Abstract
Crimean–Congo hemorrhagic fever (CCHF) is a severe tick-borne disease caused by CCHF virus (CCHFV). Ticks in the genus Hyalomma are the main vectors and reservoirs of CCHFV. In Spain, CCHFV was first detected in Hyalomma ticks from Cáceres in 2010. Subsequently, two autochthonous CCHF cases were reported in August 2016. In this study, we describe the characterization of the CCHFV genome directly from Hyalomma lusitanicum collected in Cáceres in 2014. Phylogenetic analyses reveal a close relationship with clade III strains from West Africa, with an estimated divergence time of 50 years. The results of this work suggest that CCHFV has been circulating in Spain for some time, and most likely originated from West Africa.
Keywords: : Crimean–Congo hemorrhagic fever (CCHF), emerging disease, Hyalomma tick, Spain, tick-borne disease
Introduction
Crimean–Congo hemorrhagic fever (CCHF) is a tick-borne disease that is widely distributed throughout Africa, Asia, and Southeastern Europe (Bente et al. 2013). The causative agent, CCHF virus (CCHFV) belongs to the genus Orthonairovirus, family Nairoviridae (Adams et al. 2017). The genome consists of three negative-sense RNA segments designated S (small, 1.7 kb), M (medium, 5.3 kb), and L (large, 12.1 kb). These segments encode a nucleoprotein, structural and nonstructural glycoproteins, and an RNA-dependent RNA polymerase, respectively.
The virus is maintained in vertical and horizontal transmission cycles involving ixodid ticks and a variety of wild and domestic vertebrates (Bente et al. 2013). The primary vector and reservoir of CCHFV are ticks of the genus Hyalomma; Hyalomma marginatum is the principal vector in the eastern Mediterranean basin (Bente et al. 2013).
The detection of CCHFV RNA from Hyalomma ticks in Cáceres, Spain in 2010 (Estrada-Peña et al. 2012) and subsequent autochthonous human case in the province of Ávila in 2016 (Negredo et al. 2017), highlight the emerging threat for Spain and potentially all Western Europe. The objectives of this study were to examine the genetic relationships of the CCHFV from Hyalomma lusitanicum tick, in this study referred to as Cáceres 2014, and to investigate the origin of CCHFV in Spain.
Materials and Methods
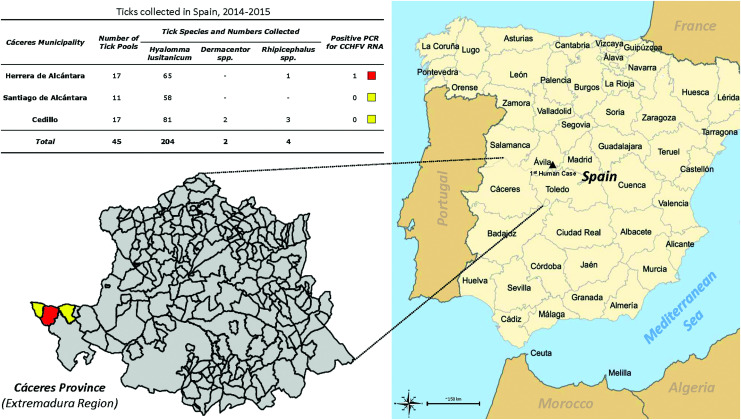
From October 2014 to January 2015, 210 ticks (204 adult H. lusitanicum, 2 adult Dermacentor spp., 4 Rhipicephalus spp. nymphs) were collected from deer at eight localities in three municipalities in Cáceres, Spain (Fig. 1). Ticks were identified and shipped on dry ice to the University of Texas Medical Branch (Galveston, Texas, USA), and stored at −80°C until processing. Ticks were divided into 45 pools, consisting of five to nine unfed adults or one semi- or engorged adult female. Each pool was washed with 3% hydrogen peroxide solution, rinsed with Hanks balanced salt solution (HBSS) containing antibiotics, minced using a pair of scissors, homogenized using a pellet pestle in a 1.5-mL tube (Kimble Chase, Vineland, NJ), and resuspended in 1-mL of tick grinding solution (HBSS with 10% fetal bovine serum and antibiotics).
FIG. 1.
Map of Spain's provinces and the location and number/species of ticks collected in 2014–2015. Ticks were collected from the municipalities of Herrera de Alcántara, Santiago de Alcántara, and Cedillo in Cáceres (highlighted in yellow or red). A total of 210 ticks were collected, divided into 45 pools, and each pool tested for CCHFV S segment RNA by nested PCR (inset table). CCHFV, Crimean–Congo hemorrhagic fever virus. Color images available online at www.liebertpub.com/vbz
Virus isolations on each tick pool were attempted in human adrenocortical carcinoma SW-13 cells (kindly provided by E. Bergeron of CDC, Atlanta, GA), and subsequently passaged two more times in SW-13, human hepatocarcinoma HuH-7, and Vero E6 (ATCC, Manassas, VA) cells as described previously (Shepherd et al. 1986, Rodriguez et al. 1997). Inoculated cultures were maintained for 14 days, harvested, and tested for CCHFV S segment RNA by real-time PCR as done previously (Atkinson et al. 2012) and by nested PCR assay (Rodriguez et al. 1997). Virus isolation on the CCHFV RNA-positive pool was attempted in 3-day-old AG129 mice following procedures described previously (Shepherd et al. 1986). Blood from mice and dams collected on day 18 postinoculation was tested for the presence of IgG antibody against CCHFV UG3010 antigen by ELISA (Rodriguez et al. 1997). All work with tick homogenization and virus isolations in cell culture and suckling mice were performed in the Biosafety Level-4 laboratories at the Galveston National Laboratory, located on the UTMB campus.
Total RNA was extracted from 0.2-mL of homogenate in TRIzol reagent (Life Technologies, Carlsbad, CA) by using the Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA). Each pool was tested for CCHFV in nested PCR assays using primers specific for the S segment as described previously (Rodriguez et al. 1997, Midilli et al. 2009, Estrada-Peña et al. 2012). First-strand cDNA was synthesized from total RNA by using SuperScript III Reverse Transcriptase (Life Technologies) and primer CCHFV-RT (5′-TCTCAAGAA-3′). The PCRs used Platinum Taq DNA polymerase (Life Technologies). Controls for each PCR included a blank (water) and a positive control RNA extracted from Vero E6 cells inoculated with CCHFV strain Turkey 200406546. The complete sequences of the S, M, and L segments were determined for the RT/PCR-positive pool. Overlapping fragments 1.0- to 2.2-kb long were amplified using Platinum Taq (Life Technologies) or HotMaster Taq DNA polymerase (Quantabio, Beverly, MA) with primers specific for the S, M, and L segments (Deyde et al. 2006, Xia et al. 2016). The amplicons overlapped by 72-nt to 220-nt, excluding primers. Both strands of each PCR product were sequenced using the Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and analyzed on an ABI PRISM® 3100-Avant™ Genetic Analyzer at the UTMB Molecular Genomics Core. The nucleotide sequences of the 5′ and 3′ termini of the S, M, and L segments were not empirically determined. The list of primers is available from the authors upon request.
The complete S, M, and L segment sequences of Cáceres 2014 were assembled using ClustalW and submitted to GenBank under accession nos. MF547415, MF547416, and MF547417, respectively. The nucleotide sequences of published CCHFV strains were obtained from the GenBank sequence database. A table listing CCHFV strain's country of origin, isolation year, and accession number is available as Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/vbz). The amino acid sequences of the S, M, and L open reading frames were aligned using ClustalW or Muscle in the MEGA version 6 package (Tamura et al. 2013), and the corresponding nucleotide sequence alignments were manually adjusted. The S, M, and L datasets consisted of 105, 75, and 64 CCHFV strains, respectively. Sequence comparisons (uncorrected p-distances) with partial S segment sequences from Spain (Estrada-Peña et al. 2012, Negredo et al. 2017) were done in MEGA6.
The maximum likelihood analyses were generated with RAxML-HPC BlackBox v8.2.10 implemented through the CIPRES Science Gateway (Miller et al. 2010). Temporal signature was validated with TempEst v1.5 (previously known as Path-O-Gen) by testing for a statistically significant linear correlation between the root-to-tip distance and isolation date of each sequence on a maximum likelihood phylogeny (Rambaut et al. 2016). Temporal signature is required to perform accurate molecular clock estimations during the generation of Bayesian phylogenies. BEAUTi v1.8.4 and BEAST v1.8.4 were used to generate XML files and conduct the Bayesian analysis, respectively (Drummond et al. 2012). The best nucleotide substitution model was determined with Jmodel Test 2 v2.1.6 (Darriba et al. 2012). Molecular clock models and tree priors were compared with path-sampling and stepping-stone approaches. Three independent trees were run for 50,000,000 generations, sampled every 5000 steps, and reviewed in tracer to ensure ESS values exceeded 200. Trees were combined in LogCombiner v1.8.4, annotated in TreeAnnotator v1.8.4, and visualized in FigTree v1.4.3.
Results
CCHFV S RNA was detected in 1 of 45 tick pools (a semi-engorged female H. lusitanicum, collected from Cervus elaphus on November 29, 2014 in Herrera de Alcántara, Cáceres). The complete S, M, and L genomic segments were sequenced from viral RNA. However, no infectious virus was recovered from the 45 homogenates inoculated on SW-13 cells or the subsequent passages in cell culture. None of the three litters of AG129 mice (n = 16) inoculated with tick homogenate showed signs of illness within a 14-day period, and none of the blood from these mice and their three dams had detectable IgG antibody to CCHFV.
Sequence comparisons of the Cáceres 2014 S segment with other Spanish sequences showed 99.4% identity with the 173-nt sequence obtained from H. lusitanicum in 2010 (Estrada-Peña et al. 2012), 99% identity with the 592-nt sequences from human cases (KY492289 and KY492290) in 2016 (Negredo et al. 2017), and 98.8% identity with the 173-nt sequences reported from H. marginatum on migratory birds in Morocco, 2011 (Palomar et al. 2013). The sequences from the human cases showed highest identity (>99.7%) with clade III strains ArD39554 from Mauritania and Daral 2012 from Mali (Zivcec et al. 2014). None of the other Spanish or Moroccan sequences includes M or L segments.
The complete S and M segments were 1673 and 5366-nt, respectively, and the L segment coding sequence was 11,838-nt. In pairwise comparisons, the full S segment coding sequences of Cáceres 2014 showed the highest identity (≥98.7%) with strains ArD39554, Daral 2012, and Sudan AB1-2009. Comparisons of the M segment coding sequences showed the highest identity (≥96.7%) with strains IbAr10200 and IbAn7620 from Nigeria; and comparisons of L segment coding sequences showed the highest identity (≥98.3%) with ArD39554 and Sudan AB1-2009.
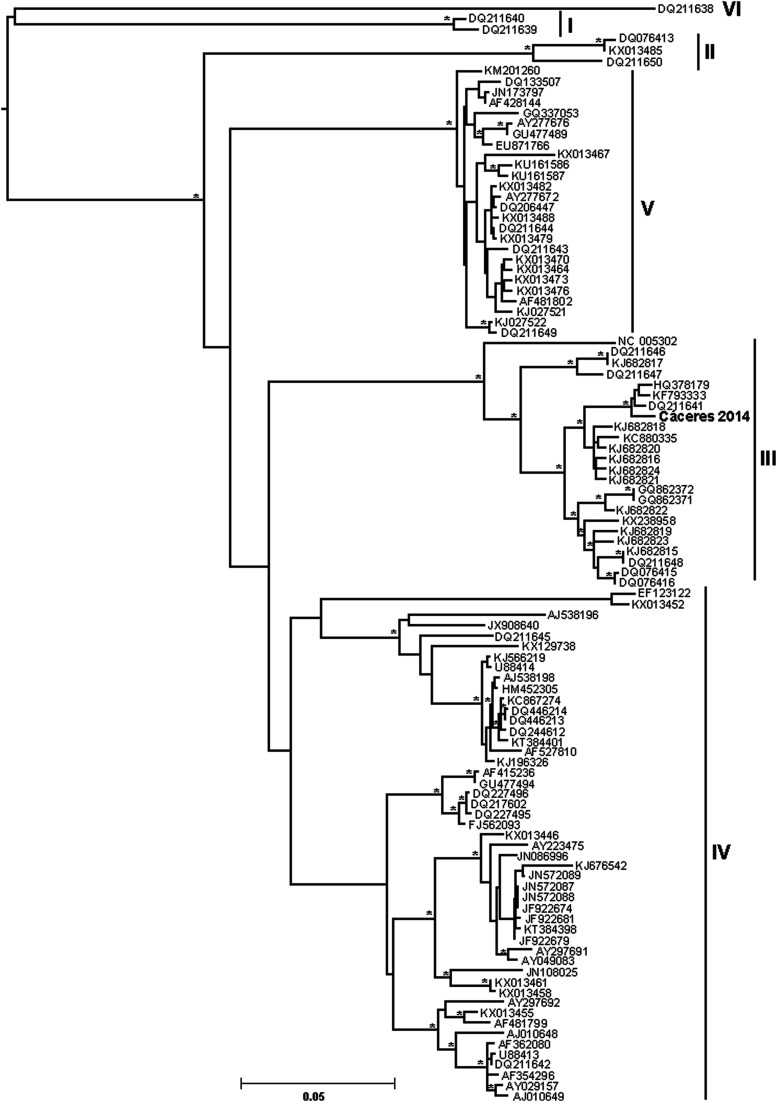
Phylogenetic analyses of the complete S segment coding sequences separated CCHFV into six well-supported groups (Fig. 2), designated clades I thru VI (Deyde et al. 2006, Bente et al. 2013). CCHFV Cáceres 2014 grouped with clade III viruses from South Africa and West Africa, among ArD39554, Daral 2012, and Sudan AB1-2009.
FIG. 2.
Evolutionary relationships among CCHFVs based on the small (S) segment. This tree and the M and L trees in Figure 4 were generated using maximum likelihood methods (RAxML-HPC BlackBox v8.2.10) implemented in CIPRES Science Gateway. CCHFV strain Cáceres 2014 from Spain is shown in bold. Scale bar indicates nucleotide substitution per site. An asterisk at a node indicates >90% bootstrap support, based on 1000 replicates. The different CCHFV clades are labeled with Roman numerals I–VI, designation by Bente et al. (2013). Branch labels are GenBank accession numbers.
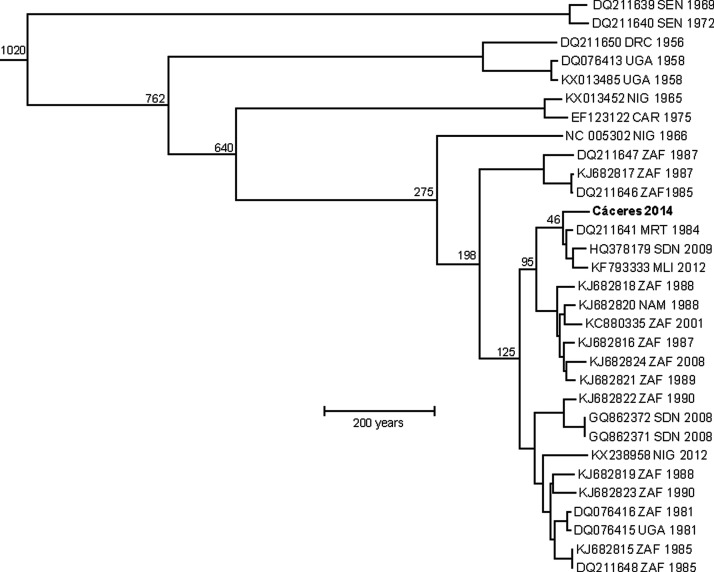
Analysis of the maximum likelihood phylogeny in TempEST revealed poor correlation between date of isolation and genetic distance, indicating poor temporal signature and preventing molecular clock analysis from being applied to the full S segment dataset. This may have been due to erroneous sampling dates, inconsistent viral mutation rates, low-quality sequencing or accumulation of mutations acquired during passage. However, strong temporal signature was identified among the African and Cáceres 2014 sequences. Comparisons of all nucleotide substitution models, clock models, and tree priors demonstrated that combination of the general time reversible model with a gamma distribution and invariable sites nucleotide substitution model, uncorrelated lognormal clock model, and exponential tree prior outperformed all other model combinations. Bayesian analyses of the African and Cáceres 2014 sequences using the above parameters reveal that Cáceres 2014 diverged from clade III viruses ∼49.14 (95%HDP 33.93–67.44) years ago (Fig. 3).
FIG. 3.
Bayesian phylogenetic tree of African (clade III) CCHFV strains based on the S segment. Bayesian analyses were conducted using BEAUTi v1.8.4 and BEAST v1.8.4. CCHFV strain Cáceres 2014 from Spain is shown in bold. Branch tips correspond to the date (year) of collection, and branch lengths correspond to length of time as measured by the scale (in years). The numbers at the nodes represent estimates (in years) of the initial divergence of the most recent common ancestors. Branch labels include GenBank accession number, country, and year of collection.
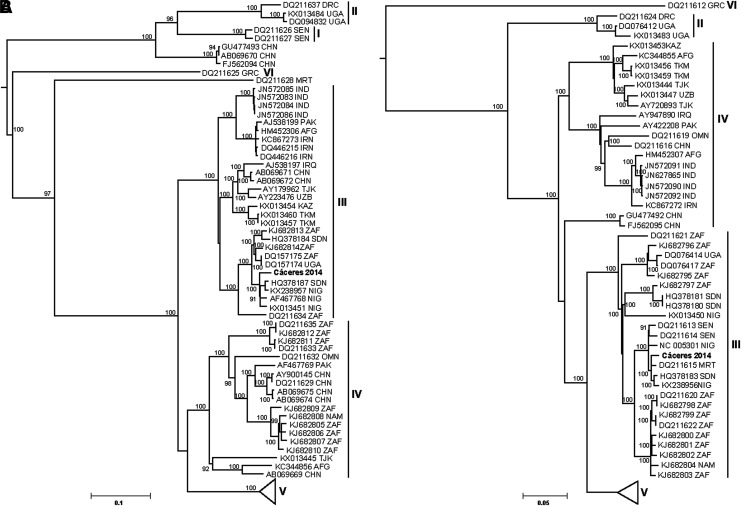
The maximum likelihood analyses of complete M segment coding sequences include Cáceres 2014 in a lineage comprised of viruses from South Africa, Nigeria, and Sudan (Fig. 4A) with IbAr10200, IbAn7620, and Borno (Nigeria) and Sudan AB1-1009. Maximum likelihood analyses of complete L segment coding sequences grouped Cáceres 2014 with clade III viruses from South Africa and West Africa (Fig. 4B) among ArD39554, Daral 2012, and Borno.
FIG. 4.
Maximum likelihood tree showing genetic relationships among CCHFVs based on the M segment (A) and L segment (B). CCHFV strain Cáceres 2014 from Spain is shown in bold. Bootstrap values >90% are shown, based on 1000 replicates. Scale bar indicates nucleotide substitution per site. The CCHFV clades are labeled I–VI and branch labels include GenBank accession number and country.
Discussion
In this study, we describe the characterization of the CCHFV genome directly from H. lusitanicum collected in Cáceres in 2014. Previous studies were limited as they only looked at a small region of the genome within the S segment. The CCHFV sequence detected in this study, and in H. lusitanicum in 2010 were genetically most closely related to clade III strains ArD39554 from Mauritania isolated from H. marginatum rufipes, and Daral 2012 from Mali detected in Hyalomma spp., supporting the notion that CCHFV in Spain was introduced from West Africa rather than from Eastern Europe. The close genetic relationship with CCHFV in H. marginatum ticks on migratory birds from Morocco in 2011 further supports this theory of northward movement into Spain (Palomar et al. 2013). Unfortunately, we were not able to isolate the virus and conduct further studies. This might be due to the fact that viral genome was detected in a semi-engorged tick, which might have imbibed neutralizing antibodies from the host.
Bayesian analyses suggest that Cáceres 2014 diverged from West African clade III viruses approximately five decades ago, and accordingly, CCHFV may have been in Spain for some time. This finding is supported by the detection of CCHFV RNA from Hyalomma ticks in four regions (Extremadura, Castilla-La Mancha, Castilla y León, and Madrid) in Spain following the 2016 CCHF cases (Ministerio de Sanidad, Servicios Sociales e Igualdad 2017). The detection of antibodies to CCHFV in persons from Southern Portugal near the border with Spain in 1980 (Filipe et al. 1985) further suggests that CCHFV has been circulating silently in the Iberian Peninsula.
The emergence of two autochthonous CCHF cases and the detection of CCHFV RNA in Hyalomma ticks in four regions of Spain highlight the need for active surveillance and ecological studies (Ministerio de Sanidad, Servicios Sociales e Igualdad 2017, Negredo et al. 2017). Since Hyalomma tick populations are widely distributed in the Mediterranean basin (Estrada-Peña et al. 2004), surveillance for the presence of CCHFV in all Western Europe is of utmost importance.
Supplementary Material
Acknowledgments
The authors thank Dr. Slobodan Paessler for providing the AG129 mice, and Ms. Mary Louise Milazzo for providing assistance with animal protocols and procedures. They also thank Mrs. Domenica Zimmerman for the help with the tick shipment, and Ms. Maricela Torres for help with Spanish literature.
Author Disclosure Statement
No competing financial interests exist.
References
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol 2017; 162:2505–2538 [DOI] [PubMed] [Google Scholar]
- Atkinson B, Chamberlain J, Logue CH, Cook N, et al. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis 2012; 12:722–726 [DOI] [PubMed] [Google Scholar]
- Bente DA, Forrester NL, Watts DM, McAuley AJ, et al. Crimean-Congo hemorrhagic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 2013; 100:159–189 [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. JModelTest 2: More models, new heuristics and parallel computing. Nat Methods 2012; 9:772. DOI: 10.1038/nmeth.2109 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, et al. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol 2006; 80:8834–8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard M, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Peña A, Bouattour A, Camicas JL, Walker AR. Ticks of Domestic Animals in the Mediterranean region: A Guide to Identification of Species, 1st ed. Zaragoza, Spain: University of Zaragoza Press, 2004 [Google Scholar]
- Estrada-Peña A, Palomar AM, Santibańez P, Sánchez N, et al. Crimean-Congo hemorrhagic fever virus in ticks, southwestern Europe, 2010. Emerg Infect Dis 2012; 18:179–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe AR, Calisher CH, Lazuick J. Antibodies to Congo-Crimean haemorrhagic fever, Dhori, Thogoto and Bhanja viruses in Southern Portugal. Acta Virol 1985; 29:324–328 [PubMed] [Google Scholar]
- Midilli K, Gargili A, Ergonul O, Eleyli M, et al. The first clinical case due to AP92 like strain of Crimean-Congo hemorrhagic fever virus and a field survey. BMC Infect Dis 2009; 9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE) IEEE; New Orleans, LA, 2010:1–8 [Google Scholar]
- Ministerio de Sanidad, Servicios Sociales e Igualdad: Informe de Situacióny Evaluación del riesgo de Transmisión de Fiebre Hemorrágica de Crimea-Congo (FHCC) en España. April 2017. (consulted 1 May 2017). Available at www.msssi.gob.es/profesionales/saludPublica/enfermedadesEmergentes/Crimea Congo/docs/ACTUALIZACION ER FHCC 20.04.2017.pdf
- Negredo A, de la Calle-Prieto F, Palencia-Herrejón E, Mora-Rillo M, et al. Autochthonous Crimean-Congo hemorrhagic fever in Spain. N Engl J Med 2017; 377:154–161 [DOI] [PubMed] [Google Scholar]
- Palomar AM, Portillo A Santibañez P, Mazuelas D, et al. Crimean-Congo hemorrhagic fever virus in ticks from migratory birds, Morocco. Emerg Infect Dis 2013; 19:260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2016; 2:vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LL, Maupin GO, Ksiazek TG, Rollin PE, et al. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am J Trop Med Hyg 1997; 57:512–518 [DOI] [PubMed] [Google Scholar]
- Shepherd AJ, Swanepoel R, Leman PA, Shepherd SP. Comparison of methods for isolation and titration of Crimean-Congo hemorrhagic fever virus. J Clin Microbiol 1986; 24:654–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Beck AS, Gargili A, Forrester N, et al. Transstadial transmission and long-term association of Crimean-Congo hemorrhagic fever virus in ticks shapes genome plasticity. Sci Rep 2016; 6:35819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M, Maïga O, Kelly A, Feldmann F, et al. Unique strain of Crimean-Congo hemorrhagic fever virus, Mali. Emerg Infect Dis 2014; 20:911–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.