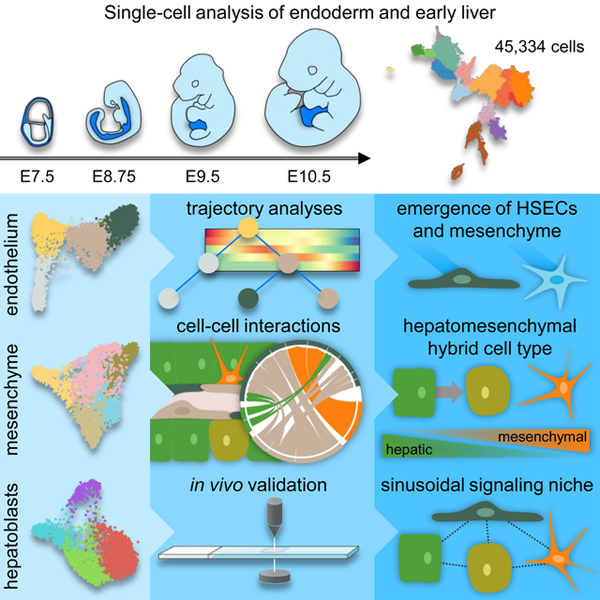
Summary
The cellular complexity and scale of the early liver have constrained analyses examining its emergence during organogenesis. To circumvent these issues, we analyzed 45,334 single-cell transcriptomes from embryonic day (E)7.5, when endoderm progenitors are specified, to E10.5 liver, when liver parenchymal and non-parenchymal cell lineages emerge. Our data detail divergence of vascular and sinusoidal endothelia, including a distinct transcriptional profile for sinusoidal endothelial specification by E8.75. We characterize two distinct mesothelial cell types as well as early hepatic stellate cells, and reveal distinct spatiotemporal distributions for these populations. We capture transcriptional profiles for hepatoblast specification and migration, including the emergence of a hepatomesenchymal cell type and evidence for hepatoblast collective cell migration. Further, we identify cell-cell interactions during the organization of the primitive sinusoid. This study provides a comprehensive atlas of liver lineage establishment from the endoderm and mesoderm through to the organization of the primitive sinusoid at single-cell resolution.
Graphical Abstract
eTOC blurb
A comprehensive atlas of mouse liver emergence is described at single-cell resolution starting at endoderm progenitor specification, including data detailing divergence of vascular and sinusoidal endothelia, hepatoblast specification, and the emergence of a distinct, migratory hepatomesenchymal cell type.
Introduction
The liver is a complex, multifunctional organ composed of cells from multiple lineages, derived from endoderm and mesoderm (Gordillo et al., 2015). How these cell types organize to form the highly elegant structure, necessary for its functions, is poorly understood. Progress has been hindered by a lack of knowledge with respect to parenchymal and non-parenchymal cell type diversity, how these cells are derived from their endodermal and mesodermal progenitors, and how these lineages interact during the formation of the liver bud.
Hepatoblasts, the bipotent progenitor for hepatocytes and cholangiocytes, develop from the definitive endoderm by embryonic day (E)8.75 (Gordillo et al., 2015; Tremblay and Zaret, 2005). Signals from the overlying cardiac mesoderm and septum transversum mesenchyme (STM) induce their transition into a pseudostratified columnar epithelium and their invasion of the STM by E9.5 (Figure 1B) (Houssaint, 1980; Tremblay and Zaret, 2005). These parenchymal progenitors begin expression of genes associated with hepatic metabolism and subsequently differentiate to form hepatocytes, the main parenchymal cell type, and cholangiocytes, the liver biliary epithelium.
Figure 1.
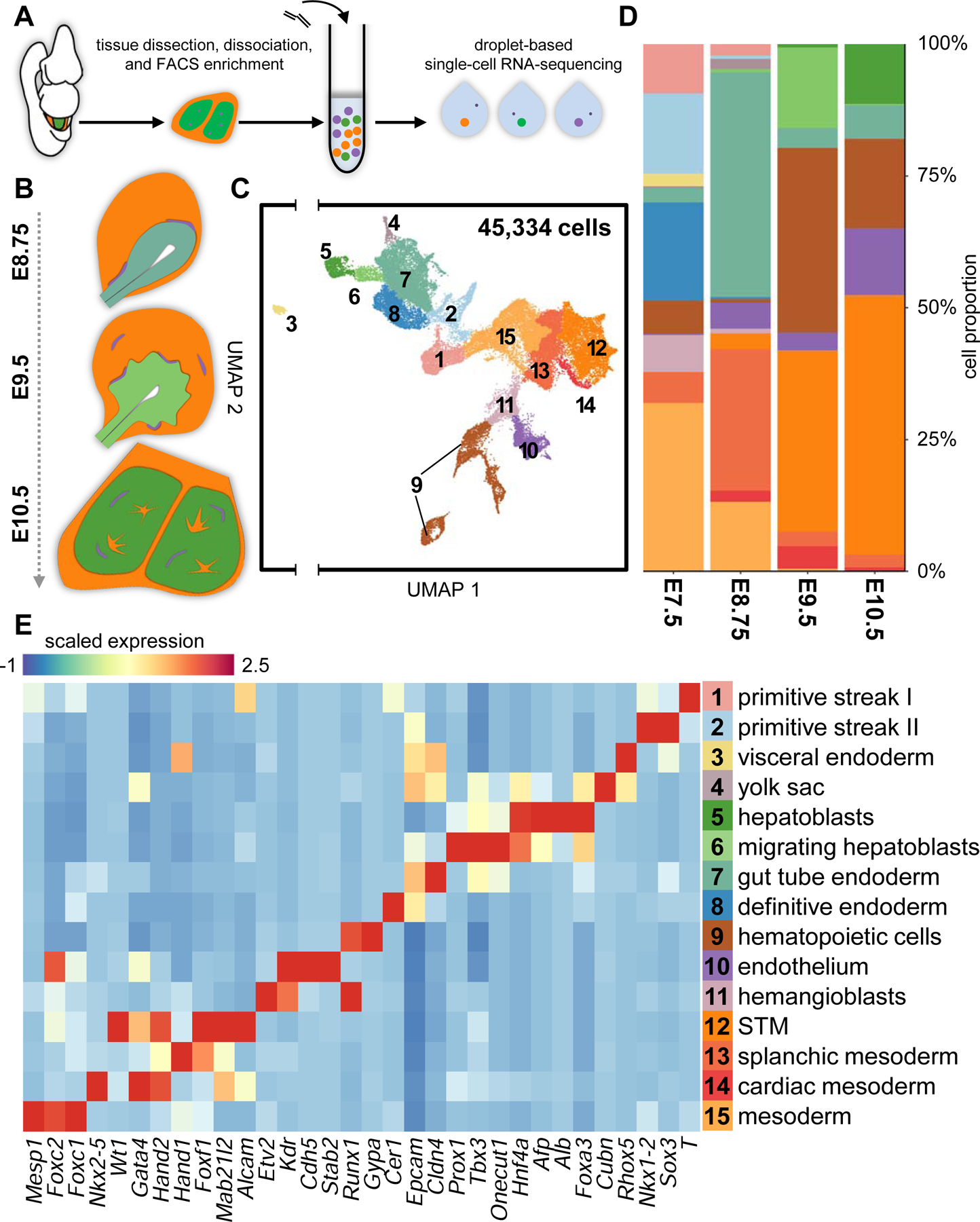
Tracking emergent parenchymal and non-parenchymal liver lineages using scRNA-seq.
(A) Schematic of experimental approach. (B) Overview of murine liver development. (C) UMAP visualization of all cells sequenced. Each dot represents a single-cell that is color-coded by cell type. (D) Cell type composition by developmental stage. (E) Heatmap depicting select differentially-expressing genes for all cell types identified. STM, septum transversum mesenchyme; FACS, fluorescently-activated cell sorting. See also Figure S1 and Table S2.
Liver non-parenchymal cells, including the endothelium, mesenchyme, blood, and nerves, play supportive roles to the liver parenchymal cells. The liver vasculature encompasses vascular and lymphatic capillaries, in addition to discontinuous, fenestrated sinusoidal capillaries, lined by hepatic sinusoidal endothelial cells (HSECs). HSECs present in the early liver are derived from the hemangioblast, from which vascular endothelial cells and early haematopoietic cells are also derived (Bollerot et al., 2005; Zovein et al., 2008), though a proportion may be derived from the endocardium and endoderm (Goldman et al., 2014; Zhang et al., 2016). This specialized endothelium plays important roles in regulating blood flow in the sinusoid, scavenging blood-born waste, cytokine secretion, and antigen presentation to liver resident immune cells (Poisson et al., 2017). The STM gives rise to the liver mesenchyme, which in the adult liver encompasses the mesothelium, the external cellular lining of the liver, and hepatic stellate cells (HSCs), the sinusoidal pericytes (Asahina et al., 2009). During liver bud formation, the STM and emergent liver mesenchyme provide signaling cues and substrate for hepatoblast specification, differentiation, and migration.
The fetal liver is also the main site of hematopoiesis during development, harboring hematopoietic stem cells from E10.5 until E16.5 when they migrate to the bone marrow (Gordillo et al., 2015). Inducible lineage tracing of early hemogenic sites with Cdh5-cre, however, does not account for all of hematopoietic cells within the embryo (Hirschi, 2012; Zovein et al., 2008). This suggests the possibility that hematopoietic stem cells may be derived de novo from hemogenic sites at later stages of embryogenesis, with some postulating the existence of hemogenic endothelium in the fetal liver (Hirschi, 2012).
By E10.5, hepatoblasts, endothelia, and HSCs begin to organize into primitive sinusoidal capillaries (Gordillo et al., 2015). The cellular interactions that contribute to the establishment of the sinusoid, including HSC migration, HSEC fenestration, basement membrane formation, and hepatoblast differentiation are poorly understood. Sinusoidal dysfunction, including defenestration and HSC activation, precedes scar formation and liver fibrosis (Poisson et al., 2017; Puche et al., 2013). Little is known about how hepatoblasts, HSCs, and HSECs organize to form the primitive sinusoids, despite the understanding that many of the pathways involved in their development are reactivated in disease states (Delgado et al., 2014; Marrone et al., 2015; Poisson et al., 2017; Puche et al., 2013).
In the present study, we have employed single-cell RNA-seq (scRNA-seq) to elucidate the lineage specification events that occur during the formation of the early liver from the definitive endoderm. We have combined 15 libraries comprising 45,334 single-cell transcriptomes enriched for E7.5 endoderm, E8.75 endoderm gut tube, E9.5 hepatic bud, and E10.5 liver (Nowotschin et al., 2019). In addition to the endodermal lineages, we compiled cells derived from the adjacent mesenchyme and endothelium and tracked their development from mesoderm to the non-parenchymal lineages of the liver. These transcriptomes represent the initial stages of liver organogenesis, including the specification of hepatoblasts from the definitive endoderm, invasion of STM by hepatoblasts, and liver sinusoid organization. We have elucidated the cell fate decisions that occur during liver cell lineage specification by combining Harmony and Palantir. Harmony constructs nearest-neighbour networks for batch correction across developmental timepoints (Nowotschin et al., 2019), whereas Palantir defines pseudotime and branch probabilities by differentiation potential as each cell type reaches its terminal state from a user-defined ‘start-cell’ (Setty et al., 2019). These methods allow for computational predictions of cell plasticity and bifurcation points as liver cell lineages are specified from their progenitors. Using these methods, we have revealed uncharacterized diversity within endothelial, mesenchymal, and hepatic cell lineages during liver development and have further generated a complete pseudotemporal map of the emergent endothelial and hepatic lineages. Together, this study provides a comprehensive single-cell atlas of liver cell lineage establishment from the endoderm and mesoderm.
Results
Tracking emergent lineages within the developing liver
To generate an overview of liver cell lineage specification at the single-cell level, we analyzed single-cell transcriptomic data from endodermal and liver tissue from E7.5-E10.5 (Figure 1A and S1A) (Nowotschin et al., 2019). To enrich for parenchymal and non-parenchymal liver progenitors, endoderm and associated mesoderm at E7.5 and E8.75, and liver buds at E9.5 and E10.5 were dissected before single-cell dissociation. In addition, the extra-embryonic endoderm- and hepatoblast-specific Afp-GFP mouse line was used to deplete for AFP-GFP+ visceral endoderm at E7.5 and delineate AFP-GFP+ liver parenchymal derivatives from AFP-GFP− non-parenchymal cells at E10.5 (Figure S1A) (Kwon et al., 2006; Wei et al., 2018). In total, 45,334 single-cell transcriptomes from 15 scRNA-seq libraries passed quality control, with a median of 4,190 genes detected per cell (Figures 1C and S1B; Table S1). We aggregated all single-cell transcriptomes and performed clustering and differential expression analyses to characterize the complete heterogeneity within our datasets (Table S2). We identified 15 clusters, which we have annotated as emergent endodermal, mesenchymal, and endothelial lineages, as well as hematopoietic cells (Figures 1C–1E). Our annotations were validated against single-cell transcriptomic data from Pijuan-Sala et al. (2019) through mutual-nearest neighbour batch correction (Figure S1C). We found overwhelming agreement between our respective cluster annotations, apart from cells annotated as surface ectoderm from Pijuan-Sala et al. (2019) (Figure S1C). While surface ectoderm is not captured in our libraries, due to expression of genes associated with epithelial identity, such as Epcam and Cdh1, we found overlap between it and our gut tube endoderm cluster (Figure S1C and data not shown).
Within our data, we further note two distinct primitive streak clusters, composed of cells from E7.5, which appear primed to either an endodermal or mesodermal cell fate, likely corresponding to the proximal and distal primitive streak (Figures 1C–E and S1D). To more clearly visualize the emergence of liver cell types during development, endodermal, mesenchymal, and endothelial lineages were computationally segregated and analyzed separately. These lineages were individually visualized using force-directed layouts and further analysed using Harmony and Palantir to reconstruct lineage relationships over developmental time (Nowotschin et al., 2019; Setty et al., 2019).
Sinusoidal and vascular endothelia diverge by E9.5 from a common endothelial progenitor
To explore the cell fate decisions required for the emergence of the sinusoidal endothelium, we analyzed 2,066 endothelial and hemangioblast transcriptomes from E7.5 to E10.5. We further included 503 hematopoietic progenitors from E7.5 to better define the divergence between endothelial and hematopoietic lineages. We identify four distinct populations based on clustering and differential gene expression analyses, representing hemangioblasts, hematopoietic progenitors, endothelium, and HSECs (Figures 2A and S2A; Table S2). The hemangioblast cluster is composed of cells predominantly from E7.5, the endothelial cluster is composed of cells from E8.75 to E10.5, and HSECs are composed predominantly of cells from E10.5, though cells from E8.75 and E9.5 are also present (Figures 2A, 2B, and S2C). While both endothelial and HSEC lineages express endothelial biomarkers, including Kdr, Pecam1, Cd34 and Cdh5 (Goncharov et al., 2017; Vestweber, 2008), the endothelial cluster is enriched for mechanosensitive ion channels and transcription factors associated with flow-induced shear stress and angiogenesis, such as Piezo1/2, Klf2/4, Foxc2, and Tbx1 (Figure 2C) (Chen et al., 2010; Sangwung et al., 2017; Seo et al., 2006). In contrast, early HSECs express transcription factors associated with sinusoidal identity, such as Sox17, Tfec, Maf, and Dab2 (Figures 2C and S2A) (Géraud et al., 2010). Surprisingly, the early HSEC transcriptome further suggests the activation of sinusoidal function, including endocytosis (Stab2, Colec12, Ehd3, and Lyve1), antigen presentation (Fcgrt, Fcer1g, and Mrc1), and signaling (Gpr182, Bmp2, Dll4 and Efna1) (Figures 2C, S2A, and S2F) (Géraud et al., 2010; Lao et al., 2017; Schmid et al., 2018). This suggests a functional role for HSECs within the developing liver starting earlier in developmental time than previously appreciated.
Figure 2.
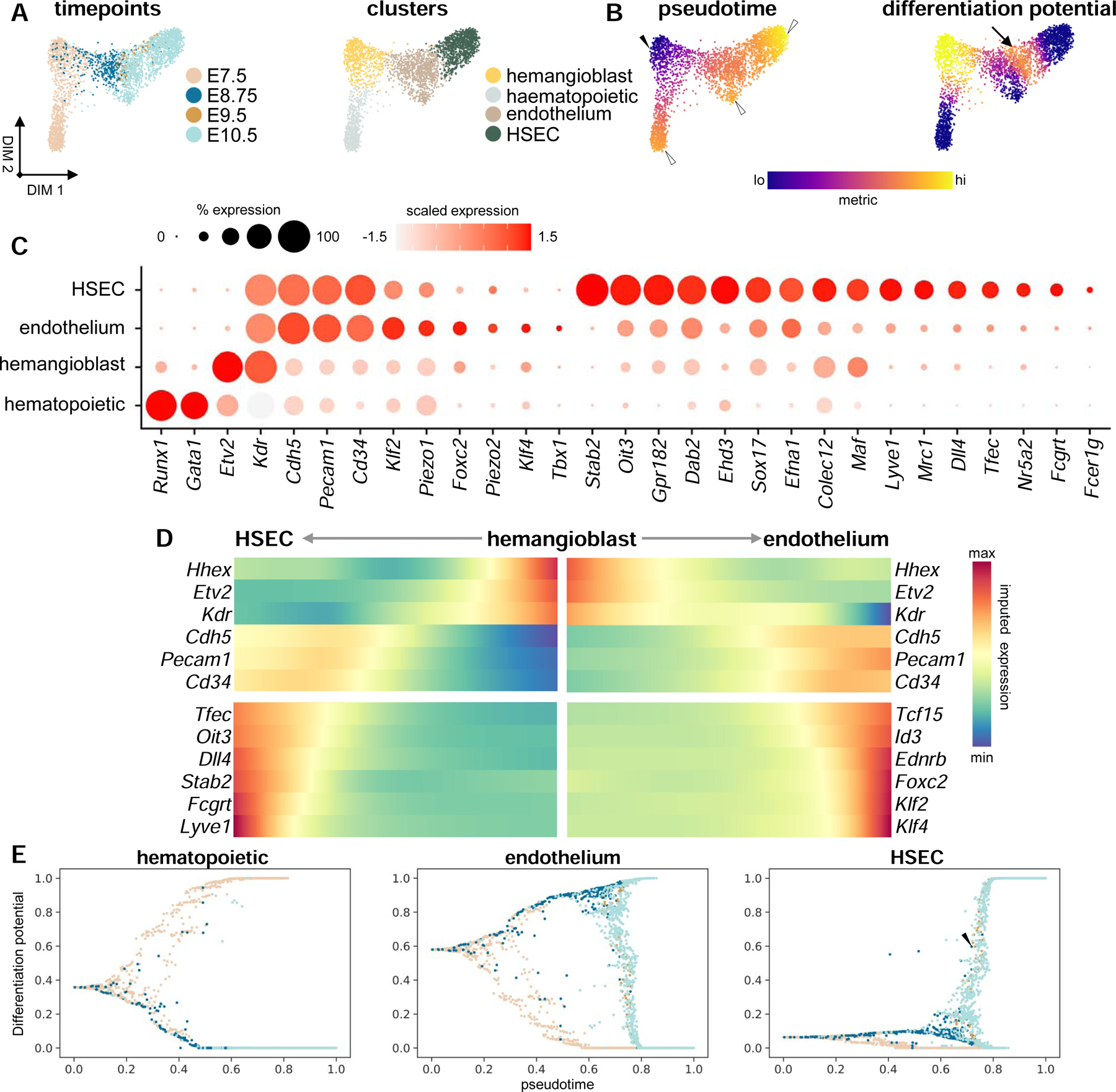
Sinusoidal and vascular endothelia diverge by E8.75 from a common endothelial progenitor.
(A) Force-directed layouts displaying timepoint and annotated cluster contribution of endothelial and E7.5 hematopoietic progenitors (n = 2569). (B) Force-directed layouts displaying pseudotime and differentiation potential in endothelial lineages from an Etv2hi ‘start cell’ (solid arrowhead) to terminal states (hollow arrowheads). Arrow indicates second local differentiation potential maximum, representing HSEC specification from the vascular endothelium. (C) Dot plot displaying known markers for hematopoietic, hemangioblast, and endothelial cell identities, including expression of immature and mature HSEC markers. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. (D) Heatmap representing gene trends over pseudotime from hemangioblast ‘start cell’ to HSEC and endothelial terminal states. (E) Differentiation potential as a function of pseudotime for hematopoietic, endothelial, and HSEC trajectories, where each dot represents a single-cell that is color-coded by embryonic timepoint as in (A). Arrowhead denotes endothelial cells at E8.75, E9.5, and E10.5 that are specified to the HSEC terminal state. HSEC, hepatic sinusoidal endothelial cell. See also Figure S2 and Table S2.
To detail the divergence between hematopoietic and endothelial cell lineages from the hemangioblast, we employed Palantir (Setty et al., 2019). We selected an E7.5 ‘start cell’ expressing high levels of the hemangioblast master regulator Etv2, which is necessary and sufficient for hemangioblast development into endothelial and hematopoietic lineages (solid arrowhead, Figure 2B) (Rasmussen et al., 2011). Further, we selected three terminal states, residing within hematopoietic, endothelial, or HSEC clusters (hollow arrowheads, Figure 2B), and calculated differentiation potential and gene expression trends along pseudotime for each trajectory as these terminal states are reached. The differentiation potential is highest within the hemangioblast cluster; however, a local maximum exists at the interface between the endothelial and HSEC clusters, suggesting the presence of a second cell fate decision in HSEC specification (arrow, Figure 2B). Emergence of hematopoietic progenitors from the hemangioblast is coincident with the activation of transcription factors required for hematopoiesis, such as Runx1 and Gata1 (Figure S2E) (Fujiwara et al., 1996; Yokomizo et al., 2008). Both the endothelial and HSEC lineages emerge initially through an unspecified endothelial progenitor, activating and maintaining expression of genes such as Cdh5, Pecam1, and Cd34, while downregulating factors associated with hemangioblast identity (Figure 2D). Cells fated for the endothelial terminal state diverge at E10.5 activating regulators of angiogenesis, such as Tbx1 and Foxc2, as well as genes associated with blood flow-associated shear stress, including Klf2 and Klf4 (Figures 2C, 2D, and S2A) (Sangwung et al., 2017). These factors are not enriched within the HSEC trajectory, where beginning as early as E8.75, HSEC-specific transcription factors are activated, including Tfec, Nr5a2, Sox17, and Oit3, as well as genes associated with HSEC identity, including Stab2, Lyve1, and Fcgrt (Figures 2C and 2D) (Géraud et al., 2010). Differentiation potential towards the HSEC terminal state further captures emergent sinusoidal endothelium as early as E8.75, expressing markers of the HSEC lineage (arrowheads, Figures 2E and S2G). Of note, while it is likely that endothelium of cardiac origin is included in this analysis, we could not deconvolute endocardial and vascular endothelial contribution to liver endothelial lineages based on transcription alone (data not shown).
Of note, the capture and sequencing of Etv2-expressing endothelial cell lineages was limited to our E7.5 and E8.75 libraries (Figures 2A and S2G). Expressed within hemangioblasts and hemogenic endothelia, the absence of Etv2 within endothelia at E9.5 and E10.5 suggests that the endothelium in the liver bud does not retain hemogenic potential (Figure S2G) (Wareing et al., 2012). We do, however, see 378 Foxa2-Kdr co-expressing cells, which may represent a population of endothelial-fated endodermal cells (Figure S2I and S2J). Re-clustering and cell population annotation of these 378 cells based on differential gene expression analysis indicates the presence of node and definitive endoderm-derived lineages, including foregut, midgut, and hindgut (Figure S2J and S2K; Table S2). We further identify a subpopulation of Foxa2-expressing endothelial cells, which express Etv2 as well as biomarkers of mature endothelium, such as Cdh5 and Fli1, (Figure S2L) (Rasmussen et al., 2011). Single-cell data from Pijuan-Sala et al. (2019) also supports the existence of these cells, where an additional 150 Foxa2-Kdr co-expressing cells can be identified (data not shown). These findings support the model where the endoderm generates endothelial cells (Goldman et al., 2014).
Septum transversum and liver mesenchymal heterogeneity in the early liver
Few studies have focused on characterizing the development of liver mesenchyme, including the STM, mesothelium, and HSCs (Asahina et al., 2009, 2011). As a result, little is known about the complete diversity and timing of emergent mesenchymal populations within the early liver. To address this shortfall, we computationally segregated 3,691 liver mesenchymal cells from our E8.75, E9.5, and E10.5 libraries and identified five distinct populations based on clustering and differential gene expression analyses (Figure 3A and S3A; Table S2). One cluster, which represents early STM (STM I), is composed predominantly of cells from E8.75 and is enriched for Hand1 and Foxf1, both of which are required for mesoderm development (Figures 3B and S3C) (Mahlapuu et al., 2001; Maska et al., 2010). A second STM cluster (STM II) is composed mostly of cells from E10.5, and has activated expression of the liver mesenchyme markers Alcam and Mab21l2 (Asahina et al., 2009; Saito et al., 2012), but does not express genes associated with specified liver mesenchyme, such as Lhx2 or Upk3b (Figures 3B and S3C) (Kanamori-Katayama et al., 2011; Kolterud et al., 2004). In contrast, the three remaining clusters represent emerging mesothelia and HSCs, including two distinct mesothelial populations. Of the mesothelial populations, one (mesothelium I) is composed of cells from E9.5 and E10.5, while the other (mesothelium II) is composed mostly of cells from E10.5, suggesting differential timing of their specification (Figures 3A and S3C). While both populations express genes associated with liver mesothelial identity, such as Wt1 and Upk3b (Chau and Hastie, 2012; Kanamori-Katayama et al., 2011), they also express distinct sets of genes, including Sox9 and Tbx18 within the mesothelium I cluster and Foxf1 and Isl1 in the mesothelium II cluster (Figures 3B and S3E). ALCAM immunostaining identifies all STM-derived populations at E9.5 and E10.5, including mesothelial progenitors and unspecified STM progenitors that lack expression of mesothelial factors (Figures 3C and 3D). These unspecified progenitors are found proximal to the hepatic bud at E9.5 and at the distal tip of the caudal lobe at E10.5 (Figures 3C and 3D). Immunostaining identifies emergent mesothelial populations expressing SOX9 or ISL1, which at E9.5 occupy the ventral and anterior portions of the tissue, respectively (Figure 3C). By E10.5, SOX9 is widespread among mesothelial cells in both caudal and rostral lobes, whereas ISL1 is limited to the rostral liver mesothelium (Figure 3D). Thus, our gene expression and immunostaining data reveal the persistence of ALCAM-expressing unspecified STM populations at the distal tip of the caudal liver lobe at E10.5 and further uncover two mesothelial populations at E10.5, with differing spatiotemporal distributions.
Figure 3.
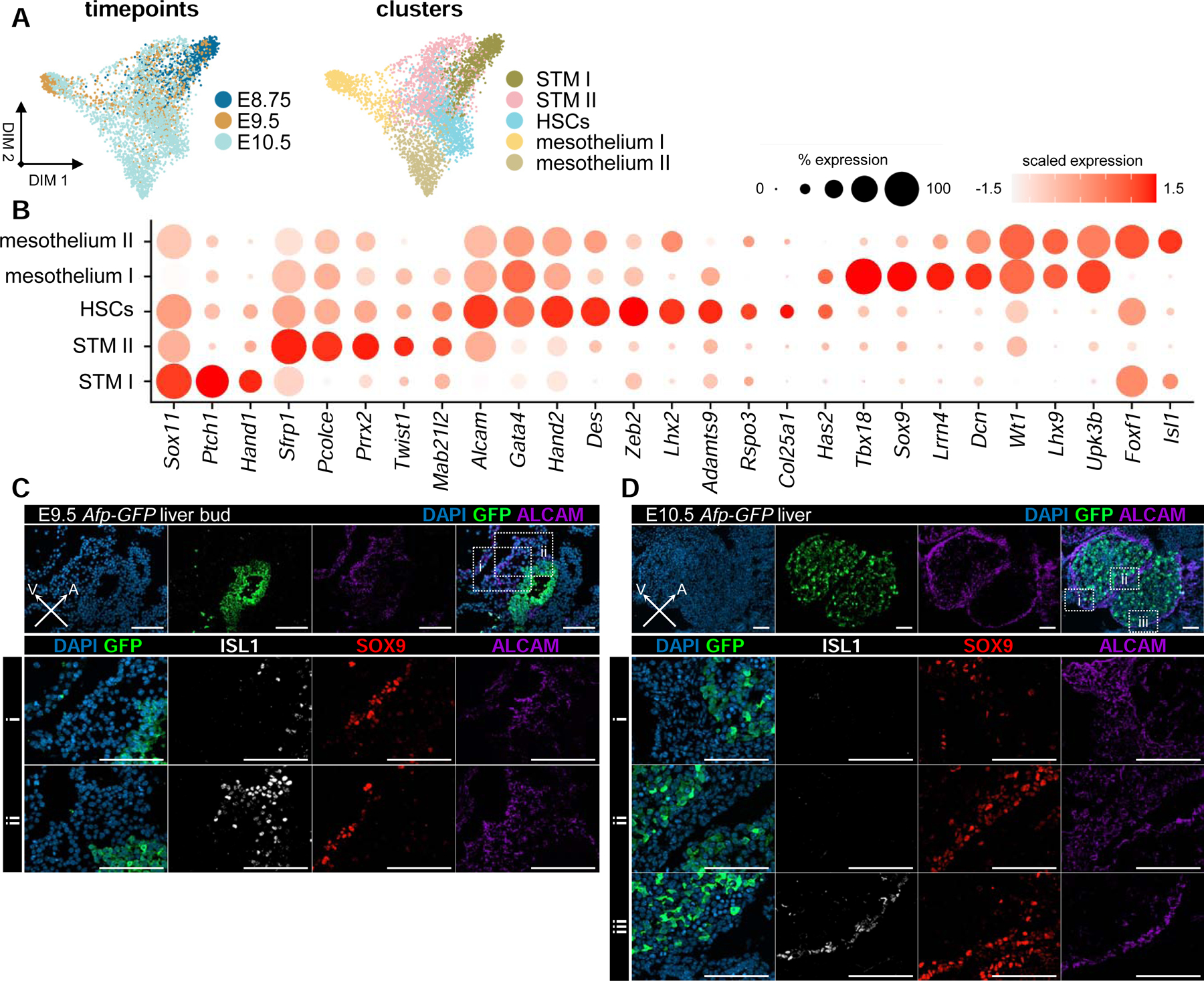
Septum transversum and liver mesenchymal heterogeneity in the early liver.
(A) Force-directed layouts displaying timepoint and annotated cluster contribution of mesenchymal progenitors (n = 3691). (B) Dot plot displaying known and previously uncharacterized markers for STM, mesothelial, and HSC identities. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. (C) ALCAM, SOX9, and ISL1 immunostaining of (i) ventral and (ii) anterior STM in E9.5 Afp-GFP livers. (D) ALCAM, SOX9, and ISL1 immunostaining of (i) mesenchyme at distal tip of caudal lobe, (ii) mesothelium separating rostral and caudal lobes, and (iii) mesothelium of rostral lobe in E10.5 Afp-GFP livers. Scale bars, 100µm. STM, septum transversum mesenchyme; HSC, hepatic stellate cells. See also Figure S3 and Table S2.
The HSC cluster, composed of cells from E9.5 and E10.5, expresses various markers of the HSC lineage, including Lhx2 and Des (Figure 3B) (Kolterud et al., 2004; Nitou et al., 2000). This population also expresses genes associated with migration, suggesting movement of HSCs into the sinusoid, where they can be identified by DES immunostaining by E10.5 in both lobes (Figures 3B and S3F). Interestingly, these emergent HSCs express genes associated with HSC activation, such as Zeb2 and Rspo3, along with collagens and other extracellular matrix proteins (Figures 3B and S3A; Table S2) (Puche et al., 2013). Cumulatively, our data demonstrate that four distinct mesenchymal populations with specific spatial distributions exist within the liver at E10.5.
A hepatomesenchymal hybrid progenitor exists within the early liver
The embryonic liver is formed from the posterior ventral foregut endoderm, which invades the STM in response to migratory cues (Gordillo et al., 2015; Tremblay and Zaret, 2005). To probe this transition, we computationally segregated 2,332 hepatic progenitors sequenced from E7.5 to E10.5 and used clustering and differential expression analyses to identify four distinct cell states (Figure 4A and S4A; Table S2). These include cells from the hepatic endoderm, composed predominantly of cells from E7.5 and E8.75 (Figure S4C). This cluster is enriched for Epcam, Cdh1, and Pyy, which are known markers of the endoderm, with Pyy being specifically expressed within the posterior foregut endoderm (Figure 4B) (Hou et al., 2007). A cluster representing hepatoblasts migrating into the STM is composed predominantly of cells from E9.5, and have activated expression of genes associated with hepatic cell fate, including Hnf4a and Foxa3 (Figure 4B) (Lee et al., 2005; Watt, 2003). This population also expresses Onecut2, Prox1, and Tbx3, along with other genes required for invasion of the STM (Figure 4B) (Lüdtke et al., 2009; Margagliotti et al., 2007; Sosa-Pineda et al., 2000). We also capture early hepatoblasts, mostly from E10.5, which robustly express Afp and Dlk1 along with genes associated with hepatic metabolism, including Alb, Apoa1, and the transcription factor Xbp1 (Figure 4B) (Lee et al., 2008; Tanimizu et al., 2003).
Figure 4.
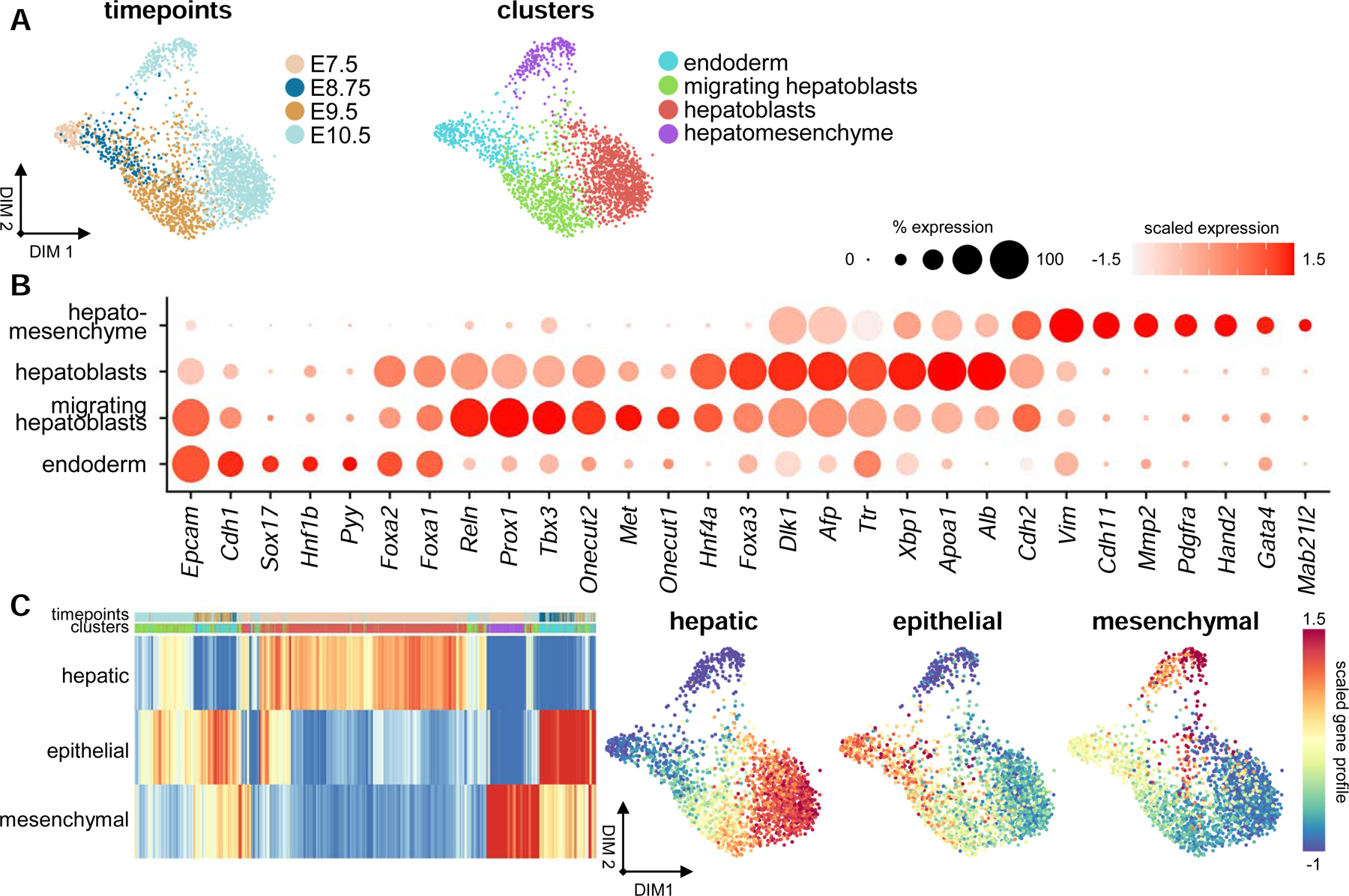
A hepatomesenchymal hybrid progenitor exists within the early liver
(A) Force-directed layouts displaying timepoint and annotated cluster contribution of hepatic parenchymal progenitors (n = 2332). (B) Dot plot displaying known and previously uncharacterized markers for endodermal, migrating hepatoblast, hepatoblast, and hepatomesenchymal cell types. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. (C) Heatmap and force-directed layouts showing average hepatic, mesenchymal, and epithelial gene profiles of all early liver parenchymal cells captured. Heatmap columns are labeled based on timepoint and cluster color schemes in (A). See also Figure S4 and Tables S2 and S3.
Our analysis further reveals a distinct hepatomesenchymal cell type, which is composed predominantly of cells from E10.5 (Figure S4C). This population maintains diminished expression of genes associated with hepatic identity, such as Afp and Dlk1, and activates genes required for mesenchymal development, such as Vim, Pdgfra, Hand2, and Gata4 (Figure 4B) (Barnes et al., 2011; Delgado et al., 2014; Saito et al., 2012). Doublet detection confirms this population is composed of single-cells (Figure S4D). VIM immunostaining of Afp-GFP livers shows presence of double-positive cells within hepatic cords at E9.75 and within the liver parenchyma at E10.5 (Figure S4I). In addition, we see GATA4-expressing GFP+ cells at E10.5 within the liver parenchyma (arrows, Figure S4I). FACS of dissociated Afp-GFP livers also shows an increase between E9.5 and E10.5 of AFP-GFP+ cells that express high levels of the liver mesenchyme-specific marker ALCAM (Figure S4F) (Asahina et al., 2011). To probe transitions in cellular identity, we calculated mean-scaled gene expression profiles for hepatic, mesenchymal, and epithelial cell identities (Figure 4C; Table S3). The epithelial profile is highest among the endoderm and decreases during hepatoblast migration, whereas the inverse is true for the hepatic profile (Figures 4C and S4G). Both of these gene profiles are low within the hepatomesenchymal cluster, where in lieu we observe the activation of a mesenchymal gene programme (Figures 4C and S4G). Further, principal component analysis of the average gene profile of hepatic and mesenchymal populations suggests that the hepatomesenchyme most closely shares transcriptional parallels with the mesothelium II cluster (Figure S4H). These results suggest the presence of an epithelial-mesenchymal hybrid progenitor cell type within the early liver parenchyma that expresses genes associated with hepatic and liver mesenchymal identity.
Hepatoblasts and hepatomesenchyme emerge via distinct migratory mechanisms
To analyze the emergence of hepatoblasts and hepatomesenchyme from the endoderm, we employed Palantir to infer differentiation trajectories from an E7.5 endodermal, Alblo ‘start cell’ (solid arrowhead, Figure 5A). Two terminal states were selected, representing emergent hepatoblasts and the hepatomesenchyme (hollow arrowheads, Figure 5A). Gene trends along pseudotime show the downregulation of endodermal markers, such as Epcam and Foxa1 (Lee et al., 2005; Sherwood et al., 2007), as well as the activation of genes associated with posterior foregut identity, such as Pyy and Onecut1 (Figure 5B) (Hou et al., 2007). Within both trajectories, we see the activation of genes associated with hepatoblast migration, including Hhex and Foxa3, as well as hepatic identity, such as Hnf4a and Afp (Figure 5B). Interestingly, within cells fated for the hepatoblast terminal state, we see the reactivation of Foxa1 along with the maintenance of genes associated with hepatic identity; however, within the hepatomesenchymal trajectory, we do not see their sustained expression (Figure 5B). Instead, genes associated with an epithelial-to-mesenchymal transition (EMT), such as Snai1, Snai2, and Zeb2, are activated (Figure 5B) (Campbell and Casanova, 2016). The expression of master regulators of mesenchymal cell fate and markers of mesenchymal identity immediately follow the activation of EMT-associated genes. These include Vim, Mab21l2, Pdgfra, and Gata4 (Figures 5B and 5C) (Asahina et al., 2011; Delgado et al., 2014; Hentsch et al., 1996; Saito et al., 2012). Differentiation potential is highest within the endoderm and migrating hepatoblasts, and low in hepatoblasts and hepatomesenchyme (Figure 5A). This suggests that the hepatomesenchyme is specified from migrating hepatoblasts between E8.75 and E9.5. Our analysis further shows two pathways to a hepatomesenchymal terminal state, which suggests their emergence may be the product of two temporally distinct migratory events (arrows, Figure 5A). Along with genes involved with mesenchymal differentiation and migration, hepatomesenchymal cells activate genes associated with the formation of actin protrusions, such as Acta2, Cnn2, Wasf1, Mfap2, and Mfap4 (Figure S4J) (Eden et al., 2002; Guo et al., 2006; Liu and Jin, 2016). Genes encoding motor proteins and cell adhesion components are also enriched, including Itgb1, Flna, Vcl, Myl4, and Myo9a (Figure S4J) (Schwartz, 2010). Together, these data suggest that hepatomesenchymal cells represent a subpopulation of hepatoblasts that exhibits epithelial-to-mesenchymal plasticity, adopting a hybrid epithelial-mesenchymal phenotype. Further, their timewise emergence as well as the expression of genes associated with migration and the generation of traction suggest that these cells may contribute to hepatoblast invasion of the STM.
Figure 5.
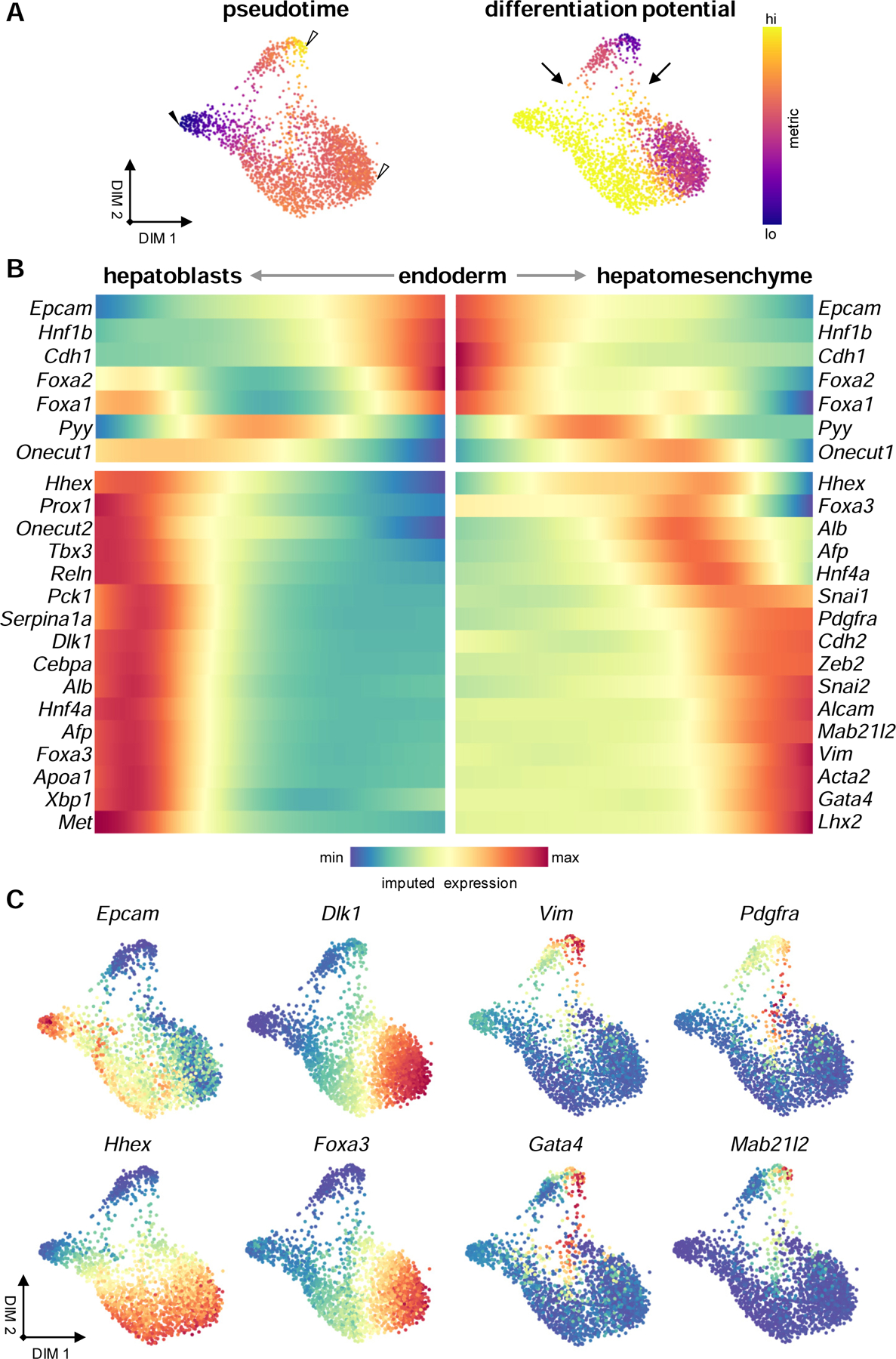
Hepatoblasts and hepatomesenchymal cells emerge via distinct migratory mechanisms
(A) Force-directed layouts displaying pseudotime and differentiation potential in hepatic parenchymal lineages from an Alblo ‘start cell’ (solid arrowhead) to terminal states (hollow arrowheads). Arrows indicate two potential trajectories to attain a hepatomesenchymal terminal state. (B) Heatmap representing gene trends over pseudotime from endodermal ‘start cell’ to hepatoblast and hepatomesenchymal terminal states. (C) Force-directed layouts displaying expression levels of cell type markers identified. See also Figure S4.
The signaling niche of the primitive sinusoid
To explore cell-cell interactions active during sinusoid formation, we computationally segregated endothelial, HSEC, hepatoblast, HSC, and hepatomesenchymal lineages from our datasets and applied CellPhoneDB to identify signaling crosstalk between these populations (Figures 6A, Table S4) (Efremova et al., 2020). CellPhoneDB assesses cell-cell interactions through the integration of single-cell expression data with a repository of ligands and receptors, which includes heteromeric complexes. As expected, we see pronounced VEGF signaling towards liver endothelia, as well as PDGF signaling between hepatoblasts and HSCs (Figure 6A) (Awuah et al., 2013; Yamane et al., 1994). Hepatomesenchymal cells are also predicted to receive signaling cues via PDGFRα. Immunostaining of E10.5 Afp-GFP livers shows robust expression of PDGFRα in HSCs as well as in a portion of GFP+ cells, likely representing the hepatomesenchyme (Figure S5C). TGFβ family signaling is also predicted to contribute to sinusoid development. BMP2 and BMP7 from endothelial and hepatic lineages, respectively, are predicted to signal towards HSC and hepatomesenchymal populations through BMPR1A/ACVR2B (Figure 6A). TGFβ1 from endothelia and TGFβ2 from HSCs and the hepatomesenchyme are predicted to signal via TGFβR3 towards all populations (Figure 6A). While not directly involved in signal transduction, here TGFβR3 may act as a reservoir for TGFβ ligand, establishing a signaling environment for later in development (Andres et al., 1989). Interestingly, though heavily implicated in hepatogenesis, WNT family signaling is not included within our top interactions, due to limited expression of WNT signaling components within sinusoidal populations.
Figure 6.
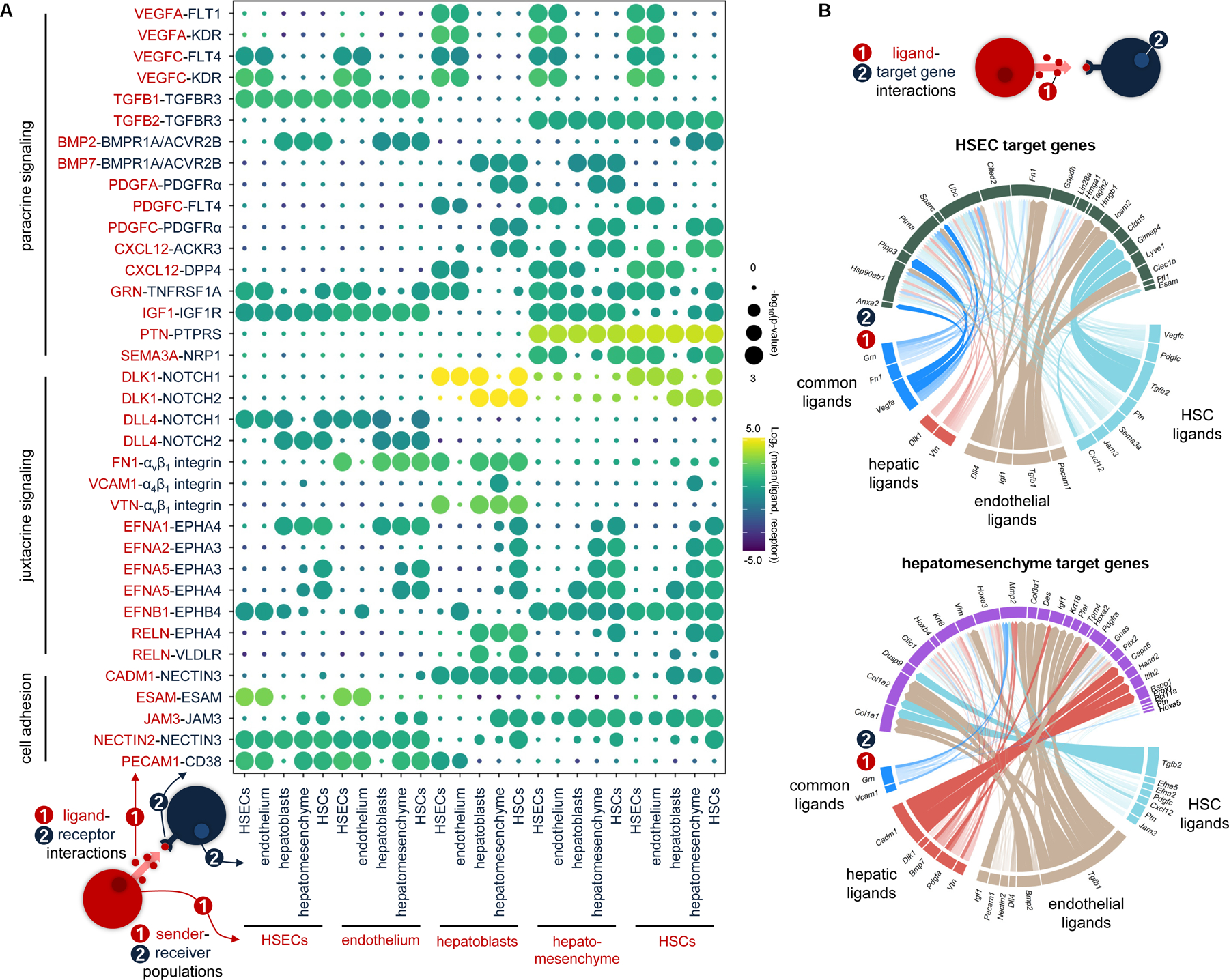
The signaling niche of the primitive sinusoid.
(A) Dotplot displaying putative ligand-receptor interactions between hepatoblast, HSEC, endothelial, hepatomesenchymal, and HSC lineages captured at E10.5. Size of the dot represents statistical significance of the indicated interactions. Color indicates the means of the average expression level of the ligand from cluster 1 and the receptor from cluster 2. (B) Circle plots depicting links between (1) ligands and (2) their predicted target genes for HSECs and hepatomesenchymal cells. Link colors indicate ligand sender populations, whereas width and opacity of links correlate to ligand-receptor interaction weights and ligand activity scores, respectively. HSCs, hepatic stellate cells; HSECs, hepatic sinusoidal endothelial cells. See also Figure S5 and Tables S4 and S5.
Identification of active ephrin, Notch, and reelin cell-cell interactions suggest an active role for juxtacrine signaling in the development of the primitive sinusoid (Figure 6A). We identify more than 40 significant ephrin-Eph interactions in our analysis, which include interactions between all sinusoidal cell populations (Figure 6A; Table S4). Dynamic NOTCH pathway activation and inhibition between sinusoidal populations is also predicted. DLL4 active in the endothelia, and enriched within HSECs, signals autocrinally via NOTCH1, whereas it further signals to other sinusoidal populations via NOTCH2 (Figure 6A). Interestingly, signaling via the non-canonical NOTCH inhibitor DLK1, expressed by non-endothelial populations, towards NOTCH receptors may compete with endothelial DLL4 for receptor occupancy (Baladrón et al., 2005; Finn et al., 2019). Reln is among the first genes expressed by hepatoblasts during hepatic endoderm specification at E8.75, and immunostaining demonstrates specific expression of RELN at E9.5 and E10.5 in AFP-GFP+ hepatoblasts (Figures 6A and S5C). Our analysis predicts canonical RELN signaling via VLDLR autocrinally as well as towards HSCs, while non-canonical RELN interactions via EPHA4 are predicted to influence HSCs and hepatomesenchymal cells. (Bouché et al., 2013; Wasser and Herz, 2017) (Figure 6A).
To assess the transcriptional influence of ligands identified above, we applied NicheNet to predict ligand-target regulatory potential during sinusoidal cell lineage development, using lineage-specific genes as potential pathway targets (Figure 6B, S5A, and S5B; Table S5). Of interest, DLL4, VEGFA, and the TGFβs are predicted to play a significant role in the regulation of genes associated with HSEC identity, including Icam2, Sparc, and Lyve1 (Figure 6B). Hepatomesenchymal target genes were predicted to be influenced by a range of ligands including BMPs, TGFβs, and IGF1. Interestingly, BMPs and TGFβs are predicted to promote a variety of genes associated with mesenchymal cell fate, including Col1a1, Des, and Mmp2, whereas IGF1 is predicted to specifically activate Pdgfra (Figure 6B). Together, these analyses suggest roles for paracrine and juxtacrine signaling in driving activation of genes associated with liver lineage development in the primitive sinusoid.
Discussion
Using single-cell transcriptomics, we have charted the emergence of diverse liver parenchymal and non-parenchymal populations from their endodermal and mesodermal ancestors. Through this analysis, we have identified an epithelial-mesenchymal hybrid population of cells that co-expresses hepatoblast and mesenchymal gene signatures. Trajectory analysis suggests that this hepatomesenchymal cell type diverges from typical hepatoblasts through a partial EMT event, though their final cell state and function within the liver parenchyma remains elusive. Our data further describe the specification events in the establishment of the non-parenchymal lineages, including the earliest stages of HSEC cell fate emergence. We further observe unexpected heterogeneity within mesenchymal populations, including emergent HSCs and two distinct mesothelial populations, which occupy distinct niches within the early liver.
The temporal emergence of hepatoblasts is in line with previous studies, with specification from the endoderm in early organogenesis (E8.75), migration at E9.5, and the beginnings of hepatoblast function at E10.5 (Gordillo et al., 2015; Si-Tayeb et al., 2010; Tremblay and Zaret, 2005). Of interest, while FGF and BMP signaling have previously been shown to coordinate the specification of anterior and posterior hepatic endodermal populations, respectively, we see no evidence of distinct hepatoblast populations based on transcriptional profiles at E9.5 (Palaria et al., 2018; Tremblay and Zaret, 2005; Wang et al., 2015). This suggests that distinct signals from the overlying mesenchyme induce a convergent transcriptional profile among developing hepatoblasts. In addition to hepatoblasts, we identify the hepatomesenchyme, a distinct cell type that expresses a gene profile reminiscent of hepatoblasts and STM progenitors. While epithelial-mesenchymal plasticity has been previously identified within hepatic tissues and cell lines (Dong et al., 2018; Goldman et al., 2016; Li et al., 2011), our analyses further delineate the mechanisms behind hepatoblast and hepatomesenchyme emergence. Within hepatoblasts, trajectory analyses reveal a gradual shift from an epithelial to a hepatic transcriptional profile alongside the activation of genes canonically associated with hepatic migration, including Hhex and Tbx3, starting at E8.75. In contrast, these analyses suggest that hepatomesenchymal cells are specified at two temporally distinct points during the epithelial-to-hepatic transition and migrate into the STM via a partial EMT, with the activation of Snai1, Snai2, and Zeb2. Generation of AFP-GFP+ and AFP-GFP− libraries at E10.5 confirms the hepatic origin of the hepatomesenchyme as the hepatoblast-specific Afp-GFP mouse line allows for the short-term lineage tracing of liver parenchymal derivatives even after Afp RNA expression has waned (Figure S4E). Gene expression analysis rules out the possibility that these cells are derivatives of other endodermal organs, which express low levels of Afp-GFP (Figure S1D).
The divergence of hepatoblast and hepatomesenchymal lineages is reminiscent of the polarization that takes place during collective cell migration (CCM). CCM is a migratory process by which a collective of cells is polarized in response to a signaling gradient into leader and follower cells, which collaborate to promote cellular movement. In zebrafish, emergence and positioning of hepatoblasts is accomplished by CCM, with ephrin signaling and hepatoblast filopodia mediating contacts between gut tube and mesodermal lineages (Cayuso et al., 2016) and in vitro studies have further shown evidence of collective migration of hepatic cell lines (Cheng et al., 2018; Han et al., 2019; Suárez-Causado et al., 2015). Our data suggest that hepatoblast migration is accomplished through CCM in mouse, where parenchymal progenitors are polarized into follower and leader cells: hepatoblasts and hepatomesenchyme, respectively. Hepatoblasts remain pseudo-epithelial; they maintain cell polarity, stable epithelial contacts, and a regular, cobblestone-like morphology (Gordillo et al., 2015). Hepatomesenchymal cells undergo a partial EMT, activating the expression of a suite of genes involved in cell migration. The signaling cues required for the polarization of these cell types are unclear; however, our signaling analysis suggests potential roles for PDGF, TGFβ, or IGF pathways. However, this remains to be explored in detail experimentally.
Differential susceptibility to perfusion between the vascular and sinusoidal capillaries may act as a driving force behind the divergence of liver endothelial lineages. Vascular endothelia express mechanosensitive ion channels along with their downstream flow-responsive genes, including Klf2 and Klf4. In contrast, our interaction analyses suggest that reciprocal interactions within HSECs through DLL4 and NOTCH receptors may drive HSEC development. Interestingly, in models of liver fibrosis, DLL4 promotes capillarization and liver scarring, whereas KLF2 maintains endothelial integrity (Chen et al., 2019; Marrone et al., 2015). In addition, the organization of the sinusoidal capillaries abuts HSECs against hepatic and mesenchymal lineages, which robustly express the Notch inhibitor Dlk1 (Falix et al., 2012; Tanaka et al., 2009). DLK1 may contribute to the lateral inhibition of Notch in HSEC progenitors, comparable to its role in the differentiation of type I from type II alveolar cells in lung repair (Finn et al., 2019). Taken together, this suggests roles for blood flow as well as competition for NOTCH receptor occupancy by Delta-like ligands during HSEC specification. Further, the expression of genes associated with sinusoidal function and identity indicate that specification of HSECs begins at E8.75, earlier than previously appreciated. Whether HSEC function, including scavenging and antigen presentation, is required for early liver development has not been explored; however, the activation of these genes in the earliest stages of liver development suggests a role prior to complete sinusoidal perfusion and fenestration, which occur at E12.5 and E15.5, respectively (Enzan et al., 1997; Lorenz et al., 2018).
Our data show diverse mesothelial and stellate cell populations with distinct spatial distributions within the developing liver by E9.5, suggesting emergence of these populations from the STM earlier than previously shown. While trajectory analysis failed to delineate lineage relationships (data not shown), likely due to incomplete capture of liver mesenchymal progenitors or complex lineage relationships between these populations, immunostaining suggests parallels between the Isl1- and Sox9-expressing anterior and ventral STM at E9.5 and distinct mesothelial populations at E10.5. We further show the earliest evidence of HSCs within the liver by E9.5. HSC progenitors in the early liver express genes associated with HSC activation, including migration and extracellular matrix deposition. This suggests the activation of HSCs during liver bud formation plays a role in sinusoidal organization and basement membrane formation. Taken together, our data demonstrate substantial, unappreciated diversity exists within the STM-derived liver mesenchymal progenitor populations.
Overall, our single-cell transcriptomic analysis of liver development reveals early emergence of highly diverse hepatic, endothelial, and mesenchymal cell lineages. We delineate distinct hepatic parenchymal populations in the liver bud, including a hepatomesenchymal cell type, which emerge via separate migratory mechanisms. We characterize exceptional diversity within spatially distinct liver mesenchymal progenitors, including distinct mesothelial populations and the earliest evidence of HSCs. We further show evidence of HSEC progenitors by E8.75, which express genes associated with sinusoidal function, and we propose a role for blood flow in their divergence from the vascular endothelium. Further analyses are required to confirm the lineage relationships proposed between these populations, as well as to determine a functional role for the hepatomesenchymal progenitors.
STAR Methods
RESOURSE AVAILABILITY
Lead Contact
Requests for resources, reagents, or further information should be directed to and will be fulfilled by the Lead Contact, Pamela A. Hoodless (hoodless@bccrc.ca).
Materials Availability
This study did not generate new or unique reagents.
Data and Code Availability
All single-cell RNA-sequencing data generated during this study have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-9334. The data can also be explored at https://singlecell.broadinstitute.org/single_cell/study/SCP1022/single-cell-transcriptomics-reveals-early-emergence-of-liver-parenchymal-and-non-parenchymal-cell-lineages. This study did not generate any unique code, and all analyses were performed in R and Python using standard protocols from previously published packages. All scripts associated with this manuscript are available by request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse husbandry
All mice were maintained in accordance with the University of British Columbia’s Animal Care Committee’s standards under specific pathogen-free conditions. Up to five mice were housed per cage and were maintained on a regular chow diet ad libitum on a 12-hour light-dark cycle. Afp-GFP (Kwon et al., 2006) and CD1 Elite mouse colonies (Charles River) were used for all animal procedures. Afp-GFP mice were maintained on a hemizygous background, and were used for the generation of E7.5 and E10.5 single-cell libraries, as well as for immunostaining. CD-1 mice were used for all other experiments (Nowotschin et al., 2019). Embryos from timed matings were considered E0.5 at noon of the day a vaginal plug was observed. Embryos were collected at noon of the experimentally-determined timepoint in ice-cold phosphate-buffered saline (PBS) under a Leica MZ6 dissecting scope. Inheritance of Afp-GFP transgene was determined based on GFP fluorescence within the yolk sac or livers of embryos, which was observed under a Zeiss AxioZoom V16. Sex-specific differences were not anticipated and embryo sex was not determined.
METHOD DETAILS
Single-cell dissociation of endodermal and hepatic tissues
Definitive and gut tube endoderm were dissected and prepared from E7.5 and E8.75 embryos as detailed in Nowotschin et al. (2019). For E7.5 definitive endoderm, embryos were washed in three drops of DMEM/F12 on ice, then incubated in pancreatin/trypsin (2.5% pancreatin/0.25% trypsin in PBS) for three minutes on ice. The embryos were subsequently washed in three drops of DMEM/F12, 10% newborn calf serum on ice, then the endoderm layer was removed using tungsten needles (FST, cat. #10130–10). Dissected endoderm was washed in cold DMEM/F12, then dissociated at 37°C for 20 minutes in accutase/0.25% trypsin (1:2). E8.75 embryos, with heads and extra-embryonic membranes removed, were washed in three drops of DMEM/F12 on ice, then incubated in pancreatin/trypsin (2.5% pancreatin.0.5% trypsin in PBS) on ice for five minutes. Embryos were washed in three drops of DMEM/F12, 10% newborn calf serum on ice, then gut tubes were removed using tungsten needles and divided into anterior and posterior portions at the foregut-midgut junction. Dissected endoderm gut tube segments were washed in cold DMEM/F12, then dissociated at 37°C for 20 minutes in accutase/0.25% trypsin (1:2). E9.5 and E10.5 single-cell suspensions were generated by dissection of liver buds, including STM, in PBS with fine dissecting forceps, ensuring removal of cardiac, yolk sac, and non-hepatic endodermal tissues. Livers were washed in PBS, and incubated for five minutes at 37°C in 1ml PBS, 10 µg/ml DNAse I, 4mM EDTA, and 2% fetal bovine serum (FBS) (PDEF) with dispase/collagenase (2.5µU/ml dispase, 0.0025% collagenase type I) to dissociate tissues into single-cells. Tissues were gently triturated using a 1000µl pipette before and after enzymatic digestion to facilitate dissociation. Single-cell suspensions were filtered through 40µm FlowMI cell strainers (Sigma BAH136800040) into 5ml FACS tubes to remove debris and incompletely dissociated cells. The strainers were washed with PBS to collect remaining cells. The filtrate was subsequently centrifuged at 1000g for four minutes to sediment single cells and the supernatant was removed.
Fluorescent-activated cell sorting
Filtered, dissociated single-cells were resuspended in DMEM or DMEM/F12 and sorted using a FACSARIA III (BD Biosciences) with a 100µm nozzle in purity mode before single-cell RNA-seq library preparation. All cell suspensions were stained with 4µM ethidium homodimer-1 to purify live cells. Cellular debris and cell doublets were excluded based on forward and side scatter. Single-cell suspensions generated from E7.5 endoderm were sorted based on AFP-GFP expression, with AFP-GFP+ fractions representing visceral endodermal derivatives and AFP-GFP− fractions representing definitive endoderm and associated mesoderm (Nowotschin et al., 2019). E10.5 livers were also sorted based on AFP-GFP expression, with AFP-GFP+ fractions representing hepatic parenchymal progenitors and AFP-GFP− fractions representing non-parenchymal progenitors (Kwon et al., 2006; Wei et al., 2018). Due to the expansion of hematopoietic populations within the liver by E10.5, we depleted these progenitors prior to library construction. Filtered, dissociated liver cells from E10.5 were resuspended in 2% FBS in PBS, and stained with APC-conjugated anti-Ter119 (1:200) anti-CD45 (1:200), and anti-cKIT (1:200) at 4°C for 15 minutes. After staining, cell suspensions were washed three times in PDEF, centrifuging at 1000g for four minutes, and were ultimately resuspended in DMEM. Subsequent APC+ fractions were not included during E10.5 library construction. For ALCAM FACS analysis, tissue suspensions were processed identically, except cells were stained with PE-conjugated anti-ALCAM (1:200) at 4°C for 15 minutes. FACS analyses and plots were generated using FlowJo Software (Becton, Dickinson and Company).
Single-cell library preparation and sequencing
Sorted single-cell suspensions were counted and diluted to a final concentration in DMEM, 10% FBS or DMEM/F12, and cellular suspensions were loaded on a Chromium Controller to generate single-cell gel bead emulsions, targeting 2,500–10,000 cells depending on tissue and embryonic stage. Three libraries at E7.5, eight at E8.75 (4 each anterior and posterior gut tube endoderm), two at E9.5, and two at E10.5 were generated individually (Figure S1A). Single-cell 3’ RNA-seq libraries were generated according to the manufacturer’s instructions (Chromium Single Cell 3’ Reagent v2 Chemistry Kit, 10X Genomics, Inc.). Libraries were sequenced to an average depth of ~200,000 reads per cell on either an Illumina Novaseq 6000 system (E7.5–9.5 libraries) or Illumina Nextseq system (E10.5 libraries). Refer to Table S1 for detailed library quality metrics.
Single-cell RNA-seq data analysis
Sequenced reads from all libraries were aligned and quantified using the Cell Ranger Single Cell Software Suite version 2.1.0 (10X Genomics, Inc.) against the 10X Genomics pre-built mm10 (GRCm38.93) reference genome. CellRanger was used to identify droplets containing cells for each library individually. CellRanger identifies cells based on the number of UMIs and gene expression profiles. Further quality metrics were calculated using the calculateQCMetrics in the scater package in R (Figures S2D, S3D, and S4D) (McCarthy et al., 2017), and cells were subsequently filtered based on unique features (>1000 genes) and mitochondrial RNA content (<20%).
Size-factor normalization, feature selection, dimensionality reduction, and doublet identification were performed using the scran and scater packages in R (Lun et al., 2016; McCarthy et al., 2017). Briefly, filtered cells were loosely clustered using the quickCluster function, then size-factor normalized using the computeSumFactors function, taking both library sizes and quickCluster ID into account. Single-cell transcriptomes were subsequently log-transformed using the normalize function in scater to correct for biases between cells, taking size factors into account. Highly variable genes were selected based on gene expression levels and variance across all cells to highlight biological differences across datasets. Ribosomal, mitochondrial, Y-chromosome genes, and Xist were removed from highly variable genes. Doublet scores were calculated using the doubletCells function in scran, which assigns each cell a score that is defined as the ratio of the density of simulated doublets to the squared density of original cells in the neighbourhood of that cell.
Principal component analysis (PCA) dimensionality reduction and mutual-nearest neighbours (Haghverdi et al., 2018) or Harmony (Nowotschin et al., 2019) batch correction were performed to correct for non-biological confounders between replicates and timepoints (Figures S2B, S3B, and S4B). PCA was completed using the RunPCA function in Seurat (Butler et al., 2018). For MNN batch correction, fastMNN from the scran package was employed, and the first 100 principal components were used as input. k=20 neighbours were used for calculation of mutual-nearest neighbours. Harmony uses timepoint metadata to construct augmented affinity matrices, which uses mutual-nearest neighbours across consecutive timepoints to correct for batch effects. Log-normalized expression-matrices, containing data for highly variable genes only, were used as inputs. A k of 20–35 was used for the mutual nearest neighbour calculation for the hepatic, endothelial, and mesenchymal lineage analyses. The augmented affinity matrices were subsequently used to calculate the force-directed layouts.
tSNE and UMAP dimensionality reduction were performed using the runTSNE and runUMAP functions in scater, respectively using the first 100 principal components as inputs. Force-directed layouts to display cell type diversity were calculated using adjusted affinity matrices generated with the Harmony package in Python (Nowotschin et al., 2019). Graph-based clustering and Wilcoxon rank-sum test for differential gene expression were performed using phenograph (Levine et al., 2015) and Seurat (Butler et al., 2018) packages in Python and R, respectively. For phenograph clustering, a k of 150 to 300 was used and the normalized expression matrix of highly variable genes served as the input. The FindAllMarkers function in Seurat was used to identify differentially-expressed genes. Globally-enriched genes within a cluster were identified based on a log fold-change of 0.25 with an expression threshold of >25% within a population using the Wilcoxon rank-sum test. Only positively enriched genes with a false-discovery rate (FDR) of 0.05 or lower are included. Gene expression profiles were calculated for each single-cell in the hepatic subset as the average log-normalized gene expression of hepatic, epithelial, or mesenchymal-enriched genes (Table S3), then mean-scaled. All heatmaps were generated in R using pheatmap with clustering done using the ward.D2 method.
Palantir single-cell differentiation trajectories, which infer pseudotime progression, cell fate decisions, and terminal cell states from a defined ‘start cell’, were calculated using diffusion components from Harmony augmented affinity matrices discussed in the previous section (Nowotschin et al., 2019; Setty et al., 2019). Emergent hepatic and endothelial lineage trajectories were calculated independently by filtering out cells of interest using annotated cell types (Figures S1A–C). ‘Start cells’ for each trajectory were selected based on current developmental paradigms, including an E7.5 Alblo definitive endodermal progenitor for the hepatic lineages, and an Etv2hi hemangioblast progenitor for the endothelial lineages. 30 nearest-neighbours and 700 to 1000 waypoints were used as inputs for the generation of Palantir differentiation trajectories. For both hepatic and endothelial trajectories, we calculated the Pearson’s correlation of Palantir results over a range of k nearest-neighbours. This demonstrates that pseudotime estimation, differentiation potential, and branch probabilities to terminal states are robust with respect to changes in the number of nearest-neighbours, k (Figures S2H and S4K). Gene expression was visualized after imputation with the MAGIC algorithm to denoise the single-cell data and fill in dropouts (van Dijk et al., 2018). Gene expression trends along pseudotime were calculated based on branch probabilities and generalized additive models, showing stepwise transcriptional progression of endodermal and endothelial lineages along pseudotime using Palantir (Setty et al., 2019).
CellPhoneDB Analysis
We applied CellPhoneDB to our data to infer ligand-receptor interactions present within the developing sinusoid (Table S4). CellPhoneDB determines cellular crosstalk based on significant enrichment of reciprocal expression of ligands and receptors between annotated cell populations. The significant enrichment is calculated by randomly permutating the labels for the annotated cell populations and determining if the mean receptor and ligand expressions of the annotated cell populations are significantly higher than those of populations of cells determined from random permutations. We took a subset of our data containing parenchymal and non-parenchymal cell types, which are present in the liver bud E10.5. This includes clusters representing cell types such as vascular endothelium, sinusoidal endothelium, hepatoblasts, hepatomesenchymal cells, nascent HSCs, septum transversum mesenchyme and hematopoietic cells as annotated in figures 1, 2A, 3A and 4A. To discern the most biologically relevant ligand-receptor pairs, we only included ligands or receptors if they had a mean combined expression of 0.7 and were expressed in over 40% of all cells in their respective clusters. The expression values for receptors and ligands were calculated from the library size normalized non-log-transformed UMI (unique molecular identifier) counts.
We added interactions involving RELN to the default v2.0.0 CellPhoneDB database of receptor-ligand interactions, specifically interactions of RELN with EPHB1, EPHB2, EPHB3, EPHA4, the α3β1 integrin complex (Bouché et al., 2013), VLDLR (Ha et al., 2017), or LRP8 (Andersen et al., 2003).
NicheNet Analysis
The transcriptional influence of ligands within the early sinusoidal niche was predicted by applying NicheNet (Browaeys et al., 2020). NicheNet predicts which ligands produced by a sender cell are the most active in affecting gene expression in a receiver cell, through the correlation of ligand activity with genes previously characterized as targets of their downstream pathways. Using the same clusters included for our CellPhoneDB analysis, we applied NicheNet to predict which ligands influence transcription during sinusoidal endothelial specification and hepatomesenchyme development. To separately define these specification events, hepatomesenchymal and sinusoidal endothelial clusters were defined as receiver populations in separate analyses, and all five primitive sinusoidal cell types were defined as potential sender cells, including vascular endothelium, sinusoidal endothelium, hepatoblasts, hepatomesenchyme, and stellate cells. Gene sets of interest were defined based on differential gene expression analysis where the top 125 differentially-enriched genes based on false-discovery rate were used (average log2 fold change > 0.25; expressed in at least 25% of population of interest) (Table S5). This includes genes specifically enriched within the hepatomesenchyme over hepatoblasts, or enriched within the sinusoidal endothelium over vascular endothelium. A background gene set includes all other genes that were not differentially expressed between these populations. NicheNet’s ligand-target model was converted from human to mouse genes using the convert_human_to_mouse_symbols function. Activities of ligands identified in our CellPhoneDB analysis were ranked for both the hepatomesenchyme and sinusoidal endothelial populations. This is done by calculating Pearson’s correlation coefficients of the ligand-target regulatory potential scores for each ligand and the target indicator vector, which defines a gene as present or absent within the gene set of interest. Z-score normalization of the Pearson coefficients allows for the comparison of ligand activities between target gene sets (Figure S5A).
The circlize package in R was used to separately represent the regulatory networks between the predicted ligands and their targets expressed in the hepatomesenchymal sinusoidal endothelial, hepatoblast, or hepatic stellate cell settings (Gu et al., 2014). Ligands were grouped and colourized based on cell of origin. Width and opacity of links were determined based on the ligand-receptor interaction weights and ligands’ activity scores, respectively.
Annotation of liver lineages
Endodermal, endothelial, and mesenchymal liver lineages were computationally segregated from each library individually (Figures S1A–C). Liver progenitors from E8.75, E9.5, and E10.5 were classified based on expression of known markers of the emergent hepatoblasts, such as Hhex, Prox1, Hnf4a, and Afp. A subset of definitive endodermal progenitors from E7.5 was also included. Pancreatic, biliary, yolk sac, lung, and gut tube cells were removed based on their expression of characteristic markers. Endothelial cells from E8.75, E9.5, and E10.5 were classified based on expression of known markers, such as Cdh5, Kdr, and Pecam1. Hemangioblast and hematopoietic progenitors from E7.5 were segregated based on their expression of Etv2, Kdr, as well as Runx1, and Gata1. Mesenchymal cells from E8.75, E9.5, and E10.5 were classified based on expression of known septum transversum and liver mesenchymal markers, such as Alcam, Mab21l2, Lhx2, and Wt1. Cardiac, stomach, and pancreatic mesenchymal progenitors were eliminated based on their expression of cell-type specific markers, including Tnnt2, Tbx20, Ryr2, Osr1, and Nkx6–1 (Figures S1A–C).
To validate our cell type annotations, we compared our annotations with the manual annotations from Pijuan-Sala et al. (2019) using mutual-nearest neighbour (MNN) batch correction. Aggregate cell type annotations between our complete dataset and a random subset of 20,000 cells from Pijuan-Sala et al. (2019) were compared using fastMNN from the scran package. The 100 first principal components were used as input, and a k of five was used to identify neighbours between datasets. MNN pairs between clusters from each dataset were then compared to approximate consistency of annotations between datasets
Immunofluorescence
Whole E9.5, E9.75, and E10.5 Afp-GFP embryos were washed with PBS and fixed with 4% paraformaldehyde (PFA) at 4°C for 4–12 hours. Embryos were washed with PBS three times, and then transferred through a sucrose gradient (15%–30%–60% sucrose in PBS, 4°C, 1–12 hours each) before embedding in TissueTek optimal cutting temperature compound and frozen at −20°C. 8µm sections were collected using a Leica CM3050S cryostat at −25°C with SuperFrost Plus slides. Sections were circumscribed with an Elite PAP pen (Diagnostic Biosystems K039) and slides were subsequently placed in an opaque humidity chamber. Sections were re-fixed for 10 minutes at room temperature in 4% PFA in PBS, washed three times with PBS for five minutes, then incubated in block solution (5% bovine serum albumin, and 0.01% Triton-X in PBS) for one hour at room temperature. Sections were subsequently incubated with the following primary antibodies in block solution overnight at 4°C: anti-GFP (1:500), anti-SOX9 (1:500), anti-ISL1 (1:10), anti-ALCAM (1:40), anti-GATA4 (1:50), anti-RELN (1:100), anti-DES (1:50), anti-VIM (1:50), and anti-PDGFRα (1:40). The next day, sections were washed with PBS three times for five minutes, then incubated with species-specific Alexa Fluor 488-, 568-, 594-, or 647-conjugated secondary antibodies (all at 1:500) in block solution for one hour at room temperature. Sections were washed with PBS three times for five minutes, incubated with 1:1000 DAPI to counterstain nuclei, and then washed again with PBS three times for five minutes. Slides were then mounted using a minimal volume (~100µl) 25mg/ml DABCO in 9:1 glycerol:PBS. Coverslips were fixed in place using clear nail polish. Images were captured using a Zeiss AxioImager M2 microscope with an Apotome.2 and a Zeiss Axiocam 503 mono or with a Nikon Instruments Eclipse Ti confocal laser microscope. Image capture or processing was done using ZEN 2.3 software, NIS Elements 5.0, and Fiji in ImageJ (Schneider et al., 2012), with brightness, contrast, pseudo-coloring adjustments, and z-stack alignments applied equally across all images in a given series.
Experimental Design
All of the libraries generated were collected at unique embryonic timepoints, i.e. E7.5, E8.75 (somite stage 13), E9.5, and E10.5. E7.5 libraries were generated in triplicate, E8.75 anterior and posterior gut tube libraries were generated in quadruplicate, E9.5 liver diverticulum libraries were generated in duplicate, and single libraries of AFP-GFP+ and AFP-GFP− single-cells were generated from E10.5 liver buds. Randomization, stratification, blinding, or sample-size estimation were not performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed and graphs generated using R and Python. scRNA-seq analyses were performed as described above. The number of cells used in each of the statistical analyses is indicated in the figures or figure legends.
Single-cell RNA-seq data processing
The Cell Ranger Single Cell Software Suite version 2.1.0 (10X Genomics, Inc.) was used for alignment, de-multiplexing, and UMI counting using the default parameters. Sequencing metrics are presented in Table S1. From 15 samples, 45,547 single cells were collected, ranging from 1413 to 7983 cells collected per sample. Median reads per cell ranged from 53,200 to 384,336 with median UMI of 9572 to 40,995 per cell. Cells with fewer than 1000 expressed genes or if mitochondrial gene expression accounted for greater than 20% of total RNA content were considered of low quality and excluded from downstream analyses.
Statistical Analyses
After data normalization, clustering, and dimensionality reduction, marker genes for each cluster were identified using the FindAllMarkers function in Seurat using the Wilcoxon rank-sum test. Genes with an FDR < 0.05 were considered statistically enriched within a cluster.
For CellPhoneDB ligand-receptor analysis, a p-value > 0.05 indicated a significant interaction. This was graphically represented as the -log10 of the p-value in Figure 6 for clarity.
Pearson’s correlation coefficients of a ligand’s target gene predictions and the observed transcriptional response in the target population was used to define ligand activity for NicheNet ligand-target gene analysis. The top tertile of scores of interactions were visualized.
Pearson’s correlation of Palantir results over a range of k nearest-neighbours demonstrates that pseudotime estimation, differentiation potential, and branch probabilities to terminal states are robust with respect to changes in the number of nearest-neighbours, k.
Supplementary Material
Figure S1. Cell-type annotation of single-cell libraries. Related to Figure 1.
(A) Schematic outlining collection of single-cells for sequencing and target cell types. Green, Afp-GFP+ cells; blue, dissected and dissociated tissue. (B) UMAP visualization and quality control parameters for all replicates sequenced. (C) Heatmap and UMAPs showing mutual-nearest neighbor (MNN) batch correction between data presented in this manuscript and data collected from whole embryos from Pijuan-Sala et al. (2019). Agreement between annotated clusters from both manuscripts validates annotation strategies. (D) Clustering and differential gene expression analyses identify 10, 17, 9, and 12 clusters at E7.5, E8.75, E9.5, and E10.5, respectively. Dotplots highlight select top differentially-expressed genes. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. Ant., anterior; ExE, extraembryonic; Def., definitive; STM, septum transversum mesenchyme; NMP, neuromuscular progenitors; PGC, primitive germ cells; post., posterior; rep., replicate.
Figure S2. Characterization of endothelial lineages captured from E7.5 to E10.5. Related to Figure 2.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of endothelial lineages before and after Harmony correction. (C) Distribution of embryonic timepoints across clusters, showing timing of emergence of endothelial lineages. (D) Quality control parameters for endothelial lineages, sorted by cluster. (E) Heatmap representing gene trends along pseudotime from Etv2hi hemangioblast ‘start cell’ to hematopoietic terminal state. (F) Force-directed layouts displaying expression of select genes enriched within the HSEC cluster. (G) Force-directed layouts, separated by timepoint, display normalized expression of genes associated with endothelial and hematopoietic lineages. Arrowheads indicated cells with early expression of HSEC markers Stab2 and Fcgrt. (H) Palantir results are robust when varying nearest-20 neighbor input, k. Plot show Pearson’s correlation for pseudotime, differentiation potential, and branch probability between pairs of Palantir runs. (I) Distribution of Foxa2-Kdr co-expressing cells (>1 logcounts) within complete UMAP and by cluster. (J) Re-clustering of Foxa2-Kdr co-expressing cells reveals definitive endodermal and endothelial cell states, as well as a transitional cells population. (K) Distribution of embryonic timepoints across clusters in Foxa2-Kdr co-expression subset. (L) Dot plot displaying transcriptional heterogeneity between clusters within the Foxa2-Kdr co-expression subset. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. DE, definitive endoderm; HSEC, hepatic sinusoidal endothelial cells; hemang., hemangioblasts; hemato., hematopoietic cells; STM, septum transversum mesenchyme; YS, yolk sac.
Figure S3. Characterization of mesenchymal lineages captured from E8.75 to E10.5. Related to Figure 3.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of mesenchymal lineages before and after Harmony correction. (C) Distribution of timepoints across clusters, showing timing of emergence of mesenchymal lineages. (D) Quality control parameters for mesenchymal lineages, sorted by cluster. (E) Volcano plot displaying top differentially-expressed genes between the mesothelial clusters identified. Genes highlighted in red represent the top 25 most differentially-expressed based on –log10(p-value). (F) Immunostaining for ALCAM and DES in E10.5 Afp-GFP livers show presence of DES+ HSCs within the (i) rostral and (ii) caudal liver parenchyma. Scale bars, 100µm - 10µm for magnified single-cells. A, anterior; HSCs, hepatic stellate cells; STM, septum transversum mesenchyme; V, ventral.
Figure S4. Characterization of the liver parenchymal progenitors captures from E7.5 to E10.5. Related to Figures 4 and 5.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of endodermal and hepatic lineages before and after Harmony correction. (C) Distribution of embryonic timepoints across clusters, showing timing of emergence of hepatic lineages. (D) Quality control parameters for hepatic lineages, sorted by cluster. (E) Distribution of AFP-GFP+ and AFP-GFP− cells sequenced at E10.5, showing contribution of AFP-GFP+ hepatic parenchymal derivatives to mesenchymal clusters. (F) FACS plots showing AFP-GFP versus the STM-enriched marker ALCAM in dissocated Afp-GFP livers from E9.5 and E10.5. Note enrichment of GFP+ALCAMhi cells at E10.5. (G) Scatterplot comparing the single-cell hepatic, epithelial, and mesenchymal profiles either by timepoint or cluster. Mesenchymal lineages included for the mesenchymal profile comparisons as a reference point. (H) Principal component analysis of average expression profiles of parenchymal and mesenchymal clusters shows close co-clustering of hepatomesenchyme with mesothelium II and STM II clusters, suggesting transcriptional parallels. (I) Immunstaining for GATA4 and VIM in E9.75 or E10.5 Afp-GFP livers shows presence of double-positive cells. Arrowheads indicate AFP-GFP+;GATA4+ or AFP-GFP+;VIM+ hepatomesenchymal cells. 100µm - 10µm for magnified single-cells. (J) Force-directed layouts displaying expression of select genes enriched within hepatomesenchymal cells. (K) Palantir results are robust when varying nearest-neighbor input, k. Plot show Pearson’s correlation for pseudotime, differentiation potential, and branch probability between pairs of Palantir runs. A, anterior; HSCs, hepatic stellate cells; R, right; STM, septum transversum mesenchyme; V, ventral.
Figure S5. Cell-cell signaling within the primitive sinusoid at E10.5. Related to Figure 6.
(A) Heatmap displaying scaled ligand-target regulatory potential for ligands highlighted in Figure 6. (B) Circle plots depicting links between (1) ligands and (2) their predicted target genes for hepatoblasts and stellate cells. Link colors indicate ligand sender populations, whereas width and opacity of links correlate to ligand-receptor interaction weights and ligand activity scores, respectively. (C) Immunostaining for RELN and PDGFRα in E9.5 or E10.5 Afp-GFP livers. RELN is shown to be an early and specific marker for emergent hepatoblasts, whereas PDGFRα is robustly expressed in stellate cell progenitors, but also weakly expressed in a subset of the AFP-GFP+ parenchymal progenitors, likely representing the hepatomesenchyme. Arrowheads indicate AFP-GFP+;RELN+ or AFP-GFP+;PDGFRα+ cells. 100µm - 10µm for magnified single-cells. A, anterior; HSECs, hepatic sinusoidal endothelial cells; V, ventral.
Table S1. Summary of single-cell libraries analyzed. Related to Figure 1.
Table S2. Differential gene expression analysis of libraries and lineage subsets. Related to Figures 1, 2, 3, and 4.
Table S3. Genes used for hepatic, epithelial, and mesenchymal gene profile analysis. Related to Figure 4.
Table S4. Complete CellPhoneDB results. Related to Figure 6.
Table S5. Target genes of interest for NicheNet analysis. Related to Figure 6.
Highlights.
Single-cell analyses provide a comprehensive atlas of liver cell lineage emergence
A distinct, migratory, hepatomesenchymal hybrid cell type is identified
Liver endothelial and mesenchymal progenitors emerge during early hepatogenesis
Candidate cell-cell interactions are identified within the primitive liver sinusoid
Acknowledgements
We thank S. Campbell, D. Lowe, T. Stephan, A. Thakur, and K. Tremblay for constructive discussions as well as H. Soliman and F. Rossi for reagents. We acknowledge funding from the Natural Sciences and Research Council of Canada (RGPIN-2018-05018 to P.A.H.), the Canadian Institutes of Health Research (FRN-153006 to P.A.H.), the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, the National Institutes of Health (R01-HD094868, R01-DK084391 and P30-CA008748 for A.-K.H.), the MSKCC Society for Special Projects and Functional Genomics Initiative (for A.-K.H. and D.P.), and 10X Genomics. J.L. is the recipient of an NSERC PGSD scholarship, S.D. is the recipient of a CIHR CGSM scholarship, and E.M.B. is the recipient of an NSERC CGSD scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
S.C.B and D.M.C. are employees and shareholders at 10x Genomics.
References
- Andersen OM, Benhayon D, Curran T, and Willnow TE (2003). Differential binding of ligands to the apolipoprotein E receptor 2. Biochemistry 42, 9355–9364. [DOI] [PubMed] [Google Scholar]
- Andres JL, Stanley K, Cheifetz S, and Massague J (1989). Membrane-anchored and soluble forms of betaglycan, a polymorphic proteoglycan that binds transforming growth factor-β. J. Cell Biol 109, 3137–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina K, Tsai SY, Li P, Ishii M, Maxson RE, Sucov HM, and Tsukamoto H (2009). Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology 49, 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina K, Zhou B, Pu WT, and Tsukamoto H (2011). Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology 53, 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awuah PK, Nejak-Bowen KN, and Monga SPS (2013). Role and regulation of PDGFRα signaling in liver development and regeneration. Am. J. Pathol 182, 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladrón V, Ruiz-Hidalgo MJ, Nueda ML, Díaz-Guerra MJM, García-Ramírez JJ, Bonvini E, Gubina E, and Laborda J (2005). Dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp. Cell Res 303, 343–359. [DOI] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, VanDusen NJ, Morikawa Y, Conway SJ, Cserjesi P, Vincentz JW, and Firulli AB (2011). Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ. Res 108, 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollerot K, Pouget C, and Jaffredo T (2005). The embryonic origins of hematopoietic stem cells: A tale of hemangioblast and hemogenic endothelium. APMIS 113, 790–803. [DOI] [PubMed] [Google Scholar]
- Bouché E, Romero-Ortega MI, Henkemeyer M, Catchpole T, Leemhuis J, Frotscher M, May P, Herz J, and Bock HH (2013). Reelin induces EphB activation. Cell Res 23, 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R, Saelens W, and Saeys Y (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, and Casanova J (2016). A common framework for EMT and collective cell migration. Development 143, 4291–4300. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Dzementsei A, Fischer JC, Karemore G, Caviglia S, Bartholdson J, Wright GJ, and Ober EA (2016). EphrinB1/EphB3b Coordinate Bidirectional Epithelial-Mesenchymal Interactions Controlling Liver Morphogenesis and Laterality. Dev. Cell 39, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau YY, and Hastie ND (2012). The role of Wt1 in regulating mesenchyme in cancer, development, and tissue homeostasis. Trends Genet 28, 515–524. [DOI] [PubMed] [Google Scholar]
- Chen L, Mupo A, Huynh T, Cioffi S, Woods M, Jin C, McKeehan W, Thompson-Snipes L, Baldini A, and Illingworth E (2010). Tbx1 regulates Vegfr3 and is required for lymphatic vessel development. J. Cell Biol 189, 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gu T, Li B, Li F, Ma Z, Zhang Q, Cai X, and Lu L (2019). Delta-like ligand 4/DLL4 regulates the capillarization of liver sinusoidal endothelial cell and liver fibrogenesis. Biochim. Biophys. Acta - Mol. Cell Res [DOI] [PubMed]
- Cheng CC, Chao WT, Liao CC, Tseng YH, Lai YCC, Lai YS, Hsu YH, and Liu YH (2018). Plectin deficiency in liver cancer cells promotes cell migration and sensitivity to sorafenib treatment. Cell Adhes. Migr 12, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I, Carrasco M, Cano E, Carmona R, García-Carbonero R, Marín-Gómez LM, Soria B, Martín F, Cano DA, Muñoz-Chápuli R, et al. (2014). GATA4 loss in the septum transversum mesenchyme promotes liver fibrosis in mice. Hepatology 59, 2358–2370. [DOI] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Hu Y, Fan X, Wu X, Mao Y, Hu B, Guo H, Wen L, and Tang F (2018). Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, and Kirschner MW (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, and Vento-Tormo R (2020). CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc 15, 1484–1506. [DOI] [PubMed] [Google Scholar]
- Enzan H, Himeno H, Hiroi M, Kiyoku H, Saibara T, and Onishi S (1997). Development of hepatic sinusoidal structure with special reference to the Ito cells. Microsc. Res. Tech 39, 336–349. [DOI] [PubMed] [Google Scholar]
- Falix FA, Aronson DC, Lamers WH, and Gaemers IC (2012). Possible roles of DLK1 in the Notch pathway during development and disease. Biochim. Biophys. Acta - Mol. Basis Dis 1822, 988–995. [DOI] [PubMed] [Google Scholar]
- Finn J, Sottoriva K, Pajcini KV, Zhang W, Malik AB, and Liu Correspondence Y (2019). Dlk1-Mediated Temporal Regulation of Notch Signaling Is Required for Differentiation of Alveolar Type II to Type I Cells during Repair. Cell Rep 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, and Orkin SH (1996). Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. U. S. A 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraud C, Schledzewski K, Demory A, Klein D, Kaus M, Peyre F, Sticht C, Evdokimov K, Lu S, Schmieder A, et al. (2010). Liver sinusoidal endothelium: A microenvironment-dependent differentiation program in rat including the novel junctional protein liver endothelial differentiation-associated protein-1. Hepatology 52, 313–326. [DOI] [PubMed] [Google Scholar]
- Goldman O, Han S, Hamou W, Jodon De Villeroche V, Uzan G, Lickert H, and Gouon-Evans V (2014). Endoderm generates endothelial cells during liver development. Stem Cell Reports 3, 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman O, Valdes VJ, Ezhkova E, and Gouon-Evans V (2016). The mesenchymal transcription factor SNAI-1 instructs human liver specification. Stem Cell Res. 17, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharov NV, Nadeev AD, Jenkins RO, and Avdonin PV (2017). Markers and Biomarkers of Endothelium: When Something Is Rotten in the State. Oxid. Med. Cell. Longev 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Evans T, and Gouon-Evans V (2015). Orchestrating liver development. Development 142, 2094–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- Guo F, Debidda M, Yang L, Williams DA, and Zheng Y (2006). Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J. Biol. Chem 281, 18652–18659. [DOI] [PubMed] [Google Scholar]
- Ha S, Tripathi PP, Mihalas AB, Hevner RF, and Beier DR (2017). C-terminal region truncation of RELN disrupts an interaction with VLDLR, causing abnormal development of the cerebral cortex and hippocampus. J. Neurosci 37, 960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Liu J, Lei Y, Lin Z, Tian D, and Yan W (2019). Netrin-1 promotes the collective cell migration of liver cancer cells in a 3D cell culture model. J. Physiol. Biochem [DOI] [PubMed]
- Hentsch B, Lyons I, Li R, Hartley L, Lints TJ, Adams JM, and Harvey RP (1996). Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev 10, 70–79. [DOI] [PubMed] [Google Scholar]
- Hirschi KK (2012). Hemogenic endothelium during development and beyond. Blood 119, 4823–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Charters AM, Lee SC, Zhao Y, Wu MK, Jones SJM, Marra MA, and Hoodless PA (2007). A systematic screen for genes expressed in definitive endoderm by Serial Analysis of Gene Expression (SAGE). BMC Dev. Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssaint E (1980). Differentiation of the mouse hepatic primordium. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Differ 9, 269–279. [DOI] [PubMed] [Google Scholar]
- Kanamori-Katayama M, Kaiho A, Ishizu Y, Okamura-Oho Y, Hino O, Abe M, Kishimoto T, Sekihara H, Nakamura Y, Suzuki H, et al. (2011). LRRN4 and UPK3B Are Markers of Primary Mesothelial Cells. PLoS One 6, e25391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud Å, Wandzioch E, and Carlsson L (2004). Lhx2 is expressed in the septum transversum mesenchyme that becomes an integral part of the liver and the formation of these cells is independent of functional Lhx2. Gene Expr. Patterns 4, 521–528. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Fraser ST, Eakin GS, Mangano M, Isern J, Sahr KE, Hadjantonakis AK, and Baron MH (2006). Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev. Dyn 235, 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y, Li Y, Hou Y, Chen H, Qiu B, Lin W, Sun A, Wei H, Jiang Y, and He F (2017). Proteomic Analysis Reveals Dab2 Mediated Receptor Endocytosis Promotes Liver Sinusoidal Endothelial Cell Dedifferentiation. Sci. Rep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, and Glimcher LH (2008). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science (80-.) 320, 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, and Kaestner KH (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir ED, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zheng YW, Sano Y, and Taniguchi H (2011). Evidence for mesenchymal-epithelial transition associated with mouse hepatic stem cell differentiation. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, and Jin J-P (2016). Calponin isoforms CNN1, CNN2 and CNN3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene 585, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz L, Axnick J, Buschmann T, Henning C, Urner S, Fang S, Nurmi H, Eichhorst N, Holtmeier R, Bódis K, et al. (2018). Mechanosensing by β1 integrin induces angiocrine signals for liver growth and survival. Nature 562, 128–132. [DOI] [PubMed] [Google Scholar]
- Lüdtke TH-W, Christoffels VM, Petry M, and Kispert A (2009). Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology 49, 969–978. [DOI] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerbäck S, and Carlsson P (2001). The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155–166. [DOI] [PubMed] [Google Scholar]
- Margagliotti S, Clotman F, Pierreux CE, Beaudry J-B, Jacquemin P, Rousseau GG, and Lemaigre FP (2007). The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev. Biol 311, 579–589. [DOI] [PubMed] [Google Scholar]
- Marrone G, Maeso-Díaz R, García-Cardena G, Abraldes JG, García-Pagán JC, Bosch J, and Gracia-Sancho J (2015). KLF2 exerts antifibrotic and vasoprotective effects in cirrhotic rat livers: Behind the molecular mechanisms of statins. Gut 64, 1434–1443. [DOI] [PubMed] [Google Scholar]
- Maska EL, Cserjesi P, Hua LL, Garstka ME, Brody HM, and Morikawa Y (2010). A Tlx2-Cre mouse line uncovers essential roles for hand1 in extraembryonic and lateral mesoderm. Genesis 48, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitou M, Ishikawa K, and Shiojiri N (2000). Immunohistochemical analysis of development of desmin-positive hepatic stellate cells in mouse liver. J. Anat 197, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Setty M, Kuo Y-Y, Liu V, Garg V, Sharma R, Simon CS, Saiz N, Gardner R, Boutet SC, et al. (2019). The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaria A, Angelo JR, Guertin TM, Mager J, and Tremblay KD (2018). Patterning of the hepato-pancreatobiliary boundary by BMP reveals heterogeneity within the murine liver bud. Hepatology 68, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, and Rautou PE (2017). Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J. Hepatol 66, 212–227. [DOI] [PubMed] [Google Scholar]
- Puche JE, Saiman Y, and Friedman SL (2013). Hepatic stellate cells and liver fibrosis. Compr. Physiol 3, 1473–1492. [DOI] [PubMed] [Google Scholar]
- Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, et al. (2011). ER71 directs mesodermal fate decisions during embryogenesis. Development 138, 4801–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kojima T, and Takahashi N (2012). Mab21l2 is essential for embryonic heart and liver development. PLoS One 7, e32991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwung P, Zhou G, Nayak L, Chan ER, Kumar S, Kang D-W, Zhang R, Liao X, Lu Y, Sugi K, et al. (2017). KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CD, Schledzewski K, Mogler C, Waldburger N, Kalna V, Marx A, Randi AM, Géraud C, Goerdt S, and Koch PS (2018). GPR182 is a novel marker for sinusoidal endothelial differentiation with distinct GPCR signaling activity in vitro. Biochem. Biophys. Res. Commun 497, 32–38. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA (2010). Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect. Biol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, and Kume T (2006). The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev. Biol 294, 458–470. [DOI] [PubMed] [Google Scholar]
- Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, and Pe’er D (2019). Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol 37, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Jitianu C, Cleaver O, Shaywitz DA, Lamenzo JO, Chen AE, Golub TR, and Melton DA (2007). Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev. Biol 304, 541–555. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, and Duncan SA (2010). Organogenesis and Development of the Liver. Dev. Cell 18, 175–189. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, and Oliver G (2000). Hepatocyte migration during liver development requires Prox1. Nat. Genet 25, 254–255. [DOI] [PubMed] [Google Scholar]
- Suárez-Causado A, Caballero-Díaz D, Bertrán E, Roncero C, Addante A, García-Álvaro M, Fernández M, Herrera B, Porras A, Fabregat I, et al. (2015). HGF/c-Met signaling promotes liver progenitor cell migration and invasion by an epithelial-mesenchymal transition-independent, phosphatidyl inositol-3 kinase-dependent pathway in an in vitro model. Biochim. Biophys. Acta - Mol. Cell Res 1853, 2453–2463. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Okabe M, Suzuki K, Kamiya Y, Tsukahara Y, Saito S, and Miyajima A (2009). Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: Drastic change of EpCAM expression during liver development. Mech. Dev 126, 665–676. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa M, Saito H, Tsujimura T, and Miyajima A (2003). Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J. Cell Sci 116, 1775–1786. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, and Zaret KS (2005). Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev. Biol 280, 87–99. [DOI] [PubMed] [Google Scholar]
- Vestweber D (2008). VE-cadherin: The major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler. Thromb. Vasc. Biol 28, 223–232. [DOI] [PubMed] [Google Scholar]
- Wang J, Rhee S, Palaria A, and Tremblay KD (2015). FGF signaling is required for anterior but not posterior specification of the murine liver bud. Dev. Dyn 244, 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing S, Eliades A, Lacaud G, and Kouskoff V (2012). ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev. Dyn 241, 1454–1464. [DOI] [PubMed] [Google Scholar]
- Wasser CR, and Herz J (2017). Reelin: Neurodevelopmental architect and homeostatic regulator of excitatory synapses. J. Biol. Chem 292, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt A (2003). HNF4: A central regulator of hepatocyte differentiation and function. Hepatology 37, 1249–1253. [DOI] [PubMed] [Google Scholar]
- Wei W, Lotto J, and Hoodless PA (2018). Expression patterns of Yes-associated protein 1 in the developing mouse liver. Gene Expr. Patterns 29. [DOI] [PubMed] [Google Scholar]
- Yamane A, Seetharam L, Yamaguchi S, Gotoh N, Takahashi T, Neufeld G, and Shibuya M (1994). A new communication system between hepatocytes and sinusoidal endothelial cells in liver through vascular endothelial growth factor and Flt tyrosine kinase receptor family (Flt-1 and KDR/Flk-1). Oncogene 9, 2683–2690. [PubMed] [Google Scholar]
- Yokomizo T, Hasegawa K, Ishitobi H, Osato M, Ema M, Ito Y, Yamamoto M, and Takahashi S (2008). Runx1 is involved in primitive erythropoiesis in the mouse. Blood 111, 4075–4080. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pu W, Tian X, Huang X, He L, Liu Q, Li Y, Zhang L, He L, Liu K, et al. (2016). Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet 48, 537–543. [DOI] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. (2008). Fate Tracing Reveals the Endothelial Origin of Hematopoietic Stem Cells. Cell Stem Cell 3, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cell-type annotation of single-cell libraries. Related to Figure 1.
(A) Schematic outlining collection of single-cells for sequencing and target cell types. Green, Afp-GFP+ cells; blue, dissected and dissociated tissue. (B) UMAP visualization and quality control parameters for all replicates sequenced. (C) Heatmap and UMAPs showing mutual-nearest neighbor (MNN) batch correction between data presented in this manuscript and data collected from whole embryos from Pijuan-Sala et al. (2019). Agreement between annotated clusters from both manuscripts validates annotation strategies. (D) Clustering and differential gene expression analyses identify 10, 17, 9, and 12 clusters at E7.5, E8.75, E9.5, and E10.5, respectively. Dotplots highlight select top differentially-expressed genes. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. Ant., anterior; ExE, extraembryonic; Def., definitive; STM, septum transversum mesenchyme; NMP, neuromuscular progenitors; PGC, primitive germ cells; post., posterior; rep., replicate.
Figure S2. Characterization of endothelial lineages captured from E7.5 to E10.5. Related to Figure 2.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of endothelial lineages before and after Harmony correction. (C) Distribution of embryonic timepoints across clusters, showing timing of emergence of endothelial lineages. (D) Quality control parameters for endothelial lineages, sorted by cluster. (E) Heatmap representing gene trends along pseudotime from Etv2hi hemangioblast ‘start cell’ to hematopoietic terminal state. (F) Force-directed layouts displaying expression of select genes enriched within the HSEC cluster. (G) Force-directed layouts, separated by timepoint, display normalized expression of genes associated with endothelial and hematopoietic lineages. Arrowheads indicated cells with early expression of HSEC markers Stab2 and Fcgrt. (H) Palantir results are robust when varying nearest-20 neighbor input, k. Plot show Pearson’s correlation for pseudotime, differentiation potential, and branch probability between pairs of Palantir runs. (I) Distribution of Foxa2-Kdr co-expressing cells (>1 logcounts) within complete UMAP and by cluster. (J) Re-clustering of Foxa2-Kdr co-expressing cells reveals definitive endodermal and endothelial cell states, as well as a transitional cells population. (K) Distribution of embryonic timepoints across clusters in Foxa2-Kdr co-expression subset. (L) Dot plot displaying transcriptional heterogeneity between clusters within the Foxa2-Kdr co-expression subset. Size of the dot represents proportion of the population that expresses each gene. Color indicates level of expression. DE, definitive endoderm; HSEC, hepatic sinusoidal endothelial cells; hemang., hemangioblasts; hemato., hematopoietic cells; STM, septum transversum mesenchyme; YS, yolk sac.
Figure S3. Characterization of mesenchymal lineages captured from E8.75 to E10.5. Related to Figure 3.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of mesenchymal lineages before and after Harmony correction. (C) Distribution of timepoints across clusters, showing timing of emergence of mesenchymal lineages. (D) Quality control parameters for mesenchymal lineages, sorted by cluster. (E) Volcano plot displaying top differentially-expressed genes between the mesothelial clusters identified. Genes highlighted in red represent the top 25 most differentially-expressed based on –log10(p-value). (F) Immunostaining for ALCAM and DES in E10.5 Afp-GFP livers show presence of DES+ HSCs within the (i) rostral and (ii) caudal liver parenchyma. Scale bars, 100µm - 10µm for magnified single-cells. A, anterior; HSCs, hepatic stellate cells; STM, septum transversum mesenchyme; V, ventral.
Figure S4. Characterization of the liver parenchymal progenitors captures from E7.5 to E10.5. Related to Figures 4 and 5.
(A) Heatmap highlighting select top differentially-expressed genes. (B) Force-directed layouts showing timepoint and cluster distribution of endodermal and hepatic lineages before and after Harmony correction. (C) Distribution of embryonic timepoints across clusters, showing timing of emergence of hepatic lineages. (D) Quality control parameters for hepatic lineages, sorted by cluster. (E) Distribution of AFP-GFP+ and AFP-GFP− cells sequenced at E10.5, showing contribution of AFP-GFP+ hepatic parenchymal derivatives to mesenchymal clusters. (F) FACS plots showing AFP-GFP versus the STM-enriched marker ALCAM in dissocated Afp-GFP livers from E9.5 and E10.5. Note enrichment of GFP+ALCAMhi cells at E10.5. (G) Scatterplot comparing the single-cell hepatic, epithelial, and mesenchymal profiles either by timepoint or cluster. Mesenchymal lineages included for the mesenchymal profile comparisons as a reference point. (H) Principal component analysis of average expression profiles of parenchymal and mesenchymal clusters shows close co-clustering of hepatomesenchyme with mesothelium II and STM II clusters, suggesting transcriptional parallels. (I) Immunstaining for GATA4 and VIM in E9.75 or E10.5 Afp-GFP livers shows presence of double-positive cells. Arrowheads indicate AFP-GFP+;GATA4+ or AFP-GFP+;VIM+ hepatomesenchymal cells. 100µm - 10µm for magnified single-cells. (J) Force-directed layouts displaying expression of select genes enriched within hepatomesenchymal cells. (K) Palantir results are robust when varying nearest-neighbor input, k. Plot show Pearson’s correlation for pseudotime, differentiation potential, and branch probability between pairs of Palantir runs. A, anterior; HSCs, hepatic stellate cells; R, right; STM, septum transversum mesenchyme; V, ventral.
Figure S5. Cell-cell signaling within the primitive sinusoid at E10.5. Related to Figure 6.
(A) Heatmap displaying scaled ligand-target regulatory potential for ligands highlighted in Figure 6. (B) Circle plots depicting links between (1) ligands and (2) their predicted target genes for hepatoblasts and stellate cells. Link colors indicate ligand sender populations, whereas width and opacity of links correlate to ligand-receptor interaction weights and ligand activity scores, respectively. (C) Immunostaining for RELN and PDGFRα in E9.5 or E10.5 Afp-GFP livers. RELN is shown to be an early and specific marker for emergent hepatoblasts, whereas PDGFRα is robustly expressed in stellate cell progenitors, but also weakly expressed in a subset of the AFP-GFP+ parenchymal progenitors, likely representing the hepatomesenchyme. Arrowheads indicate AFP-GFP+;RELN+ or AFP-GFP+;PDGFRα+ cells. 100µm - 10µm for magnified single-cells. A, anterior; HSECs, hepatic sinusoidal endothelial cells; V, ventral.
Table S1. Summary of single-cell libraries analyzed. Related to Figure 1.
Table S2. Differential gene expression analysis of libraries and lineage subsets. Related to Figures 1, 2, 3, and 4.
Table S3. Genes used for hepatic, epithelial, and mesenchymal gene profile analysis. Related to Figure 4.
Table S4. Complete CellPhoneDB results. Related to Figure 6.
Table S5. Target genes of interest for NicheNet analysis. Related to Figure 6.
Data Availability Statement
All single-cell RNA-sequencing data generated during this study have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-9334. The data can also be explored at https://singlecell.broadinstitute.org/single_cell/study/SCP1022/single-cell-transcriptomics-reveals-early-emergence-of-liver-parenchymal-and-non-parenchymal-cell-lineages. This study did not generate any unique code, and all analyses were performed in R and Python using standard protocols from previously published packages. All scripts associated with this manuscript are available by request.


