Abstract
Social bonds such as parent-infant attachment or pair bonds can be critical for mental and physical well-being. The monogamous prairie vole (Microtus ochrogaster) has proven useful for examining the neural substrates regulating social behaviors, including social bonding. Oxytocin (OXT) and oxytocin receptor (OXTR) play critical roles in alloparental care, pair bonding and consoling behavior in prairie voles. While OXTR in a few regions, such as the nucleus accumbnes (NAcc), prefrontal cortex (PFC) and anterior cingulate cortex (ACC), have been implicated in regulating these behaviors, the extent to which other OXT sensitive areas modulate social behaviors has not been investigated. The NAcc is a central hub for modulating OXTR dependent social behaviors. To identify neurons expressing Oxtr in prairie vole brain, we generated gene knock-in voles expressing Cre recombinase in tandem with Oxtr (Oxtr-ires-Cre) using CRISPR/Cas9 genome editing. We confirmed Oxtr and Cre mRNA co-localization in NAcc, validating this model. Next, we identified putative Oxtr-expressing neurons projecting to NAcc by infusing retrograde CRE-dependent EGFP AAV into NAcc and visualizing fluorescence. We found EGFP positive neurons in anterior olfactory nucleus, PFC, ACC, insular cortex, paraventricular thalamus, basolateral amygdala, amygdalohippocampal area, and cortical amygdaloid area. The ACC to NAcc OXTR projection may represent a species-specific circuit since Oxtr-expressing neurons in the ACC of mice were reported not to project to the NAcc. This is the first delineation of Oxtr-expressing neural circuits in the prairie vole, and demonstrates the utility of this novel genetically modified organism for characterizing OXTR circuits involved in social behaviors.
Keywords: Oxytocin receptor, genome editing, nucleus accumbens, social bonding, affiliation, neural circuits, CRISPR
Introduction
Social bonds such as the close and enduring attachments between parents and infants or between opposite sex mates in monogamous mammals can play a critical role in promoting mental and physical well-being (Feldman, 2012). Disruption of social bonds can have devastating effects on mental and physical health (Bosch and Young, 2018; Pohl et al., 2019). Recent research has focused on understanding how genes, neuronal modulators, receptors, and circuits regulate social bond formation and maintenance, which may have translational implications for improving quality of life in certain psychiatric disorders such as autism, PTSD, and depression (Modi et al., 2015; Arai et al., 2016; Bosch et al., 2016; King et al., 2016; Amadei et al., 2017; Hirota et al., 2020).
The prairie vole (Microtus ochrogaster) is a monogamous rodent species showing enduring pair bonding between mates, biparental care, and the consoling behavior toward a distressed partner (Young and Wang, 2004; Aragona et al., 2006; Gobrogge et al., 2009; Young et al., 2011; Johnson and Young, 2015; Burkett et al., 2016; Walum and Young, 2018). Prairie voles have proven to be an ideal model for investigating brain mechanisms that regulate social bonding though studying the pair bond (McGraw and Young, 2010; Walum and Young, 2018). Oxytocin (OXT) and oxytocin receptor (OXTR) are one of the key neuromodulators and receptors mediating pair bond formation and maintenance in prairie voles (Donaldson and Young, 2008; Smith and Wang, 2014; Barrett et al., 2015; Bosch, et al., 2016; Johnson et al., 2016; Johnson et al., 2017; Walum and Young, 2018). OXT is produced in neurons in the paraventricular nucleus (PVN) and supraoptic nucleus of the hypothalamus and their projections course throughout the forebrain (Ross et al., 2009; Johnson and Young, 2017; Rogers et al., 2018). OXT binds to OXTR, which is a G protein coupled receptor coupling with Gαq/11 and transducing intracellular signaling(Jurek and Neumann, 2018). OXT/OXTR signaling acts as a neuromodulator affecting social and sexual behavior, stress coping, and the processing of social stimuli, in many species (Knobloch et al., 2012; Dolen et al., 2013; Choe et al., 2015; Marlin et al., 2015; Wei et al., 2015; Oettl et al., 2016; Yokoi et al., 2020). It has been proposed that OXT acts in multiple brain regions to enhance the neural processing of social signals, increase the salience and reinforcing value of social cues, and facilitate the formation of social memories, processes critical for social bond formation. OXTR signaling in the nucleus accumbens (NAcc) plays a particularly important role in pair bond formation and maintenance in prairie voles (Young et al., 2001; Keebaugh et al., 2015). However, it is increasingly clear that OXTR signaling in a distributed social salience network is critical for many aspects of social cognition involved in pair bonding, and the NAcc is a central hub of that network (Johnson, et al., 2017). Indeed OXTR signaling appears to coordinate neural activity across the social salience network during mating (Johnson, et al., 2016) which is hypothesized to facilitate the flow of social information across the network, ultimately leading to synaptic plasticity in the NAc that enhances the reinforcing value of the partner’s cues (Walum and Young, 2018). However, the extent to which OXTR expressing neurons in regions of the social salience network, such as the anterior olfactory nucleus, prefrontal cortex, anterior cingulate cortex, and basolateral amygdala directly project to the NAcc is unknown. Although some circuits projecting to NAcc are known to regulate reward processing in rodents (Zhu et al., 2016; Otis et al., 2019; Lafferty et al., 2020), only a few studies have explored the function and anatomy of these circuits in prairie voles (Amadei, et al., 2017) and the relationships between these circuits and the OXT/OXTR system remains to be elucidated. To begin to explore the interconnected nature of OXTR circuits with the NAcc that may be involved in social bonding in prairie voles, we generated Oxtr-internal ribosomal entry site (ires)-Cre prairie voles using clustered regularly interspaced short palindromic repeat (CRISPR)/ CRISPR associated protein (Cas) genome editing.
Adeno associated virus (AAV) has been proven useful as a tracer to visualize afferent and efferent neuronal circuits in rodents (Chamberlin et al., 1998; Tervo et al., 2016). Anterograde AAV carrying fluorescent proteins like enhanced green fluorescent protein (EGFP) can be expressed in cell bodies of neurons exclusively, and by tracing fibers filled with EGFP in the whole brain, efferent projections can be determined (Bosch, et al., 2016). Retrograde AAV can transduce axon terminals in the injection site, be transported to the cell body, and express in neurons that project to injection site and afferent projections of the injection site can be determined. To our knowledge, retrograde AAV tracing studies have yet to be used in prairie voles to identify cell-type specific afferent projections in prairie voles.
Although the expression of Oxtr in brains of prairie voles has shown by autoradiography (Witt et al., 1991; Insel and Shapiro, 1992), it has not been possible to visualize Oxtr expressing neurons at a cellular level, or examine their projections in prairie voles. To examine neural circuits expressing Oxtr that project to NAcc specifically, a knock-in prairie vole that expresses Cre recombinase (Cre) under the control of Oxtr would be useful, however embryonic stem cells- or induced pluripotent stem cells-based homologous recombination has not been achieved in prairie voles. Alternatively, the CRISPR/Cas9 system has been proven to be useful for generating gene knock in animals because double strand break, generated by site-specific endonuclease-like activity by CRISPR/Cas9, stimulates and accelerate homologous recombination (Aida et al., 2015; Boender and Young, 2020). The CRISPR/Cas system is an RNA-based adaptive immune system of archaea and bacteria against bacteriophage or foreign nucleic acids (Barrangou et al., 2007; Wiedenheft et al., 2012). CRISPR/Cas9 system derived from Streptococcus pyogenes is in the Type II - A CRISPR/Cas system consisting of Cas9 nuclease and two short RNAs, crispr RNA (crRNA) and trans activating crRNA (tracrRNA)(Deltcheva et al., 2011). crRNA containing 20 nt of sequence homologous with the target genome, tracrRNA, and Cas9 protein bind to the target region adjacent to the 5’-NGG-3’ palindromic adjacent motif (PAM) sequence. Cas9 induces a double strand break into the target sequence and insertions or deletions are induced into the target sequence while it is repaired by endogenous DNA repair pathways (Cho et al., 2013; Cong et al., 2013; Mali et al., 2013; Wang et al., 2013). At that time, if there are homologous templates like knock-in plasmids or single strand DNAs, and exogenous DNA sequence can be inserted in the targeted region (Wang, et al., 2013; Aida, et al., 2015; Boender and Young, 2020).
In this study, we first generated a knock-in prairie vole carrying ires - Cre in the Oxtr gene by using CRISPR/Cas9. This is the first reported knock-in prairie vole, demonstrating the potential of this strategy for diversifying the genetic tools available for this model organism. By combining the Oxtr-ires-Cre knock-in prairie vole with the injection of retrograde AAV carrying CRE-dependent EGFP, we mapped the Oxtr-expressing neurons that project to NAcc (afferents) and report evidence that prairie voles may have novel OXT sensitive circuits relevant to their species-specific social behavioral repertoire.
Experimental procedures
Animals
Prairie vole colonies were maintained first at Tohoku University in Japan and then at Emory University in Atlanta as described previously (Horie et al., 2019). The Tohoku colony originated from the Emory University colony, which were originally derived from field caught specimens from Illinois, USA. Opposite sex pairs were housed together to breed. After weaning at postnatal day (P) 20–23, the offspring were separated from their parents and housed in groups of up to three same-sex siblings per cage. All animals were housed using alternating 12-h periods of light and dark at 25 °C and were allowed ad libitum access to food (Labo MR Stock, NOSAN, Japan, or Laboratory Rabbit Diet HF 5326, LabDiet) and water. All animal experiments were performed according to a protocol approved by Tohoku University Guidelines for Animal Experimentation or by the Institutional Animal Care and Use Committee at Emory University.
Construction of crRNA and the knock-in plasmid
The crRNA targeting the 3’ untranslated region (UTR) of the prairie vole Oxtr was designed using Benchling (https://www.benchling.com). The crRNA with the desired spacer sequence (5’- TTCAGCATGAGCCACCTGTCTGG −3’) and tracrRNA were synthesized by GenomeCraft service of FASMAC (GE-001, FASMAC, Japan). The efficiency of the crRNA was confirmed by in vitro digestion assay (IDA) as described previously (Aida, et al., 2015) (Supplementary Figure 1).
A knock-in plasmid containing ires-Cre sequence flanked by homology arms were made with In-Fusion HD Cloning kit (638909, Clontech, Japan). Ires-Cre sequence was placed 7 bp downstream from the stop codon of Oxtr gene. Seven hundred fifty base pairs of 5’ and 3’ homology arms were amplified with a forward primer 5’ - CCGCGGGAATTCGATCAGGCAGGTTGGAAGTTG - 3’ and a reverse primer 5’ - CGCGGTGGGCCAGACAGGTGGCTCATGCTGAAG - 3’, and a forward primer 5’ - GTCTGGCCCACCGCGCCT - 3’ and a reverse primer 5’ - GAATTCACTAGTGATCAGCTCCAGAGTGTGTGT - 3’ respectively. The pGEM-T-easy vector (Promega, Madison WI, USA) was linearized by PCR with a forward primer 5’ - ATCACTAGTGAATTCGCGGC - 3’ and 5’ - ATCGAATTCCCGCGGCCGCC - 3’ and homology arms were cloned into linearized pGEM T-easy vector with In-Fusion HD Cloning kit (638909, Clontech, Japan) (pGEM-arm). The Ires-Cre sequence was amplified from the genome of Oxtr-ires-Cre knock-in mouse (Hidema et al., 2016) by PCR with a forward primer 5’ - CAGCATGAGCCACCTAGATGGGGGTCCTGGGCC - 3’ and a reverse primer 5’ - CGCGGTGGGCCAGACCTAATCGCCATCTTCCAGC - 3’ and cloned into pGEM-arm linearized by PCR with a forward primer 5’ - GTCTGGCCCACCGCGCCTGC - 3’ and 5’ - AGGTGGCTCATGCTGAAGAT - 3’. Every PCR was performed with PrimeSTAR GXL DNA Polymerase (R050B, Takara, Japan) and T100 thermal cycler (Promega, Madison WI, USA).
Embryo manipulations
Superovulation and embryo collection
Superovulation and embryo transfer were performed as described previously (Horie et al., 2015; Horie, et al., 2019). Briefly, 48 h after 60 IU pregnant mare serum gonadotropin was administered to the female prairie vole, 60 IU human chorionic gonadotropin was administered and the subject was allowed to mate with the stud male for 16 h. Then embryos were collected from oviducts of the female and cultured in G-1 PLUS medium (10128, Vitrolife, Sweden) at 37 °C with 5 % CO2 until they were used for the microinjection.
Microinjection of crRNA, tracrRAN, and Cas9 protein
Microinjecting Cas9 nuclease protein, synthetic crRNA, and tracrRNA to the nucleus and cytoplasm of embryos instead of microinjecting Cas9 mRNA and single guide RNA dramatically improved knock-in efficiency in mouse (Aida, et al., 2015). Thus, we used this strategy to generate Oxtr-ires-Cre knock-in prairie voles.
Thirty ng/μl Cas9 nuclease protein (M0386S, NEB, Ipswich, MA USA), 0.61 pmol/μl crRNA, 0.61 pmol/μl tracrRNA, and 10 ng/μl knock-in plasmid were prepared in Nuclease-Free water (AM9932, Ambion, Austin TX, USA). The mixture was injected into the nucleus and cytoplasm of embryos with a microinjector (IM-11–2、NARISHIGE, Japan) and a micromanipulator (MN-4、M0–202U、NARISHIGE, Japan) under an inverted microscope (Ts2R, Nikon, Japan). Injected embryos were cultured in G-1 Plus medium for 16–20 hours until they developed to the 2-cell stage.
Embryo transfer
Injected embryos were transferred to the oviducts of pseudopregnant female voles as described previously (Horie, et al., 2019). Briefly, up to 15 injected embryos were transferred into the oviducts of pseudopregnant female voles with a glass capillary. Twenty days later, the subjects gave birth pups.
Genotyping and Sanger sequencing of Oxtr in newborns
Tissues were collected from their ear punches or toe clips and the target regions were amplified with 3 primer pairs (5’ arm PCR, F1 and R1, 5’ - GCCAGGCAGAGAGTTTTGTC - 3’ and 5’ - CAGAGCAGCCCTGTACATCA - 3’; insert PCR, F2 and R2, 5’ - ATGGTCCCAGAGTGACCCTTTG - 3’ and 5’ - TTGTCCTAGCCCACTGGGTTTC - 3’; 3’ arm PCR, F3 and R3, 5’ - TGGAAGATGGCGATTAGGTC - 3’ and 5’ - CGAGGTTACAAACGCACTGA - 3’) and analyzed by gel electrophoresis as shown in Figure 1. Each PCR amplicon from cre1 vole was purified and cloned into pGEM-T easy vector, and 10 of each plasmid were sequenced using Sanger sequencing.
Figure 1. Generation of Oxtr-ires-Cre knock-in prairie voles.
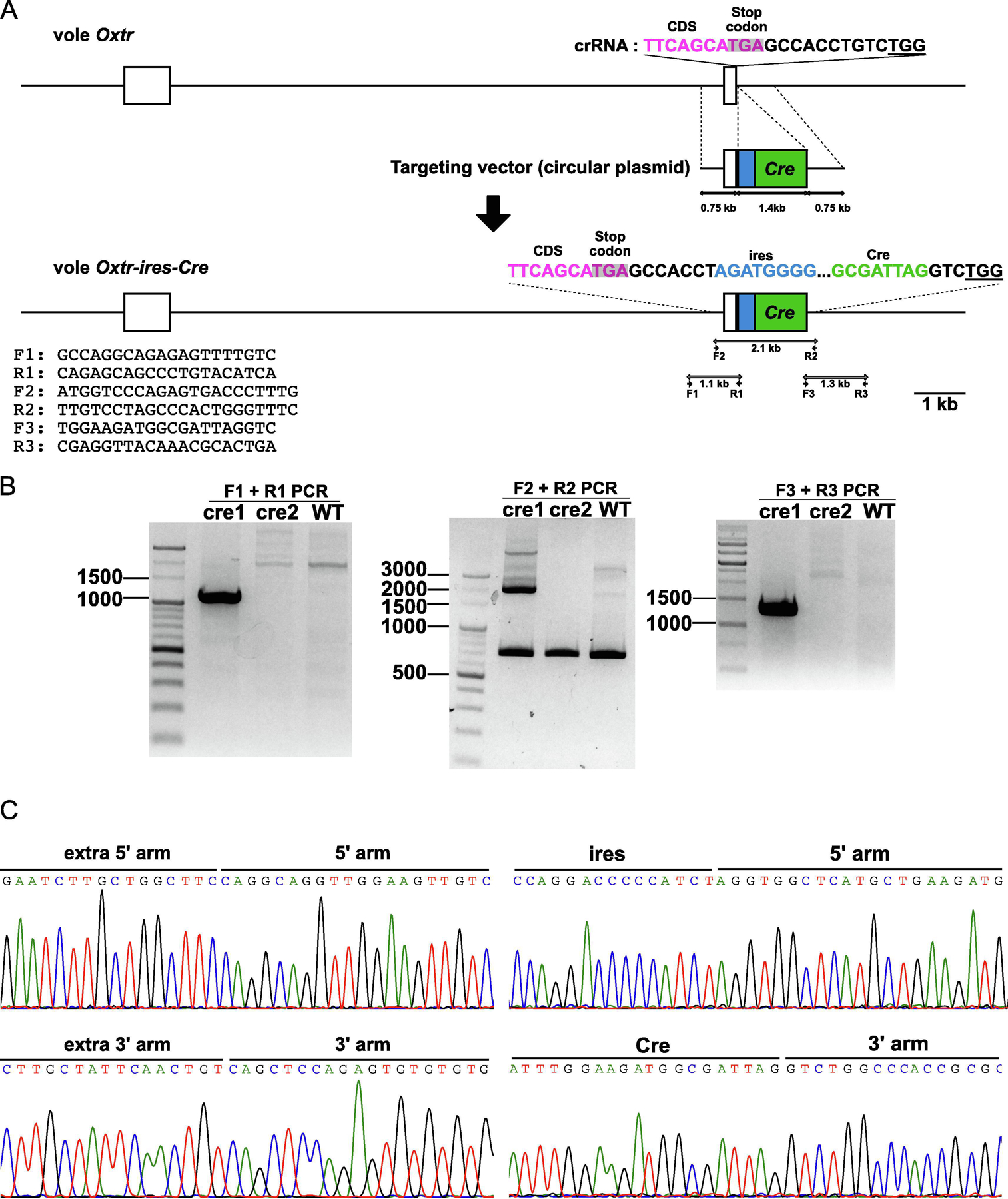
A. The Oxtr gene was targeted by a crRNA targeting TTCAGCATGAGCCACCTGTC. The underline indicates the PAM sequence. The ires-Cre sequence was placed at the 7 bp downstream from the stop codon of Oxtr. Coding sequence (CDS), magenta; ires, blue; Cre, green; stop codon, gray highlight. B. Cre1 was confirmed as a knock-in vole by the PCR analysis. C. The target region of Cre1 was sequenced and confirmed as a knock-in vole. The boundary sequences between endogenous sequences and knock-in arms, and arms and ires-Cre sequence were shown as representative figures to show the precise knock-in.
Off target analysis
Potential off-target sequences were predicted by Benchling. The off-target site having the score higher than 0.8 was regarded as a potential off-target site and 9 potential off-target sites were detected. Each potential off-target was amplified from genomes of 14 voles that were the breeding pairs of F3 generation by PCR with each primer set (OffT1: 5’ - CTTACCAGATGGTCAACAGC - 3’ and 5’ - TCTAGGACAGTGATCTTGCC - 3’; OffT2: 5’ - TTATGTCTGTGTACTACCTA - 3’ and 5’ - TACATATATAGACATAGTTA - 3’, or 5’ - GAGGCTCAGTGCCCACACCT - 3’ and 5’ - AAGTGGCTCATACCTACCTG - 3’; OffT3: 5’ - ATTCACGGTTATATGGGCAT - 3’ and 5’ - TCTCAATAAAATAACAAGGG - 3’; OffT4: 5’ - TCACACTTGGGGATTCTGTG - 3’ and 5’ - TTTGCACAATAAAGGGGATC - 3’; OffT5: 5’ - AGGGCCCTGCACACAGTGAC - 3’ and 5’ - GTATGTCAGTGCATGCATGT - 3’; OffT6: 5’ - TAGAATTTTGTCTGGAAGTT - 3’ and 5’ - AATACAAGTAAAATATCACT - 3’; OffT7: 5’ - GAATTGGCAATGTTGTTGCT - 3’ and 5’ - ACTTATCCTCAACCACTGCC - 3’; OffT8: 5’ - GCCTGCCTTTGCTTCTCAGG - 3’ and 5’ - AGACAGTAAAGGGAATAATG - 3’; OffT9: 5’ - TGGAAACTTTAAACTCTGCA - 3’ and 5’ - TTTTGCAGAGCTGTAAGAGT - 3’) and directly sequenced by the sanger sequencing with each sequencing primer (OffT1: 5’ - CTTACCAGATGGTCAACAGC - 3’ or 5’ - AACACAGGCTCTTTAAGTCC - 3’; OffT2: 5’ - GTCTGTGTACTACCTATGTA - 3’ or 5’ - ACACAGTACCCGGAGACTCCAG - 3’; OffT3: 5’ - ATTCACGGTTATATGGGCAT - 3’; OffT4: 5’ - TCACACTTGGGGATTCTGTG - 3’; OffT5: 5’ - AGGGCCCTGCACACAGTGAC - 3’; OffT6; 5’ - TAGAATTTTGTCTGGAAGTT - 3’; OffT7; 5’ - GAATTGGCAATGTTGTTGCT - 3’, 5’ - CATCTTCATTTCTGACTCAT - 3’, 5’ - ACTTATCCTCAACCACTGCC - 3’, or 5’ - CCTCAACCACTGCCTTCCCG - 3’; OffT8: 5’ - GCCTGCCTTTGCTTCTCAGG - 3’, or 5’ - CTTAACTATCTGGGCCACCG - 3’; OffT9: 5’ - TGGAAACTTTAAACTCTGCA - 3’).
RNAscope in situ hybridization (ISH)
Prairie voles were deeply anesthetized by Isoflurane and euthanized by decapitation and their brains were immediately removed and frozen on dry ice and stored at −80 °C until they were sliced. Twenty μm thick brain slices were prepared on a cryostat (CryoStar™ NX-70, ThermoFisher Scientific, USA).
A prairie vole specific Oxtr probe (422561-C3, Advanced Cell Diagnostics, Inc, USA) and a Cre probe (312281-C2, Advanced Cell Diagnostics, Inc, USA) were used for fluorescent RNAscope ISH. Brain slices mounted on the glass slides (Superfrost Plus Microscope Slides, Fisherbrand, USA) were placed in 4% paraformaldehyde (PFA) for 15 min at 4°C. Slices were dehydrated serially in 50% ethanol for 5 min, 70% ethanol for 5min, and 100% ethanol for 5 min at room temperature, and then incubated in 100% ethanol at −20°C over a night. Slides were dried for 5 min and barriers were drawn with an Immedge™ hydrophobic barrier pen (H-4000, Vector Laboratory, USA) around each section. Protease IV (Advanced Cell Diagnostics, Inc, USA) was applied and slides were incubated for 30 min at room temperature followed by two washes in PBS. Oxtr and Cre probes were diluted in C1 probe diluent (Advanced Cell Diagnostics, Inc, USA) in 3:1:100 ratios, respectively. The mixture was applied to slides and incubated for 2 hrs at 40°C. Slides were washed in 1x wash buffer (Advanced Cell Diagnostics, Inc, USA) twice for 2 min each at room temperature and Amp 1-FL was put on the sections and the slides were incubated for 30min at 40°C. The slides were then washed in 1x wash buffer for 2 min at room temperature twice and Amp 2-FL was applied to the sections and they were incubated for 15min at 40°C. The slides were then washed in 1x wash buffer for 2 min at room temperature twice and Amp 3-FL was applied to the sections and they are incubated for 30min at 40°C. Slides were then washed in 1x wash buffer for 2 min at room temperature twice and Amp 4-FL was applied to the sections and they are incubated for 15min at 40°C. Slides were washed in 1x wash buffer for 2 min at room temperature twice, and then DAPI (Advanced Cell Diagnostics, Inc, USA) was applied to the sections and incubated for 30 sec at room temperature. Excess liquid was removed and ProLong Gold Antifade Mountant (P36930, Thermo Fisher Scientific, USA) was applied onto each section and cover slips were placed on slides.
Images were taken using the Keyence microscope (BZ-X700, Keyence, Japan) with x40 lens.
OXTR receptor autoradiography
Brain sections were prepared according to the methods described in the RNAscope ISH section. Oxytocin receptor autoradiography was performed as described previously (Horie, et al., 2019).
Colony maintenance and genotyping
For daily genotyping, a primer pair of 5’ - AAATGCTTCTGTCCGTTTGC - 3’, 5’ - TGCCTTCATCATCGCCATGC - 3’, and 5’ - GAGCACTCAGGTTGGGGTAG - 3’ were used for PCR. It is performed using KAPA2G Robust PCR Kits, HotStart ReadyMix (KK5701, Roche, Switzerland) with touchdown PCR. PCR mixtures were denatured at 94 °C for 3 min, then denatured at 94 °C for 10 sec. They were annealed with primers at 65 °C for 30 sec, and extendedat 68 °C for 1min, and then, this cycle was repeated ten times with decreasing annealing temperature 0.5 °C per cycle. Then 28 cycles of denature at 94 °C for 10 sec, annealing at 60 °C for 30 sec, and extension at 72 °C for 1 min were performed and amplicons were analyzed by the gel electrophoresis in 2% Agarose gel.
Stereotaxic injection
Subjects were anesthetized with 3% Isoflurane and anesthesia was maintained with 1–3 % Isoflurane during surgery. They were administered with 2 mg/kg Meloxicam as a pre-surgical analgesic. The head was fixed with ear bars coated with Lidocaine and a small incision was made on the head. To visualize projections of Oxtr-expressing neurons to NAcc, 200–1000 nl of AAVrg- the human synapsin promoter (hSyn)-DIO-EGFP (7.6×1012 gc/ml, 50457-AAVrg, Addgene, USA) was injected into NAcc (anteroposterior: +1.7 mm; mediolateral: 0.85 mm; dorsoventral; 5.00 mm) with a NanoFil syringe (NANOFIL, WORLD PRECISION INSTRUMENTS, USA) with the NanoFil needle (NF33BL-2, WORLD PRECISION INSTRUMENTS, USA) at 100 nl/min by using Micro 4 microinfusion pump (WPI, USA). Subjects were administered 2 mg/kg Meloxicam on the following day and recovered for 2 weeks.
Immunostaining and imaging
Subjects injected with AAVrg-hSyn-DIO-EGFP were perfused with 4% PFA and brains were removed from their skulls. Brains were incubated in 4% PFA for 16 h and then placed in 2% Agarose in PBS. Fifty μm thick slices were prepared for using a vibratome (VT1200, Leica, USA) and served for immunohistochemistry.
Sections were washed in PBS three times at room temperature. They were incubated in 0.05% Triton X-100 in PBS (PBST) for 30 min for penetrating and then incubated in 5% normal horse serum (16050122, Thermo Fisher Scientific) in PBST (NPBST) for 30 min for blocking. Sections were incubated with rabbit anti-GFP antibody (598, MBL, Japan) diluted 1/1000 in NPBST for 16 h at 4 °C. Then, the sections were washed in PBS three times at room temperature and incubated with goat anti-rabbit IgG secondary antibody conjugated with Alexa Fluor 488 diluted 1/400 in NPBST (A-11008, Thermo Fisher Scientific) with DAPI diluted 1/1000 for 1 h at room temperature. Sections were washed in PBS three times at room temperature and mounted on the glass and dried for 20 min. VECTASHIELD® Antifade Mounting Media (H-1400, VCTOR LABORATORIES, USA) was dropped on slides and covered with cover glasses. Images were taken using a confocal microscope (LSM800, ZEISS, Germany) or a fluorescent microscope (BZ-X710 ,Keyence, Japan).
Statistical analysis
Tukey’s multiple comparison test was performed using Prism for the comparison of the mean of the intensity of OXTR binding in the autoradiography experiments.
Results
Generating Oxtr-ires-Cre knock-in prairie vole using CRISPR/Cas9
To express Cre under the control of Oxtr gene expression, we generated Oxtr-ires-Cre knock-in prairie vole using CRISPR/Cas9. We designed the knock-in plasmid vector containing ires-Cre flanked on either side by 750 bp arms having homology to the target site, with the ires place 7 bp downstream of the Oxtr stop codon (Figure 1A). We microinjected crRNA, tracrRNA, and Cas9 protein into fifty four embryos and 23 embryos developed to the 2-cell stage and were transferred into the oviducts of pseudopregnant female voles (the efficiency of development = injected embryos/two-cells, 43.0%). Two candidate pups (cre1 and cre2) were obtained (Birth rate = new borns/transferred embryos, 8.7%) and one (cre1) was confirmed as a knock-in prairie vole by genotyping of tail tissues (Figure1 B). As further confirmation, the Sanger sequencing around the target region and the inserted sequence showed the knock-in construct was precisely inserted in the target site as planned (Figure 1B and C) (the efficiency of knock-in = knocked-in voles/newborns, 50%). Based on these results, we concluded that we successfully generated an Oxtr-ires-Cre knock-in prairie vole.
Off-target analysis
Results of Sanger sequencing of 14 breeders at the F3 generation showed that a few potential off-target sites had single nucleotide polymorphisms, as to be expected in outbred voles, but no deletions or insertions (indels) indicative of CRISPR editing were detected in any target for any animal (Supplementary figure 2). In the analyses of off-target 4, a G/T polymorphism in the PAM sequence was detected in animals #3 and 6, and a G/A polymorphism in the 3’ region adjacent to the PAM sequence was detected in animal #3, 6, and 9, indicated by the red boxes in FigS2. In the off-target 5 analyses, an A/C polymorphism outside of the seed sequence in the protospacer sequence was detected in animals #1 and 8. In the analyses of off-target 8, a G/T polymorphism outside of the seed sequence in the protospacer sequence was detected in animal #13. In the analyses of off-target 9, a C/A polymorphism in the protospacer sequence was detected in animal #3, 8, 9, and 13. Such polymorphism in non-coding regions are expected in the outbred vole colony as previously described (Okhovat et al., 2015; King, et al., 2016). We conclude that no detectable off-target effects were induced by CRISPR/Cas9 genome editing at the most probable off-target sites in the Oxtr-ires-Cre line.
Confirmation of Co-expression of Oxtr and Cre mRNA
To confirm whether Cre co-localizes in cells expressing Oxtr in the brain, Oxtr and Cre mRNA were detected by in situ hybridization with RNAscope (Figure 2; male: +/+, +/Cre, Cre/Cre, each N = 3; female: +/+, +/Cre, Cre/Cre, each N =3). In WT brains, Oxtr mRNA was detected but no Cre mRNA was detected in NAcc. In contrast, Cre mRNA was detected in the cells expressing Oxtr in NAcc of +/Cre and Cre/Cre animals. Although no probe controls gave us little background noise, the signal with the Cre probe in +/Cre and Cre/Cre brains were much stronger than the background of WT. From these results, we concluded Cre mRNAs were transcribed exclusively in Oxtr-expressing cells in the brain. Interestingly Cre mRNA signal was highly restricted to one or two puncta per cell in +/Cre tissue while Oxtr mRNA signal was widely distributed across the cell body. By contrast, in Cre/Cre animals, both Cre mRNA and Oxtr mRNA were both highly restricted and colocalized in their distribution as puncta, suggesting altered processing of Oxtr-ires-Cre tandem mRNA produced by the knock-in allele.
Figure 2. Co-localization of Oxtr and Cre mRNA in brains of Oxtr-ires-Cre knock-in priaire voles.
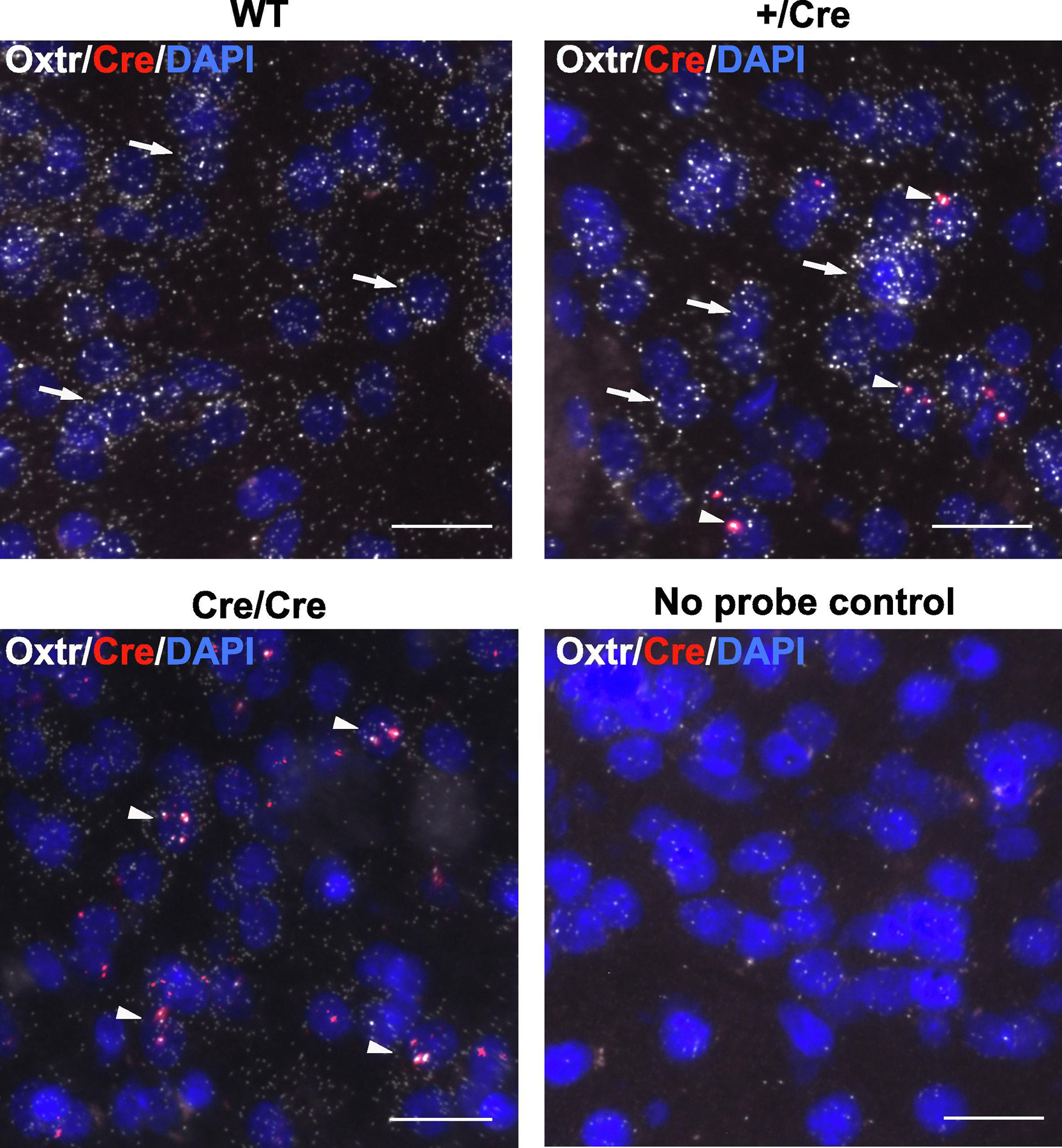
Oxtr and Cre mRNA localization in NAcc were analyzed using in situ hybridization with RNAscope in brains of wildtype (WT), heterozygous +/Cre, and homozygous Cre/Cre prairie voles (male, each N = 3; female, each N = 3). Note the restriction and co-localization of Oxtr and Cre signal in aggregates in the Cre/Cre brain.
Detection of OXTR proteins by autoradiography
The insertion of ires sequence is known to affect the expression level of the gene before and after the ires sequence (Cheng et al., 2019). We used receptor autoradiography to detect OXTR protein in the brain and analyzed binding density in the NAcc. Quantitative autoradiography showed that OXTR protein levels did not differ between WT and +/Cre (Figure 3; male: +/+, +/Cre, Cre/Cre, each N = 3; female: +/+, +/Cre, Cre/Cre, each N =3). However, OXTR in the brain of Cre/Cre was significantly decreased. These data suggested the insertion of ires-Cre sequence significantly reduced expression or translation of the Oxtr mRNA. Because of these results, we used heterozygous +/Cre knock-in prairie voles for further experiments.
Figure 3. Detection of OXTR proteins by autoradiography.
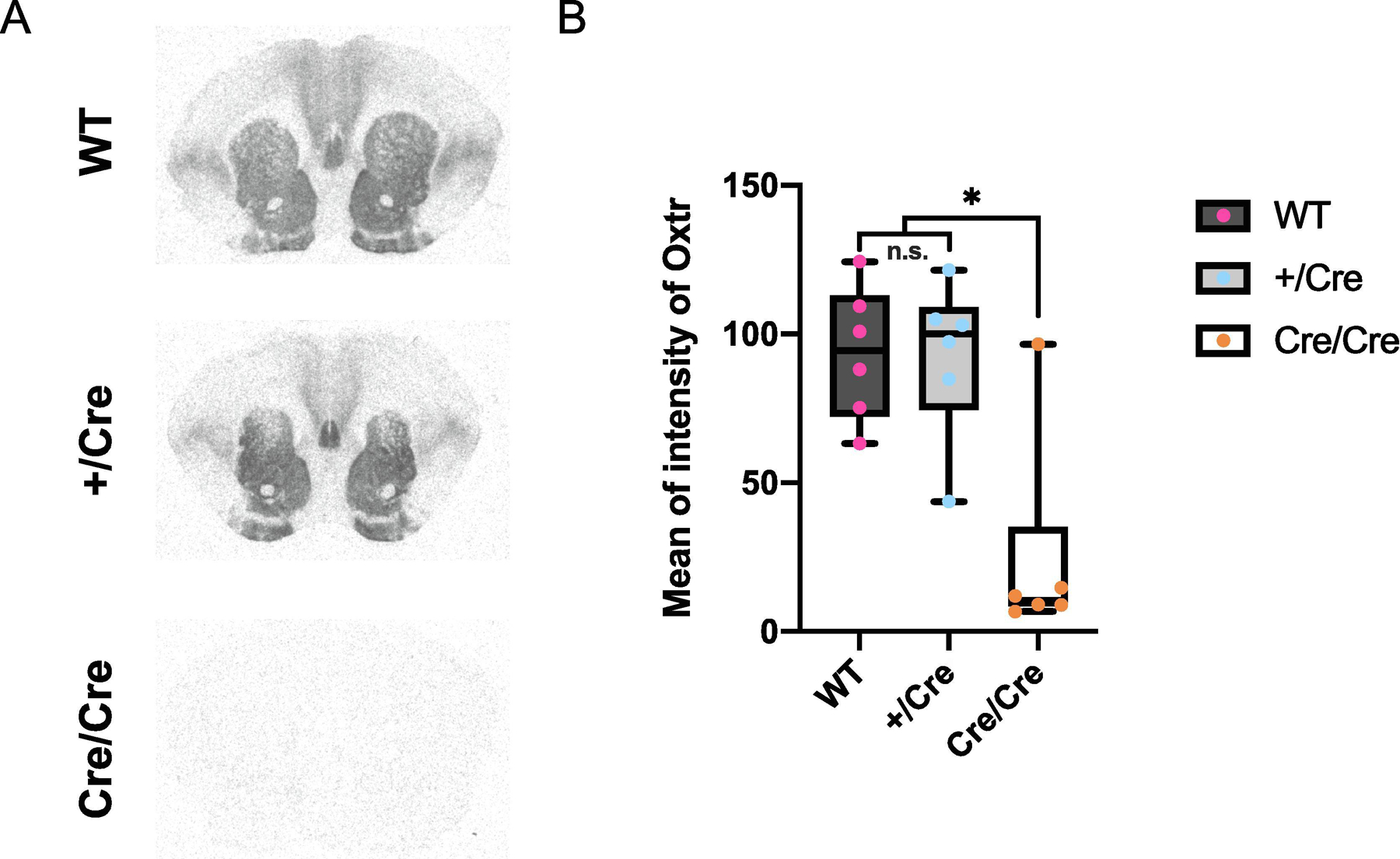
A. Representative images of autoradiography of brains of WT, heterozygous +/Cre, and homozygous Cre/Cre (male, each N = 3; female, each N = 3). B. Mean of intensity of Oxtr in NAcc of brains of Cre/Cre was significantly lower than WT and +/Cre (WT, 93.5 ± 9.20; +/Cre, 92.5 ± 10.9; Cre/Cre, 24.6 ± 14.4; WT vs . +/Cre, P = 0.998; WT vs. Cre/Cre, P = 0.0023; +/Cre vs. Cre/Cre, P = 0.0026). The box plot depicts the median and the 25th and 75th quartiles and the whisker shows the 5th and 95th percentile.
Retrograde tracing of Oxtr-expressing neurons projecting to NAcc
To reveal Oxtr-expressing neurons that project to the NAcc in prairie voles, AAVrg-hSyn-DIO-EGFP was injected into the NAcc of heterozygous +/Cre knock-in prairie voles and the expression of EGFP was mapped in the whole brain (Figure 4A; male +/Cre , N = 3; female +/Cre, N =3). The AAVrg is taken up by terminals in the NAcc and transported back to the nucleus and if Cre is present (i.e. only in Oxtr expressing neurons), the neuron will express EGFP. EGFP was detected in anterior olfactory nucleus (AON), prelimbic cortex (PrL), anterior cingulate cortex (Cg2), insular cortex (IC), paraventricular thalamus (PVT), basolateral amygdala (BLA), and posteromedial and posterolateral cortical amygdaloid area (PMCo, PLCo) (Figure 4 B–H).
Figure 4. Retrograde tracing of Oxtr-expressing neurons projecting NAcc.
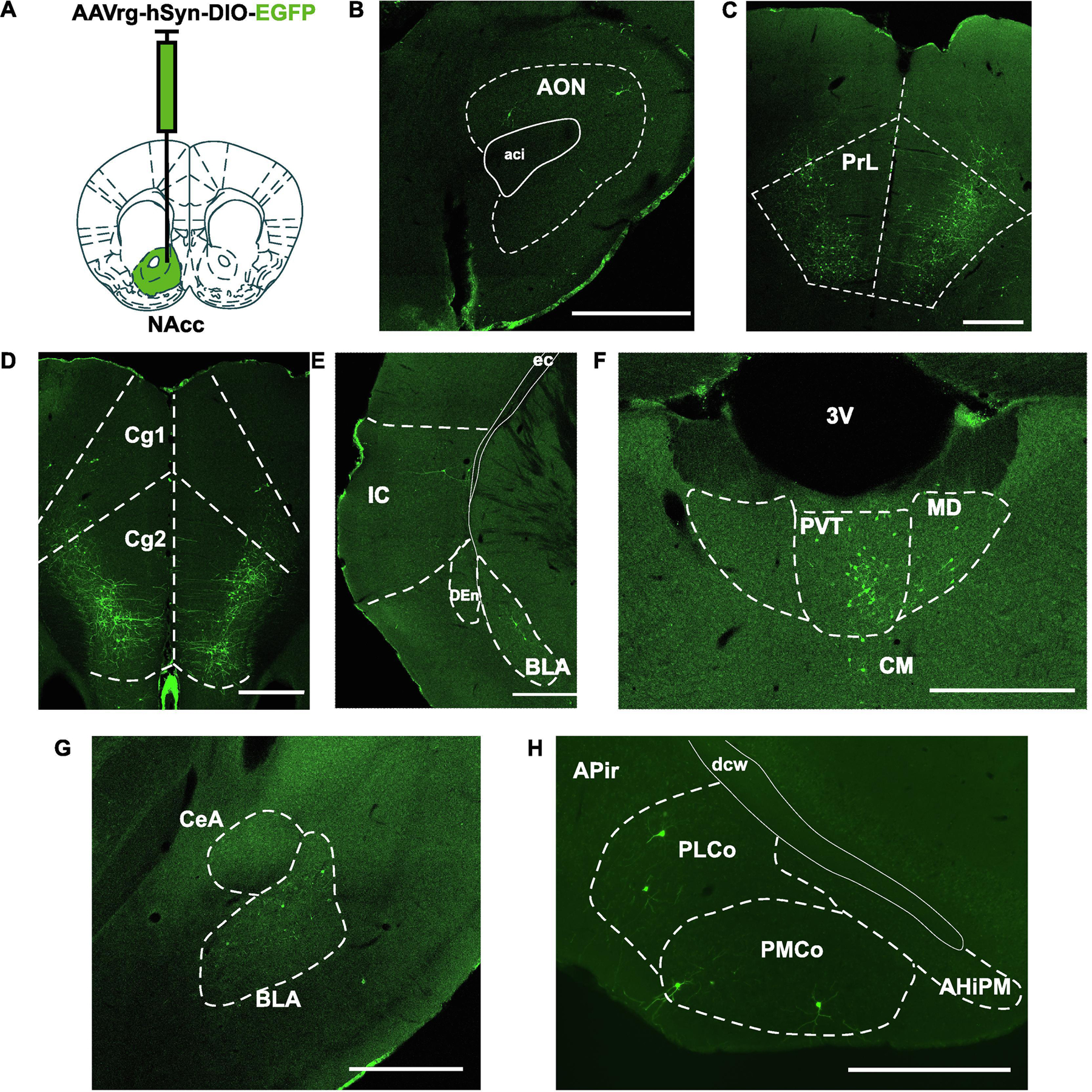
A. AAVrg-hSyn-DIO-EGFP was injected into NAcc of Oxtr-ires-Cre knock-in prairie voles (male, N = 3; female, N = 3). B-I. EGFP signals were detected in anterior olfactory nucleus (AON), prelimbic cortex (PrL), anterior cingulate cortex (Cg2), insular cortex (IC), paraventricular thalamus (PVT), basolateral amygdala (BLA), and posteromedial and posterolateral cortical amygdaloid area (PMCo, PLCo). Abbreviations: anterior commissure (aci), anterior cingulate cortex (Cg1), dorsal endopiriform nucleus (DEn), external capsule (ec), 3rd ventricle (3V), mediodorsal thalamic nucleus (MD), central medial thalamic nucleus (CM), central amygdala (CeA), posterior amygdalohippocampal area (AHiPM), amygdalopiriform transition area (APir), deep cerebral white matter (dcw). Scale bar = 500 μm.
Discussion
Here we report that the first Oxtr-ires-Cre knock-in prairie vole was efficiently generated using CRISPR/Cas9 and use Cre-dependent AAVrg to document for the first time several populations of Oxtr-expressing neurons that provide direct projections to the prairie vole NAcc. In the prairie vole, we achieved efficient (50%) knock-in of the knock-in plasmid into Oxtr gene by using the microinjection of crRNA, tracrRNA, and Cas9 protein, which is consistent with the efficiency showed in the previous report generating knock-in mice(Aida, et al., 2015). This technique should be widely applicable for generating many mutant lines that can greatly accelerate the elucidation of neural circuit mechanisms underlying social behavior in this model organism.
In previous studies of prairie voles, GFP expressing transgenic voles were generated by lentiviral mediated transgenesis (Donaldson et al., 2009; Keebaugh et al., 2012). Also, prairie vole induced pluripotent cells (iPSCs) have been established by two different groups (Manoli et al., 2012; Katayama et al., 2016). Although those studies suggested the possibility of generating knock-out or knock-in prairie voles using homologous recombination, it was not achieved because of technical difficulties of maintenance of quality of iPSCs and modification of the targeted gene. To overcome the difficulties, we used CRISPR/Cas9 to edit the Oxtr gene and successfully generated Oxtr knock-out prairie voles in a previous study (Horie, et al., 2019). By applying those methods described in the previous paper combined with the high-efficiency method for knocking-in, we successfully generated Oxtr-ires-Cre knock-in prairie voles. Sequencing from genome of knock-in prairie voles indicated homologous recombination (HR) was induced in the prairie vole embryos, which means that HR pathway could be conserved in prairie voles.
In the in situ hybridization experiments, colocalization of Oxtr and Cre RNA in the heterozygous and homozygous knock-in prairie voles indicated that the expression of Cre is restricted to Oxtr-expressing cells. Although Oxtr signals in WT brains were detected as sparse, distributed signals, Cre mRNA in brains of heterozygous and homozygous knock-in prairie voles seemed to aggregate and signals were more dense than Oxtr signals in the WT brains, and Oxtr signals also aggregated. These data suggested that the insertion of Cre sequence into the Oxtr gene might disturb the localization or conformation of Oxtr mRNA. Additionally, in the autoradiography experiments, ligand binding was not detected in the homozygous knock-in prairie voles, which suggested the insertion of Cre sequence suppressed the translation of OXTR as well. This problem does not appear to be brain specific as a female Cre/Cre animals delivered pups but they did not survive presumably due to starvation because of a defective milk ejection reflex in the mother (The successful rate of weaning (weaned pups/new borns * 100) = 0%), similar to that observe in Oxtr KO voles and mice. Although OXTR might not be efficiently translated from the Cre inserted allele, there was no difference in the expression of Oxtr between WT and heterozygous knock-in voles in autoradiography and CRE-dependent virus expressed well in brains of heterozygous knock-in prairie voles. Those data suggested that heterozygous knock-in prairie voles should be used for further experiments, such as chemogenetic or optogenetic manipulation of behaviors, to avoid negative effects of deficits of OXTR proteins to results of behavioral experiments. However, CRE expression may be limited to the highest OXTR expressing neurons due to the abnormal localization of the mRNA. Inserting the ires-Cre further downstream of the Oxtr stop coding deeper into the 3’ flanking region, or the use of a P2A cleavage sequence might eliminate the aggregation and result in more efficient translation of both the OXTR and CRE.
Neural circuits of Oxtr-expressing neurons projecting to NAcc found in the current study are regarded as key components for regulating social cue processing (Walum and Young, 2018). In AON, Oxtr-expressing neurons facilitates social recognition by increasing the signal to noise ratio in olfactory bulb (Oettl, et al., 2016). Although the function of Oxtr-expressing neurons from AON to NAcc has yet to be reported, this circuit might regulate processing odorant cues of the partner in prairie voles. PrL is a region that has the projection to NAcc, and this circuit enhances females’ affiliative behavior during the cohabitation in prairie voles (Amadei, et al., 2017). Oxtr-expressing neurons in PrL has been proven to regulate the reward processing, social behavior, anxiety, and affiliation behaviors (Young, et al., 2001; Sabihi et al., 2014; Everett et al., 2019; He et al., 2019). Combining those finding, we presume that Oxtr-expressing neurons in PrL projecting to NAcc may regulate the pair bonding in prairie voles. The BLA has been proposed as a key brain region regulating pair bonding as well (Numan and Young, 2016; Walum and Young, 2018). The BLA to NAcc circuit regulates reward seeking in mice(Ambroggi et al., 2008) and OXT in the amygdala plays a role in regulating social decision-making in non-human primates (Chang et al., 2015). These observations suggest that Oxtr-expressing neurons in BLA projecting to NAcc might regulate social reward processing in animals and might facilitate pair bonding in prairie voles. Also, PVT has been emerging as one of the key brain regions regulating the reward processing (Zhu, et al., 2016; Do-Monte et al., 2017; Zhu et al., 2018). The blockade of OXTR in PVT decreased the maternal crouching behavior toward pups in mice (Watarai et al., 2020). Although the function of PVT in regulating social behaviors has yet to be investigated, these findings and the projection that was found in the current study suggested that Oxtr-expressing neurons in PVT projecting to NAcc might regulate the pair bonding behavior in prairie voles, and their role should be investigated in future studies.
A previous mouse study showed that NAcc core had afferent inputs of Oxtr-expressing neurons from PVN, AON, PVT, BLA, central amygdala (CeA), cortex of the amygdala (CoA), ventral tegmental area (VTA), dorsal raphe nucleus (dRph), ventral subiculum, and ventral hippocampal CA1 (vCA1) by combining the immunostaining of Oxtr-Venus knock-in mice (Yoshida et al., 2009) and the retrograde tracing from NAcc core by rabies virus (RBV) carrying tdTomato (Dolen, et al., 2013). Another paper showed that Oxtr-expressing neurons in VTA had projections to NAcc in mice by the injection of CRE-dependent AAV-chanelrhodopsin2 (ChR2) into VTA of Oxtr-Cre transgenic mice (Peris et al., 2017). By the injection of CRE-dependent AAV-ChR2 into PFC of Oxtr-Cre transgenic mice, the projection of Oxtr-expressing neurons from PFC to NAcc were shown as well (Tan et al., 2019). Thus, we detected a more limited distribution of Oxtr-expressing neurons projecting to the NAcc, which suggests that due to the reduced expression/translation of the Oxtr-ires-Cre transcript, we may only be detecting the neurons with the most robust Oxtr expression. However, we did detect one set of Oxtr neurons with robust signal that was not detected in mice, the ACC. Although we targeted the entire NAcc and didn’t distinguish between core and shell, we found that Oxtr-expressing neurons in ACC had projections to NAcc in prairie voles, which was not found in previous studies of mice (Dolen, et al., 2013). In prairie voles, inhibition of OXTR in ACC impaired consolation behaviors toward the stressed partners in prairie voles (Burkett, et al., 2016). Based on these studies, we propose the hypothesis that Oxtr-expressing neurons from ACC to NAcc found in the current study might be a prairie vole specific circuit that regulates partner specific affiliation or consolation behaviors that is not present in mice. Although further confirmation to make sure the specificity of the neural circuits of Oxtr-expressing neurons between NAcc core and shell are required, this circuit was recognized only in prairie voles, not in mice and rats.
Regarding IC, although the circuit of Oxtr-expressing neurons from IC to NAcc was not confirmed in mice, we found it in prairie voles, but there were only a few labeled neurons in IC of prairie voles (Figure 4E). IC to NAcc circuits have been regarded as the key brain region that regulates social approach behaviors to the stressed juvenile in rats (Rogers-Carter et al., 2019). Blockade of OXTR in IC disrupted social approach behaviors in rats (Rogers-Carter et al., 2018). Based on these studies, we hypothesis that rats and prairie vole share a common circuit of Oxtr-expressing neurons from IC projecting to NAcc which is absent in mice. Oxtr-expressing neurons in vCA1 projecting to NAcc were not found in brains of prairie voles. The vCA1 to NAcc circuit is known as a region storing social memory in mice (Okuyama et al., 2016). In PVN, CeA, VTA, and dRph of prairie voles, outputs of Oxtr-expressing neurons to NAcc were not detected. Due to the altered mRNA localization and translation of the Oxtr-ires-Cre mRNA, we cannot be confident that a lack of signal in our study is strong evidence of a lack of connectivity in prairie voles. Alternate strategies of generating Oxtr-cre prairie voles are needed to have confidence of the absence of Oxtr-neuron connections with NAcc in vCA1, VTA, dRph and other regions.
As a limitation of discussion, these differences of projections between mice, rats, and prairie voles might come from differences in neurotropism between those two retrograde viruses used for the retrograde tracing (Sun et al., 2019). For the further comparison, Oxtr-ires-Cre voles should be injected with CRE-dependent RBV and the projections must be compared with the current data in the future study. Also, lower level of the expression of Cre because of the aggregation of Cre mRNA might affect results of retrograde tracing. RNAscope experiments showed that Cre containing RNAs aggregated, which means translation of Cre RNA might be suppressed. Also we were not able to detect CRE proteins on fixed brain slices by immunostaining with anti-CRE antibodies (data not shown). From those data, we presumed that it was difficult to detect CRE-dependent EGFP in some brain regions that express less Oxtr. Because of these limitations, our analysis was limited to a qualitative description in adults and we were not able to reliably determine whether there are sex differences or developmental changes in Oxtr neurons projecting to the NAcc in the current study.
Oxtr-ires-Cre knock-in prairie voles generated in the current study is the first knock-in prairie vole that is an unprecedented tool to investigate Oxtr-expressing neural circuits related to the regulation of social affiliation, albeit with unfortunate limitations. Combining Oxtr-ires-Cre knock-in prairie voles with using CRE-dependent hM3Dq/hM4Di expression, or ChR2/halorhodopsin enable researchers to manipulate Oxtr-expressing neural circuits during partner preference tests (Boender and Young, 2020). It will be possible to monitor the activity of Oxtr-expressing neurons using CRE-dependent GCaMP in vivo as well. We believe that those future experiments using Oxtr-ire-Cre knock-in voles will greatly accelerate our elucidation of the neural circuit mechanisms underlying social bonding.
Supplementary Material
Supplementary figure 1. In vitro digestion assay (IDA) of crRNA targeting to Oxtr gene. The crRNA that we used digested the PCR amplicon that contains the target sequence of crRNA (Middle lane), but not without the crRNA (Right lane). M, 100 bp ladder marker.
Supplementary figure 2. Off-target effects were analyzed by Sanger sequencing. Off-target effects were predicted by Benchling Target sequences were amplified by PCR with each primer set and read by Sanger sequencing. Sites of polymorphisms are indicated by a red box.
Highlights.
Prairie voles are a model species for studying neural mechanisms of social behavior
We generated the first CRISPR generated Oxtr-ires-Cre knock-in prairie vole.
Cre mRNA was restricted to Oxtr cells, but aggregated, reducing translation.
CRE-dependent retrograde viruses were used to identify Oxtr neurons projecting to nucleus accumbens.
Oxtr neurons in the ACC project to nucleus accumbens, and may represent a species-specific circuit.
Acknowledgements
We thank Lorra Julian for help maintaining the vole colony at Emory. The research performed in Japan was funded by the Strategic Research Program for Brain Sciences from Japan Agency for Medical Research and Development (AMED; 18dm0107076h0003, 2016–2020) and JSPS Grant-in-Aid for Scientific Research (15H02442, 2015–2018) to KN, and Grant-in-Aid for JSPS Fellows (16J05070, 2016–2019) to KH. The work performed at Emory in the USA was supported by NIH grants R01MH112788 and P50MH100023 to LJY and P51OD11132 to YNPRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, et al. (2015), Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadei EA, Johnson ZV, Jun Kwon Y, Shpiner AC, Saravanan V, Mays WD, Ryan SJ, Walum H, et al. (2017), Dynamic corticostriatal activity biases social bonding in monogamous female prairie voles. Nature 546:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggi F, Ishikawa A, Fields HL, Nicola SM (2008), Basolateral amygdala neurons facilitate reward-seeking behavior by exciting nucleus accumbens neurons. Neuron 59:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Yu YJ, Curtis JT, Detwiler JM, Insel TR, Wang Z (2006), Nucleus accumbens dopamine differentially mediates the formation and maintenance of monogamous pair bonds. Nat Neurosci 9:133–139. [DOI] [PubMed] [Google Scholar]
- Arai A, Hirota Y, Miyase N, Miyata S, Young LJ, Osako Y, Yuri K, Mitsui S (2016), A single prolonged stress paradigm produces enduring impairments in social bonding in monogamous prairie voles. Behav Brain Res 315:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007), CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- Barrett CE, Arambula SE, Young LJ (2015), The oxytocin system promotes resilience to the effects of neonatal isolation on adult social attachment in female prairie voles. Transl Psychiatry 5:e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boender AJ, Young LJ (2020), Oxytocin, vasopressin and social behavior in the age of genome editing: A comparative perspective. Horm Behav 124:104780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Dabrowska J, Modi ME, Johnson ZV, Keebaugh AC, Barrett CE, Ahern TH, Guo J, et al. (2016), Oxytocin in the nucleus accumbens shell reverses CRFR2-evoked passive stress-coping after partner loss in monogamous male prairie voles. Psychoneuroendocrinology 64:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Young LJ (2018), Oxytocin and Social Relationships: From Attachment to Bond Disruption. Curr Top Behav Neurosci 35:97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett JP, Andari E, Johnson ZV, Curry DC, de Waal FB, Young LJ (2016), Oxytocin-dependent consolation behavior in rodents. Science 351:375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin NL, Du B, de Lacalle S, Saper CB (1998), Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain Res 793:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SW, Fagan NA, Toda K, Utevsky AV, Pearson JM, Platt ML (2015), Neural mechanisms of social decision-making in the primate amygdala. Proc Natl Acad Sci U S A 112:16012–16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AH, Fung SW, Cheng HM (2019), Limitations of the Avp-IRES2-Cre (JAX #023530) and Vip-IRES-Cre (JAX #010908) Models for Chronobiological Investigations. J Biol Rhythms 34:634–644. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS (2013), Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. [DOI] [PubMed] [Google Scholar]
- Choe HK, Reed MD, Benavidez N, Montgomery D, Soares N, Yim YS, Choi GB (2015), Oxytocin Mediates Entrainment of Sensory Stimuli to Social Cues of Opposing Valence. Neuron 87:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, et al. (2013), Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, et al. (2011), CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do-Monte FH, Minier-Toribio A, Quinones-Laracuente K, Medina-Colon EM, Quirk GJ (2017), Thalamic Regulation of Sucrose Seeking during Unexpected Reward Omission. Neuron 94:388–400 e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Darvishzadeh A, Huang KW, Malenka RC (2013), Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature 501:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Yang SH, Chan AW, Young LJ (2009), Production of germline transgenic prairie voles (Microtus ochrogaster) using lentiviral vectors. Biol Reprod 81:1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Young LJ (2008), Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322:900–904. [DOI] [PubMed] [Google Scholar]
- Everett N, Baracz S, Cornish J (2019), Oxytocin treatment in the prelimbic cortex reduces relapse to methamphetamine-seeking and is associated with reduced activity in the rostral nucleus accumbens core. Pharmacol Biochem Behav 183:64–71. [DOI] [PubMed] [Google Scholar]
- Feldman R (2012), Oxytocin and social affiliation in humans. Horm Behav 61:380–391. [DOI] [PubMed] [Google Scholar]
- Gobrogge KL, Liu Y, Young LJ, Wang Z (2009), Anterior hypothalamic vasopressin regulates pair-bonding and drug-induced aggression in a monogamous rodent. Proc Natl Acad Sci U S A 106:19144–19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Young L, Ma XM, Guo Q, Wang L, Yang Y, Luo L, Yuan W, et al. (2019), Increased anxiety and decreased sociability induced by paternal deprivation involve the PVN-PrL OTergic pathway. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidema S, Fukuda T, Hiraoka Y, Mizukami H, Hayashi R, Otsuka A, Suzuki S, Miyazaki S, et al. (2016), Generation of Oxtr cDNA(HA)-Ires-Cre Mice for Gene Expression in an Oxytocin Receptor Specific Manner. J Cell Biochem 117:1099–1111. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Arai A, Young LJ, Osako Y, Yuri K, Mitsui S (2020), Oxytocin receptor antagonist reverses the blunting effect of pair bonding on fear learning in monogamous prairie voles. Horm Behav 120:104685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Hidema S, Hirayama T, Nishimori K (2015), In vitro culture and in vitro fertilization techniques for prairie voles (Microtus ochrogaster). Biochem Biophys Res Commun 463:907–911. [DOI] [PubMed] [Google Scholar]
- Horie K, Inoue K, Suzuki S, Adachi S, Yada S, Hirayama T, Hidema S, Young LJ, et al. (2019), Oxytocin receptor knockout prairie voles generated by CRISPR/Cas9 editing show reduced preference for social novelty and exaggerated repetitive behaviors. Horm Behav 111:60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Shapiro LE (1992), Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proc Natl Acad Sci U S A 89:5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Walum H, Jamal YA, Xiao Y, Keebaugh AC, Inoue K, Young LJ (2016), Central oxytocin receptors mediate mating-induced partner preferences and enhance correlated activation across forebrain nuclei in male prairie voles. Horm Behav 79:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Walum H, Xiao Y, Riefkohl PC, Young LJ (2017), Oxytocin receptors modulate a social salience neural network in male prairie voles. Horm Behav 87:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Young LJ (2015), Neurobiological mechanisms of social attachment and pair bonding. Curr Opin Behav Sci 3:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Young LJ (2017), Oxytocin and vasopressin neural networks: Implications for social behavioral diversity and translational neuroscience. Neurosci Biobehav Rev 76:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurek B, Neumann ID (2018), The Oxytocin Receptor: From Intracellular Signaling to Behavior. Physiol Rev 98:1805–1908. [DOI] [PubMed] [Google Scholar]
- Katayama M, Hirayama T, Horie K, Kiyono T, Donai K, Takeda S, Nishimori K, Fukuda T (2016), Induced Pluripotent Stem Cells With Six Reprogramming Factors From Prairie Vole, Which Is an Animal Model for Social Behaviors. Cell Transplant 25:783–796. [DOI] [PubMed] [Google Scholar]
- Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, Young LJ (2015), RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Soc Neurosci 10:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh AC, Modi ME, Barrett CE, Jin C, Young LJ (2012), Identification of variables contributing to superovulation efficiency for production of transgenic prairie voles (Microtus ochrogaster). Reprod Biol Endocrinol 10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King LB, Walum H, Inoue K, Eyrich NW, Young LJ (2016), Variation in the Oxytocin Receptor Gene Predicts Brain Region-Specific Expression and Social Attachment. Biol Psychiatry 80:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch HS, Charlet A, Hoffmann LC, Eliava M, Khrulev S, Cetin AH, Osten P, Schwarz MK, et al. (2012), Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73:553–566. [DOI] [PubMed] [Google Scholar]
- Lafferty CK, Yang AK, Mendoza JA, Britt JP (2020), Nucleus Accumbens Cell Type- and Input-Specific Suppression of Unproductive Reward Seeking. Cell Rep 30:3729–3742 e3723. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013), RNA-guided human genome engineering via Cas9. Science 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli DS, Subramanyam D, Carey C, Sudin E, Van Westerhuyzen JA, Bales KL, Blelloch R, Shah NM (2012), Generation of induced pluripotent stem cells from the prairie vole. PLoS One 7:e38119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin BJ, Mitre M, D’Amour JA, Chao MV, Froemke RC (2015), Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature 520:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Young LJ (2010), The prairie vole: an emerging model organism for understanding the social brain. Trends Neurosci 33:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi ME, Inoue K, Barrett CE, Kittelberger KA, Smith DG, Landgraf R, Young LJ (2015), Melanocortin Receptor Agonists Facilitate Oxytocin-Dependent Partner Preference Formation in the Prairie Vole. Neuropsychopharmacology 40:1856–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan M, Young LJ (2016), Neural mechanisms of mother-infant bonding and pair bonding: Similarities, differences, and broader implications. Horm Behav 77:98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettl LL, Ravi N, Schneider M, Scheller MF, Schneider P, Mitre M, da Silva Gouveia M, Froemke RC, et al. (2016), Oxytocin Enhances Social Recognition by Modulating Cortical Control of Early Olfactory Processing. Neuron 90:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okhovat M, Berrio A, Wallace G, Ophir AG, Phelps SM (2015), Sexual fidelity trade-offs promote regulatory variation in the prairie vole brain. Science 350:1371–1374. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Kitamura T, Roy DS, Itohara S, Tonegawa S (2016), Ventral CA1 neurons store social memory. Science 353:1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otis JM, Zhu M, Namboodiri VMK, Cook CA, Kosyk O, Matan AM, Ying R, Hashikawa Y, et al. (2019), Paraventricular Thalamus Projection Neurons Integrate Cortical and Hypothalamic Signals for Cue-Reward Processing. Neuron 103:423–431 e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris J, MacFadyen K, Smith JA, de Kloet AD, Wang L, Krause EG (2017), Oxytocin receptors are expressed on dopamine and glutamate neurons in the mouse ventral tegmental area that project to nucleus accumbens and other mesolimbic targets. J Comp Neurol 525:1094–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl TT, Young LJ, Bosch OJ (2019), Lost connections: Oxytocin and the neural, physiological, and behavioral consequences of disrupted relationships. Int J Psychophysiol 136:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CN, Ross AP, Sahu SP, Siegel ER, Dooyema JM, Cree MA, Stopa EG, Young LJ, et al. (2018), Oxytocin- and arginine vasopressin-containing fibers in the cortex of humans, chimpanzees, and rhesus macaques. Am J Primatol 80:e22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers-Carter MM, Djerdjaj A, Gribbons KB, Varela JA, Christianson JP (2019), Insular Cortex Projections to Nucleus Accumbens Core Mediate Social Approach to Stressed Juvenile Rats. J Neurosci 39:8717–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers-Carter MM, Varela JA, Gribbons KB, Pierce AF, McGoey MT, Ritchey M, Christianson JP (2018), Insular cortex mediates approach and avoidance responses to social affective stimuli. Nat Neurosci 21:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HE, Cole CD, Smith Y, Neumann ID, Landgraf R, Murphy AZ, Young LJ (2009), Characterization of the oxytocin system regulating affiliative behavior in female prairie voles. Neuroscience 162:892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabihi S, Durosko NE, Dong SM, Leuner B (2014), Oxytocin in the prelimbic medial prefrontal cortex reduces anxiety-like behavior in female and male rats. Psychoneuroendocrinology 45:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AS, Wang Z (2014), Hypothalamic oxytocin mediates social buffering of the stress response. Biol Psychiatry 76:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Tang Y, Yan K, Yu J, Zou Y, Xu W, Xiao K, Zhang Z, et al. (2019), Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol Neurodegener 14:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Singhal SM, Harden SW, Cahill KM, Nguyen DM, Colon-Perez LM, Sahagian TJ, Thinschmidt JS, et al. (2019), Oxytocin Receptors Are Expressed by Glutamatergic Prefrontal Cortical Neurons That Selectively Modulate Social Recognition. J Neurosci 39:3249–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, et al. (2016), A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walum H, Young LJ (2018), The neural mechanisms and circuitry of the pair bond. Nat Rev Neurosci 19:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013), One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai A, Tsutaki S, Nishimori K, Okuyama T, Mogi K, Kikusui T (2020), The blockade of oxytocin receptors in the paraventricular thalamus reduces maternal crouching behavior over pups in lactating mice. Neurosci Lett 720:134761. [DOI] [PubMed] [Google Scholar]
- Wei D, Lee D, Cox CD, Karsten CA, Penagarikano O, Geschwind DH, Gall CM, Piomelli D (2015), Endocannabinoid signaling mediates oxytocin-driven social reward. Proc Natl Acad Sci U S A 112:14084–14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA (2012), RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338. [DOI] [PubMed] [Google Scholar]
- Witt DM, Carter CS, Lnsel TR (1991), Oxytocin receptor binding in female prairie voles: endogenous and exogenous oestradiol stimulation. J Neuroendocrinol 3:155–161. [DOI] [PubMed] [Google Scholar]
- Yokoi S, Naruse K, Kamei Y, Ansai S, Kinoshita M, Mito M, Iwasaki S, Inoue S, et al. (2020), Sexually dimorphic role of oxytocin in medaka mate choice. Proc Natl Acad Sci U S A 117:4802–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Takayanagi Y, Inoue K, Kimura T, Young LJ, Onaka T, Nishimori K (2009), Evidence that oxytocin exerts anxiolytic effects via oxytocin receptor expressed in serotonergic neurons in mice. J Neurosci 29:2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KA, Gobrogge KL, Liu Y, Wang Z (2011), The neurobiology of pair bonding: insights from a socially monogamous rodent. Front Neuroendocrinol 32:53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Lim MM, Gingrich B, Insel TR (2001), Cellular mechanisms of social attachment. Horm Behav 40:133–138. [DOI] [PubMed] [Google Scholar]
- Young LJ, Wang Z (2004), The neurobiology of pair bonding. Nat Neurosci 7:1048–1054. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nachtrab G, Keyes PC, Allen WE, Luo L, Chen X (2018), Dynamic salience processing in paraventricular thalamus gates associative learning. Science 362:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wienecke CF, Nachtrab G, Chen X (2016), A thalamic input to the nucleus accumbens mediates opiate dependence. Nature 530:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. In vitro digestion assay (IDA) of crRNA targeting to Oxtr gene. The crRNA that we used digested the PCR amplicon that contains the target sequence of crRNA (Middle lane), but not without the crRNA (Right lane). M, 100 bp ladder marker.
Supplementary figure 2. Off-target effects were analyzed by Sanger sequencing. Off-target effects were predicted by Benchling Target sequences were amplified by PCR with each primer set and read by Sanger sequencing. Sites of polymorphisms are indicated by a red box.


