Abstract
To facilitate scalable profiling of single cells, we developed Split Pool Ligation-based Transcriptome sequencing (SPLiT-seq), a single-cell RNA-seq (scRNA-seq) method that labels the cellular origin of RNA through combinatorial barcoding. SPLiT-seq is compatible with fixed cells or nuclei, allows efficient sample multiplexing and requires no customized equipment. We used SPLiT-seq to analyze 156,049 single-nucleus transcriptomes from postnatal day 2 and 11 mouse brains and spinal cords. Over 100 cell types were identified, with gene expression patterns corresponding to cellular function, regional specificity, and stage of differentiation. Pseudotime analysis revealed transcriptional programs driving four developmental lineages, providing a snapshot of early postnatal development in the murine central nervous system. SPLiT-seq provides a path towards comprehensive single-cell transcriptomic analysis of other similarly complex multicellular systems.
One Sentence Summary:
Here we use SPLiT-seq, a scalable method for single-cell or single-nucleus sequencing, to profile 156,049 single nuclei from the developing mouse brain and spinal cord.
Introduction
Over three hundred years have passed since Leeuwenhoek first described living cells, yet we still do not have a complete catalogue of cell types or their functions. Recently, transcriptomic profiling of individual cells has emerged as an essential tool for characterizing cellular diversity (1–3). Single cell RNA-seq (scRNA-seq) methods have profiled tens of thousands of individual cells (4–6), revealing new insights about cell types within both healthy (7–14) and diseased tissues (15–18). Unfortunately, since these methods require cell sorters, custom microfluidics, or microwells, throughput is still limited and experiments are costly. We introduce SPLiT-seq, a low-cost, scRNA-seq method that enables transcriptional profiling of hundreds of thousands of fixed cells or nuclei in a single experiment. SPLiT-seq does not require partitioning single cells into individual compartments (droplets, microwells or wells), but relies on the cells themselves as compartments. The entire workflow before sequencing consists just of pipetting steps and no complex instruments are needed.
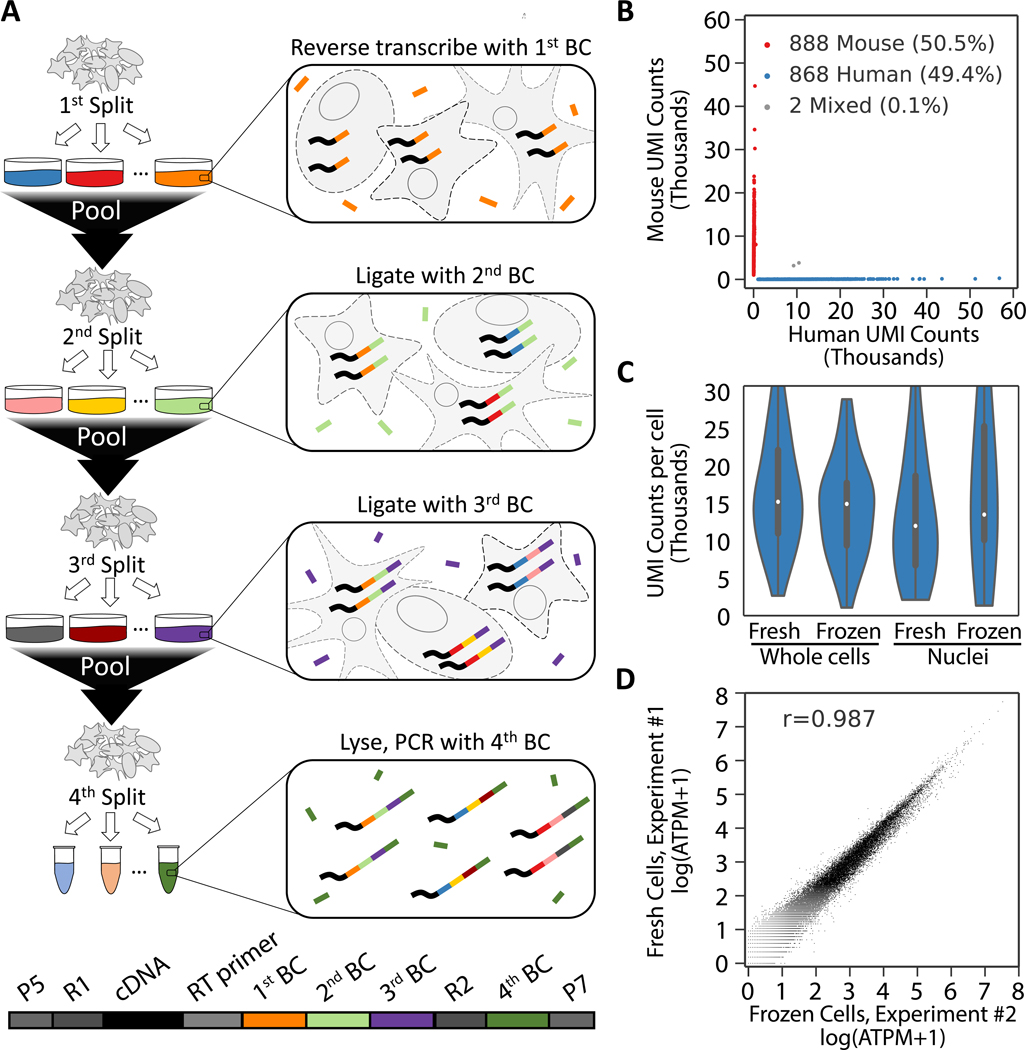
In SPLiT-seq, individual transcriptomes are uniquely labeled by passing a suspension of formaldehyde fixed cells or nuclei through four rounds of combinatorial barcoding. In the first round of barcoding, cells are distributed into a 96-well plate and cDNA is generated with an in-cell reverse transcription (RT) reaction using well-specific barcoded primers. Each well can contain a different biological sample – thereby enabling multiplexing of up to 96 samples in a single experiment. After this step, cells from all wells are pooled and redistributed into a new 96-well plate, where an in-cell ligation reaction appends a second well-specific barcode to the cDNA. The third-round barcode, which also contains a unique molecular identifier (UMI), is then appended with another round of pooling, splitting, and ligation. After three rounds of barcoding, the cells are pooled, split into sublibraries, and sequencing barcodes are introduced by PCR. This final step provides a fourth barcode, while also making it possible to sequence different numbers of cells in each sublibrary. After sequencing, each transcriptome is assembled by combining reads containing the same four-barcode combination (Fig. 1A, Fig. S1A).
Fig. 1. Overview of SPLiT-seq.
(A) Labeling transcriptomes with split-pool barcoding. In each split-pool round, fixed cells or nuclei are randomly distributed into wells and transcripts are labeled with well-specific barcodes. Barcoded RT primers are used in the first round. Second and third round barcodes are appended to cDNA through ligation. A fourth barcode is added to cDNA molecules by PCR during sequencing library preparation. The bottom scheme shows the final barcoded cDNA molecule. (B) Species mixing experiment with a library prepared from 1,758 whole cells. Human UBCs are blue, mouse UBCs are red, and mixed-species UBCs are gray. The estimated barcode collision rate is 0.2%, whereas species purity is >99%. (C) UMI counts from mixing experiments performed with fresh and frozen (stored at −80°C for 2 weeks) cells and nuclei. Median human UMI counts for fresh cells: 15,365; frozen cells: 15,078; nuclei: 12,113; frozen nuclei: 13,636. (D) Measured gene expression by SPLiT-seq is highly correlated between frozen cells and cells processed immediately (Pearson-r: 0.987). Frozen and fresh cells were processed in two different SPLiT-seq experiments.
Four rounds of combinatorial barcoding can yield 21,233,664 barcode combinations (three rounds of barcoding in 96-well plates followed by a fourth round with 24 PCR reactions) - enough to uniquely label over 1 million cells. Even larger numbers of barcode combinations can be achieved by performing experiments in 384-well plates or through additional rounds of barcoding (Fig. S1B). In addition, by performing the first step in a 384-well plate, up to 384 different biological samples could be combined in a single experiment.
SPLiT-seq validation
To test SPLiT-seq’s ability to generate uniquely barcoded cells (UBCs), we performed a species-mixing experiment. We mixed cells from one mouse and two human cell lines (NIH/3T3, HEK293, and Hela-S3), fixed them, and used SPLiT-seq to generate a scRNA-seq library with 1,758 UBCs. The library was sequenced and reads were aligned to a combined mouse-human genome. 99.9% of the UBCs were unambiguously assigned to a single species (>90% of reads aligned to a single genome) with the remaining 0.1% of UBCs representing barcode collisions between mouse and human cells (Fig. 1B). At saturating read coverage (>500,000 reads per cell), we identified a median of 15,365 UMIs and 5,498 genes per human cell and 12,243 UMIs and 4,497 genes per mouse cell. The species purity in both human and mouse UBCs was high: 99.6% of reads in human UBCs and 99.0% of reads in mouse UBCs aligned to their respective genomes. We repeated SPLiT-seq experiments with freshly prepared nuclei as well as nuclei and cells that had been preserved at −80°C for two weeks. In all samples, we detected similar numbers of transcripts and genes per cell (Fig. 1C, Fig. S2, Table S1). Gene expression was highly correlated between preserved and freshly prepared cells (Fig. 1D, Fig. S2, Pearson-r: 0.987) as well as between cells and nuclei (Fig. S2, Pearson-r: 0.952). We also examined gene and UMI detection at different sequencing depths and found that the sensitivity of SPLiT-seq is comparable to droplet-based scRNA-seq methods (Fig. S3).
snRNA-seq of developing mouse brain and spinal cord
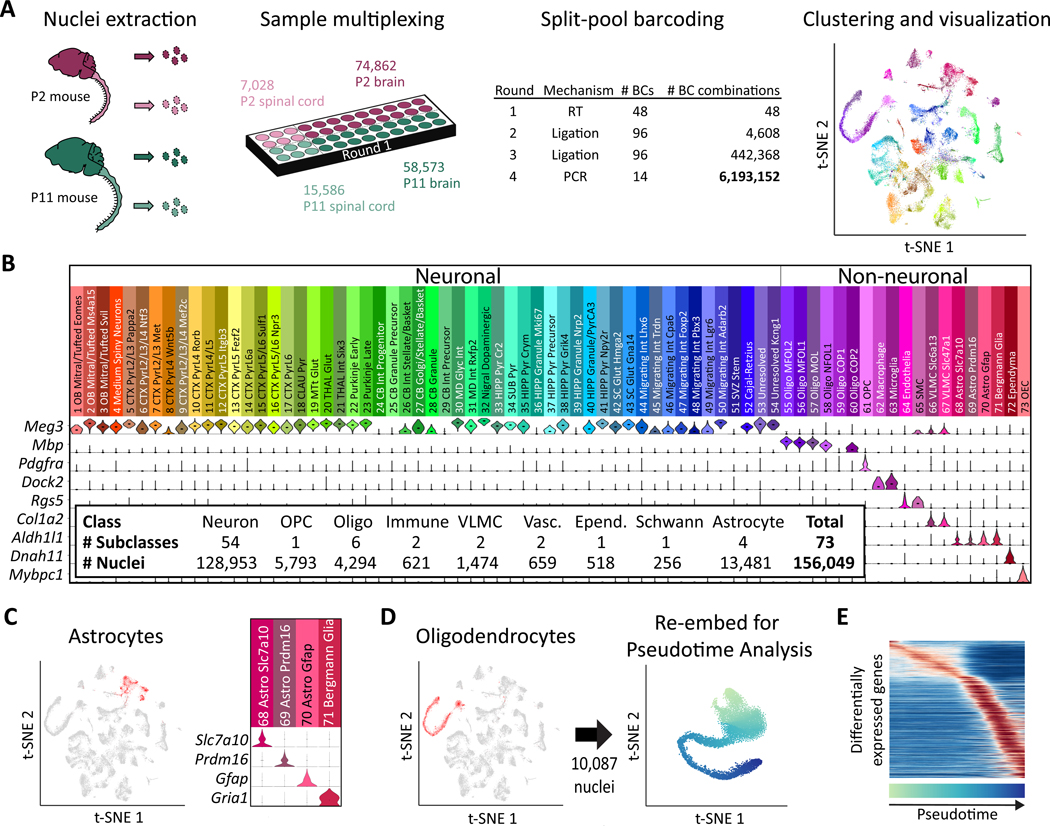
We used SPLiT-seq to profile nuclei from the developing brain and spinal cord of postnatal day 2 and 11 (P2 and P11) mice. The first round of barcoding assigned identifiers for the P2 brain, P2 spinal cord, P11 brain, and P11 spinal cord samples (Fig. 2A, Fig. S4). In total, four rounds of barcoding (48 × 96 × 96 × 14) generated over 6 million distinct barcode combinations, making it possible to process hundreds of thousands of nuclei in a single experiment with minimal barcode collisions (2.5% expected collisions for 150,000 nuclei).
Fig. 2. Single-cell transcriptome landscape of postnatal brain and spinal cord development by SPLiT-seq.
(A) Over 150,000 nuclei from P2 and P11 mouse brains and spinal cords were profiled in a single experiment employing over six million barcode combinations. Transcriptomes were clustered and then visualized using t-SNE. Cells are colored according to cell type. Each cluster was downsampled to 1,000 cells for visualization. (B) A total of 73 distinct clusters were assigned to nine cell classes based on expression of established markers. The violin plots show marker gene expression in each cluster. (C) Astrocyte clusters are highlighted in red in the t-SNE. The violin plots show markers that are differentially expressed between astrocyte subtypes. (D) Seven OPC and oligodendrocyte clusters (containing 10,087 nuclei) colocalized in the original t-SNE (highlighted in red), forming a lineage. Cells from these clusters were re-embedded with t-SNE. (E) The heatmap shows genes expressed differentially across pseudotime in the oligodendrocyte lineage.
To test how many transcripts SPLiT-seq detects within nuclei from the central nervous system, we performed deep sequencing on a sublibrary containing only 131 nuclei. We detected 4,943 UMIs and 2,055 genes per nucleus (UMI duplication: 95%). We then sequenced the rest of the library at lower depth, resulting in a median of 677 genes and 1,022 UMIs per nucleus (UMI duplication: 58%) (Table S2). Low-quality transcriptomes were removed from analysis (19), yielding 156,049 single-nucleus transcriptomes (74,862 P2 brain; 7,028 P2 spinal cord; 58,573 P11 brain; 15,586 P11 spinal cord).
Unsupervised clustering grouped transcriptomes into 73 distinct clusters (19) (Tables S3–S5), which were visualized by t-Distributed Stochastic Neighbor Embedding (t-SNE, Fig. 2A). Each of these 73 clusters was assigned to a cell class on the basis of expression of established marker genes (Fig. 2B). Neurons accounted for 79% of the profiled transcriptomes (54 clusters), with most clusters expressing Meg3.
The 27,096 non-neuronal transcriptomes spanned 19 different clusters, each assigned to a specific cell type. Four astrocyte types (Fig. 2C) accounted for 50% of all non-neuronal nuclei (n=13,481). Oligodendrocytes (6 types, n=4,294) and oligodendrocyte precursor cells (OPC, 1 type, n=5,793) formed the second most abundant population. We further identified two vascular and leptomeningeal cell (VLMC) types (Fig. S5A), endothelial cells, smooth muscle cells (Fig. S5B), microglia, macrophages (Fig. S5C) (20, 21), ependymal cells, and olfactory ensheathing cells.
Previous work has observed that t-SNE can order cells in 2D space according to stages of differentiation (9). Moving through t-SNE space along the path of differentiation can then be viewed as moving through “pseudotime”(22). As oligogenesis spans the first two postnatal weeks of murine development (23), we asked whether the oligodendrocyte and OPC clusters might reflect a continuous developmental trajectory. When we examined the oligodendrocyte clusters, we found that they formed an overlapping elongated shape in the t-SNE visualization. OPCs and oligodendrocytes from the P2 mouse, were enriched at one end of the structure while oligodendrocytes from the P11 mouse were enriched at the opposite end (Fig. S6), indicative of a lineage (19, 22).
We then performed a more thorough analysis of this putative lineage. To ensure that our ordering of oligodendrocytes was determined exclusively by their relationship to other oligodendrocytes, rather than all cells, we re-embedded only transcriptomes within these seven clusters with t-SNE (Fig. 2D, Fig. S7A). We calculated the moving average of gene expression in the resulting pseudotime ordering (Fig. 2E, Fig. S8). Analysis of these expression patterns confirmed that proliferating OPCs segregated to one end of the t-SNE, whereas mature oligodendrocytes segregated to the opposite end (Fig. S7B). We also detected previously reported intermediate stages of oligodendrocyte development, with the order of gene expression across pseudotime nearly identical to the one defined previously (9) (Fig. S7C, Spearman-r: 0.94). When analyzing spinal cord and brain derived cells separately, we found more mature oligodendrocytes in the spinal cord than in the brain (Fig. S7D), indicating that oligodendrocyte maturation occurs earlier in the spinal cord.
Neuronal Cell Types
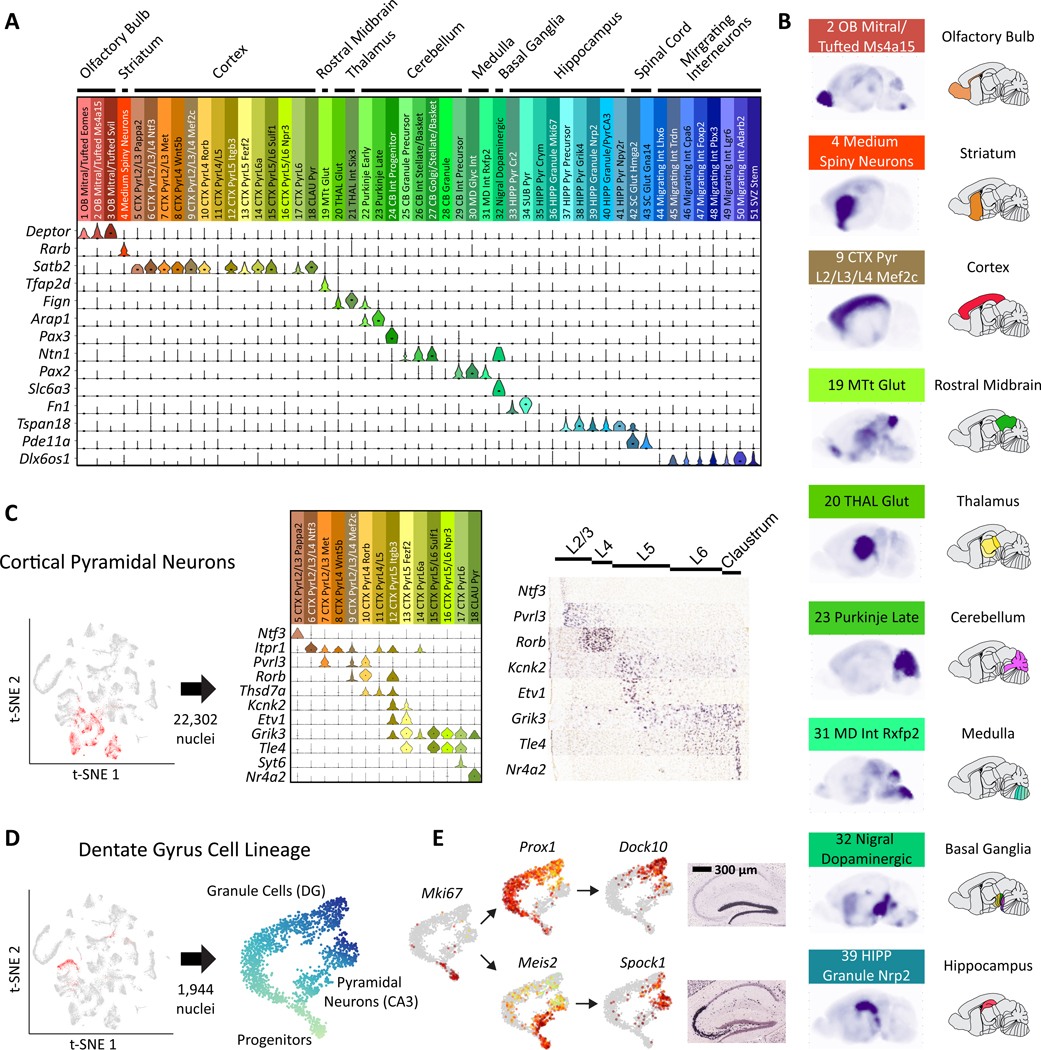
Using known gene markers, we were able to assign most neuronal clusters to specific cell types (19). While some clusters corresponded to abundant cell types, such as cerebellar granule cells (CGCs), others mapped to rare and often less characterized cell types, such as mitral/tufted cells. Previously characterized regional markers were used to assign the majority of clusters to a specific region of the brain (24) (Fig. 3A). Regional assignments were validated with RNA in situ hybridization (ISH) from the Allen Institute’s Developing Mouse Brain Atlas (Allen DMBA) (25). Specifically, we generated composite ISH maps by averaging across the five most highly enriched genes from each of our clusters (Tables S6, S7). For clusters primarily containing P2 or P11 nuclei, we used the P4 or P14 atlases, respectively. The resulting composite maps confirmed the high regional specificity of most types (Figs. 3B, S9 and S10). Cortical pyramidal neuronal types could be further assigned to specific layers using marker genes (Fig. 3C) (7, 8).
Fig. 3. Neuronal clusters exhibit regional specificity.
(A) Marker gene expression was used to map neuronal clusters to specific brain regions. (B) Sagittal composite RNA ISH maps for nine representative clusters from distinct areas. For each cell type, we averaged ISH intensities from the Allen DMBA across the top five differentially expressed genes. (C) Types of pyramidal neurons in the cortex display layer-specific enrichments according to marker genes: cortical pyramidal neurons are highlighted in red in the t-SNE. Expression of example marker genes in pyramidal clusters is shown in the middle and corresponding available RNA ISH results on the right. (D) Three clusters constitute a developmental trajectory in the hippocampus. Re-embedding these clusters highlights the branching of the two differentiation trajectories in pseudotime. (E) Expression of differentiation marker genes is overlaid on the t-SNE. RNA ISH maps (Allen DMBA) show the regional specificity of granule cell and pyramidal neuron markers.
Granule cell fate in the hippocampus
In the hippocampus, immature granule cells in the dentate gyrus give rise not only to mature granule cells, but also to pyramidal neurons (26). This process is one of two instances of neurogenesis that continues into adulthood (27), but little is known about the underlying transcriptional program. We determined that three neuronal cell types from the hippocampus likely constituted a developmental trajectory (19). Analysis of only these transcriptomes with t-SNE revealed a clear branching structure (Fig. 3D, Fig. S11A). The transcription factor Prox1, suspected to be necessary for granule cell identity (28), was exclusively expressed in one branch, whereas genes known to be specific to CA3 pyramidal neurons such as Spock1 (29) were expressed exclusively in the other branch. Markers of dividing neuronal progenitors were expressed before the branching point and genes in the Slit-Robo signaling pathway were differentially expressed between the two lineages (Fig. S11B). We used these data to identify specific temporal dynamics of transcription factors across the two lineages, with Meis2 as a candidate marker of early pyramidal cell differentiation (Fig. 3E, Fig. S12).
Profiling cells in the developing cerebellum
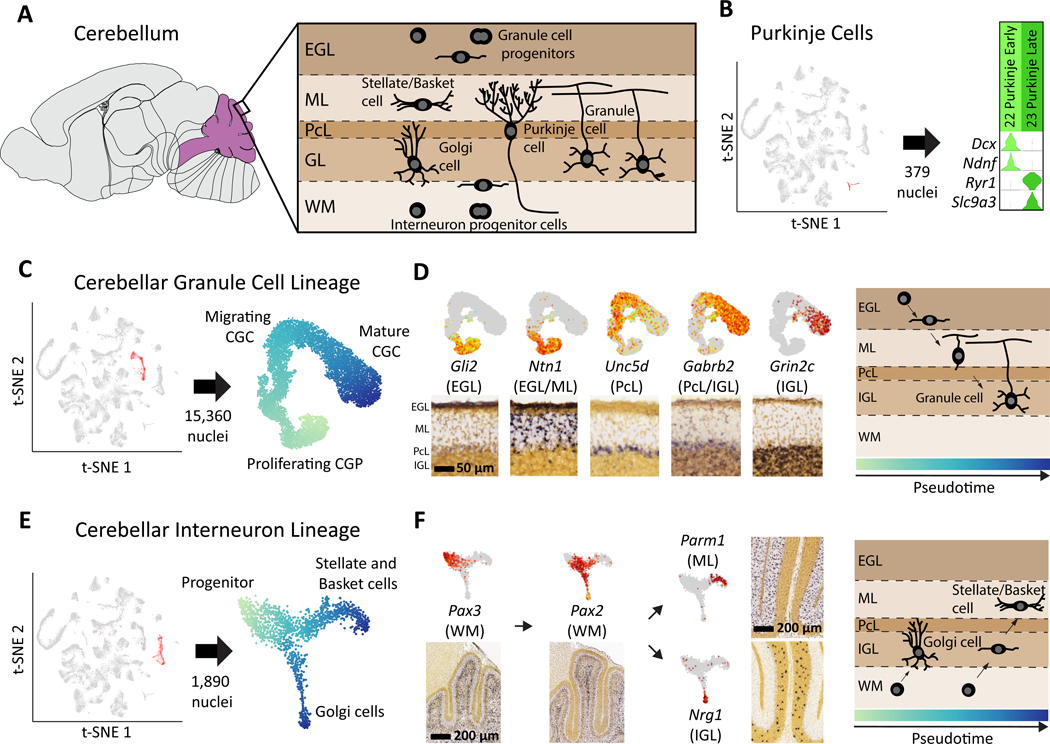
The cerebellum accounts for only 9% of the brain mass in adult mice, but contains nearly 85% of all neurons (30). Despite the wide range of functions performed by the cerebellum, many of the gene expression programs driving development of cerebellar cell types remain unknown. We identified the four main cerebellar neuronal types (Fig. 4A): Purkinje cells, Golgi cells, stellate/basket cells, and CGCs. Two types of Purkinje cells (Fig. 4B) were segregated primarily by age (P2 vs P11), and did not form a continuous trajectory in t-SNE but rather two clearly segregated clusters. The absence of cells at intermediate stages of maturation suggests that Purkinje cell development may be more synchronous than other processes of neurogenesis captured by our dataset.
Fig. 4. Neuronal differentiation trajectories in the cerebellum revealed by SPLiT-seq.
(A) Major cell types and their locations in the cerebellum. (B) Two types of Purkinje cells with distinct gene expression programs were identified. Early Purkinje cells are primarily found in the P2 brain and late Purkinje cells in the P11 brain. (C) t-SNE re-embedding of 15,360 nuclei suggests a pseudotime ordering from proliferating, to migrating, to mature CGCs. (D) Expression of marker genes is overlaid on the t-SNE, and the corresponding RNA ISH from Allen DMBA is shown below. Marker genes associated with different layers of the cerebellum are expressed at different points in pseudotime. Gene expression order is consistent with ordering of the physical layers. RNA ISH maps confirm regional specificity of marker genes. (E) t-SNE re-embedding of 1,890 nuclei reveals a branching differentiation trajectory. Progenitors can either become Golgi cells or stellate/basket cells. (F) Markers for progenitors and mature cell types are expressed at different points in pseudotime and have layer specificity.
CGCs, the most numerous type of neuron in the brain (31), drive the postnatal foliation of the cerebellar cortex by migrating from the external granule layer (EGL), through the molecular layer (ML) and the Purkinje cell layer (PcL) to the internal granule layer (IGL) (32, 33). We created a pseudotime ordering of 15,360 CGCs (Fig. 4C, Fig. S13) and measured gene expression across this lineage. We defined genes with specific expression at different points in pseudotime (Fig. S14), and then used RNA ISH to map these genes to layers of the developing cerebellar cortex. Genes ordered from early-to-late in pseudotime were progressively expressed from outer-to-inner layers, consistent with the known direction of CGC migration (Fig. 4D). Our analysis revealed previously unknown pseudotime and layer specific gene expression patterns within pathways related to axonal development and neuronal migration (Fig. S15).
Origins of cerebellar inhibitory interneurons
The question of whether all cerebellar inhibitory interneurons arise from the same progenitor population has been a point of contention (34). Early hypotheses proposed that stellate/basket cells originated from precursors in the EGL, whereas Golgi cell precursors resided in the ventricular epithelium (35). Later evidence indicated that these two interneurons shared a common precursor in the cerebellar white matter (36, 37). However, the molecular profile of the inhibitory neuron lineage in the cerebellum remains largely unknown.
We found a cerebellar inhibitory interneuron lineage (1,517 cells, Fig. 4E, Fig. S16A) with a shared progenitor branching into either Golgi or stellate/basket cells (Fig. S17). This lineage includes a known precursor cell type expressing Pax2 (36), but also a previously unknown, earlier precursor expressing Pax3 (Fig. 4F). RNA ISH analysis suggests that this Pax3+ precursor is located deep within the cerebellar white matter. Moreover, we found that stellate/basket cells expressed genes specific to the molecular layer, whereas Golgi cells expressed genes specific to the granule cell layer (Fig. 4F, Fig. S18). The distribution of P2 and P11 nuclei within the lineage clearly demonstrated that the maturation of Golgi cells was well underway by P2 and complete by P11 (Fig. S16B). In contrast, stellate/basket cells had not begun to differentiate at P2 and were still not fully mature by P11. These results indicate that the same molecularly defined precursor gives rise to two distinct interneurons at different stages of development.
Cell types in the developing spinal cord
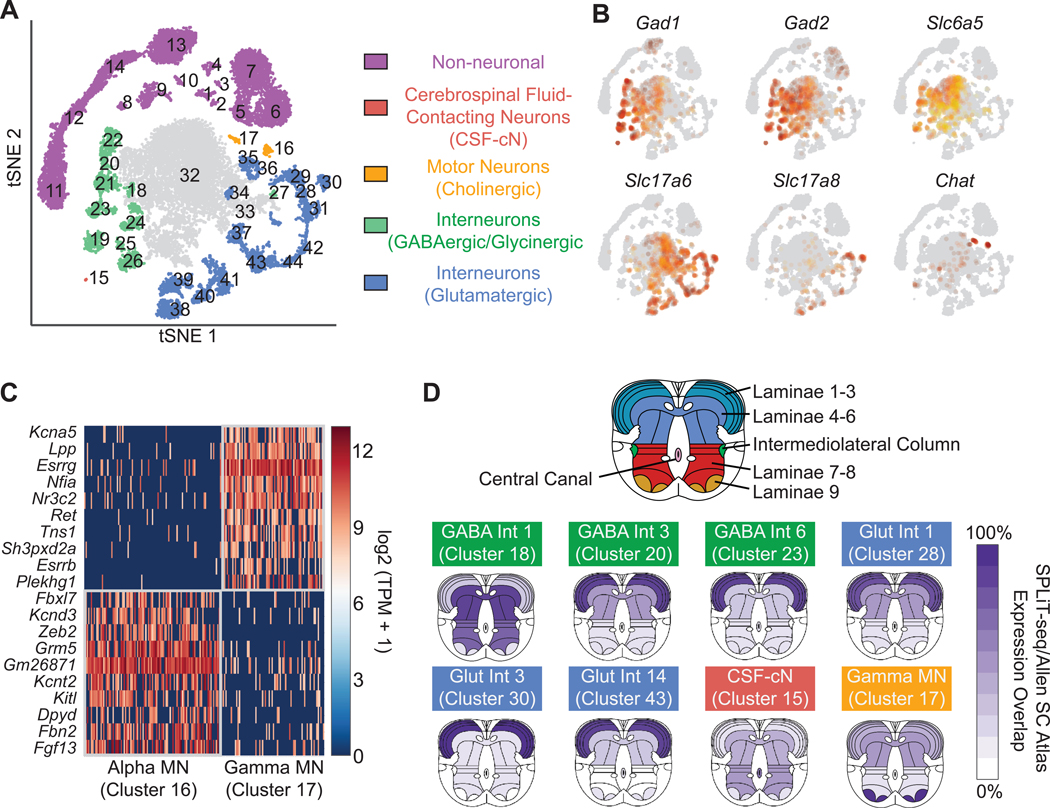
The original clustering was dominated by cells in the brain, and many spinal cord cells did not segregate into well-defined clusters (Fig. S19). To resolve more cell types in the spinal cord, we selected all the nuclei originating from the spinal cord and re-clustered them (19), resulting in 44 clusters: 14 non-neuronal types (12 of which were also found in the brain) and 30 neuronal types (Fig. 5A and Tables S8–S10). We identified 11 different types of GABAergic neurons, of which several were also glycinergic (Fig. 5B). One GABAergic type was identified as cerebrospinal fluid-contacting neurons (CSF-cNs) (38), with the other ten types corresponding to inhibitory interneurons. Glutamatergic interneurons accounted for 15 additional types. We also identified two clusters of cholinergic motor neuron types (alpha and gamma) (39). To date, known markers exist only for gamma motor neurons (Esrrg and Esrrb) (40), however, we identified specific markers for both alpha and gamma neurons (Fig. 5C).
Fig. 5. Gene expression patterns and spatial origin of cell types in the spinal cord.
(A) Re-clustering spinal cord nuclei resulted in 30 neuronal and 14 non-neuronal clusters. (B) GABAergic neurons were defined by expression of Gad1 and Gad2. A subset of GABAergic neurons are also glycinergic, based on expression of Slc6a5. Glutamatergic neurons were defined by expression of VGLUT1 (Slc17a6), whereas cholinergic motor neurons express Chat. (C) Novel gene markers distinguish gamma motor neurons from alpha motor neurons. (D) Inferred spatial origin of neuronal clusters within the spinal cord. We analyzed the Allen Spinal Cord Atlas expression patterns of the top ten enriched genes in each cluster. Dark purple indicates expression of all ten genes in the given region, while white indicates none of the ten genes were expressed in the given region.
To infer the spatial origin of neuronal types in the spinal cord, we identified the ten most enriched genes in each type according to our snRNA-seq data and created composite ISH maps based on the Allen Mouse Spinal Cord Atlas (41) (Fig. 5D, Fig. S20). Some interneuron subtypes appeared to originate primarily from laminae 1–3, with others originating from laminae 4–6. We found both inhibitory and excitatory neurons in each region. Motor neurons expressed genes found in laminae 9, while CSF-cNs were the only neuronal type expressing genes found in the central canal. These data allowed us to create an atlas of gene expression in the early spinal cord, providing a rich resource for further understanding development of the central nervous system.
Discussion
In this work, we profiled hundreds of thousands of cells using only basic laboratory equipment with a library preparation cost of ~$0.01 per cell (Fig. S21, Table S11). In our analysis of more than 150,000 single-nucleus transcriptomes from two early postnatal stages, we identified 69 types of cells in the brain and 44 types in the spinal cord. We defined many new molecular markers for specific cell types, and explored gene expression in four different developmental lineages.
SPLiT-seq’s compatibility with fixed cells and fixed nuclei overcomes challenges faced by other scRNA-seq methods. Fixation can reduce perturbations to endogenous gene expression during cell handling (42) and makes it possible to store cells for future experiments. Moreover, the use of nuclei bypasses the need to obtain intact single cells, which can be challenging for many complex tissues. SPLiT-seq’s compatibility with formaldehyde-fixed nuclei suggests it may be used to profile single nuclei from formalin-fixed, paraffin-embedded tissue (43).
SPLiT-seq enables flexible and scalable cell and sample multiplexing. The use of the first-round barcode as a sample identifier makes it possible to profile a large number and variety of samples in parallel, thus minimizing batch effects. As the number of unique barcodes grows exponentially with the number of barcoding rounds, larger numbers of cells than presented here could be processed by adding a fifth barcoding round or by switching to a 384-well plate format. Although for such large cell numbers sequencing cost may currently be forbidding, it is easy to imagine extended applications, such as targeted sequencing of gene panels, which would even now benefit from very large cell numbers and only require shallow sequencing depth.
Our hope is that the increased scale and accessibility provided by the low cost and minimal equipment requirements of SPLiT-seq will further accelerate the widespread adoption of scRNA-seq.
Supplementary Material
Acknowledgments:
We would like to thank Thuc Nghi Nguyen for help with cluster identity assignment. All relevant sequencing files will be deposited to Sequence Read Archive (SRA). Processed data will be submitted to GEO. This work was supported by NIH R01CA207029 and NSF CCF-1317653 to GS and NIH R01NS064404 and R21NS086500 to SHP. ZY, LG and BT were supported by the Allen Institute for Brain Science. CR was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number TL1 TR002318.
References and Notes:
- 1.Picelli S et al. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Hashimshony T, Wagner F, Sher N, Yanai I, CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Jaitin DA et al. , Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 343, 776–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macosko EZ et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein AM et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 161, 1187–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng GXY et al. , Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasic B et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci 19, 335–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeisel A et al. , Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347, 1138–42 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Marques S et al. , Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 352, 1326–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darmanis S et al. , A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U. S. A 112, 7285–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lake BB et al. , Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 352, 1586–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalek AK et al. , Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 498, 236–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grün D et al. , Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 525, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Moignard V et al. , Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat. Biotechnol 33, 269–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venteicher AS et al. , Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science (80-. ). 355 (2017) (available at http://science.sciencemag.org/content/355/6332/eaai8478/tab-pdf). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirosh I et al. , Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352, 189–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang L et al. , Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 321, 1095–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng C et al. , Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169, 1342–1356.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Materials and methods are provided as supplementary materials.
- 20.Hickman SE et al. , The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci 16, 1896–905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matcovitch-Natan O et al. , Science, in press, doi: 10.1126/science.aad8670. [DOI] [Google Scholar]
- 22.Trapnell C et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levison SW, Goldman JE, Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 10, 201–12 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Lein ES et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Allen Institute for Brain Science, Developing Mouse Brain Atlas (2008), (available at http://developingmouse.brain-map.org/).
- 26.Iwano T, Masuda A, Kiyonari H, Enomoto H, Matsuzaki F, Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development. 139, 3051–62 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Zhao C, Deng W, Gage FH, Mechanisms and Functional Implications of Adult Neurogenesis. Cell. 132 (2008), pp. 645–660. [DOI] [PubMed] [Google Scholar]
- 28.Lavado A, Lagutin OV, Chow LML, Baker SJ, Oliver G, Prox1 Is Required for Granule Cell Maturation and Intermediate Progenitor Maintenance During Brain Neurogenesis. PLoS Biol. 8, e1000460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnet F et al. , Structure and cellular distribution of mouse brain testican. Association with the postsynaptic area of hippocampus pyramidal cells. J. Biol. Chem 271, 4373–80 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Herculano-Houzel S, The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci 3, 31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakashima K, Umeshima H, Kengaku M, Cerebellar granule cells are predominantly generated by terminal symmetric divisions of granule cell precursors. Dev. Dyn 244, 748–758 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Sudarov A, Joyner AL, Cerebellum morphogenesis: the foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2, 26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JC et al. , Mitotic Events in Cerebellar Granule Progenitor Cells That Expand Cerebellar Surface Area Are Critical for Normal Cerebellar Cortical Lamination in Mice. J. Neuropathol. Exp. Neurol 74, 261–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schilling K, Oberdick J, Rossi F, Baader SL, Besides Purkinje cells and granule neurons: an appraisal of the cell biology of the interneurons of the cerebellar cortex. Histochem. Cell Biol. 130, 601–615 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Altman J, Bayer SA, Development of the precerebellar nuclei in the rat: I. The precerebellar neuroepithelium of the rhombencephalon. J. Comp. Neurol 257, 477–489 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Maricich SM, Herrup K, Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J. Neurobiol 41, 281–94 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Weisheit G et al. , Postnatal development of the murine cerebellar cortex: formation and early dispersal of basket, stellate and Golgi neurons. Eur. J. Neurosci 24, 466–478 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Petracca YL et al. , The late and dual origin of cerebrospinal fluid-contacting neurons in the mouse spinal cord. Development. 143, 880–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enjin A et al. , Identification of novel spinal cholinergic genetic subtypes disclose Chodl and Pitx2 as markers for fast motor neurons and partition cells. J. Comp. Neurol 518, 2284–2304 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Lalancette-Hebert M, Sharma A, Lyashchenko AK, Shneider NA, Gamma motor neurons survive and exacerbate alpha motor neuron degeneration in ALS. Proc. Natl. Acad. Sci. U. S. A 113, E8316–E8325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen Institute for Brain Science, Allen Mouse Spinal Cord Atlas (2008), (available at http://mousespinal.brain-map.org/imageseries/showref.html).
- 42.Lacar B et al. , Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun 7, 11022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen ER et al. , Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods 13, 87–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.