Abstract
Mutations in leucine-rich repeat kinase 2 (lrrk2) are the most common genetic cause of Parkinson’s disease. Difficulty in elucidating the pathogenic mechanisms resulting from disease-associated Lrrk2 variants stems from the complexity of Lrrk2 function and activities. Lrrk2 contains multiple protein-protein interacting domains, a GTPase domain, and a kinase domain. Lrrk2 is implicated in many cellular processes including vesicular trafficking, autophagy, cytoskeleton dynamics, and Wnt signaling. Here, we generated a zebrafish lrrk2 allelic series to study the requirements for Lrrk2 during development and to dissect the importance of its various domains. The alleles are predicted to encode proteins that either lack all functional domains (lrrk2sbu304), the GTPase, and kinase domains (lrrk2sbu71) or the kinase domain (lrrk2sbu96). All three lrrk2 mutants are viable, morphologically normal, and display wild-type-like locomotion. Because Lrrk2 modulates Wnt signaling in some contexts, we assessed Wnt signaling in all three mutant lines. Analysis of Wnt signaling by studying the expression of target genes using whole mount RNA in situ hybridization and a transgenic Wnt reporter revealed wild-type domains of Wnt activity in each of the mutants. However, we found that Wnt pathway activation is attenuated in lrrk2sbu304/sbu304, which lacks both scaffolding and catalytic domains, but not in the other alleles during late embryogenesis. This supports a model in which Lrrk2 scaffolding functions are key to a context-dependent role in promoting canonical Wnt signaling.
Keywords: CRISPR-Cas9, Lrrk2, Parkinson’s disease, Wnt, zebrafish
1 |. INTRODUCTION
Parkinson’s disease (PD) is a debilitating, progressive, neurodegenerative disorder marked by tremors, rigidity, and cognitive impairment. PD affects approximately 2% of the population over the age of 65 in the United States (Davie, 2008; Lill, 2016; Thomas & Beal, 2007). PD is defined by progressive degradation and dysfunction of dopaminergic neurons (DA) in the substantia nigra pars compacta (SNpc) (Atashrazm & Dzamko, 2016; Gasser, 2009; Parkinson, 2002). Although the majority of PD cases are idiopathic (Larsen, Hanss, & Kruger, 2018), a few commonly mutated genes have been identified (Houlden & Singleton, 2012; Iwaki et al., 2019; Kitada et al., 1998). Among these, leucine-rich repeat kinase 2 (lrrk2) is the most commonly mutated autosomal dominant PD gene (Dachsel & Farrer, 2010). Mutations in the lrrk2 gene account for 5%−13% of familial inheritance cases and 1%−5% of all sporadic PD cases (Iwaki et al., 2019; Li et al., 2014).
Lrrk2 encodes a ~280 kDa protein that contains multiple enzymatic domains and protein-protein interacting domains (Figure 1). These include armadillo-like repeats (ARM), ankyrin repeats (ANK), leucine-rich repeats (LRR), and a WD40 domain, which all function as sites for protein-protein interactions (Berwick, Heaton, Azeggagh, & Harvey, 2019). The enzymatic domains of Lrrk2 include the kinase domain and the GTPase domain, which is comprised of the Roc (Ras of complex protein) and COR (C-terminal of Roc) domains (Langston, Rudenko, & Cookson, 2016).
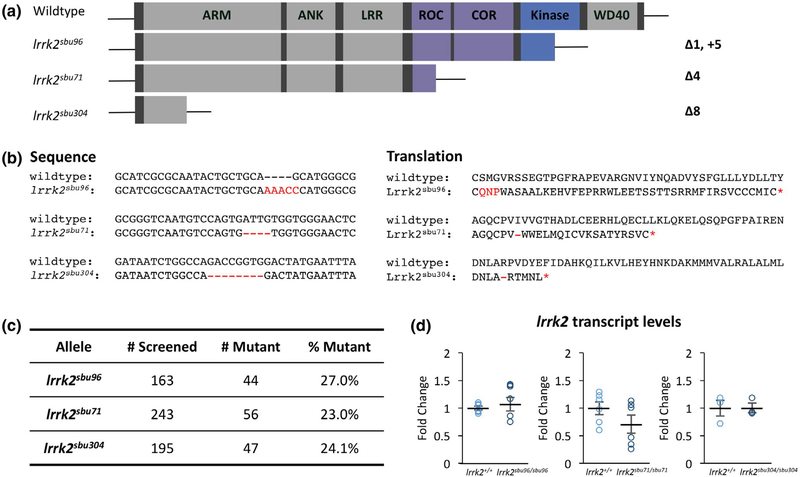
FIGURE 1.
Zebrafish lrrk2 Loss-of-Function Allelic Series. (a) Schematics of lrrk2 loss-of-function mutants created with CRISPR-Cas9. (b) lrrk2sbu96 encodes a 5bp insertion and 1bp deletion in the kinase domain, lrrk2sbu71 encodes a 4bp deletion in the ROC domain, and lrrk2sbu304 encodes an 8bp deletion in the armadillo repeat domain. All deletions in the lrrk2 zebrafish lines create a premature stop codon indicated by a red asterisk. b (Left): Nucleotide sequences of the loss-of-function mutants and b (Right): Predicted amino acid translations of the loss-of-function mutants. (c) Ratio of adults recovered for each lrrk2 allele from heterozygous intercross. (d) lrrk2 mRNA expression in each of the lrrk2 mutants Student’s t test, (lrrk2sbu96, n = 6, p = 0.23), (lrrk2sbu71, n = 6, p = 0.17), and (lrrk2sbu304, n = 3, p = 0.97)
Many PD-associated lrrk2 mutations fall within the GTPase or kinase domains (Cookson, 2010). The most common mutation is G2019S located in the kinase domain, which results in increased kinase activity (Dachsel & Farrer, 2010). The GTPase domain modulates kinase domain function, and some mutations within the GTPase domain lower GTPase activity and enhance kinase activity (Cookson, 2015). Lrrk2 G2019S mutation results in a three- to fourfold increase in kinase activity (Chen et al., 2012). BAC transgenic mice harboring the Lrrk2 G2019S mutation display some features of PD including reduced locomotor activity, impaired motor performance, and a decrease in striatal dopamine release (Chen et al., 2012). However, Lrrk2 function is multifaceted and a reduced kinase activity has also been reported in other PD-associated mutations, such as Lrrk2 G2385R (Rudenko, Chia, & Cookson, 2012). A thorough understanding of the range of wild-type Lrrk2 activity is essential to interpreting the consequences of disease-associated Lrrk2 variants.
Wnt signaling is important for many developmental processes; and it is also linked to Lrrk2 pathogenic mechanisms (Berwick & Harvey, 2012; Nixon-Abell et al., 2016). The relationship between Lrrk2 and Wnt signaling is complex and context dependent. Lrrk2 interacts with multiple components of the β-catenin destruction complex, including Disheveled family proteins, and binds to the intracellular domain of LRP 5/6 (Berwick et al., 2017; Sancho, Law, & Harvey, 2009). In addition, Lrrk2 associates with Wnt/PCP components PRICKLE1 and CELSR1 (Salasova et al., 2017). Lrrk2 knockout mice display enhanced basal levels of Wnt activation and Lrrk2 knockout fibroblasts also show enhanced Wnt responsiveness (Berwick et al., 2017). However, some pharmacological antagonists of Lrrk2 have opposite effects on Wnt signaling (Berwick et al., 2017; Deng et al., 2011). In addition, the non-canonical Wnt pathway component, PRICKLE1, converts Lrrk2 to an activator of canonical Wnt signaling in cell culture (Salasova et al., 2017). Based on these data and additional analyses, alterations in Wnt signaling are proposed to contribute to the etiology of PD (Harvey & Outeiro, 2019; Le Grand, Gonzalez-Cano, Pavlou, & Schwamborn, 2015).
Lrrk2 and Lrrk1 double-knockout mice display age-dependent loss of dopaminergic neurons in the substantia nigra, impairment of protein degradation pathways, alpha-synuclein accumulation, and increased apoptosis (Giaime et al., 2017). In zebrafish, seemingly contradictory effects of Lrrk2 disruption on DA neurons have been described. Morpholino-mediated deletion of the WD40 domain, or knockdown of the entire protein, was reported to result in significant loss of DA neurons and an overall reduction in neuron numbers during early embryonic development (Prabhudesai et al., 2016; Sheng et al., 2010). However, in contrast Ren et al, found that disruption of zebrafish Lrrk2 WD40 domain did not lead to loss of DA neurons (Ren, Xin, Li, Zhong, & Lin, 2011). Lrrk2 knockdown also produced additional effects including axial defects and edema in the eyes, lens, and otic vesicles (Prabhudesai et al., 2016).
To circumvent limitations associated with knockdown studies and assess the role of specific domains of Lrrk2 in modulating locomotor behavior, dopaminergic neuron specification, and Wnt signaling, we generated a series of zebrafish lrrk2 alleles that truncate the protein near the N-terminus, in the GTPase domain or in the kinase domain. All three lrrk2 mutants were viable. Mutant larvae did not display locomotor defects, alterations in dopaminergic neurons, or alterations in the basal levels of canonical Wnt target genes. However, upon activation of Wnt signaling using the GSK3p inhibitor BIO, we observed attenuated activation of some Wnt target genes in the strongest allele, which eliminates scaffolding and catalytic functions. Together, these findings suggest that Lrrk2 is not required for zebrafish viability but instead acts as a context specific activator of Wnt signaling.
2 |. METHODS
2.1 |. Fish husbandry
Zebrafish embryos were obtained from natural crosses and maintained at 28.5°C under 13/11 hr light/dark cycle as previously described (Moravec, Samuel, Weng, Wood, & Sirotkin, 2016). Fish were fed either artemia twice daily or Gemma micropellets (Skrettering). Adult fish were genotyped and generally separated by sex into 1.8 L tanks at approximately 3 months of age. The wild-type strain used for all experiments was a hybrid wild-type background consisting of Tubingen long-fin crossed to Brian’s wild type. For all experiments, embryos and larvae were maintained in egg water (0.3 g/L Instant Ocean, 0.001 g/L methylene blue). This work was approved and conducted in accordance with the Stony Brook University Institutional Animal Care and Use Committee. As sex in zebrafish is indeterminate during embryonic and larval stages, we were unable to consider sex as a variable.
2.2 |. Generation of the Lrrk2 mutations
An allelic series was created for the Lrrk2 gene using the CRISPR/Cas9 system. gRNAs were generated using Ambion MegaScript T7 or the Integrated DNA Technologies Alt-R single guide RNA system. The target sequences for the guide RNAs were lrrk2sbu71 5′-GTCAAT GTCCAGTGATTG-3′, lrrk2sbu96 5′-GCGCAGTACTGCTGCAGCAT-3′, and lrrk2sbu304 5′-GGATAATCTGGCCAGACCGG-3′. Guide RNA (75 pg) was co-injected with 150 pg of Cas9 protein (PNA Biosystems) into the cell of one-cell stage wild-type embryos. Fish were genotyped using the following primers: lrrk2sbu71 forward 5′-TTTGTGGATCTATTAAACCTTGCTC-3′, lrrk2sbu71 reverse 5′-CGCTCTTCACACAAATCTGC-3′, lrrk2sbu96 forward 5′-GGAGGATTATGGGTAAAT GAGC-3′, lrrk2sbu96 reverse 5′-ACGCTGAGATCGCTAAA-3′, and lrrk2sbu304 forward 5′-CTCTGCTGTGTCGGCTGAT-3′, lrrk2 sbu304 reverse 5′-CACTCCAACTTACTTGTGTGCAT-3′.
2.3 |. Quantitative RT-PCR
Total RNA was extracted from pools of six embryos at 4 hr postfertilization (hpf) or pools of four embryos at 56 hpf using Trizol (Invitrogen). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Quantitative real-time PCR (qPCR) was performed with a Light Cycler 480 (Roche) using Quanta SYBR Green (Quanta Bioscience). Transcript levels from each sample were normalized top-actin. Each experiment consisted of three pools of embryos run in duplicate. The primer pairs used include: lef1 forward 5′-TCGCCCATGAAAACTCTACTG-3′ and reverse 5′-TGGACCAAAAGTGACGAGC-3′, dkk1 forward 5′-ACCCACAGGTGAAACAGGAG-3′ and reverse 5′-CAGCATGAAAGCGTTTAGAGG-3′, sp5 forward 5′-GCTTGAGGAACTCGAGGAAG-3’ and reverse 5′-TGTTTTCCGGAGAGGAG TTC-3′, sp5l forward 5′-CGGACAATTTCCTCCACAAT-3′ and reverse 5′-CTTCCTGCCAAGCCTGACT-3′, and lrrk2 forward 5′-ATGAGGAGGAATGGGATGTG and reverse 5′-CTCCGCCAGGTGATACTCAG-3′.
2.4 |. Whole mount RNA in situ hybridization
The following probes have all been previously described dkk1 (Hashimoto et al., 2000), lef1 (Dorsky et al., 1999), sp and sp5l (Weidinger, Thorpe, Wuennenberg-Stapleton, Ngai, & Moon, 2005). Embryos were fixed at 14, 27, 48, and 72 hpf with 4% PFA at 4°C overnight, washed with PBT, then stored in 100% methanol at −20°C (~15–20 embryos per tube). Whole mount RNA in situ hybridization protocol was adapted from Thisse, Thisse, Schilling, and Postlethwait (1993).
2.5 |. Behavioral assays
Behaviors of 6 dpf larvae were recorded using a Zebrabox imaging system (Viewpoint Life Sciences, France) with constant illumination by infrared light and tracking with automated video-tracking software (Zebralab; Viewpoint Life Sciences, France). All experiments were conducted between 12 and 6 pm The visual-motor behavior paradigm consisted of 20 min of acclimation, 15 min in the light followed by a stimulus (light change), and 15 min in the dark. Beginning at 1 dpf, fish were grown in 24-well plates, with one fish per well. We tracked behavioral parameters such as number, distance, and duration, of movements as well as spatial preference and stimulus-evoked movements upon a change from light to dark. Data were assessed in 1-min bins for the analysis of spontaneous movement and 1-s bins to capture acute response to stimulus.
2.6 |. Statistics
Data analysis was analyzed using SPSS, GraphPad, and R Suite statistical language programming. Significance was denoted at a p value less than or equal to 0.05 for all tests, with error bars representing standard error of mean. Outliers were detected using the Grubbs’ test and removed from analysis. To analyze the larval behavioral data a Student’s t test was used for comparison between wild type and mutant with values presented as ±SEM. In instances where we were interested in how multiple groups varied from each other, we used a two-way ANOVA and followed with a post hoc Tukey’s test. Statistical significance was set to p < 0.05.
2.7 |. Pharmacological treatments
Working solutions of 6-Bromoindirubin-3′-oxime (BIO, Cayman Chemical) 2.5 and 0.5 μM were diluted in 1% DMSO E3 embryo media. For the qPCR experiments, zebrafish were treated at 36 hpf with BIO and the embryos were collected after 20 hr. The zebrafish were divided into six groups: wild-type control group (DMSO control), lrrk2 allele mutant control group (DMSO control), 0.5 μM BIO-treated wild-type group, 0.5 μM BIO-treated lrrk2 allele mutant group, 2.5 μM BIO-treated wild-type group, and 2.5 μM BIO-treated lrrk2 allele mutant group. In these experiments, lrrk2 mutants were obtained from crosses of homozygous parents. Control wild types were generated from crosses of wild-type siblings of the lrrk2 mutants.
2.8 |. Fluorescent imaging
The irrk2sbu71/+, irrk2sbu96/+, and irrk2sbu304/+ alleles were crossed with a siam: GFP transgenic line (Moro et al., 2012). Embryos from the heterozygous cross were placed in 1% 1-phenyl 2-thiourea during segmentation to inhibit pigmentation. Embryos were anesthetized with MS-222 (Tricaine) and mounted on slides using 3% methyl cellulose. Images of 12–15 fish were acquired at each stage during one imaging session using a Zeiss Axioplan2 microscope and genotyped post hoc.
3 |. RESULTS
3.1 |. Generation of a lrrk2 allelic series
Zebrafish Lrrk2 has a domain structure very similar to human Lrrk2 as both are comprised of armadillo-like repeats, ankyrin repeats, leucine-rich repeat, a Roc and COR GTPase, kinase, and WD40 domains (Sheng et al., 2010) (Figure 1). To study the function of Lrrk2 in zebrafish development we created an allelic series of mutants. The lrrk2 mutants were generated by microinjecting single guide RNAs with CAS9 mRNA or protein into one-cell zebrafish embryos to truncate the protein in distinct regions (Figure 1a). Germline transmission was achieved for lesions produced by each gRNA. Lesions were amplified by PCR and sequenced. The alleles we chose to investigate further included: irrk2sbu96, a net 4 nucleotide insertion in the kinase domain; irrk2sbu71, a 4-base pair deletion in the ROC domain; and irrk2sbu304 an 8 nucleotide deletion in the armadillo repeats that is predicted to remove all of the key functional domains (Figure 1b).
Homozygous lrrk2 mutants for each of the alleles appeared morphologically normal throughout embryonic and larval development (not shown). Mutant adults were recovered at the expected Mendelian ratios (Figure 1c). Offspring of mutant females appeared wild type suggesting that there is no requirement for maternal lrrk2 mRNA. Because nonsense-mediated decay of mutant mRNA can cause upregulation of related genes and transcriptional adaptation (El-Brolosy et al., 2019), we measured lrrk2 transcript levels by qPCR in each mutant line. In each case, no evidence for nonsense-mediated decay of mutant lrrk2 transcripts was found as lrrk2 mRNA was comparable in mutants and control embryos (Figure 1d).
3.2 |. lrrk2 mutants display wild-type locomotor behavior
Because disruption of PD-associated genes and alterations in the dopaminergic system result in locomotor defects in zebrafish (Godoy, Noble, Yoon, Anisman, & Ekker, 2015; Lambert, Bonkowsky, & Masino, 2012; Xi et al., 2010) we evaluated the locomotor behavior of each of the lrrk2 mutants. We studied spontaneous and illumination-evoked changes in locomotor behavior of all three lrrk2 alleles at 6 dpf. Offspring of heterozygous parents was assayed in 24-well dishes at 6 dpf using the Zebrabox system (Viewpoint). Following a 20-min acclimation period, 15 min of spontaneous movements in the light were recorded. The lighting was then extinguished and swimming behavior in the dark was recorded for 15 min. Post hoc genotyping was performed to identify lrrk2 mutants and wild-type siblings. The number of movements, distance traveled, swim time, swim length, and speed were compared between lrrk2 mutants and sibling controls (Figure 2 and Figure S1). We found that the lrrk2sbu96/sbu96, irrk2sbu71/sbu71, and irrk2sbu304/sbu304 larvae all made similar numbers of movements, when compared to their respective sibling controls, in both light and dark conditions (Figure 2b,c,e,f,h,i). No differences between lrrk2 mutants and controls were observed when other parameters such as duration of movements, distance of movements, swim length, and speed were assessed (Figure S1).
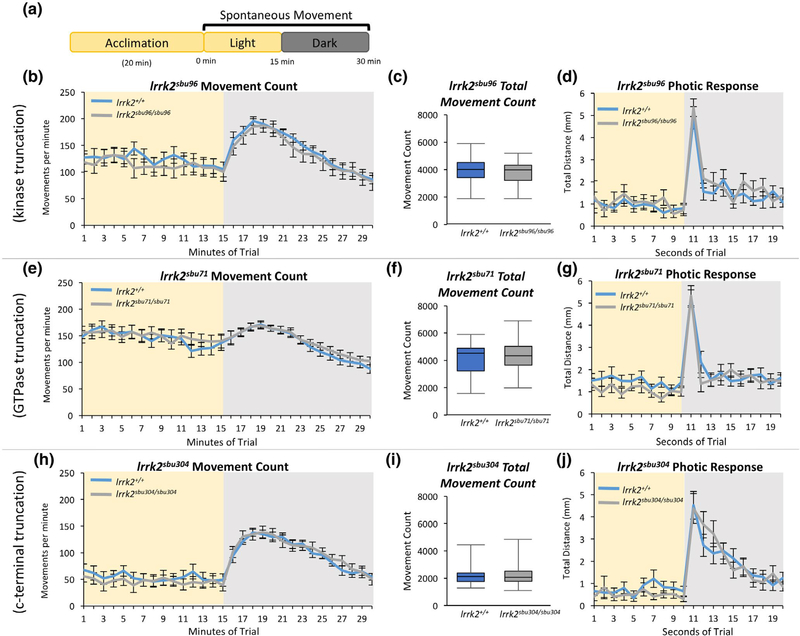
FIGURE 2.
Assessment of Locomotor Behavior of lrrk2 Mutants. (a) Visual-motor paradigm for assessing spontaneous locomotor behavior in 6 dpf zebrafish. (b,e,h) Spontaneous movement count per minute in the light and dark (illumination is removed at minute 15 of the trial) for (b) lrrk2+/+ (n = 28) and lrrk2sbu96/sbu96 (n = 21), (e) lrrk2+/+ (n = 34) and lrrk2sbu71/sbu71 (n = 35), (h) lrrk2+/+ (n = 24) and lrrk2sbu304/sbu304 (n = 24). (c,f,i) The average total number of movements for each genotype recorded during the 15 min in the light (p = 0.41, p = 0.65, p = 0.82, Student’s t test, for lrrk2sbu96, lrrk2sbu71, lrrk2sbu304 respectively). (d,g,j) Distance traveled per second upon light change in response to the removal of illumination (photic response) at second 11 of the trial
Shifting zebrafish larvae from light to dark produces an abrupt swimming burst (dark flash photokinesis) (Burgess & Granato, 2007a; Wolman, Jain, Liss, & Granato, 2011) followed by a characteristic increase in movement (visual-motor response) (Burgess & Granato, 2007b; Emran et al., 2007; Liu et al., 2015) for several minutes following the light change. Both of these behaviors were indistinguishable in lrrk2 mutants and sibling controls (Figure 2b,d,e,g,h,j). Together, these results demonstrate that locomotor behaviors are not disrupted in any of the lrrk2 mutants in our assays.
3.3 |. Dopaminergic neuron numbers are unaltered in lrrk2 mutant larvae
Loss of dopaminergic neurons is a defining feature of PD (Poewe et al., 2017). Previous Lrrk2 knockdown studies in zebrafish have come to opposing conclusions on the requirement for Lrrk2 in dopaminergic neuron specification (Prabhudesai et al., 2016; Ren et al., 2011; Sheng et al., 2010). To assess the requirement for Lrrk2 in early dopaminergic neuron development in our lrrk2 mutants, we performed whole mount RNA in situ hybridization using two markers of dopaminergic neurons, tyrosine hydroxylase (th1) and d-alanine aminotransferase (dat) at 72 hpf on the lrrk2sbu304/sbu304 mutants (Figure 3 and Figure S2), which are predicted to disrupt all of the key domains. The pattern of staining was unaltered in lrrk2sbu304/sbu304 mutants and the number of dopaminergic neurons was comparable between mutants and wild-type siblings (Figure 3c) suggesting that Lrrk2 is not required in zebrafish for dopaminergic neuron specification.
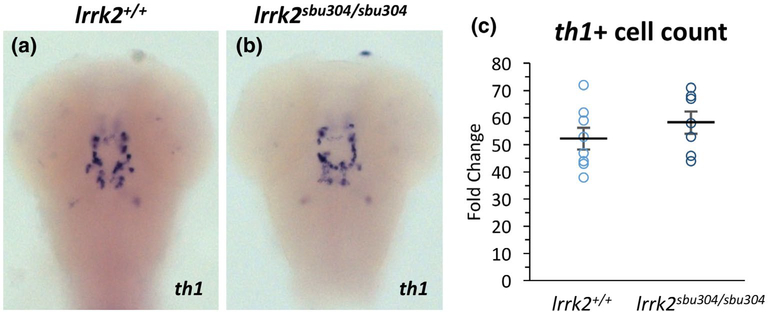
FIGURE 3.
th1 mRNA Expression and cell count for lrrk2sbu304 Mutant. (a,b) RNA in situ hybridization of th1 on 72 hpf wild type and lrrk2sbu304/sbu304 mutant embryos from heterozygous intercrosses. Images taken on a dorsal view, focused on the midbrain. (c) Cell counts of th1 positive cells in wild types and lrrk2sbu304/sbu304 mutants. Student’s t test, lrrk2+/+ (n = 8) and lrrk2sbu304/sbu304 (n = 7), p = 0.32
3.4 |. Regulation of Wnt target gene expression by Lrrk2
Alterations in Wnt signaling have been implicated in PD and several lines of evidence support roles for Lrrk2 within this pathway (Berwick & Harvey, 2012; Berwick et al., 2017; Harvey & Marchetti, 2014; Le Grand et al., 2015; Nixon-Abell et al., 2016; Sancho et al., 2009). Zebrafish lrrk2 is expressed throughout embryonic and larval development (Sheng et al., 2010). To examine the zebrafish requirement for Lrrk2 in modulating Wnt signaling, we first evaluated Wnt signaling in lrrk2 mutants using whole mount RNA in situ hybridization and a transgenic Wnt reporter, Tg(7xTCF-Xla.Siam:GFP)ia4 (Moro et al., 2012).
Wnt target genes studied by RNA in situ hybridization included sp5l, dkk1, sp5, and lef1 (Dorsky et al., 1999; Hashimoto et al., 2000; Weidinger et al., 2005). Embryos were collected from heterozygous intercrosses and genotypes established post hoc. sp5l expression in the tailbud region of 10-somite embryos (14 hpf) was similar between the lrrk2 mutants and their wild-type siblings (Figure 4a–f). dkk1 expression at 27 hpf in the pharyngeal pouches was comparable in lrrk2 mutants and their wild-type siblings (Figure 4g–l). Lef1 and sp5 are both expressed in the brain at 48 and 72 hpf respectively and were unaltered in lrrk2 mutants compared to controls (figure 4m–x).
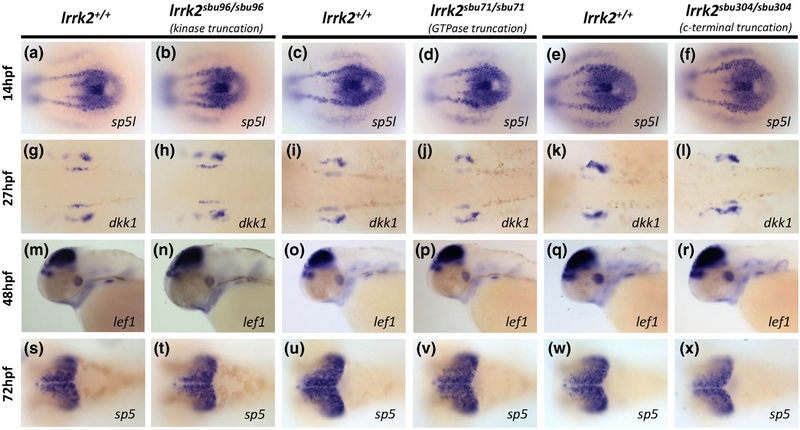
FIGURE 4.
Wnt Target Gene Expression in lrrk2 Mutants. (a-f) RNA in situ hybridization of sp5l in 10-somite embryos from lrrk2sbu96, lrrk2sbu71, lrrk2sbu304 heterozygous intercrosses, dorsal view of the tailbud region. (g-l) RNA in situ hybridization of dkk1 in 27 hpf embryos from lrrk2sbu96, lrrk2sbu71, lrrk2sbu304 heterozygous intercrosses; dorsal view of the hindbrain expression. (m-r) RNA in situ hybridization of lef1 in 48 hpf embryos from lrrk2sbu96, lrrk2sbu71, lrrk2sbu304 heterozygous intercrosses; lateral view of the head. (s-x) RNA in situ hybridization of sp5 in 72 hpf embryos from lrrk2sbu96, lrrk2sbu71, lrrk2sbu304 heterozygous crosses; dorsal view of the brain
The transgenic reporter line Tg(7xTCF-Xla.Siam:GFP)ia4 (Moro et al., 2012) was used to generate dynamic readouts of Wnt activity. Each of the three lrrk2 alleles were crossed into the Tg(7xTCF-Xla. Siam:GFP)ia4 background to evaluate the effects of Lrrk2 disruption on canonical Wnt signaling. The pattern of expression of the reporter was assessed by fluorescent microscopy in living embryos at 27, 36, and 56 hpf (Figure 5a,b, Figure S3 and data not shown). Spatial Wnt reporter expression was found to be the same for each of the mutants and wild-type siblings. Together, these findings suggest that Lrrk2 disruption does not dramatically alter basal Wnt signaling during early zebrafish development.
FIGURE 5.
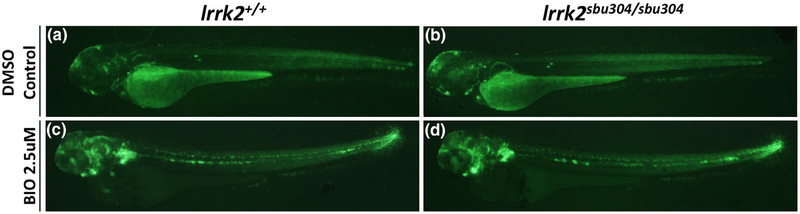
Wnt Reporter Tg(7xTCF-Xla.Siam:GFP)ia4 in lrrk2sbu304 Mutants with Wnt Activator BIO Treatment. (a-d) Fluorescent images of lrrk2sbu304 mutants in a Siam:GFP background at 56 hpf. Response to Wnt activator: (a,b) DMSO control 56 hpf lrrk2+/+ and lrrk2sbu304 sbu304. (c,d) Treated with BIO (2.5 μM) 56 hpf lrrk2+/+ and irrk2sbu304/sbu304. Images taken in lateral view
3.5 |. lrrk2 disruption attenuates response to the Wnt activator BIO
We reasoned that subtle effects of Lrrk2 on canonical Wnt signaling might be most apparent under conditions when Wnt signaling is enhanced. Therefore, to assess the impacts of Lrrk2 on Wnt signaling, we challenged lrrk2 mutants with the Wnt activator BIO (Cayman Chemical) and assessed signaling using the transgenic Wnt reporter line and by qPCR. Treatment of the lrrk2 mutants with 2.5 μM BIO at 56 hpf did not result in appreciable spatial changes in fluorescence of the reporter (Figure 5c,d and data not shown).
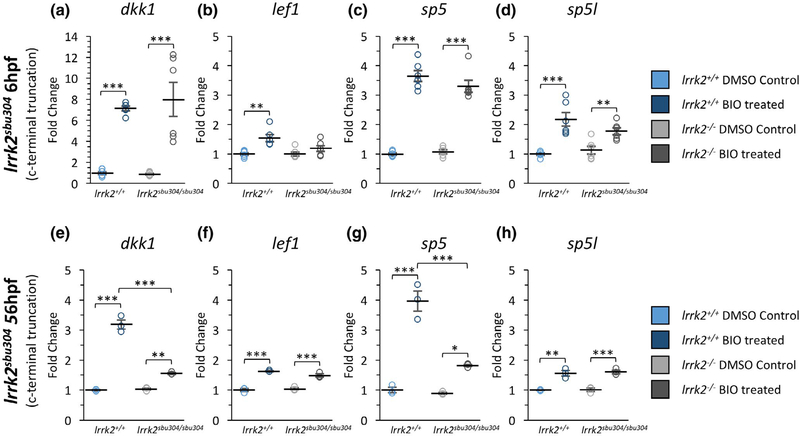
We next monitored the effects of Lrrk2 dysfunction on levels of Wnt target gene expression following treatment with BIO during early gastrula (6 hpf) and late embryogenesis (56 hpf) using qPCR. We first studied the lrrk2sbu304 allele because it lacks all of the protein interaction and catalytic domains. Basal levels of the Wnt target genes in untreated control and lrrk2sbu304/sbu304 mutants were similar at 6 and 56 hpf. As expected, levels of dkk1, lef1, sp5, and sp5l were increased in wild-type embryos challenged with 2.5 μM BIO (Figure 6), with induction of dkk1 and sp5 more robust than lef1 and sp51 at both 6 and 56 hpf. lrrk2sbu304/sbu304 embryos and controls had comparable increases in expression levels of target genes in response to BIO treatment at 6 hpf (Figure 6a–d). However, in contrast to controls, at 56 hpf, BIO-mediated induction of dkk1 and sp5 was attenuated in the lrrk2sbu304/sbu304 mutant, and this reduction in response was also apparent at a lower BIO concentration (0.5 μM) (Figure S4). These findings suggest that Lrrk2 functions as an activator of Wnt signaling at late embryogenesis, but not during the early gastrula stage.
FIGURE 6.
mRNA Expression of Wnt Target Genes in lrrk2sbu304 Mutants. (a-d) Real-time qPCR showing the expression level of (a) dkk1 (n = 6), (b) lef1 (n = 6), (c) sp5 (n = 6) and (d) sp5l (n = 6) for lrrk2+/+ and lrrk2sbu304/sbu304 at 6 hpf for vehicle (DMSO)-and BIO (2.5uM)-treated embryos. (a) Expression levels of dkk1 comparing lrrk2+/+ control and treated (p = 0.00022) and lrrk2sbu304/sbu304 control and treated (p = 0.000034). (b) Expression levels of lef1 comparing lrrk2+/+ control and treated (p = 0.0019). (c) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 6.7e-8) and lrrk2sbu304/sbu304 control and treated (p = 1.2e-6). (d) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.000082) and lrrk2sbu304/sbu304 control and treated (p = 0.028). (e-h) Real-time qPCR showing the expression of (e) dkk1(n = 3), (f) lef1(n = 3), (g) sp5 (n = 3), and (h) sp5l (n = 3) for lrrk2+/+ and lrrk2sbu304/sbu304 at 56 hpf for vehicle (DMSO)- and BIO (2.5uM)-treated embryos. (e) Expression levels of dkk1 comparing lrrk2+/+ control and treated (p = 3.1e-7), lrrk2sbu304/sbu304 control and treated (p = 0.0082), and lrrk2+/+ treated and lrrk2sbu304/sbu304 treated (p = 2.8e-6). (f) Expression levels of lef1 comparing lrrk2+/+ control and treated (p = 0.000025) and lrrk2sbu304/sbu304 control and treated (p = 0.00032). (g) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 0.00001) and lrrk2sbu304/sbu304 control and treated (p = 0.023), and lrrk2+/+ treated and lrrk2sbu304/sbu304 treated (p = 0.00012). (h) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.0012) and lrrk2sbu304/sbu304 control and treated (p = 0.00076). Statistical analysis for pairwise comparisons was done with a post hoc Tukey’s test. A two-way analysis of variance was conducted to test if there was a significant interaction between genotype and treatment. We found a significant interaction between genotype and treatment for (e) dkk1(F(1,8) = 105.7, n = 3, p = 6.9e-6)) and (g) sp5 (F(18) = 33.89, n = 3, p = 0.00039) at 56 hpf. p < 0.05 (*), p < 0.01 (**), p < 0.001 (***)
3.6 |. Robust activation of Wnt target genes requires Lrrk2 scaffolding domains
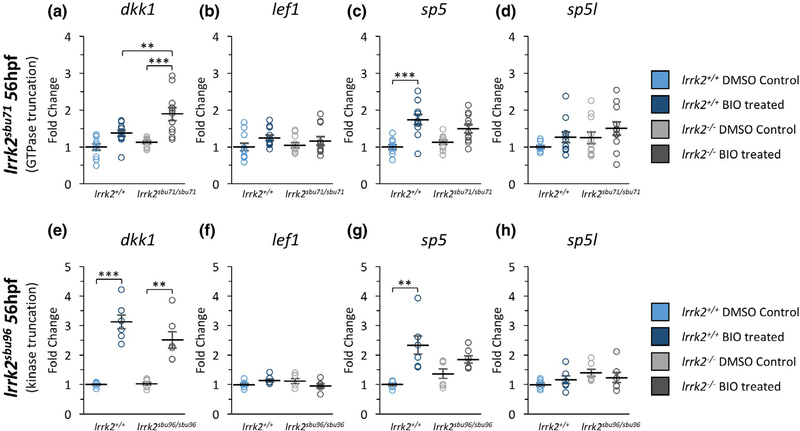
The lrrk2sbu304 allele is predicted to truncate the protein in the third exon, eliminating the majority of the protein including most of the protein-protein interaction domains as well as the ROC GTPase domain and the kinase domain (Figure 1). Effects of Lrrk2 on Wnt signaling could be dependent on scaffolding functions (mediated by the armadillo, Ank, and LRR domains) or by the catalytic domains. To distinguish between these possibilities, we challenged the mutants that are predicted to spare the main scaffolding domains, lrrk2sbu71/sbu71 and lrrk2sbu96/sbu96, with 2.5 μM BIO at 56 hpf and measured levels of Wnt target genes by qPCR (Figure 7). We failed to observe reduced dkk1 or sp5 induction in response to BIO treatment in either lrrk2sbu71/sbu71, which lacks the GTPase and kinase domains, or lrrk2sbu96/sbu96, which lacks only the kinase domain, when compared to related wild-type controls (Figure 7a,c,e,g). In fact, dkk1 induction in lrrk2sbu71/sbu71 is slightly enhanced compared to levels in BIO-treated wild type (Figure 7a). Lrrk2 function is complex, therefore, domain specific effects on the Wnt pathway may be highly context dependent. Nevertheless, the dependency on Lrrk2 for robust activation of dkk1 and sp5 by BIO observed in the lrrk2sbu304/sbu304 experiments (Figure 6) was not replicated in either lrrk2sbu71/sbu71 or lrrk2sbu96/sbu96. While we cannot exclude catalytic domain functions in modulation of Wnt signaling (and some data may imply a role), our data are most consistent with a requirement for the scaffolding functions of Lrrk2 for robust activation of Wnt signaling in zebrafish during late embryogenesis.
FIGURE 7.
mRNA Expression of Wnt Target Genes in lrrk2sbu71 and lrrk2sbu96 Mutants. (a-d) Real-time qPCR showing the expression level of (a) dkk1 (n = 11), (b) lef1 (n = 11), (c) sp5 (n = 11), and (d) sp5l (n = 111) for lrrk2+/+ and lrrk2sbu71/sbu71 56 hpf for vehicle (DMSO)- and BIO (2.5uM)-treated embryos. (a) Expression levels of dkk1 comparing lrrk2+/+ treated and lrrk2sbu71/sbu71 treated (p = 0.0073) and lrrk2sbu71/sbu71 control and treated (p = 0.000067). (c) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 0.000025). (e-h) Real-time qPCR showing the expression of (e) dkk1 (n = 6), (f) lef1(n = 6), (g) sp5 (n = 6) and (h) sp5l (n = 6) for lrrk2+/+ and lrrk2sbu96/sbu96 at 56 hpf for vehicle (DMSO)- and BIO (2.5 μM)-treated embryos. (e) Expression levels of dkk1 comparing lrrk2+/+ control and treated (p = 0.000041), lrrk2sbu96/ sbu96 control and treated (p = 0.0023). (g) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 0.0016). Statistical analysis for pairwise comparisons was done with a post hoc Tukey’s test. p < 0.05 (*), p < 0.01 (**), p < 0.001 (***)
4 |. DISCUSSION
Challenges in elucidating the pathogenic mechanisms and wild-type functions of Lrrk2 stem from the complex structure of the protein, which contains multiple protein interaction and catalytic domains. To study the requirement for Lrrk2 during zebrafish development and parse functions of regions of the protein, we generated a lrrk2 allelic series that encodes truncations in the armadillo repeats (which likely encodes a null allele), GTPase domain (which removes both the GTPase and kinase domain) or the kinase domain (Figure 1). All of these lrrk2 mutants are viable. Although, overexpression of Lrrk2 produces convergence-extension defects in Xenopus (Salasova et al., 2017), we did not observe effects on convergence–extension or morphological defects in any of our lrrk2 mutants.
As PD is associated with difficulty in initiating movement, movement kinetics are a convenient means to assess the rudimentary nervous system function of larval zebrafish. Previous knockdown studies yielded conflicting conclusions on the role of Lrrk2 in controlling larval zebrafish locomotion (Ren et al., 2011; Sheng et al., 2010). Furthermore, a lrrk2 mutation that disrupted the C-terminal WD40 domain produced clutch-dependent effects on locomotion making it difficult to interpret this finding (Sheng et al., 2018). In our studies, normal locomotor behavior was observed for each of the three mutant lines (Figure 2 and Figure S1) suggesting that either Lrrk2 does not play a significant role in larval locomotion or that compensatory mechanisms sometimes obscure the role. Although off-target effects are sometimes associated with morpholinos, compensatory mechanisms can occur in mutants. The lack of nonsense-mediated decay in our mutants does not rule out additional compensatory mechanisms.
One of the principal features of PD is a reduction in dopaminergic neurons in the substantia nigra pars compacta (Surmeier, 2018). These neurons use dopamine to promote motor activity by reducing inhibition (Poewe et al., 2017). In PD patients, greater exertion of effort is needed to create any given movement (Tolosa, Wenning, & Poewe, 2006). Analysis of the strongest allele, lrrk2sbu304, did not reveal any differences in the number and pattern of DA neurons (Figure 3 and Figure S2). Previous assessments of DA neurons in larval zebrafish following lrrk2 disruption yielded conflicting findings (Prabhudesai et al., 2016; Ren et al., 2011; Sheng et al., 2010), and our data support conclusion that Lrrk2 does not impact early DA specification or survival. Because our alleles are loss of function (LOF) and PD-associated mutations tend to enhance kinase activity (although GTPase activity is often reduced), it is not surprising that larval DA neuron populations are intact.
Impacts on Wnt signaling have been proposed to be central to Lrrk2 pathogenic mechanisms in PD (Berwick et al., 2017; Le Grand et al., 2015). Lrrk2 associates with multiple canonical and non-canonical/PCP Wnt pathway components including Dvl 1–3, LRP5/6, GSKβ3, CELSR1, and PRICKLE1 (Berwick & Harvey, 2012; Salasova et al., 2017; Sancho et al., 2009). Because of the myriad interactions and the intricate cross talk between canonical Wnt signaling components and PCP components, the impacts of Lrrk2 on Wnt activity is highly context dependent and Lrrk2 has been proposed to play a role in balancing canonical and non-canonical/PCP Wnt signaling (Berwick et al., 2019). We did not detect any difference in the basal output of canonical Wnt signaling in our lrrk2 mutants (Figures 5 and 6). This contrasts in vitro studies and assays of mouse bone development (Berwick et al., 2017) that shows Lrrk2 acts to repress Wnt activity. Our results suggest that during early zebrafish development Lrrk2 impacts on basal Wnt signaling are either subtle or absent. However, we observed a Lrrk2-dependent change when the Wnt pathway was activated with a small molecule, BIO (Figures 6 and 7). Because the strongest lrrk2 allele (sbu304), which is predicted to eliminate both the domains involved in scaffolding and catalytic function, reduced the magnitude of Wnt pathway activation by BIO at 56 hpf, we conclude that in this context, Lrrk2 promotes the Wnt response (Figure 6). However, no effect was observed in the same assay at 6 hpf (early gastrula). The observation that Lrrk2 functions as an activator of Wnt signaling is consistent with the finding that PRICKLE1 and Lrrk2 together enhance Wnt activity in mammalian cell culture (Salasova et al., 2017).
We investigated whether the Lrrk2 alleles that encoded predicted proteins lacking the kinase domain (lrrk2sbu96) or the kinase and GTPase domains (lrrk2sbu71) were able to foster robust activation of the Wnt pathway at 56 hpf (Figure 7). In neither case did we observe clear reductions in the levels of Wnt targets as we did in the lrrk2sbu304 allele, which lacks the protein interaction domains. While induction of Wnt target genes by BIO was comparable in the control-related wild types for the lrrk2sbu304 and lrrk2sbu96 lines, induction was less robust in the lrrk2sbu71 controls. This effect is unrelated to lrrk2 function and likely stems from subtle genetic background differences.
We did not observe nonsense-mediated decay of lrrk2 mutant transcripts (Figure 1d) and were unable to directly assess mutant protein levels or biochemical properties. However, many Lrrk2 fragments and isolated domains function in in vitro assays (Berwick & Harvey, 2012; Salasova et al., 2017; Sancho et al., 2009). While we cannot entirely rule out effects of the Lrrk2 catalytic domains in modulation of Wnt, our data support a model in which Lrrk2 scaffolding functions are paramount. Many Lrrk1 functional properties differ from Lrrk2, but Lrrk1 also impacts Wnt signaling in some contexts (Berwick & Harvey, 2012). Future studies should address potential compensatory activities of Lrrk1 in the lrrk2 mutants.
In conclusion, we discovered that Lrrk2 is able to facilitate robust activation of the Wnt signaling pathway during late zebrafish embryogenesis. However, lrrk2 mutants display normal morphology and locomotor behavior and are fully viable. Pathogenic Lrrk2 mutations are generally gain of function and may take decades to produce discernable phenotypes. We have not detected overt abnormalities in adult lrrk2 mutants, but future studies to investigate roles for Lrrk2 during later life stages are warranted. Together, these findings suggest that Lrrk2 has subtle influences and a highly context-dependent impact on Wnt signaling.
Supplementary Material
FIGURE S1 Assessment of Locomotor Behavior of lrrk2 Mutants. (a,e,i) The duration of movements per minute in the light and dark for (a) lrrk2+/+ (n = 28) and /rrk2sbu96/sbu96 (n = 21), (e) lrrk2+/+ (n = 34) and lrrk2sbu71/sbu71 (n = 35), (i) lrrk2+/+ (n = 24) and lrrk2sbu304/sbu304 (n = 24). (b,f,j) The distance traveled per minute for each genotype tested in the light and dark periods. (c,g,k) The average swim length (distance traveled per movement) for each genotype. (d,h,l) The average speed per movement for each genotype
FIGURE S2 dat mRNA Expression and Cell Count /rrk2sbu304 Mutants. (a,b) RNA in situ hybridization for dat in 72 hpf wild type and lrrk2sbu304 mutant embryos from heterozygous crosses. Images are dorsal view focused on the midbrain
FIGURE S3 Wnt Reporter Tg(7xTCF-X/a.Siam:GFP)ia4in lrrk2 Mutants. (a-f) Fluorescent images of lrrk2sbu96, lrrk2sbu71 and lrrk2sbu304 mutants in a Siam:GFP background at 56 hpf. Images taken in lateral view
FIGURE S4 mRNA Expression of Wnt Target Genes in lrrk2sbu304 Mutants. (a-d) Real-time qPCR showing the expression level of (a) dkk1 (n = 3), (b) lef1 (n = 3), (c) sp5 (n = 3) and (d) sp5l (n = 3) for lrrk2+/+ and lrrk2sbu304/sbu304 56 hpf for vehicle (DMSO)- and BIO (0.5 μM)-treated embryos. (a) Expression levels of dkk1 comparing lrrk2+/+ treated and lrrk2sbu304/sbu304 treated (p = 0.0094), lrrk2+/+ control and treated (p = 0.00076), and lrrk2sbu304/sbu304 control and treated (p = 0.018). (c) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 0.00084) and lrrk2+/+ treated lrrk2sbu304/sbu304 treated (p = 0.0035). (d) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.023). (e-h) Real-time qPCR showing the expression of (e) dkk1 (n = 6), (f) lef1 (n = 6), (g) sp5 (n = 6), and (h) sp5l (n = 6) for lrrk2+/+ and lrrk2sbu304/sbu304 at 6 hpf for vehicle (DMSO)- and BIO (0.5 μM)-treated embryos. (e) Expression levels of dkk1 comparing lrrk2+/+ control and treated (p = 4.8e-5), and lrrk2sbu304/sbu304 control and treated (p = 6.2e-5). (f) Expression levels of lef1 comparing lrrk2+/+ control and treated (p = 0.022). (g) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 1.9e-6) and lrrk2sbu304/sbu304 control and treated (p = 0.00002). (h) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.006). Statistical analysis for pairwise comparisons was done with a post hoc Tukey’s test. A two-way analysis of variance was conducted between genotype and treatment. There was a significant interaction between genotype and treatment for (c) sp5 (F(1,8) = 10.889, n = 3, p = 0.0108). p < 0.05 (*), p < 0.01 (**), p < 0.001 (***)
Significance.
Leucine-rich repeat kinase 2 (Lrrk2) is a complex protein with multiple enzymatic and protein interaction domains. The pathways by which altered Lrrk2 function leads to Parkinson’s disease are unknown. As a means to better understand the actions of Lrrk2 during neural development, we generated a series of truncation mutants in zebrafish and assessed the impacts on larval locomotion, dopaminergic neurons, and Wnt signaling. We did not find significant changes in locomotion or dopaminergic neurons in any of the lrrk2 mutants. However, enhanced responsiveness to Wnt activation was observed, which may over time produce more pronounced impacts.
ACKNOWLEDGMENTS
We thank the many members of the zebrafish community who provided probes and fish lines, in particular Dr. Richard Dorsky who provided several reagents to assay Wnt signaling. The groundwork for these studies was laid by Cara Moravec, Alexandra DellaPenna, and Ejike Okoye. We are grateful to Dr. Bernadette Holdener, Amalia Napoli, and Josiah Zoodsma for comments on the manuscript, Josiah Zoodsma for assistance with statistical analyses and to Neal Bhattacharji, Stephanie Flanagan and the undergraduate fish room aides for fish care. This work was supported by Turner Fellowship (JW), IMSD Merge support (JW), R03NS102949 (HS) and Hartman Foundation support (HS).
Funding information
National Institutes of Health; Thomas Hartman Center for Parkinson’ Research, Stony Brook University
Footnotes
CONFLICT OF INTEREST
We declare no financial conflict of interest.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Atashrazm F, & Dzamko N (2016). LRRK2 inhibitors and their potential in the treatment of Parkinson’s disease: Current perspectives. Journal of Clinical Pharmacology, 8, 177–189. 10.2147/CPAA.S102191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, & Harvey K (2012). LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Human Molecular Genetics, 21(22), 4966–4979. 10.1093/hmg/dds342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, Heaton GR, Azeggagh S, & Harvey K (2019). LRRK2 Biology from structure to dysfunction: Research progresses, but the themes remain the same. Molecular Neurodegeneration, 14(1), 49 10.1186/s13024-019-0344-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, Javaheri B, Wetzel A, Hopkinson M, Nixon-Abell J, Granno S, … Harvey K (2017). Pathogenic LRRK2 variants are gain-of-function mutations that enhance LRRK2-mediated repression of beta-catenin signaling. Molecular Neurodegeneration, 12(1), 9 10.1186/s13024-017-0153-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HA, & Granato M (2007a). Modulation of locomotor activity in larval zebrafish during light adaptation. Journal of Experimental Biology, 210(Pt 14), 2526–2539. 10.1242/jeb.003939 [DOI] [PubMed] [Google Scholar]
- Burgess HA, & Granato M (2007b). Sensorimotor gating in larval zebrafish. Journal of Neuroscience, 27(18), 4984–4994. 10.1523/JNEUROSCI.0615-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Weng Y-H, Chien K-Y, Lin K-J, Yeh T-H, Cheng Y-P, … Wang H-L (2012). (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death and Differentiation, 19(10), 1623–1633. 10.1038/cdd.2012.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR (2010). The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nature Reviews Neuroscience, 11(12), 791–797. 10.1038/nrn2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR (2015). LRRK2 pathways leading to neurodegeneration. Current Neurology and Neuroscience Reports, 15(7), 42 10.1007/s11910-015-0564-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachsel JC, & Farrer MJ (2010). LRRK2 and Parkinson disease. Archives of Neurology, 67(5), 542–547. 10.1001/archneurol.2010.79 [DOI] [PubMed] [Google Scholar]
- Davie CA (2008). A review of Parkinson’s disease. British Medical Bulletin, 86, 109–127. 10.1093/bmb/ldn013 [DOI] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, … Gray NS (2011). Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nature Chemical Biology, 7(4), 203–205. 10.1038/nchembio.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Snyder A, Cretekos CJ, Grunwald DJ, Geisler R, Haffter P, … Raible DW (1999). Maternal and embryonic expression of zebrafish lef1. Mechanisms of Development, 86(1–2), 147–150. 10.1016/s0925-4773(99)00101-x [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, … Stainier DYR (2019). Genetic compensation trig gered by mutant mRNA degradation. Nature, 568(7751), 193–197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran F, Rihel J, Adolph AR, Wong KY, Kraves S & Dowling JE (2007). OFF ganglion cells cannot drive the optokinetic reflex in zebrafish. Proceedings of the National Academy of Sciences, 104, 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T (2009). Mendelian forms of Parkinson’s disease. Biochimica et Biophysica Acta, 1792(7), 587–596. 10.1016/j.bbadis.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Giaime E, Tong Y, Wagner LK, Yuan Y, Huang G, & Shen J (2017). Age-dependent dopaminergic neurodegeneration and impairment of the autophagy-lysosomal pathway in LRRK-deficient mice. Neuron, 96(4), 796–807.e796. https://doi.org/10.1016Zj.neuron.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy R, Noble S, Yoon K, Anisman H, & Ekker M (2015). Chemogenetic ablation of dopaminergic neurons leads to transient locomotor impairments in zebrafish larvae. Journal of Neurochemistry, 135(2), 249–260. 10.1111/jnc.13214 [DOI] [PubMed] [Google Scholar]
- Harvey K, & Marchetti B (2014). Regulating Wnt signaling: A strategy to prevent neurodegeneration and induce regeneration. Journal of Molecular Cell Biology, 6(1), 1–2. 10.1093/jmcb/mju002 [DOI] [PubMed] [Google Scholar]
- Harvey K, & Outeiro TF (2019). The role of LRRK2 in cell signalling. Biochemical Society Transactions, 47(1), 197–207. 10.1042/BST2018046 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Itoh M, Yamanaka Y, Yamashita S, Shimizu T, Solnica-Krezel L, … Hirano T (2000). Zebrafish Dkk1 functions in forebrain specification and axial mesendoderm formation. Developmental Biology, 217(1), 138–152. 10.1006/dbio.1999.9537 [DOI] [PubMed] [Google Scholar]
- Houlden H, & Singleton AB (2012). The genetics and neuropathology of Parkinson’s disease. Acta Neuropathologica, 124(3), 325–338. 10.1007/s00401-012-1013-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki H, Blauwendraat C, Leonard HL, Kim JJ, Liu G, Maple-Grødem J, … Nalls MA (2019). Genomewide association study of Parkinson’s disease clinical biomarkers in 12 longitudinal patients’ cohorts. Movement Disorders, 34, 1839–1850. 10.1002/mds.27845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, … Shimizu N (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature, 392(6676), 605–608. 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Lambert AM, Bonkowsky JL, & Masino MA (2012). The conserved dopaminergic diencephalospinal tract mediates vertebrate locomotor development in zebrafish larvae. Journal of Neuroscience, 32(39), 13488–13500. 10.1523/JNEUROSCI.1638-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston RG, Rudenko IN, & Cookson MR (2016). The function of orthologues of the human Parkinson’s disease gene LRRK2 across species: Implications for disease modelling in preclinical research. The Biochemical Journal, 473(3), 221–232. 10.1042/BJ20150985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SB, Hanss Z, & Kruger R (2018). The genetic architecture of mitochondrial dysfunction in Parkinson’s disease. Cell and Tissue Research, 373(1), 21–37. 10.1007/s00441-017-2768-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand JN, Gonzalez-Cano L, Pavlou MA, & Schwamborn JC (2015). Neural stem cells in Parkinson’s disease: A role for neurogenesis defects in onset and progression. Cellular and Molecular Life Sciences, 72(4), 773–797. 10.1007/s00018-014-1774-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang D, Zhong S, Thomas JM, Xue F, Liu J, … Smith WW (2014). Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson’s disease cell and mouse models. Human Molecular Genetics, 23(23), 6212–6222. 10.1093/hmg/ddu341 [DOI] [PubMed] [Google Scholar]
- Lill CM (2016). Genetics of Parkinson’s disease. Molecular and Cellular Probes, 30, 386–396. 10.1016/j.mcp.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Liu G, Sgobio C, Gu X, Sun L, Lin X, Yu J, … Cai H (2015). Selective expression of Parkinson’s disease-related Leucine-rich repeat kinase 2 G2019S missense mutation in midbrain dopaminergic neurons impairs dopamine release and dopaminergic gene expression. Human Molecular Genetics, 24(18), 5299–5312. 10.1093/hmg/ddv249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec CE, Samuel J, Weng W, Wood IC, & Sirotkin HI (2016). Maternal rest/Nrsf regulates zebrafish behavior through snap25a/b. Journal of Neuroscience, 36(36), 9407–9419. 10.1523/JNEUROSCI.1246-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, … Argenton F (2012). In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Developmental Biology, 366(2), 327–340. 10.1016/j.ydbio.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J, Berwick DC, Granno S, Spain VA, Blackstone C, & Harvey K (2016). Protective LRRK2 R1398H variant enhances GTPase and Wnt signaling activity. Frontiers in Molecular Neuroscience, 9, 18 10.3389/fnmol.2016.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J (2002). An essay on the shaking palsy. Journal of Neuropsychiatry and Clinical Neurosciences, 14(2), 223–236; discussion 222. 10.1176/jnp.14.2.223 [DOI] [PubMed] [Google Scholar]
- Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, … Lang AE (2017). Parkinson disease. Nature Reviews Disease Primers, 3, 17013 10.1038/nrdp.2017.13 [DOI] [PubMed] [Google Scholar]
- Prabhudesai S, Bensabeur FZ, Abdullah R, Basak I, Baez S, Alves G, … Moller SG (2016). LRRK2 knockdown in zebrafish causes developmental defects, neuronal loss, and synuclein aggregation. Journal of Neuroscience Research, 94(8), 717–735. 10.1002/jnr.23754 [DOI] [PubMed] [Google Scholar]
- Ren G, Xin S, Li S, Zhong H, & Lin S (2011). Disruption of LRRK2 does not cause specific loss of dopaminergic neurons in zebrafish. PLoS One, 6(6), e20630 10.1371/journal.pone.0020630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko IN, Chia R, & Cookson MR (2012). Is inhibition of kinase activity the only therapeutic strategy for LRRK2- associated Parkinson’s disease? BMC Medicine, 10, 20 10.1186/1741-7015-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salasova A, Yokota C, Potesil D, Zdrahal Z, Bryja V, & Arenas E (2017). A proteomic analysis of LRRK2 binding partners reveals interactions with multiple signaling components of the WNT/ PCP pathway. Molecular Neurodegeneration, 12(1), 54 10.1186/s13024-017-0193-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho RM, Law BM, & Harvey K (2009). Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signalling pathways. Human Molecular Genetics, 18(20), 3955–3968. 10.1093/hmg/ddp337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng D, Qu D, Kwok KHH, Ng SS, Lim AYM, Aw SS, … Liu J (2010). Deletion of the WD40 domain of LRRK2 in zebrafish causes Parkinsonism-like loss of neurons and locomotive defect. PLoS Genetics, 6(4), e1000914 10.1371/journal.pgen.1000914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng D, See K, Hu XU, Yu D, Wang Y, Liu Q Liu, J. (2018). Disruption of LRRK2 in zebrafish leads to hyperactivity and weakened antibacterial response. Biochemical and Biophysical Research Communications, 497(4), 1104–1109. 10.1016/j.bbrc.2018.02.186 [DOI] [PubMed] [Google Scholar]
- Surmeier DJ (2018). Determinants of dopaminergic neuron loss in Parkinson’s disease. FEBS Journal, 285(19), 3657–3668. 10.1111/febs.14607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, & Postlethwait JH (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development, 119(4), 1203–1215. [DOI] [PubMed] [Google Scholar]
- Thomas B, & Beal MF (2007). Parkinson’s disease. Human Molecular Genetics, 16(Spec No. 2), R183–R194. 10.1093/hmg/ddm159 [DOI] [PubMed] [Google Scholar]
- Tolosa E, Wenning G, & Poewe W (2006). The diagnosis of Parkinson’s disease. The Lancet Neurology, 5(1), 75–86. 10.1016/S1474-4422(05)70285-4 [DOI] [PubMed] [Google Scholar]
- Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, & Moon RT (2005). The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Current Biology, 15(6), 489–500. https://doi.org/10.1016Zj.cub.2005.01.041 [DOI] [PubMed] [Google Scholar]
- Wolman MA, Jain RA, Liss L, & Granato M (2011). Chemical modulation of memory formation in larval zebrafish. Proceedings of the National Academy of Sciences of the United States of America, 108(37), 15468–15473. 10.1073/pnas.1107156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Ryan J, Noble S, Yu M, Yilbas AE, & Ekker M (2010). Impaired dopaminergic neuron development and locomotor function in zebrafish with loss of pink1 function. European Journal of Neuroscience, 31(4), 623–633. 10.1111/j.1460-9568.2010.07091.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Assessment of Locomotor Behavior of lrrk2 Mutants. (a,e,i) The duration of movements per minute in the light and dark for (a) lrrk2+/+ (n = 28) and /rrk2sbu96/sbu96 (n = 21), (e) lrrk2+/+ (n = 34) and lrrk2sbu71/sbu71 (n = 35), (i) lrrk2+/+ (n = 24) and lrrk2sbu304/sbu304 (n = 24). (b,f,j) The distance traveled per minute for each genotype tested in the light and dark periods. (c,g,k) The average swim length (distance traveled per movement) for each genotype. (d,h,l) The average speed per movement for each genotype
FIGURE S2 dat mRNA Expression and Cell Count /rrk2sbu304 Mutants. (a,b) RNA in situ hybridization for dat in 72 hpf wild type and lrrk2sbu304 mutant embryos from heterozygous crosses. Images are dorsal view focused on the midbrain
FIGURE S3 Wnt Reporter Tg(7xTCF-X/a.Siam:GFP)ia4in lrrk2 Mutants. (a-f) Fluorescent images of lrrk2sbu96, lrrk2sbu71 and lrrk2sbu304 mutants in a Siam:GFP background at 56 hpf. Images taken in lateral view
FIGURE S4 mRNA Expression of Wnt Target Genes in lrrk2sbu304 Mutants. (a-d) Real-time qPCR showing the expression level of (a) dkk1 (n = 3), (b) lef1 (n = 3), (c) sp5 (n = 3) and (d) sp5l (n = 3) for lrrk2+/+ and lrrk2sbu304/sbu304 56 hpf for vehicle (DMSO)- and BIO (0.5 μM)-treated embryos. (a) Expression levels of dkk1 comparing lrrk2+/+ treated and lrrk2sbu304/sbu304 treated (p = 0.0094), lrrk2+/+ control and treated (p = 0.00076), and lrrk2sbu304/sbu304 control and treated (p = 0.018). (c) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 0.00084) and lrrk2+/+ treated lrrk2sbu304/sbu304 treated (p = 0.0035). (d) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.023). (e-h) Real-time qPCR showing the expression of (e) dkk1 (n = 6), (f) lef1 (n = 6), (g) sp5 (n = 6), and (h) sp5l (n = 6) for lrrk2+/+ and lrrk2sbu304/sbu304 at 6 hpf for vehicle (DMSO)- and BIO (0.5 μM)-treated embryos. (e) Expression levels of dkk1 comparing lrrk2+/+ control and treated (p = 4.8e-5), and lrrk2sbu304/sbu304 control and treated (p = 6.2e-5). (f) Expression levels of lef1 comparing lrrk2+/+ control and treated (p = 0.022). (g) Expression levels of sp5 comparing lrrk2+/+ control and treated (p = 1.9e-6) and lrrk2sbu304/sbu304 control and treated (p = 0.00002). (h) Expression levels of sp5l comparing lrrk2+/+ control and treated (p = 0.006). Statistical analysis for pairwise comparisons was done with a post hoc Tukey’s test. A two-way analysis of variance was conducted between genotype and treatment. There was a significant interaction between genotype and treatment for (c) sp5 (F(1,8) = 10.889, n = 3, p = 0.0108). p < 0.05 (*), p < 0.01 (**), p < 0.001 (***)