly on this dataset, we reported the normalized metrics that were computed from , where ρ (·) denotes a metric function, x denotes a reconstruction, and x′ denotes a reference · image. Higher metric values produce better image quality for all metrics. We also investigated the accuracy of the gradient guidance estimate in terms of edge localization error that was computed from , where g is the gradient guidance, and g′ is the gradients computed from the reference image.
Baseline Methods.
We employed a CNN-based learning model as a baseline, which is similar to [8], trained on the same data, with the same network architecture as our model except that the output is the HR slice. The HR reconstruction was obtained from the fusion over the HR slice stacks estimated in different planes. We denoted this baseline by deepSlice. We compared our approach to deepSlice to demonstrate that learning a gradient guidance is superior to learning an end-to-end mapping for HR slices in volumetric SRR. We also compared our approach to the plain gradient guidance regularization method [17], denoted by GGR, to demonstrate that our approach led to a more accurate gradient guidance estimate, and in turn resulted in better HR reconstruction. We denoted our approach by deepGG, that stands for deep gradient guidance.
3. Results
We compared the accuracy of the gradient guidance estimates obtained from GGR and deepGG on the AC dataset in terms of edge localization errors. As shown in Fig. 2(a), deepGG yielded much smaller errors than GGR on all 14 HR reconstructions. The representative slices of the learned gradient guidance obtained from deepGG are shown in Fig. 2(b).
Fig. 2.
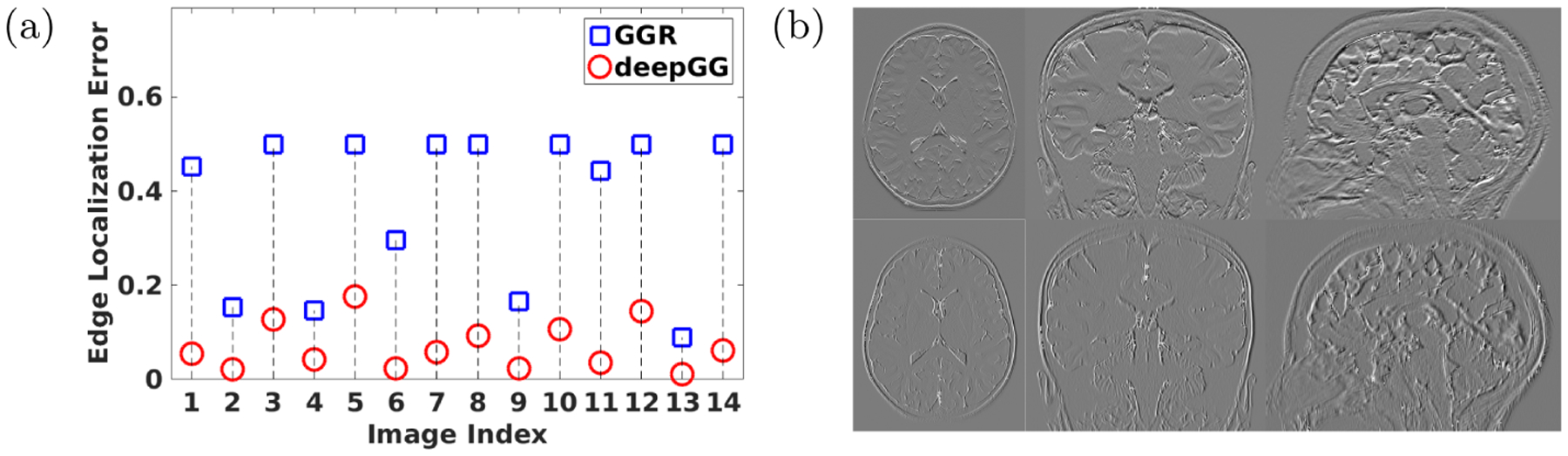
Evaluation results of the gradient guidance estimation on the AC dataset. (a) Comparisons between GGR and deepGG in the edge localization errors. (b) Representative slices of the learned gradient guidance obtained from deepGG.
Figure 3 shows the evaluation results of deepSlice, GGR, and deepGG on the simulated dataset. It is evident that deepGG consistently outperformed the two baselines by large margins at all upscale factors in terms of all metrics.
Fig. 3.
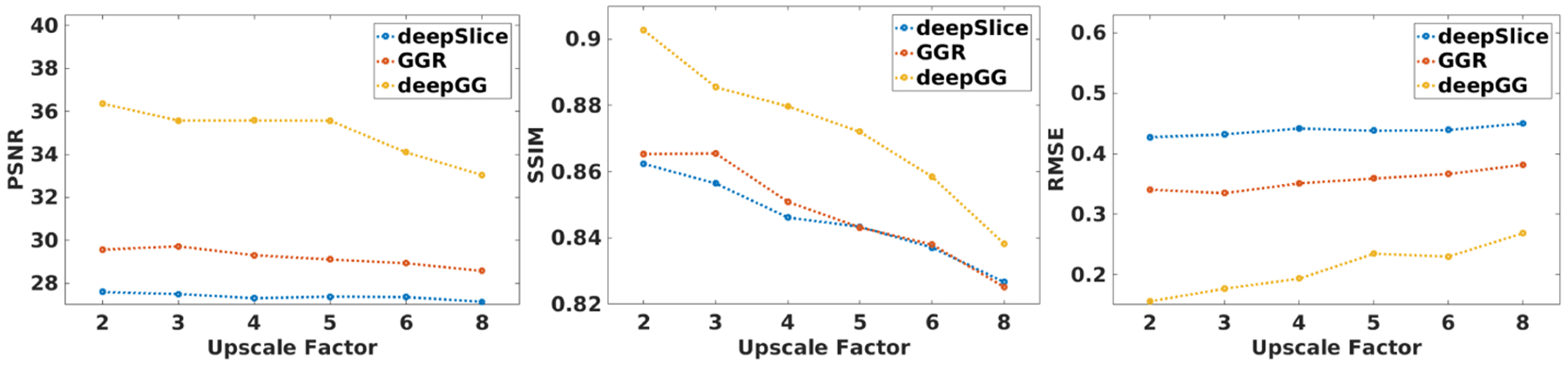
Evaluation results obtained from the three methods on the simulated dataset.
On the OSA dataset, the quantitative evaluation results were in terms of 1) PSNR: deepSlice = 35.01, GGR = 34.45, deepGG = 35.71; 2) SSIM: deepSlice = 0.912, GGR = 0.910, deepGG = 0.921; and 3) sharpness: deepSlice = 0.75, GGR = 0.78, deepGG = 0.86. Figure 4 shows the representative slices of an LR image and the reconstructed images obtained from deepSlice, GGR, and deepGG. The deepGG method achieved the sharpest images and the best contrast, which was consistent with the quantitative assessments, in particular with the sharpness metric.
Fig. 4.
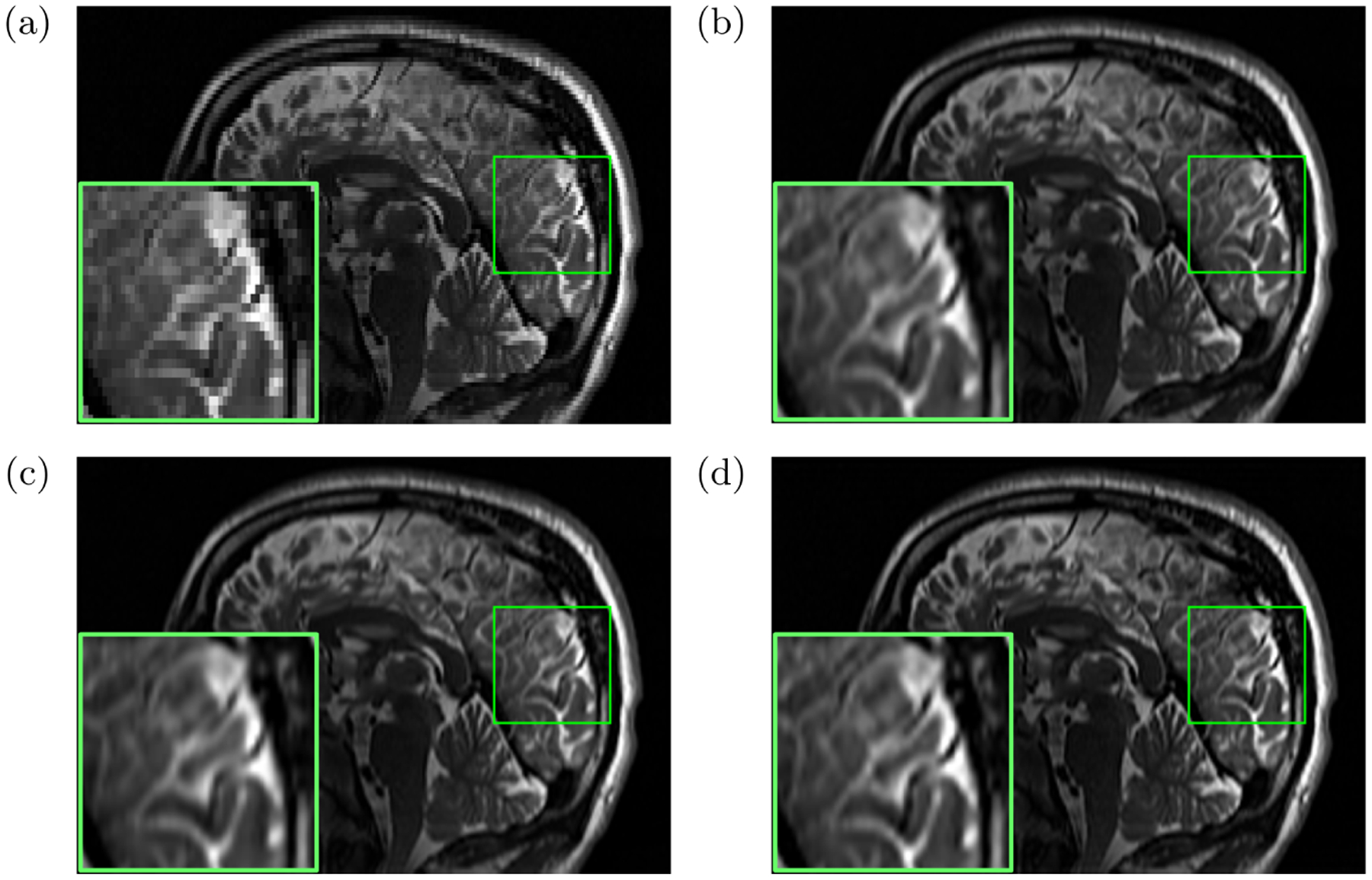
Representative slices of (a) an LR image and the reconstructed images obtained from (b) deepSlice, (c) GGR, and (d) deepGG (ours), respectively, on the OSA dataset.
Figure 5 plots the evaluation results of the three methods on the AC dataset. It is evident that deepGG considerably outperformed the two baselines. Figure 6 shows the representative slices of an LR image, and the HR images reconstructed by the three methods. It is shown that deepGG achieved more image details than the two baselines, and obtained the best contrast and the sharpest images.
Fig. 5.
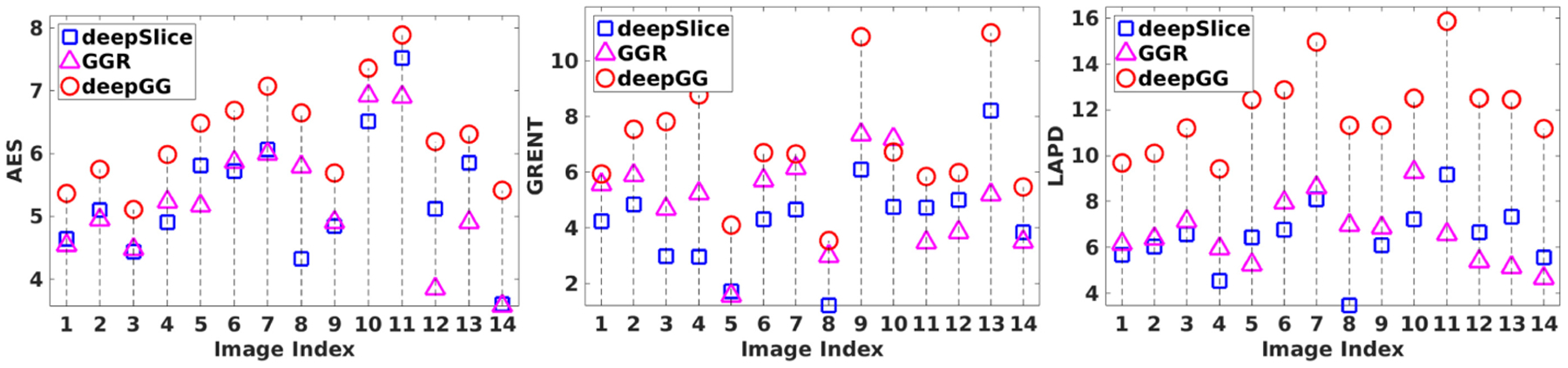
Evaluation results on the AC datasets in terms of average edge strength (AES), gradient entropy (GRENT), and diagonal XSML (LAPD), respectively.
Fig. 6.
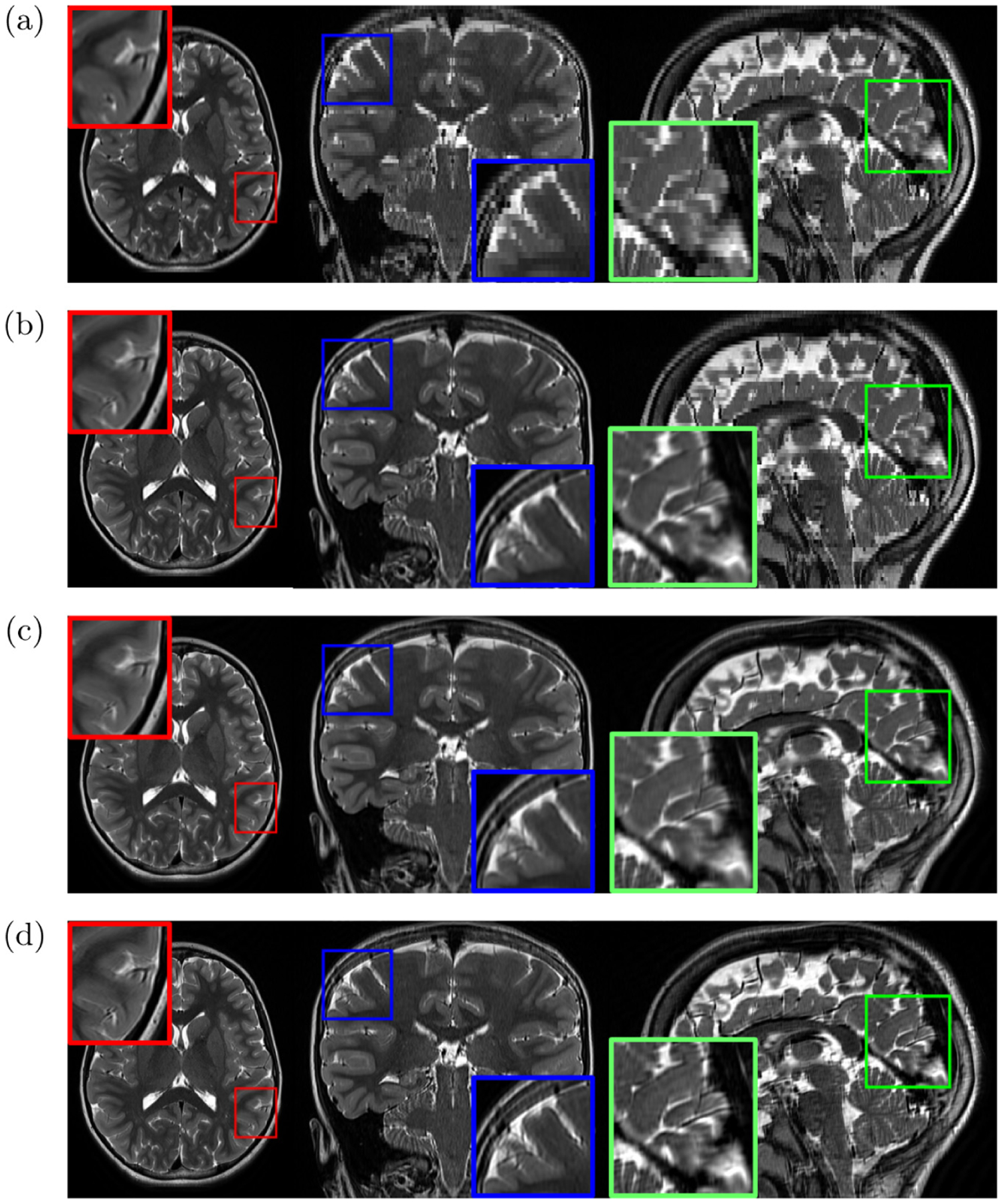
Representative slices of (a) an LR image, and the reconstructed images obtained from (b) deepSlice, (c) GGR, and (d) deepGG (ours), respectively, on the AC dataset.
4. Conclusions
We have developed a learning-based method to estimate the gradient guidance prior for MRI SRR. The learning tasks have been decomposed from 3D volumes onto 2D slices. The training dataset was constructed by exploiting the correspondences of in-plane HR and through-plane LR slices when incorporating the anisotropic acquisition schemes. Therefore, the learning model has been trained over the LR images themselves only. The learned gradient guidance, as prior knowledge, has been integrated into a forward model-based SRR framework. A closed-form solution to the SRR model, which is globally optimal, has been developed for obtaining the HR reconstruction. Extensive experimental results on both simulated and real data have demonstrated that our approach led to superior SRR over state-of-the-art methods, and obtained better images at lower or the same cost in scan time than direct HR acquisition.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) under grants R01 NS079788, R01 EB019483, R01 EB018988, R01 NS106030, IDDRC U54 HD090255; by a research grant from the Boston Children’s Hospital Translational Research Program; by a Technological Innovations in Neuroscience Award from the McKnight Foundation; by a research grant from the Thrasher Research Fund; and by a pilot grant from National Multiple Sclerosis Society under Award Number PP-1905-34002.
References
- 1.Brown RW, Cheng YCN, Haacke EM, Thompson MR, Venkatesan R: Magnetic Resonance Imaging: Physical Principles and Sequence Design, 2nd edn Wiley, Hoboken: (2014) [Google Scholar]
- 2.Chaudhari A, et al. : Super-resolution musculoskeletal MRI using deep learning. Magn. Reson. Med 80(5), 2139–2154 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Xie Y, Zhou Z, Shi F, Christodoulou AG, Li D: Brain MRI super resolution using 3D deep densely connected neural networks. In: International Symposium on Biomedical Imaging, pp. 739–742 (2018) [Google Scholar]
- 4.Chen Y, Shi F, Christodoulou AG, Xie Y, Zhou Z, Li D: Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 91–99. Springer, Cham: (2018). 10.1007/978-3-030-00928-1_11 [DOI] [Google Scholar]
- 5.Dong C, Loy CC, He K, Tang X: Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell 38(2), 295–307 (2016) [DOI] [PubMed] [Google Scholar]
- 6.Gholipour A, Estroff JA, Warfield SK: Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE Trans. Med. Imaging 29(10), 1739–1758 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson H, Patz S: The Rician distribution of noisy MRI data. Magn. Reson. Med 34, 910–914 (1995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurek J, Kocinski M, Materka A, Elgalal M, Majos A: CNN-based superreso-lution reconstruction of 3D MR images using thick-slice scans. Biocybern. Biomed. Eng 40(1), 111–125 (2020) [Google Scholar]
- 9.Loktyushin A, Nickisch H, Pohmann R, Schölkopf B: Blind multirigid retrospective motion correction of MR images. Magn. Reson. Med 73(4), 1457–1468 (2015) [DOI] [PubMed] [Google Scholar]
- 10.Lusebrink F, Sciarra A, Mattern H, Yakupov R, Speck O: T1-weighted in vivo human whole brain MRI dataset with an ultrahigh isotropic resolution of 250 μm. Sci. Data 4, 170032 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham C, Ducournau A, Fablet R, Rousseau F: Brain MRI super-resolution using deep 3D convolutional networks. In: International Symposium on Biomedical Imaging, pp. 197–200 (2017) [Google Scholar]
- 12.Pipe J: Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn. Reson. Med 42, 963–969 (1999) [DOI] [PubMed] [Google Scholar]
- 13.Plenge E, et al. : Super-resolution methods in MRI: can they improve the trade-off between resolution, signal-to-noise ratio, and acquisition time? Magn. Reson. Med 68, 1983–1993 (2012) [DOI] [PubMed] [Google Scholar]
- 14.Poot DHJ, Van Meir V, Sijbers J: General and efficient super-resolution method for multi-slice MRI In: Jiang T, Navab N, Pluim JPW, Viergever MA (eds.) MICCAI 2010. LNCS, vol. 6361, pp. 615–622. Springer, Heidelberg: (2010). 10.1007/978-3-642-15705-9_75 [DOI] [PubMed] [Google Scholar]
- 15.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P: SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med 42, 952–962 (1999) [PubMed] [Google Scholar]
- 16.Shilling RZ, Robbie TQ, Bailloeul T, Mewes K, Mersereau RM, Brum- mer ME: A super-resolution framework for 3-D high-resolution and high-contrast imaging using 2-D multislice MRI. IEEE Trans. Med. Imaging 28(5), 633–644 (2009) [DOI] [PubMed] [Google Scholar]
- 17.Sui Y, Afacan O, Gholipour A, Warfield SK: Isotropic MRI super-resolution reconstruction with multi-scale gradient field prior In: Shen D, et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 3–11. Springer, Cham: (2019). 10.1007/978-3-030-32248-9_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thelen A, Frey S, Hirsch S, Hering P: Improvements in shape-from-focus for holographic reconstructions with regard to focus operators, neighborhood-size, and height value interpolation. IEEE Trans. Image Process 18(1), 151–157 (2009) [DOI] [PubMed] [Google Scholar]
- 19.Tsai RY, Huang T: Multi-frame image restoration and registration. In: Advances in Computer Vision and Image Processing (1984) [Google Scholar]
- 20.Wang J, Chen Y, Wu Y, Shi J, Gee J: Enhanced generative adversarial network for 3D brain MRI super-resolution. In: IEEE Winter Conference on Applications of Computer Vision, pp. 3627–3636 (2020) [Google Scholar]
- 21.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP: Image quality assessment: from error measurement to structural similarity. IEEE Trans. Image Process 13(1), 600–612 (2004) [DOI] [PubMed] [Google Scholar]
- 22.Zaca D, Hasson U, Minati L, Jovicich J: A method for retrospective estimation of natural head movement during structural MRI. J. Magn. Reson. Imaging 48(4), 927–937 (2018) [DOI] [PubMed] [Google Scholar]
- 23.Zhao C, Carass A, Dewey BE, Prince JL: Self super-resolution for magnetic resonance images using deep networks. In: International Symposium on Biomedical Imaging, pp. 365–368 (2018) [Google Scholar]


