Abstract
Oriented sample solid state NMR (OS-ssNMR) spectroscopy allows direct determination of the structure and topology of membrane proteins reconstituted into aligned lipid bilayers. While OS-ssNMR theoretically has no upper size limit, its application to multi-span membrane proteins has not been established since most studies have been restricted to single or dual span proteins and peptides. Here, we present a critical assessment of the application of this method to multi-span membrane proteins. We used molecular dynamics simulations to back-calculate [15N-1H] separated local field (SLF) spectra from a G protein-coupled receptor (GPCR) and show that fully resolved spectra can be obtained theoretically for a multi-span membrane protein with currently achievable resonance linewidths.
Solid-state NMR (ssNMR) spectroscopy is the method of choice for obtaining high-resolution structural and dynamical information of membrane proteins embedded in native-like liquid crystalline lipid bilayers.[1] Directionally-dependent dipolar couplings (DCs) and chemical shift anisotropy (CSA), which broaden signals in conventional solution NMR methods, are either averaged out by magic angle spinning (MAS) techniques or directly measured using oriented sample (OS) methods for structural restraints.[2–6] MAS-ssNMR mostly removes these interactions by mechanically spinning samples in a rotor about an angle of 54.74° at high frequency (currently over 100 kHz) to resolve isotropic chemical shifts (CSs) with solution-like resolution. While increasingly popular, the application of MAS-ssNMR to membrane proteins has been limited primarily by low sensitivity as only a few microliters can be packed into high-speed rotors, much of which is composed by water and lipid. Furthermore, poorly-dispersed isotropic chemical shifts arising from transmembrane (TM) α-helical residues – a common issue for membrane protein NMR in general – often leads to poor resolution. OS-ssNMR, on the other hand, achieves comparable resolution by homogenously orienting membrane proteins by either reconstitution into supported lipid bilayers (SLBs) or into systems that spontaneously align with magnetic fields such as high-q bicelles[7] or macrodiscs.[8–9] Additionally, the anisotropic chemical shift tensor[10] greatly enhances CS dispersion (and resolution), while sample volumes upwards of 100 μL allows for milligram quantities of protein to be studied under physiological relevant lipid-protein ratios and hydration levels. An outstanding example was the use of OS-ssNMR spectroscopy to characterize the β-helical antibiotic channel-forming peptide Gramicidin A in lipid bilayers by Separovic and Cornell[11–13] and later on the determination of its high-resolution structure by Cross and coworkers.[14–15] OS-ssNMR has since advanced with improvements in theory, software and hardware such as the introduction of multidimensional separated local field (SLF) experiments,[16–20] sensitivity-enhanced pulse sequences,[21–22] simultaneous acquisition schemes,[23] low-E probes,[24] paramagnetic relaxation enhancement,[25] structural calculation methods,[26–33] higher magnetic fields[34] and dynamic nuclear polarization.[35–38] As a result, OS-ssNMR has been central to several notable structural studies including the M2 segments of the acetylcholine and N-methyl-D-aspartate (NMDA) receptors;[39] the filamentous bacteriophage fd coat protein;[40] the HIV channel-forming Vpu protein;[41–42]; the bacterial MerF mercury transporter;[43–45] the Influenza A M2 proton channel;[46] the β-barrel outer membrane protein OmpX from Escherichia coli;[47] membrane-anchored cytochrome P450 metabolic enzymes;[48] the cardiac regulatory protein phospholamban;[49–50] the tuberculosis CrgA protein involved in cell division;[51] the TM domains of the adaptive immune system’s MHC II proteins;[52–53] the TM domain of the p24 protein involved in vesicle sorting and transport;[54] the Huntington 1–17 membrane anchor;[55] cell-penetrating peptides;[56–57] and recently an in-depth characterization into the dynamics of the dimeric triple-pass bacterial EmrE drug transporter[58]. Despite its potential, difficult sample preparation and low resolution have generally limited OS-ssNMR to studies of peptides and small proteins. Recently, however, linewidths as narrow as 50 Hz are starting to be reported with the introduction of macrodiscs.[9] Prompted by these advancements, we have devised a new method to test the feasibility of OS-ssNMR spectroscopy for multi-pass membrane proteins using computational approaches.
To assess this, we carried out molecular dynamics (MD) simulations and calculated the theoretical [15N-1H] SLF spectrum of the seven transmembrane (TM) β2-adrenergic G-protein-coupled receptor (GPCR; PDB 3P0G)[59] from an MD trajectory. SLF spectra correlate 15N CSs with 15N-1H DCs of amide residues and are commonly acquired on uniformly 15N-labelled protein using PISEMA or SAMPI4 pulse sequences.[16, 18, 22] Unlike isotropic CSs observed in solution NMR and MAS-ssNMR, which are positioned according to electronic shielding or deshielding of nuclei induced by the surrounding chemical environment, anisotropic CSs in OS-ssNMR directly relate to time-averaged orientations of the chemical shift tensor components (, , and ) with respect to the magnetic field vector () and can be analyzed over an ensemble of MD snapshots similar to previously reported methods.[60] For each frame, the tensor components were projected onto the amide plane of each residue (see Figure 1A), with and lying in-plane and a rotation of by 17° (polar angle β) away from the N-H bond vector ().[61] The normalized time-averaged orientations of each component with respect to (Z-axis of MD simulation) were used to find the 15N CS (δMD) (Eqn 1):
| (1) |
where δ11, δ22 and δ33 were 57.3, 81.2 and 228.1 ppm, respectively.[62] Angular brackets denote time-averages. Correct determination of these values,[63] along with proper chemical shift calibration,[64] will strongly impact the agreement between experimental and MD-predicted spectra. Experimentally, however, the observed CS (δ) is also scaled by per-residue order parameters (SNH), the order of the alignment medium (Smemb),[41] rigid-body dynamics of the protein (Sprotein),[65–67] and the angle of the membrane normal with respect to (θmemb). The 15N CS may then be adjusted according to Eqn 2:
| (2) |
where δiso = (δ11 + δ22 + δ33)/3. Salign is a user-defined generalised order parameter to account for scaling due to Sprotein and Smemb, which may not be adequately sampled by the simulation. Like lipid C-2H order parameters,[68] the SNH parameter for each residue was estimated from Z-axis fluctuation of the amide N-H bond vector () away from its time-averaged orientation, or director axis (), and assuming fast-rotational diffusion, according to Eqn 3:
| (3) |
Consequently, disorder due to Sprotein is implicit in SNH if rigid-body (rocking) motions of the membrane protein are unrestrained during the MD simulation. These motions, however, are usually slow on MD simulation timescales (100–1000s ns) and restraining them, or aligning trajectories to starting coordinates, is recommended for faster convergence of CS and DC values.
Figure 1:
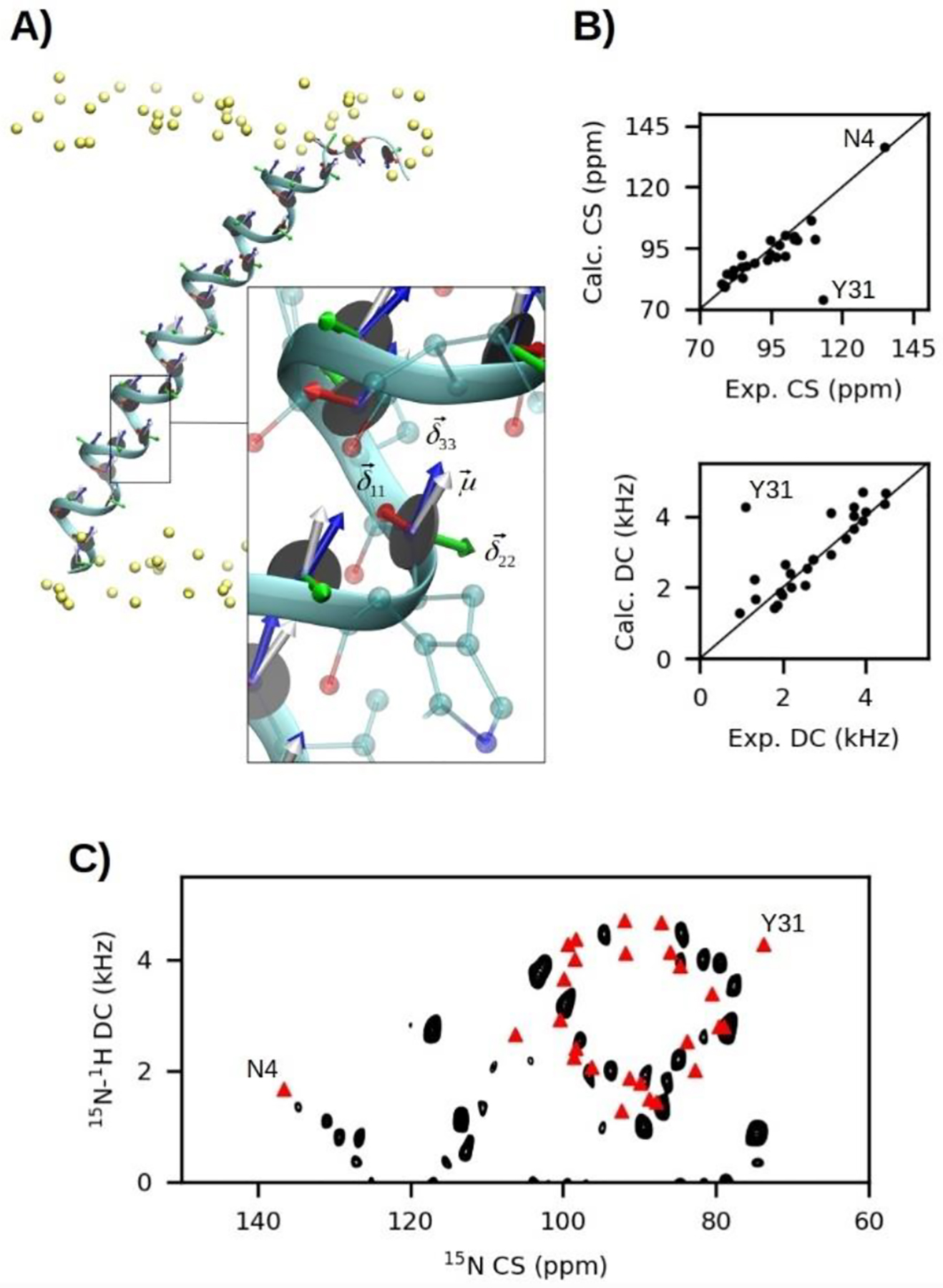
A) MD simulation snapshot of SLN embedded in a lipid bilayer with CS tensor components and the DC vectors projected onto the amide plane of each residue. B) Correlation plots between MD-derived and experimental CS and DC values. C) MD-derived CS-DC correlations (red triangles) overlaid onto an experimental SE-SAMPI4 spectrum of SLN in unflipped bicelles, where the normal of the bilayer is perpendicular to the direction of the static field (B0).[25]
Lastly, the 15N-1H DC (νMD) was determined from the normalized time-average orientation of the N-H bond vector () by Eqn 4:
| (4) |
where ν‖ is the maximal DC and set to 10.735 kHz.[27] The experimentally observable DC (ν) was then determined by Eqn 5:
| (5) |
The analysis was implemented using custom Tcl code built on VMD 1.9.3 functions[69] and available at https://github.com/weberdak/md2slf (accessed June 9th 2019) along with Python scripts to convert peak lists to NMR spectra in Sparky[70] format. The approach is analogous to restrained[26] and unrestrained[71] MD methods reported previously, but it is primarily intended as a post-processing method of MD trajectories to predict SLF spectra from pre-existing structural data rather than de novo structure calculation. Therefore, this method was developed for convenience in early stages of spectral assignment and experimental design. It is applicable to trajectories obtained from any simulation package and forcefield and does not require advanced accelerated sampling methods. In addition, it is easy to implement using freely available software, customizable parameters, and can generate data that can be directly compared to experimental results from magnetically aligned systems. For simplicity, CSs and DCs were computed only from the final ensemble averages of , , and , with SNH and user-defined parameters δ11, δ22, δ33, ν‖, Salign and θmemb, rather than at every frame of the trajectory as done previously.[26, 71] While CS and DC distributions computed from these past studies provided some insight into the broad line shapes observed in SLF spectra from SLBs, they appear negligible for relatively dynamic magnetically aligned systems where structural and topological fluctuations observed on MD timescales are completely averaged out on NMR timescales to yield narrow linewidths.[71] For this work, MD simulations were set up using the CHARMM-GUI Membrane Builder webserver[72–73] and run using CHARMM36 forcefield parameters[74–75] and the NAMD 2.12 simulation package.[76] Proteins were embedded in a bilayer containing 75% DMPC and 25% POPC lipids to be consistent with experimental bicelle compositions used for magnetic alignment at room temperature.[77]
To test the accuracy of MD-predicted CSs and DCs, their back-calculated average values from a 150 ns trajectory of SLN (PDB 1JDM[78]) were compared against experimental SE-SAMPI4[22] spectra using unflipped (θmemb = 90.0°) bicelles[25] (Figure 1B). A remarkable correlation was observed, with the largest deviation attributed to the terminal residue Y31. R2 values of 0.59 and 0.62 were determined for CSs and DCs, respectively, which increases to 0.89 and 0.87 respectively if Y31 outliers are removed. The tilt angle of SLN was restrained in the simulation to the experimentally determined angle[77] of 24° using 100 kcal/mol tilt collective variable restraints available in NAMD 2.12,[76] while azimuthal fluctuations were left unrestrained. No adjustments of Salign (i.e., set to 1.0) was required, suggesting that the MD simulation accounted for the primary sources of disorder. Experimental and calculated 15N CS and 15N-1H DC cross-peaks are overlaid in Figure 1C and produce PISA (Polar Index Slant Angle) wheel patterns that reflect the periodicity of residues in an α-helix.[30] The ability of MD-derived SLF spectra to predict deviations from idealized wheels is striking, especially the far-removed peak assigned to Asn4. These results demonstrate that MD-based predictions of SLF spectra may be a powerful alternative to PISA wheel simulation methods[27] commonly used to assign and rationalize experimental data.
With the accuracy of MD-derived CSs and DCs established, the method was then applied to predict an SLF spectrum from a crystal structure of the β2-adrenergic GPCR in the active state (PDB 3P0G).[59] The simulated GPCR structure was prepared as per previous MD studies,[79–80] with the stabilizing T4 lysozyme residues and G-protein mimetic Nb80 nanobody removed to save computation time. Drifts to the inactive state and slow-timescale rocking motions were suppressed by applying 10 kcal/mol tilt and spinAngle collective variable restraints on backbone atoms of all TM α-helices. MD-predicted peak positions were then used to simulate SLF spectra (Figure 2A) into Sparky[70] format using the NMRGlue[81] Python library. The spectral width of the simulated spectrum corresponded to the 15N frequency on a 14.1 T magnet (60.8 MHz) and peaks were represented as Gaussians of 50 Hz FWHM linewidth in CS and DC dimensions to approximate the narrowest linewidths currently reported.[9] Nearly all of 276 backbone peaks simulated were fully resolved, demonstrating the potential for OS-ssNMR to capture high-resolution structural and topological information of large membrane proteins. More realistically, however, linewidths are often on the order of 100 to 200 Hz due to factors accompanying the complexity of larger systems, such as sample and structural inhomogeneity, slow-timescale motions,[82] mosaic spread[83] and multiple conformational states[84], for which spectroscopists generally benefit from employing higher field strengths and selective labelling and unlabelling strategies.[85–86] Additionally, simulated GPCR spectra were in the form of a flipped system achieved using either SLBs or bicelles and macrodiscs doped with lanthanides[87] to obtain a parallel orientation of the normal of the bilayer with respect to the static magnetic field (B0). This essentially doubles resolution by increasing dispersion of CSs and DCs and eliminates line broadening arising from slow-timescale uniaxial rotational diffusion common of larger proteins.[88–89]
Figure 2:
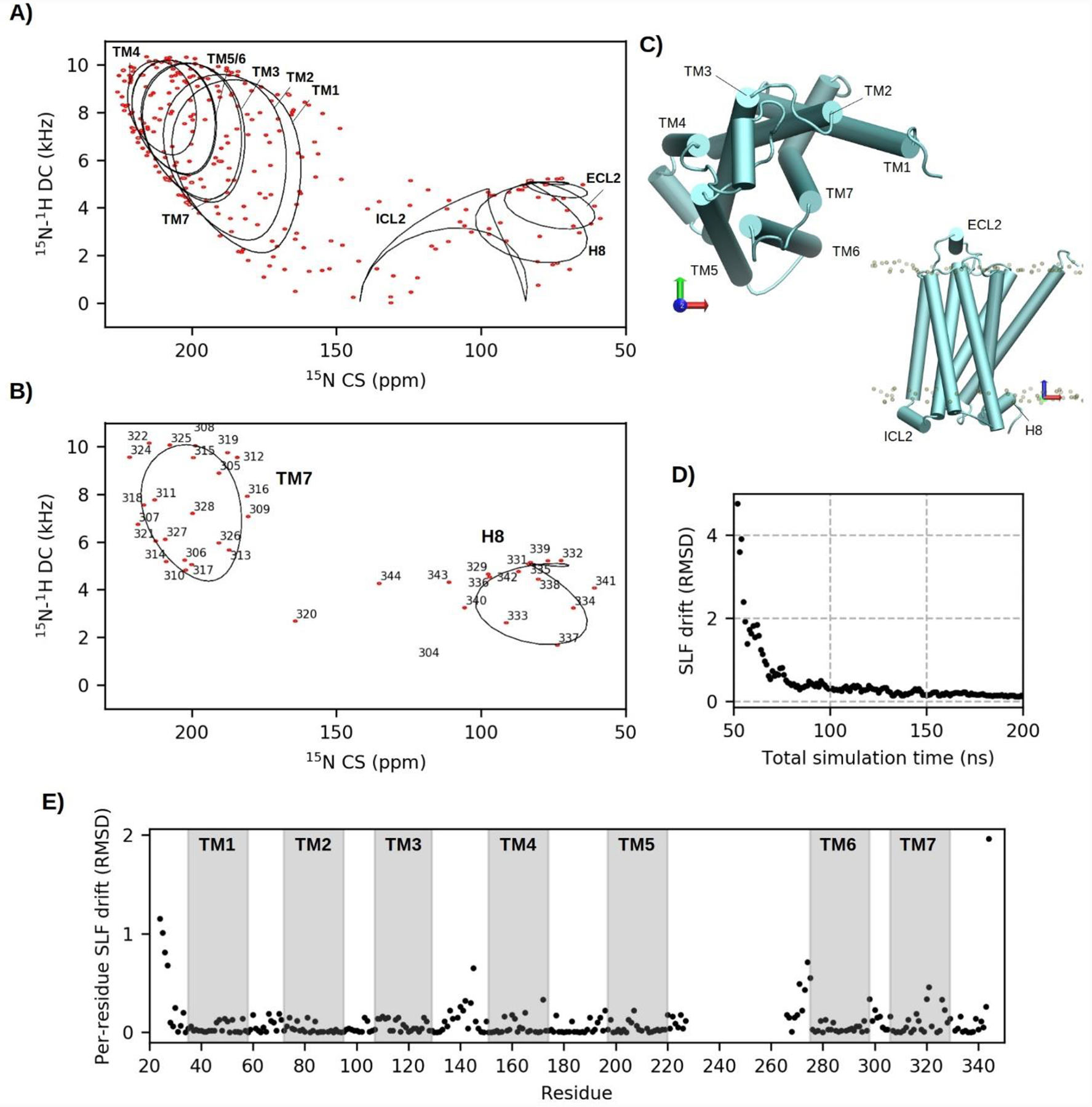
A) Simulated 15N-1H SLF spectrum (14.1 T; 50 Hz linewidths) of the β2-adrenergic GPCR back-calculated from 150 ns of equilibrated MD trajectory (red contours). Ideal PISA wheels fit to helical segments are overlaid as black lines. B) Extraction of TM7 and H8 residues for clear comparison to PISA fits. C) Nomenclature of helical segments. D) Convergence of SLF peak positions with time after 50 ns of initial equilibration. E) Per-residue contributions to the final SLF drift score after 200 ns of simulation.
Unlike SLN, PISA wheels are no longer clearly distinguishable in spectra of the GPCR due to crowding and peaks appear dispersed across the range of allowed combinations of CSs and DCs. To further rationalize the spectrum, peaks associated with residues central to each of the 10 helices were fit to ideal PISA models[27] (Figure 2A) by an exhaustive search[90] factoring in a general order parameter of 0.95 (typical of SNH) and idealized backbone structural parameters (i.e., bonds and dihedral angles) averaged from the MD trajectory. As expected, TM helices oriented parallel to the magnetic field clustered downfield of the spectrum, while perpendicular intra- (ICL) and extra-cellular (ECL) helices occupied the upfield end. Several peaks clustered in the 110 to 130 ppm range, away from ideal PISA positions, were mostly from flexible and unstructured residues at juxtamembrane positions. Helical residues of TM7 and H8 were isolated in Figure 2B (see Figure 2C for nomenclature) and closely resemble two PISA wheels. It is noteworthy that at 3.5 Å the resolution of the GPCR crystal structure (PDB 3P0G) would not allow accurate computation of SLF peaks directly from deposited PDB coordinates as non-refined backbone atom positions produce very large errors in CS and DC values[91] and do not reflect the time-averaged nature of the measurement. The power of MD-simulation to refine these errors was assessed using the convergence plot in Figure 2D. Here, the final 150 ns of equilibrated simulation was broken into 150 blocks. From the first equilibrated frame (at 50 ns), blocks of frames were successively added until the end of the trajectory (200 ns), with DC and CS values recomputed after the addition of each block. The average drift in cross-peak positions, over n cross-peaks, with each successive block (+b) was determined by Eqn 6:
| (6) |
where DCs were scaled up by 10 to match CS dispersion. From this plot, convergence of SLF values was mostly complete from just a 50 ns ensemble. Per-residue contributions to the final SLF drift (Figure 2E) show that at 200 ns the best approximations are made for structured regions, while peak positions of dynamic residues will generally require longer timescales, or accelerated sampling schemes, to converge.
In conclusion, we demonstrated the potential for OS-ssNMR to obtain site-resolved topological and structural information for large multi-pass membrane proteins through MD-predicted [15N-1H] SLF spectra of a GPCR. Realizing this potential in practice will hinge on further harnessing bicelles and macrodiscs as alignment media, which have been especially crucial for obtaining narrow linewidths,[9] as well as providing hydration levels, lipid ratios and preparative routes more amendable to maintaining larger and more-complex proteins in a functional state. The successful use of these systems, however, remains challenging as variations in temperature, pH, lipid composition and the presence of membrane proteins can have unforeseen consequences on magnetic alignment properties. While these limitations are currently being addressed,[92–93] traditionally used SLBs are still utilized as a robust method to mechanically align proteins.[52, 94] Furthermore, the striking ability of MD simulations to accurately predict SLF spectra from existing structural data sets, even at low resolution, may position OS-ssNMR as a powerful complement to cryo-electron microscopy, which has proven highly successful for solving membrane protein structures in ground states. We envision an interplay between these two techniques may involve streamlined assignment of OS-ssNMR experiments to provide crucial insights into the function of membrane proteins embedded in fluid-phase lipid bilayers at physiological temperature.
Acknowledgements
This work was partially supported by the National Institutes of Health (GM64742 and HL144130 to G.V.) and the American Heart Association (19POST34420009 to D.W.).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Chipot C; Dehez F; Schnell JR; Zitzmann N; Pebay-Peyroula E; Catoire LJ; Miroux B; Kunji ERS; Veglia G; Cross TA; Schanda P, Chem. Rev 2018, 118, 3559. doi: 10.1021/acs.chemrev.7b00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladizhansky V, Biochim. Biophys. Acta 2017, 1865, 1577. doi: 10.1016/j.bbapap.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 3.Radoicic J; Lu GJ; Opella SJ, Q. Rev. Biophys 2014, 47, 249. doi: 10.1017/S0033583514000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Wel PCA, Emerg. Top. Life Sci 2018. doi: 10.1042/ETLS20170088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wylie BJ; Do HQ; Borcik CG; Hardy EP, Mol. Phys 2016, 114, 3598. doi: 10.1080/00268976.2016.1252470 [DOI] [Google Scholar]
- 6.Opella SJ, Biomed. Spectrosc. Imaging 2014, 3, 81. doi: 10.3233/BSI-140080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders CR; Landis GC, Biochemistry. 1995, 34, 4030. doi: 10.1021/bi00012a022 [DOI] [PubMed] [Google Scholar]
- 8.Park SH; Berkamp S; Cook GA; Chan MK; Viadiu H; Opella SJ, Biochemistry. 2011, 50, 8983. doi: 10.1021/bi201289c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radoicic J; Park SH; Opella SJ, Biophys. J 2018, 115, 22. doi: 10.1016/j.bpj.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitô H; Ando I; Ramamoorthy A, Prog. Nucl. Magn. Reson. Spectrosc 2010, 57, 181. doi: 10.1016/j.pnmrs.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornell BA; Separovic F; Baldassi AJ; Smith R, Biophys. J 1988, 53, 67. doi: 10.1016/S0006-3495(88)83066-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornell BA; Weir LE; Separovic F, Eur. Biophys. J 1988, 16, 113. doi: 10.1007/BF00255521 [DOI] [PubMed] [Google Scholar]
- 13.Smith R; Thomas DE; Separovic F; Atkins AR; Cornell BA, Biophys. J 1989, 56, 307. doi: 10.1016/S0006-3495(89)82677-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketchem R; Hu W; Cross T, Science. 1993, 261, 1457. doi: 10.1126/science.7690158 [DOI] [PubMed] [Google Scholar]
- 15.Ketchem RR; Roux B; Cross TA, Structure. 1997, 5, 1655. doi: 10.1016/S0969-2126(97)00312-2 [DOI] [PubMed] [Google Scholar]
- 16.Wu CH; Ramamoorthy A; Opella SJ, J. Magn. Reson., Ser A 1994, 109, 270. doi: 10.1006/jmra.1994.1169 [DOI] [Google Scholar]
- 17.Nevzorov AA; Opella SJ, J. Magn. Reson 2003, 164, 182. doi: 10.1016/S1090-7807(03)00240-4 [DOI] [PubMed] [Google Scholar]
- 18.Nevzorov AA; Opella SJ, J. Magn. Reson 2007, 185, 59. doi: 10.1016/j.jmr.2006.09.006 [DOI] [PubMed] [Google Scholar]
- 19.Marassi FM; Gesell JJ; Valente AP; Kim Y; Oblatt-Montal M; Montal M; Opella SJ, J. Biomol. NMR 1999, 14, 141. doi: 10.1023/A:1008391823293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mote KR; Gopinath T; Traaseth NJ; Kitchen J; Gor’kov PL; Brey WW; Veglia G, J. Biomol. NMR 2011, 51, 339. doi: 10.1007/s10858-011-9571-8 [DOI] [PubMed] [Google Scholar]
- 21.Gopinath T; Mote KR; Veglia G, Prog. Nucl. Magn. Reson. Spectrosc 2013, 75, 50. doi: 10.1016/j.pnmrs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopinath T; Veglia G, J. Am. Chem. Soc 2009, 131, 5754. doi: 10.1021/ja900096d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopinath T; Mote KR; Veglia G, J. Biomol. NMR 2015, 62, 53. doi: 10.1007/s10858-015-9916-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gor’kov PL; Chekmenev EY; Li C; Cotten M; Buffy JJ; Traaseth NJ; Veglia G; Brey WW, J. Magn. Reson 2007, 185, 77. doi: 10.1016/j.jmr.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 25.Wang S; Gopinath T; Veglia G, Methods. 2018, 138–139, 54. doi: 10.1016/j.ymeth.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Simone A; Mote KR; Veglia G, Biophys. J 2014, 106, 2566. doi: 10.1016/j.bpj.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denny JK; Wang J; Cross TA; Quine JR, J. Magn. Reson 2001, 152, 217. doi: 10.1006/jmre.2001.2405 [DOI] [PubMed] [Google Scholar]
- 28.Lapin J; Nevzorov AA, J. Magn. Reson 2018, 293, 104. doi: 10.1016/j.jmr.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 29.Lapin J; Nevzorov AA, J. Biomol. NMR 2019, 73, 229. doi: 10.1007/s10858-019-00251-7 [DOI] [PubMed] [Google Scholar]
- 30.Marassi FM; Opella SJ, J. Magn. Reson 2000, 144, 150. doi: 10.1006/jmre.2000.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mascioni A; Eggimann BL; Veglia G, Chem. Phys. Lipids 2004, 132, 133. doi: 10.1016/j.chemphyslip.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 32.Nevzorov AA; Opella SJ, J. Magn. Reson 2003, 160, 33. doi: 10.1016/S1090-7807(02)00138-6 [DOI] [PubMed] [Google Scholar]
- 33.Shi L; Traaseth NJ; Verardi R; Cembran A; Gao J; Veglia G, J. Biomol. NMR 2009, 44, 195. doi: 10.1007/s10858-009-9328-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan Z; Hung I; Wang X; Paulino J; Wu G; Litvak IM; Gor’kov PL; Brey WW; Lendi P; Schiano JL; Bird MD; Dixon IR; Toth J; Boebinger GS; Cross TA, J. Magn. Reson 2017, 284, 125. doi: 10.1016/j.jmr.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechinger B, eMagRes 2018, 7, 25. doi: 10.1002/9780470034590.emrstm1558 [DOI] [Google Scholar]
- 36.Salnikov E; Rosay M; Pawsey S; Ouari O; Tordo P; Bechinger B, J. Am. Chem. Soc 2010, 132, 5940. doi: 10.1021/ja1007646 [DOI] [PubMed] [Google Scholar]
- 37.Salnikov ES; Abel S; Karthikeyan G; Karoui H; Aussenac F; Tordo P; Bechinger B; Ouari O, ChemPhysChem 2017, 18, 2103. doi: 10.1002/cphc.201700389 [DOI] [PubMed] [Google Scholar]
- 38.Salnikov ES; Sarrouj H; Reiter C; Aisenbrey C; Purea A; Aussenac F; Ouari O; Tordo P; Fedotenko I; Engelke F; Bechinger B, J. Phys. Chem. B 2015, 119, 14574. doi: 10.1021/acs.jpcb.5b07341 [DOI] [PubMed] [Google Scholar]
- 39.Opella SJ; Marassi FM; Gesell JJ; Valente AP; Kim Y; Oblatt-Montal M; Montal M, Nat. Struct. Biol 1999, 6, 374. doi: 10.1038/7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeri AC; Mesleh MF; Nevzorov A; Opella SJ, Proc. Natl. Acad. Sci 2003, 100, 6458. doi: 10.1073/pnas.1132059100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SH; De Angelis AA; Nevzorov AA; Wu CH; Opella SJ, Biophys. J 2006, 91, 3032. doi: 10.1529/biophysj.106.087106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SH; Mrse AA; Nevzorov AA; Mesleh MF; Oblatt-Montal M; Montal M; Opella SJ, J. Mol. Biol 2003, 333, 409. doi: 10.1016/j.jmb.2003.08.048 [DOI] [PubMed] [Google Scholar]
- 43.Das BB; Nothnagel HJ; Lu GJ; Son WS; Tian Y; Marassi FM; Opella SJ, J. Am. Chem. Soc 2012, 134, 2047. doi: 10.1021/ja209464f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Angelis AA; Howell SC; Nevzorov AA; Opella SJ, J. Am. Chem. Soc 2006, 128, 12256. doi: 10.1021/ja063640w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian Y; Lu GJ; Marassi FM; Opella SJ, J. Biomol. NMR 2014, 60, 67. doi: 10.1007/s10858-014-9852-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma M; Yi M; Dong H; Qin H; Peterson E; Busath DD; Zhou HX; Cross TA, Science. 2010, 330, 509. doi: 10.1126/science.1191750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahalakshmi R; Marassi FM, Biochemistry. 2008, 47, 6531. doi: 10.1021/bi800362b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dürr UHN; Waskell L; Ramamoorthy A, Biochim. Biophys. Acta 2007, 1768, 3235. doi: 10.1016/j.bbamem.2007.08.007 [DOI] [PubMed] [Google Scholar]
- 49.Traaseth NJ; Shi L; Verardi R; Mullen DG; Barany G; Veglia G, Proc. Natl. Acad. Sci 2009, 106, 10165. doi: 10.1073/pnas.0904290106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verardi R; Shi L; Traaseth NJ; Walsh N; Veglia G, Proc. Natl. Acad. Sci 2011, 108, 9101. doi: 10.1073/pnas.1016535108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das N; Dai J; Hung I; Rajagopalan M; Zhou H-X; Cross TA, Proc. Natl. Acad. Sci 2015, 112, E119. doi: 10.1073/pnas.1415908112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aisenbrey C; Salnikov ES; Bechinger B, J. Membr. Biol 2019. doi: 10.1007/s00232-019-00071-8 [DOI] [PubMed] [Google Scholar]
- 53.Salnikov ES; Aisenbrey C; Anantharamaiah GM; Bechinger B, Chem. Phys. Lipids 2019, 219, 58. doi: 10.1016/j.chemphyslip.2019.01.012 [DOI] [PubMed] [Google Scholar]
- 54.Aisenbrey C; Kemayo-Koumkoua P; Salnikov ES; Glattard E; Bechinger B, Biochemistry. 2019, 58, 2782. doi: 10.1021/acs.biochem.9b00375 [DOI] [PubMed] [Google Scholar]
- 55.Michalek M; Salnikov ES; Bechinger B, Biophys. J 2013, 105, 699. doi: 10.1016/j.bpj.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douat C; Aisenbrey C; Antunes S; Decossas M; Lambert O; Bechinger B; Kichler A; Guichard G, Angewandte Chemie - International Edition 2015, 54, 11133. doi: 10.1002/anie.201504884 [DOI] [PubMed] [Google Scholar]
- 57.Wolf J; Aisenbrey C; Harmouche N; Raya J; Bertani P; Voievoda N; Süss R; Bechinger B, Biophys. J 2017, 113, 1290. doi: 10.1016/j.bpj.2017.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gayen A; Leninger M; Traaseth NJ, Nat. Chem. Biol 2016, 12, 141. doi: 10.1038/nchembio.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen SGF; Choi HJ; Fung JJ; Pardon E; Casarosa P; Chae PS; Devree BT; Rosenbaum DM; Thian FS; Kobilka TS; Schnapp A; Konetzki I; Sunahara RK; Gellman SH; Pautsch A; Steyaert J; Weis WI; Kobilka BK, Nature. 2011, 469, 175. doi: 10.1038/nature09648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber DK; Sani MA; Downton MT; Separovic F; Keene FR; Collins JG, J. Am. Chem. Soc 2016, 138, 15267. doi: 10.1021/jacs.6b09996 [DOI] [PubMed] [Google Scholar]
- 61.Wang J; Denny J; Tian C; Kim S; Mo Y; Kovacs F; Song Z; Nishimura K; Gan Z; Fu R; Quine JR; Cross TA, J. Magn. Reson 2000, 144, 162. doi: 10.1006/jmre.2000.2037 [DOI] [PubMed] [Google Scholar]
- 62.Murray DT; Hung I; Cross TA, J. Magn. Reson 2014, 240, 34. doi: 10.1016/j.jmr.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salnikov E; Bertani P; Raap J; Bechinger B, J. Biomol. NMR 2009, 45, 373. doi: 10.1007/s10858-009-9380-5 [DOI] [PubMed] [Google Scholar]
- 64.Bertani P; Raya J; Bechinger B, Solid State Nucl. Magn. Reson 2014, 61–62, 15. doi: 10.1016/j.ssnmr.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 65.Holt A; Rougier L; Réat V; Jolibois F; Saurel O; Czaplicki J; Antoinette Killian J; Milon A, Biophys. J 2010, 98, 1864. doi: 10.1016/j.bpj.2010.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Separovic F; Pax R; Cornell B, Mol. Phys 1993, 78, 357. doi: 10.1080/00268979300100281 [DOI] [Google Scholar]
- 67.Esteban-Martín S; Strandberg E; Fuertes G; Ulrich AS; Salgado J, Biophys. J 2009, 96, 3233. doi: 10.1016/j.bpj.2008.12.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molugu TR; Xu X; Leftin A; Lope-Piedrafita S; Martinez GV; Petrache HI; Brown MF, Solid-State Deuterium NMR Spectroscopy of Membranes In Modern Magnetic Resonance, Webb GA, Ed. Springer International Publishing: Cham, 2018; pp 581–603. [Google Scholar]
- 69.Humphrey W; Dalke A; Schulten K, J. Mol. Graphics 1996, 14, 33. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 70.Lee W; Tonelli M; Markley JL, Bioinformatics. 2015, 31, 1325. doi: 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi L; Cembran A; Gao J; Veglia G, Biophys. J 2009, 96, 3648. doi: 10.1016/j.bpj.2009.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jo S; Kim T; Iyer VG; Im W, J. Comput. Chem 2008, 29, 1859. doi: 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 73.Wu EL; Cheng X; Jo S; Rui H; Song KC; Davila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM; Klauda JB; Im W, J. Comput. Chem 2014, 35, 1997. doi: 10.1002/jcc.23702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang J; MacKerell AD, J. Comput. Chem 2013, 34, 2135. doi: 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD; Pastor RW, J. Phys. Chem. B 2010, 114, 7830. doi: 10.1021/jp101759q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kalé L; Schulten K, J. Comput. Chem 2005, 26, 1781. doi: 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mote KR; Gopinath T; Veglia G, J. Biomol. NMR 2013, 57, 91. doi: 10.1007/s10858-013-9766-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mascioni A; Karim C; Barany G; Thomas DD; Veglia G, Biochemistry. 2002, 41, 475. doi: 10.1021/bi011243m [DOI] [PubMed] [Google Scholar]
- 79.Rosenbaum DM; Zhang C; Lyons JA; Holl R; Aragao D; Arlow DH; Rasmussen S. r. G. F.; Choi HJ; Devree BT; Sunahara RK; Chae PS; Gellman SH; Dror RO; Shaw DE; Weis WI; Caffrey M; Gmeiner P; Kobilka BK, Nature. 2011, 469, 236. doi: 10.1038/nature09665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dror RO; Arlow DH; Maragakis P; Mildorf TJ; Pan AC; Xu H; Borhani DW; Shaw DE, Proc. Natl. Acad. Sci 2011, 108, 18684. doi: 10.1073/pnas.1110499108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Helmus JJ; Jaroniec CP, J. Biomol. NMR 2013, 55, 355. doi: 10.1007/s10858-013-9718-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itkin A; Salnikov ES; Aisenbrey C; Raya J; Glattard E; Raussens V; Ruysschaert J-M; Bechinger B, ACS Omega 2017, 2. doi: 10.1021/acsomega.7b00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quine JR; Achuthan S; Asbury T; Bertram R; Chapman MS; Hu J; Cross TA, J. Magn. Reson 2006, 179, 190. doi: 10.1016/j.jmr.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 84.Staus DP; Strachan RT; Manglik A; Pani B; Kahsai AW; Kim TH; Wingler LM; Ahn S; Chatterjee A; Masoudi A; Kruse AC; Pardon E; Steyaert J; Weis WI; Prosser RS; Kobilka BK; Costa T; Lefkowitz RJ, Nature. 2016, 535, 448. doi: 10.1038/nature18636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lacabanne D; Meier BH; Böckmann A, J. Biomol. NMR 2018, 71, 141. doi: 10.1007/s10858-017-0156-z [DOI] [PubMed] [Google Scholar]
- 86.Verardi R; Traaseth NJ; Masterson LR; Vostrikov VV; Veglia G, Isotope Labeling for Solution and Solid-State NMR Spectroscopy of Membrane Proteins In Isotope labeling in Biomolecular NMR, Atreya HS, Ed. Springer; Netherlands: Dordrecht, 2012; Vol. 992, pp 35–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prosser RS; Hunt SA; DiNatale JA; Vold RR, J. Am. Chem. Soc 1996, 118, 269. doi: 10.1021/ja953598x [DOI] [Google Scholar]
- 88.Park SH; Mrse AA; Nevzorov AA; De Angelis AA; Opella SJ, J. Magn. Reson 2006, 178, 162. doi: 10.1016/j.jmr.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 89.Tesch DM; Pourmoazzen Z; Awosanya EO; Nevzorov AA, Appl. Magn. Reson 2018, 49, 1335. doi: 10.1007/s00723-018-1056-4 [DOI] [Google Scholar]
- 90.Buffy JJ; Traaseth NJ; Mascioni A; Gor’kov PL; Chekmenev EY; Brey WW; Veglia G, Biochemistry. 2006, 45, 10939. doi: 10.1021/bi060728d [DOI] [PubMed] [Google Scholar]
- 91.Kim S; Cross TA, Biophys. J 2002, 83, 2084. doi: 10.1016/S0006-3495(02)73969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ravula T; Hardin NZ; Ramadugu SK; Cox SJ; Ramamoorthy A, Angew. Chem. Int. Ed 2018, 57, 1342. doi: 10.1002/anie.201712017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 93.Ravula T; Hardin NZ; Ramamoorthy A, Chem. Phys. Lipids 2019, 219, 45. doi: 10.1016/j.chemphyslip.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harmouche N; Bechinger B, Biophys. J 2018, 115, 1033. doi: 10.1016/j.bpj.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]


