Abstract

Comprehensive two-dimensional gas chromatography (GC×GC) is a powerful analytical tool for both nontargeted and targeted analyses. However, there is a need for more integrated workflows for processing and managing the resultant high-complexity datasets. End-to-end workflows for processing GC×GC data are challenging and often require multiple tools or software to process a single dataset. We describe a new approach, which uses an existing underutilized interface within commercial software to integrate free and open-source/external scripts and tools, tailoring the workflow to the needs of the individual researcher within a single software environment. To demonstrate the concept, the interface was successfully used to complete a first-pass alignment on a large-scale GC×GC metabolomics dataset. The analysis was performed by interfacing bespoke and published external algorithms within a commercial software environment to automatically correct the variation in retention times captured by a routine reference standard. Variation in 1tR and 2tR was reduced on average from 8 and 16% CV prealignment to less than 1 and 2% post alignment, respectively. The interface enables automation and creation of new functions and increases the interconnectivity between chemometric tools, providing a window for integrating data-processing software with larger informatics-based data management platforms.
Introduction
Advanced analytical technologies are revealing new chemical complexities in our environment (e.g., wastewaters and air quality), our bodies (e.g., metabolome and microbiome), and our food and commodities (e.g., packaging, materials, and petrochemicals). Comprehensive two-dimensional gas chromatography (GC×GC) is a technique that (theoretically) affords unparalleled separation of volatile and semivolatile matrices, providing a powerful analytical tool for both nontargeted and targeted analyses.1−3 The increased peak capacity and separation of individual compounds within complex mixtures can benefit studies focused on biomarker discovery and signature profiling, such as global metabolomics.4−7 Targeted methods for screening, such as biomonitoring persistent organic pollutants, benefit from less-extensive sample preparations and increased confidence in chemical assignment.8,9
Much work has been done to advance the hardware to make the technique more accessible and affordable and reach new chromatographic optima.1,10,11 However, the complexity of the resultant datasets makes it difficult to automate, reproduce, and share data and data workflows. Increased adoption of GC×GC for routine analyses and for larger scale discovery analysis is at risk from the lagging development of more integrated platforms for data processing, management, and storage.
Commercially available pieces of software for processing GC×GC data (coupled to both univariate, e.g., flame ionization detectors, and multivariate detectors, e.g., mass spectrometric detectors, herein described collectively as “GC×GC” data) are powerful. They have high functionality and advanced graphical user interfaces (GUIs) (Table S1). However, despite efforts to make the software user friendly (e.g., by introducing guided workflows), users can require extensive periods of learning (with one or two dedicated experienced users per group). They are also expensive and often instrument-specific, restricting analysts to one type of software, and the data output may be incompatible with other software or require time-consuming export steps.
Furthermore, commercial or original equipment manufacturer (OEM) software for processing GC×GC data is not flexible enough for the different workflows that exist within the various applications of GC×GC. Although it is not economically viable for vendors to create custom functions on an individual basis, researchers need tools that maintain a level of flexibility to meet the demands of different projects and experimental designs. The locked-down nature of proprietary systems, although some are scriptable, makes it difficult to embed them in custom data-processing pipelines.
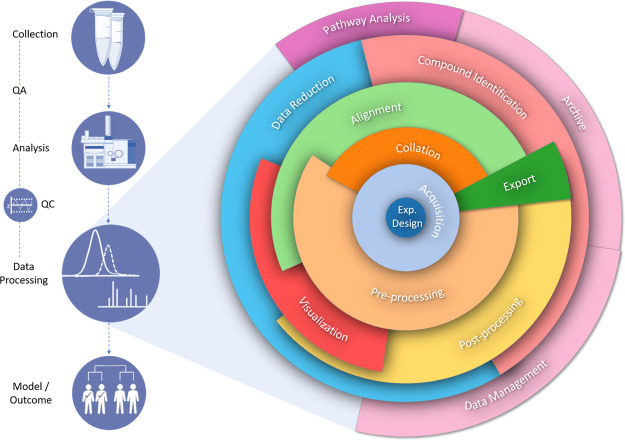
For instance, the processing of chromatographic data for the generation of a peak table (i.e., data matrix), including baseline correction, peak detection, and the critical step of alignment, is often only one part of a wider workflow (Figure 1). These steps can be preceded by sample collection and quality control steps, followed by multivariate analysis and compound identification, finally uploading the data to a data management platform or repository (Figure 1).
Figure 1.
Flow diagram depicting the complexity and layers of GC×GC data processing within a wider data workflow (preprocessing includes steps such as baseline correction and signal smoothing. Postprocessing includes steps such as feature extraction/peak detection. Data reduction can include application of multivariate analysis and machine learning techniques).
Several chemometric methods using free and open-source software (FOSS) or bespoke tools and scripts have been developed for processing GC×GC data (Table S1).12 Such tools overcome issues of accessibility and cost; they are often compatible with multiple file types for wider adoption; and the open-source programming language allows modification by the user.
Nonetheless, poor-quality supporting documentation can make it very time-consuming to determine how to use functions owing to a lack of user-friendly GUIs. They can also be a niche for certain application areas or address one task or specific part of a workflow, for example, an algorithm for performing alignment or peak detection. These tools are a great resource, which enhance the analyst’s tool kit; however, this only increases the complexity of the task faced by the analysts in choosing the optimum workflow for their data. This means that the user might have to swap between multiple software to process a single dataset. In addition, they are often specific to one programming language (unless produced as libraries with wrappers in different languages); they all are not readily accessible (i.e., published as “in-house”), and although the package or script may be made freely available, the environment/platform may not be freely available.
Consequently, analysts report unique combinations of both commercial software and FOSS. The powerful core functions of commercial software are used as a foundation (acquiring and converting raw data files at minimum) and then expanded on using external FOSS alongside manual editing to produce a data workflow.4,5,13−19 This is done in discrete laborious steps, which are difficult to report and reproduce.
Herein, we provide the primary description for interfacing open-source and commercial software for processing GC×GC data. The objectives were to demonstrate the use of an underused interface that exists within commercial software (i) for automating steps, which were not previously possible within the software GUI, (ii) for creating new functions, (iii) for integrating open-source and bespoke code with commercial software functions, and (iv) to highlight its future potential for increasing interconnectivity between chemometric tools.
To keep the proof of concept simple, the command-line interface was used in a popular piece of software for GC×GC data processing. We illustrate its use for automating the alignment of a metabolomics dataset as an example to provide analysts with the knowledge needed to accelerate and automate their own workflows. The new method makes it possible to combine the powerful functionality of commercial software and the flexibility of FOSS to produce a custom data workflow within a single software environment, tailoring the commercial software and workflow to the needs of the individual researcher.
Experimental Details
Analysis by GC×GC was conducted as described previously using an Agilent 7890A gas chromatogram, fitted with a G3486A CFT flow modulator.20 The instrument was coupled to a TD-100xr thermal desorption autosampler (Markes International Ltd, Llantrisant, UK). Sample tubes were placed in trays, typically six sample tubes per tray along with a tube loaded with the n-alkane and aromatic mixture and in every four trays another tube loaded with a reference indoor air mixture.20
Exhaled volatile organic compounds (VOCs) were collected from breath using the ReCIVA device (Owlstone Medical, Cambridge, UK) within a prospective real-world observational study involving adults presenting with self-reported acute breathlessness.20,21 Written informed consent was obtained from all participants. The study protocol was approved by the National Research Ethics Service Committee East Midlands (REC number: 16/LO/1747) IRAS 198921.
Data were acquired in MassHunter GC–MS Acquisition B.07.04.2260 (Agilent Technologies Ltd, Stockport, UK) and the data were processed using the command-line interface within the GC Image v2.8 suite (GC Image, LLC. Lincoln, NE, US). Step-by-step examples of the data analysis are described in further detail. The files used in the exemplar methods have been made available with supporting documentation via a repository at https://github.com/rcfgroup/gc-automation.
Results and Discussion
Current Software and Tools
A plethora of software tools exist for chemometric processing of GC×GC data (Table S1). These methods and tools have been reviewed previously.12,22,23 In addition, a vast number of commercial and open-source data-processing tools that are not specific for GC×GC data exist, usually for applications in metabolomics, and these have been reviewed previously.24,25 The aim, herein, is not to provide users with a new algorithm (e.g., for alignment or peak detection) or new dedicated workflow for a specific application. This method is also not to be confused with automated compound classification based on scripting, such as Microsoft VBScript in ChromaTOF18,26 and the CLIC expression tool in GC Image,27−29 for filtering features based on advanced mass spectral fragmentation pattern matching. The approach herein automates analyses by integrating commercial software as a module into a FOSS-based workflow, bringing together multiple steps and data transitions into a single input and output.
Inclusion of commercial software within a data workflow avoids repeated development; these software have full-featured GUIs for examining results and are based on proven methods capable of completing all the essential chromatographic processing and feature extraction steps. These typically include at least one method for baseline correction, peak detection (including deconvolution for MS data), alignment, data visualization (e.g., 2D and 3D rendering of chromatograms), compound identification (e.g., MS library search), and at least some basic descriptive statistics (Table S1). Only very recently have a small number of FOSS GC×GC data tools been published that attempt to offer an end-to-end workflow or pipeline that competes with commercial software functionality for large-scale datasets (Table S1).30−35 However, these tools often still require preprocessing within the vendor software before being imported. The user also does not benefit from the powerful proprietary functions developed within commercial software. One of the benefits of the approach described herein is that it allows workflows that have been developed within commercial software to be scaled up for larger or multiple datasets.
Interfacing Open-Source and Commercial Software
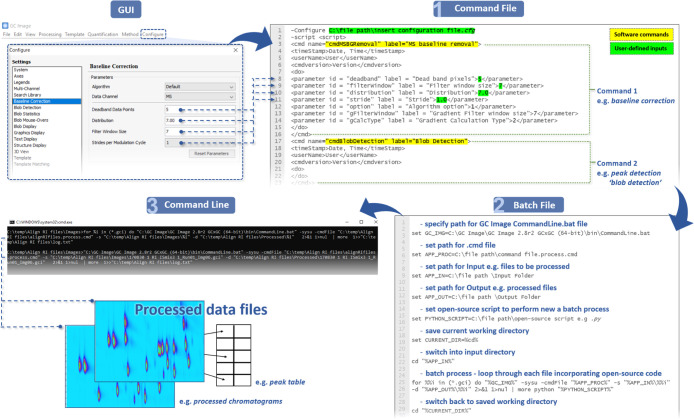
The steps for interfacing FOSS and commercial software are shown in Figure 2. To demonstrate the concept, we describe the use of the command-line interface accessible within the GC Image software. However, we emphasize that this is a proof of principle and not limited to the software used herein. Indeed, any software which has defined inputs/outputs and a command-line interface could be used (other examples are detailed in the Supporting Information). Additionally, any software which has a documented programming library or has a software development kit could be embedded into data processing scripts, for example, through Python.
Figure 2.
Flow diagram showing steps for using the command-line interface, extending beyond a commercial software GUI with the creation of a command and batch file where open-source code can be integrated, increasing the interconnectivity of chemometric tools.
Step 1—Generate a Command File
Using the command-line interface is reliant on creating two files: a command file (.cmd) and a batch file (.bat).36 The command file is the list of processing steps the user would like to perform on the data files (e.g., baseline correction and peak detection), and this can utilize the main functions in the commercial software (Figure 2; step 1).
There are two ways to generate the command file. A list of commands can be directly exported from the software, or the individual commands can be located in the program directory. The XML script format is easy to follow and modify, even for those with minimal coding experience. Each “command” or data-processing step is contained between the tag operators, <cmd name => and </cmd> (Figure 2; step 1). Details on how to generate a command file are available in the Supporting Information and example.cmd files have been made available.
Step 2—Generate a Batch File
The batch file (.bat) then tells the command-line interface to execute these steps and provides the user an opportunity to integrate their open-source code. The batch file applies the steps outlined in the command file to all the files in the input folder. In the batch file, the user specifies the file path for the interface (viz., the command line in GC Image), the path for the command file, and the paths for the input and output folders (Figure 2; step 2). The user can then also integrate their open-source code to either extend the batch processing capability of the commercial software, perform new functions, or link the data once processed with other chemometric tools (see the example batch files in the Supporting Information and repository).
Step 3—Command-Line Interface
After the command and batch files have been created, to execute the analysis, the user simply adds the sample files to the input folder and double clicks the batch file. When using the command line herein, the terminal window opens, showing the files being automatically processed (Figure 2; step 3). Once this is complete, the processed files appear in the output folder. If additional outputs are specified in the code, such as the creation of a report or another type of data export, a file path to another directory can be added.
A folder containing the abovementioned can then be shared for another user to repeat the analysis, simply changing the data in the input folder and double clicking the .bat file to perform a new analysis, making reproducing the analysis simple.
Example Applications
To demonstrate the use of the command-line interface, four examples are described for (i) automating steps which were not previously possible to batch process within the software, (ii) creating new functions, (iii) integrating open-source code with commercial software processing, (iv) and sharing workflows. These examples were then combined in a “real-world” example to demonstrate the use of the command line in the processing of GC×GC data collected within a clinical study focused on the analysis of exhaled breath metabolites. Example data, files, and guided instructions for all steps have been made available.
Example 1. Reprocessing Data
To batch process data, a “method” is created, which is then applied to all files. In this instance, the method was created in GC Image and executed in Project, during which processed chromatograms are saved as .gci files. A common step after batch processing is to perform a quick review of the processed data to check if specific peaks have been detected or to identify overloaded peaks and anomalous samples. This is often still quicker than processing each sample individually. However, once processed, each “image” file can be edited individually but a batch-wide change cannot be made (in Project) without reprocessing from the raw data again under “Runs”. This voids any incremental changes made during the review.
The command-line interface overcomes this issue (Figure 2) as it is not bound by the file type. A command file was generated, which contained the steps the user wanted to amend, such as changing a peak template, changing the matching thresholds in the configuration file, or changing the visualization. The previously processed and reviewed chromatograms were placed in the input folder as .gci files (with corresponding .bin files) and the batch file was used to perform the amendments, making the changes without having to reprocess again from the raw data.
Example 2. New Functions
A common function within commercial software is the use of a “template” (also referred to as stencils or graphics). A template is a graphical layer, which denotes the position of a peak or group of peaks of interest using a marker or outliner.
The layer retains descriptive information about the peak(s), such as the compound name and structure, retention positions, and mass spectra. The template can then be loaded and matched onto other chromatograms, identifying the same peak(s) across multiple samples, as a form of targeted analysis.
The “distance” or similarity between the template marker and the position of the peak in a new sample is reported, and the vector representing this difference can be exported as a “match” file. This vector can be used in other processes, such as alignment based on a transformation.37−41
However, the function for exporting match files from a batch is not available in the software GUI. The user would have to export each match file individually. Therefore, a new function was created using the command-line interface. A simple Python script was added to the batch file. This script extracted the retention times of the peaks of interest from a summary report (a command available in the software, added to the command file) and the retention times of the reference peaks from the template file (.bt). The script then combined the two sets of retention time data to produce the exported match files containing the vectors for the entire dataset. This example expanded the core functionality of the commercial software (e.g., templates) by integrating open-source code (providing new automation), within a single software environment.
Example 3. Integrating Open-Source Tools
This example demonstrates the integration of one new and one published open-source tool for performing alignment. The vector in the match file can be used to align chromatograms based on a transformation. Types of transformation available can be extended through plug-ins (in GC Image). For example, Gros et al.38 published a new algorithm based on the alignment of local regions, determined by a Voronoi decomposition, followed by Sibson natural neighbor interpolation. The original open-source algorithm was developed in MATLAB (Table S1) and made into a plug-in. It is possible to utilize such plug-ins or FOSS scripts within the command-line interface, incorporating them within the command file.
Batch alignment in the commercial software, as described above, can only be performed using one match file at a time. Although the files can be organized into “batches” in Project, it simply organizes the files into subfolders and does not make the workflow scalable. However, using the command-line interface, it is possible to automatically align subsets of data using multiple vectors. This is applicable during routine analyses involving the regular analysis of a reference mixture to monitor retention shifts or determine retention indices to align data on a more frequent basis. Using the command-line interface, it was possible to integrate a bespoke Python script (made available), which automatically identified a date and batch number at the beginning of each filename of the QC/reference chromatograms (e.g., n-alkane mixtures). The script then automatically found the samples which had the corresponding date and batch number and performed the alignment using the corresponding match file. In doing so, the entire dataset underwent first-pass alignment through the commercial software using the open-source alignment algorithm, in multiple discrete batches with a minimal user input. In this example, the workflow benefitted from the powerful proprietary functions within the commercial software (e.g., a platform for performing alignment), a published open-source algorithm (developed in MATLAB), and a new function for smart automation (bespoke Python code), within a single software environment (command-line interface).
Example 4. Workflow and Data Sharing
Creating a new workflow using the command-line interface still requires user input and method development by an experienced analyst. This is only as challenging as using the commercial and FOSS already being employed. However, a major advantage of integrating the steps through the interface, within a single environment, is the increased reproducibility, enabling other users, independent of experience, to process similar data.
A workflow can be readily shared in a code repository such as Github as demonstrated herein or by sending the folder containing the two files and folder tree. For other users to repeat the data-processing steps (expert or nonexpert), they add their unprocessed data files into the input folder and double click the .bat file to generate the same output. The protocol for repeating an analysis is greatly simplified, reducing the potential error in reproducing the data analysis and swapping between multiple pieces of software. In the current example, it is dependent on the other user having access to the same commercial software, which is inherent in the replication of most workflows. However, if the commercial software functions are well modularized, with a well-defined common input and output, then, it could be replaced by another commercial software as long as the software could be evoked to work in a similar way.
Interfacing Commercial and Open-Source Software for Processing Clinical Breath Data
Metabolomics is a research area where GC×GC has demonstrated promising results, especially in studies concerned with the analysis of the human volatilome.3 The enhanced separation of complex biofluids collected using noninvasive methods places GC×GC at the forefront of biomarker discovery platforms alongside LC–MS assays (e.g., for lipidomics) and high-resolution mass spectrometric methods (e.g., SESI-Orbitrap MS and MALDI- and SIFT-MS).6,42 In particular, GC×GC has proven effective for the analysis of the exhaled breath metabolome (“breathome”), which contains hundreds to thousands of VOCs. Breath analysis is being widely adopted in clinical studies for the discovery of breath-based biomarkers for a variety of diseases.43 For these studies to produce meaningful data, large patient cohorts and sample sizes are required, which means scalable data workflows for processing the GC×GC datasets collected must be developed.
A subset of data was taken from a clinical study collecting breath samples from patients suffering with acute respiratory diseases. Each patient was sampled at two time points: within 24 h of admission and later during a period of convalescence. At each time point, breath, room air, and air supply samples were collected. Continuous recruitment was planned over two years, with up to 20 patient visits/week (∼200 tubes per month). In addition, despite the clinical advantages of breath, the matrix poses unique analytical challenges. Offline analyses based on sorbent tube sampling mean short storage times, longer preparation times, and higher consumable costs (cf. practicalities of collecting breath on sorbent tubes with serum in vials).
Common approaches for analyzing samples by GC×GC are designed to minimize the variance introduced by instrumentation over time, such as retention shift and signal response, and as such reduce the burden of ensuring that the extracted features are comparable across all samples during data processing. Such approaches can include analyzing the majority of samples by GC–MS and a subset by GC×GC; or to wait for all samples to be collected and analyze in “one go” by GC×GC; or to collect samples in 2–3 batches and align per batch. Furthermore, sampling is often from vials by liquid injection or SPME. However, for studies with continuous sample collection over extended periods of time or large sample sizes based on sorbent tube sampling, such as the clinical study described (vide supra), none of the previous approaches were amenable. Therefore, this sample set provided an opportunity to demonstrate a novel use of interfacing commercial and open-source software to help address the issue of alignment.
Samples were analyzed in discrete analytical batches, six per tray with a reference mixture of n-alkanes and aromatics analyzed per tray. These discrete analytical batches reflected the continuous nature of the sampling campaign and analysis.20 However, the guided workflow in the commercial software was not optimized to automate the processing of multiple small batches. For the iterative extraction of all features across all samples as a part of a discovery workflow, many pieces of software rely on user-defined windows (often only one window per parameter for the whole chromatogram) based on retention time and/or mass spectral match, in order to identify the same chromatographic features across all samples. This can result in a high number of errors when implemented across a large sample size that has been collected and analyzed over a long timeframe.
To reduce the potential for errors herein, a command-line approach was used to automate a first-pass alignment on all the samples; performed every six runs using the reference n-alkane and aromatic mixture at the beginning of each tray (Figure S1). Little to no variation in primary and secondary retention times (1tR and 2tR) was observed within a tray, less than 0.1 and 1.3% RSD, respectively. First, a template of thirty compounds in the reference mixture, which covered the 2D chromatographic space, was applied to 414 reference chromatograms using the commercial software GUI. This captured the variation in the retention times of the n-alkane and aromatic standards over the period of recruitment. Next, an “alignment template” was then made from a subset of the thirty compounds based on the reference mixture from the first tray. It was important not to use all 30 compounds as reference points, as this can result in overfitting and spurious results for the algorithm used (optimum number is algorithm-dependent). The compounds not included in the subset provided an internal validation for assessing the effects of the alignment on the peak area and 1tR and 2tR.
Using the command-line interface, a match file (i.e., the vector representing the difference between the template and the peaks) was generated for each reference chromatogram labelled with the date and tray number (see examples 1 and 2). Next, 677 files comprising breath and corresponding environmental air samples (representing a large dataset), analyzed at different times throughout the recruitment period, were transferred to the input folder and a new command and batch file was produced. This command and batch file (as described in example 3) instructed the software to find the date and tray number at the beginning of each sample (e.g., a new function added by the user) and align the chromatogram (e.g., an existing function within the commercial software) using the algorithm developed by Gros et al.38 (e.g., integrating published open-source code) based on the corresponding match file automatically generated from the reference chromatogram with the same date and tray number. In effect, alignment vectors were automatically exported and then used to align all the samples based on the batch and tray number, correcting variation in the retention position captured by the reference chromatograms every six samples (Figure S1).
The remaining compounds not included in the template were used to assess the effect of the alignment on the peaks. Across the 414 reference chromatograms, the peak areas of all compounds pre- and post alignment had coefficients of variation (CV) of less than 1.5%, and the postalignment variation in 1tR and 2tR was less than 1.0 and 2.0%, respectively. This was an improvement for both 1tR and 2tR, which was on average 8 and 16% CV pre-alignment (dependent on compound 2tR and time since column change). This demonstrated that the first-pass alignment based on the local transformation by Gros et al.38 executed and automated through the command-line interface successfully reduced the variation in retention time observed in the samples across the recruitment period while having a minimal effect on the extracted peak areas. Further verification was provided using an external reference mixture, which contained VOCs from additional chemical classes (aldehydes, ketones, aromatics, branched- and straight-chain hydrocarbons, and terpenoids), independent of that included in the alignment template, run every four trays. The peak areas, across 120 chromatograms analyzed at different times throughout the recruitment period, had a pre- and post alignment CV of less than 3.5%. The processing time for a large dataset such as this was reduced from several weeks by manual processing of hundreds of files (including individual exports and numerous opening and closing of files and programs) to less than 2 h.
This example demonstrates how interfacing commercial and open-source software through the use of the command-line interface automated a multistep process for iterative batch processing of a large-scale GC×GC dataset within a single software environment. However, as mentioned previously, the aim herein was not to provide users with a new workflow for a specific application. The purpose was to provide analysts with the knowledge to accelerate and automate their own workflows through the use of the interface.
After the users become familiar with the basics of the method (see the Supporting Information), it is expected that they can build on the concept of using an interface in their own workflows. The number of applications for which this approach can be used is large as it is capable of automating tasks of high complexity. For example, the interface could be used to compensate for specific tasks a commercial software might lack (e.g., PARAFAC deconvolution) or new tasks which have not been developed as they lacked a method of implementation. Workflows still benefit from the powerful functionality of commercial software but are no longer bound by the GUI but that of the programming language its interfaced with.
Future Development
The method herein can be expanded in the future across many applications. However, in order to be as effective as possible, in addition to the command line and programming libraries, software manufacturers should provide and support a full application programming interface (API). This would allow different functions to run independently of the GUI (e.g., in Python scripts), with the ability to move data in and out. Depending on its final implementation, vendor-specific, the method would then also be computationally optimized. The alternative is for software manufacturers to commercialize popular methods developed in FOSS, such as the integration of Tile-based alignment and Fisher ratios into LECO’s ChromaTOF software.44
Calls for standardization in research areas such as metabolomics have highlighted a need for the development of integrated informatics platforms (e.g., LabPipe, tranSMART, and eCRF) covering end-to-end workflows from sample collection to data archiving. These platforms are used to streamline the management of metabolomics data and metadata and ensure that users abide by reported guidelines (e.g., MSI, ISA tools, and mwTab). The type of interface described herein is a window of opportunity for integrating GC×GC workflows within larger data management platforms. It allows communication between the commercial software and other FOSS platforms. Compliance with guidelines and the use of informatics-based platforms are becoming increasingly pertinent, and soon, only the software which can demonstrate interconnectivity, capable of interfacing with other chemometric tools, will be practical for large-scale discovery studies.
Conclusions
GC×GC workflows are challenging and often cannot be done in a single software environment. Through the use of an interface, such as the command line, users have the ability to create more powerful and flexible workflows by being able to mold commercial software with their own and others’ scripts and tools developed in FOSS.
The successful interface of open-source and commercial software for processing GC×GC data has been demonstrated. Advantages of the method include the automation of multisoftware steps within a single software environment and the addition of new functionalities within commercial software using open-source scripts. Applications include the development of more user-friendly tools; automated batch processing tailored for and by the individual researcher; the sharing of reproducible workflows; and the integration of data-processing tools and workflows with data standardization platforms (e.g., ISA), repositories, and sample collection platforms (e.g., LabPipe)45 for the seamless collection, analysis, and processing of high-fidelity datasets.
Acknowledgments
Research funding was provided by the MRC and EPSRC Stratified Medicine Grant for Molecular Pathology Nodes (grant no. MR/N005880/1). The research was supported by the NIHR Leicester Biomedical Research Centre, the NIHR Leicester Clinical Research Facility, and the Leicester Wellcome Trust ISSF (award no. 204801/Z/16/Z) to whom we are extremely grateful. The authors would like to acknowledge the contributions of Dr Qingping Tao, GC Image LLC, and Scott Campbell, SpectralWorks Ltd, for their guidance in helping us access the command-line interface within the commercial software and Dr Jonas Gros for his guidance on using the local alignment algorithm. We thank the anonymous reviewers for their input, particularly on shaping future directions. We would also like to acknowledge the invaluable efforts of the research nurses responsible for the in-clinic sample collection and the input from the wider EMBER consortium (https://ember.le.ac.uk/web). The views expressed are those of the author(s) and not necessarily those of the NHS and NIHR or the Department of Health.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c02844.
Instructions for using the example folders and for creating command and batch files. Files used in the exemplar methods and supporting documentation available via a repository at https://github.com/rcfgroup/gc-automation (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Amaral M. S. S.; Nolvachai Y.; Marriott P. J. Comprehensive Two-Dimensional Gas Chromatography Advances in Technology and Applications: Biennial Update. Anal. Chem. 2020, 92, 85–104. 10.1021/acs.analchem.9b05412. [DOI] [PubMed] [Google Scholar]
- Hashimoto S.; Takazawa Y.; Fushimi A.; Tanabe K.; Shibata Y.; Ieda T.; Ochiai N.; Kanda H.; Ohura T.; Tao Q.; Reichenbach S. E. Global and selective detection of organohalogens in environmental samples by comprehensive two-dimensional gas chromatography-tandem mass spectrometry and high-resolution time-of-flight mass spectrometry. J. Chromatogr. A 2011, 1218, 3799–3810. 10.1016/j.chroma.2011.04.042. [DOI] [PubMed] [Google Scholar]
- Higgins Keppler E. A.; Jenkins C. L.; Davis T. J.; Bean H. D. Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. Trends Anal. Chem. 2018, 109, 275–286. 10.1016/j.trac.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risticevic S.; Souza-Silva E. A.; DeEll J. R.; Cochran J.; Pawliszyn J. Capturing Plant Metabolome with Direct-Immersion in Vivo Solid Phase Microextraction of Plant Tissues. Anal. Chem. 2016, 88, 1266–1274. 10.1021/acs.analchem.5b03684. [DOI] [PubMed] [Google Scholar]
- Humston E. M.; Dombek K. M.; Tu B. P.; Young E. T.; Synovec R. E. Toward a global analysis of metabolites in regulatory mutants of yeast. Anal. Bioanal. Chem. 2011, 401, 2387–2402. 10.1007/s00216-011-4800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orešič M.; Hyötyläinen T.; Herukka S. K.; Sysi-Aho M.; Mattila I.; Seppänan-Laakso T.; Julkunen V.; Gopalacharyulu P. V.; Hallikainen M.; Koikkalainen J.; Kivipelto M.; Helisalmi S.; Lötjönen J.; Soininen H. Metabolome in progression to Alzheimer’s disease. Transl. Psychiatry 2011, 1, e57 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Yin X.; Prodhan M. A. I.; Zhang X.; Zhong Z.; Kato I. Global Plasma Profiling for Colorectal Cancer-Associated Volatile Organic Compounds: a Proof-of-Principle Study. J. Chromatogr. Sci. 2019, 57, 385–396. 10.1093/chromsci/bmz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez M. J.; Herrera S.; Solé D.; García-Calvo E.; Fernández-Alba A. R. Automatic Searching and Evaluation of Priority and Emerging Contaminants in Wastewater and River Water by Stir Bar Sorptive Extraction followed by Comprehensive Two-Dimensional Gas Chromatography-Time-of-Flight Mass Spectrometry. Anal. Chem. 2011, 83, 2638–2647. 10.1021/ac102909g. [DOI] [PubMed] [Google Scholar]
- Megson D.; Brown T. A.; Johnson G. W.; O’Sullivan G.; Bicknell A. W. J.; Votier S. C.; Lohan M. C.; Comber S.; Kalin R.; Worsfold P. J. Identifying the provenance of Leach’s storm petrels in the North Atlantic using polychlorinated biphenyl signatures derived from comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry. Chemosphere 2014, 114, 195–202. 10.1016/j.chemosphere.2014.04.061. [DOI] [PubMed] [Google Scholar]
- Bahaghighat H. D.; Freye C. E.; Synovec R. E. Recent advances in modulator technology for comprehensive two dimensional gas chromatography. Trends Anal. Chem. 2019, 113, 379–391. 10.1016/j.trac.2018.04.016. [DOI] [Google Scholar]
- Pollo B. J.; Alexandrino G. L.; Augusto F.; Hantao L. W. The impact of comprehensive two-dimensional gas chromatography on oil & gas analysis: Recent advances and applications in petroleum industry. Trends Anal. Chem. 2018, 105, 202–217. 10.1016/j.trac.2018.05.007. [DOI] [Google Scholar]
- Reichenbach S. E.; Tian X.; Cordero C.; Tao Q. Features for non-targeted cross-sample analysis with comprehensive two-dimensional chromatography. J. Chromatogr. A 2012, 1226, 140–148. 10.1016/j.chroma.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Veenaas C.; Bignert A.; Liljelind P.; Haglund P. Nontarget Screening and Time-Trend Analysis of Sewage Sludge Contaminants via Two-Dimensional Gas Chromatography-High Resolution Mass Spectrometry. Environ. Sci. Technol. 2018, 52, 7813–7822. 10.1021/acs.est.8b01126. [DOI] [PubMed] [Google Scholar]
- Almstetter M. F.; Oefner P. J.; Dettmer K. Comprehensive two-dimensional gas chromatography in metabolomics. Anal. Bioanal. Chem. 2012, 402, 1993–2013. 10.1007/s00216-011-5630-y. [DOI] [PubMed] [Google Scholar]
- Barcaru A.; Vivó-Truyols G. Use of Bayesian Statistics for Pairwise Comparison of Megavariate Data Sets: Extracting Meaningful Differences between GCxGC-MS Chromatograms Using Jensen-Shannon Divergence. Anal. Chem. 2016, 88, 2096–2104. 10.1021/acs.analchem.5b03506. [DOI] [PubMed] [Google Scholar]
- Harvey P. M.; Shellie R. A. Data Reduction in Comprehensive Two-Dimensional Gas Chromatography for Rapid and Repeatable Automated Data Analysis. Anal. Chem. 2012, 84, 6501–6507. 10.1021/ac300664h. [DOI] [PubMed] [Google Scholar]
- Beccaria M.; Mellors T. R.; Petion J. S.; Rees C. A.; Nasir M.; Systrom H. K.; Sairistil J. W.; Jean-Juste M.-A.; Rivera V.; Lavoile K.; Severe P.; Pape J. W.; Wright P. F.; Hill J. E. Preliminary investigation of human exhaled breath for tuberculosis diagnosis by multidimensional gas chromatography - Time of flight mass spectrometry and machine learning. J. Chromatogr. B 2018, 1074–1075, 46–50. 10.1016/j.jchromb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Purcaro G.; Stefanuto P.-H.; Franchina F. A.; Beccaria M.; Wieland-Alter W. F.; Wright P. F.; Hill J. E. SPME-GC×GC-TOF MS fingerprint of virally-infected cell culture: Sample preparation optimization and data processing evaluation. Anal. Chim. Acta 2018, 1027, 158–167. 10.1016/j.aca.2018.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo A.; Forbes S. L.; Stuart B. H.; Ueland M. Profiling the seasonal variability of decomposition odour from human remains in a temperate Australian environment. Aust. J. Forensic Sci. 2019, 51, 1–11. 10.1080/00450618.2019.1637938. [DOI] [Google Scholar]
- Wilde M. J.; Cordell R. L.; Salman D.; Zhao B.; Ibrahim W.; Bryant L.; Ruszkiewicz D.; Singapuri A.; Free R. C.; Gaillard E. A.; Beardsmore C.; Thomas C. L. P.; Brightling C. E.; Siddiqui S.; Monks P. S. Breath analysis by two-dimensional gas chromatography with dual flame ionisation and mass spectrometric detection - Method optimisation and integration within a large-scale clinical study. J. Chromatogr. A 2019, 1594, 160–172. 10.1016/j.chroma.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim W.; Wilde M.; Cordell R.; Salman D.; Ruszkiewicz D.; Bryant L.; Richardson M.; Free R. C.; Zhao B.; Yousuf A.; White C.; Russell R.; Jones S.; Patel B.; Awal A.; Phillips R.; Fowkes G.; McNally T.; Foxon C.; Bhatt H.; Peltrini R.; Singapuri A.; Hargadon B.; Suzuki T.; Ng L. L.; Gaillard E.; Beardsmore C.; Ryanna K.; Pandya H.; Coates T.; Monks P. S.; Greening N.; Brightling C. E.; Thomas P.; Siddiqui S. Assessment of breath volatile organic compounds in acute cardiorespiratory breathlessness: a protocol describing a prospective real-world observational study. BMJ Open 2019, 9, e025486 10.1136/bmjopen-2018-025486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce K. M.; Mohler R. E. A Review of Chemometrics Applied to Comprehensive Two-dimensional Separations from 2008-2010. Separ. Purif. Rev. 2012, 41, 143–168. 10.1080/15422119.2011.591868. [DOI] [Google Scholar]
- Zeng Z.; Li J.; Hugel H. M.; Xu G.; Marriott P. J. Interpretation of comprehensive two-dimensional gas chromatography data using advanced chemometrics. Trends Anal. Chem. 2014, 53, 150–166. 10.1016/j.trac.2013.08.009. [DOI] [Google Scholar]
- Johnson C. H.; Ivanisevic J.; Benton H. P.; Siuzdak G. Bioinformatics: The Next Frontier of Metabolomics. Anal. Chem. 2015, 87, 147–156. 10.1021/ac5040693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R.; Salek R. M.; Moreno P.; Cañueto D.; Steinbeck C. Navigating freely-available software tools for metabolomics analysis. Metabolomics 2017, 13, 106. 10.1007/s11306-017-1242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weggler B. A.; Gröger T.; Zimmermann R. Advanced scripting for the automated profiling of two-dimensional gas chromatography-time-of-flight mass spectrometry data from combustion aerosol. J. Chromatogr. A 2014, 1364, 241–248. 10.1016/j.chroma.2014.08.091. [DOI] [PubMed] [Google Scholar]
- Reichenbach S. E.; Kottapalli V.; Ni M.; Visvanathan A. Computer language for identifying chemicals with comprehensive two-dimensional gas chromatography and mass spectrometry. J. Chromatogr. A 2005, 1071, 263–269. 10.1016/j.chroma.2004.08.125. [DOI] [PubMed] [Google Scholar]
- Wilde M. J.; Rowland S. J. Structural Identification of Petroleum Acids by Conversion to Hydrocarbons and Multidimensional Gas Chromatography-Mass Spectrometry. Anal. Chem. 2015, 87, 8457–8465. 10.1021/acs.analchem.5b01865. [DOI] [PubMed] [Google Scholar]
- Wilde M. J.; Rowland S. J. Naphthenic acids in oil sands process waters: Identification by conversion of the acids or esters to hydrocarbons. Org. Geochem. 2018, 115, 188–196. 10.1016/j.orggeochem.2017.09.004. [DOI] [Google Scholar]
- Tian T.-F.; Wang S.-Y.; Kuo T.-C.; Tan C.-E.; Chen G.-Y.; Kuo C.-H.; Chen C.-H. S.; Chan C.-C.; Lin O. A.; Tseng Y. J. Web Server for Peak Detection, Baseline Correction, and Alignment in Two-Dimensional Gas Chromatography Mass Spectrometry-Based Metabolomics Data. Anal. Chem. 2016, 88, 10395–10403. 10.1021/acs.analchem.6b00755. [DOI] [PubMed] [Google Scholar]
- Titaley I. A.; Ogba O. M.; Chibwe L.; Hoh E.; Cheong P. H.-Y.; Simonich S. L. M. Automating data analysis for two-dimensional gas chromatography/time-of-flight mass spectrometry non-targeted analysis of comparative samples. J. Chromatogr. A 2018, 1541, 57–62. 10.1016/j.chroma.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo S.; Mattila I.; Miettinen J.; Orešič M.; Hyötyläinen T. Data Analysis Tool for Comprehensive Two-Dimensional Gas Chromatography/Time-of-Flight Mass Spectrometry. Anal. Chem. 2011, 83, 3058–3067. 10.1021/ac103308x. [DOI] [PubMed] [Google Scholar]
- Wei X.; Shi X.; Koo I.; Kim S.; Schmidt R. H.; Arteel G. E.; Watson W. H.; McClain C.; Zhang X. MetPP: a computational platform for comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry-based metabolomics. Bioinformatics 2013, 29, 1786–1792. 10.1093/bioinformatics/btt275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayedpour S.; Parastar H. RMet: An automated R based software for analyzing GC-MS and GC×GC-MS untargeted metabolomic data. Chemom. Intell. Lab. Syst. 2019, 194, 103866. 10.1016/j.chemolab.2019.103866. [DOI] [Google Scholar]
- Quiroz-Moreno C.; Furlan M. F.; Belinato J. R.; Augusto F.; Alexandrino G. L.; Mogollón N. G. S. RGCxGC toolbox: An R-package for data processing in comprehensive two-dimensional gas chromatography-mass spectrometry. Microchem. J. 2020, 156, 104830. 10.1016/j.microc.2020.104830. [DOI] [Google Scholar]
- Shah Y. I.; Paradkar A. R.; Dhayagude M. G.. Introduction to Biostatistics & Computer Science; Nirali Prakashan, 2008. [Google Scholar]
- Wang B.; Fang A.; Heim J.; Bogdanov B.; Pugh S.; Libardoni M.; Zhang X. DISCO: distance and spectrum correlation optimization alignment for two dimensional gas chromatography time-of-fligh mass spectrometry-based metabolomics. Anal. Chem. 2010, 82, 5069. 10.1021/ac100064b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J.; Nabi D.; Dimitriou-Christidis P.; Rutler R.; Arey J. S. Robust Algorithm for Aligning Two-Dimensional Chromatograms. Anal. Chem. 2012, 84, 9033–9040. 10.1021/ac301367s. [DOI] [PubMed] [Google Scholar]
- Pierce K. M.; Wood L. F.; Wright B. W.; Synovec R. E. A Comprehensive Two-Dimensional Retention Time Alignment Algorithm To Enhance Chemometric Analysis of Comprehensive Two-Dimensional Separation Data. Anal. Chem. 2005, 77, 7735–7743. 10.1021/ac0511142. [DOI] [PubMed] [Google Scholar]
- Vial J.; Noçairi H.; Sassiat P.; Mallipatu S.; Cognon G.; Thiébaut D.; Teillet B.; Rutledge D. N. Combination of dynamic time warping and multivariate analysis for the comparison of comprehensive two-dimensional gas chromatograms. J. Chromatogr. A 2009, 1216, 2866–2872. 10.1016/j.chroma.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Rempe D. W.; Reichenbach S. E.; Tao Q.; Cordero C.; Rathbun W. E.; Zini C. A. Effectiveness of Global, Low-Degree Polynomial Transformations for GCxGC Data Alignment. Anal. Chem. 2016, 88, 10028–10035. 10.1021/acs.analchem.6b02254. [DOI] [PubMed] [Google Scholar]
- Schleich F. N.; Zanella D.; Stefanuto P.-H.; Bessonov K.; Smolinska A.; Dallinga J. W.; Henket M.; Paulus V.; Guissard F.; Graff S.; Moermans C.; Wouters E. F. M.; Van Steen K.; van Schooten F.-J.; Focant J.-F.; Louis R. Exhaled Volatile Organic Compounds Are Able to Discriminate between Neutrophilic and Eosinophilic Asthma. Am. J. Respir. Crit. Care Med. 2019, 200, 444–453. 10.1164/rccm.201811-2210oc. [DOI] [PubMed] [Google Scholar]
- Wallace M. A. G.; Pleil J. D. Evolution of clinical and environmental health applications of exhaled breath research: Review of methods and instrumentation for gas-phase, condensate, and aerosols. Anal. Chim. Acta 2018, 1024, 18–38. 10.1016/j.aca.2018.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons B. A.; Marney L. C.; Siegler W. C.; Hoggard J. C.; Wright B. W.; Synovec R. E. Tile-Based Fisher Ratio Analysis of Comprehensive Two-Dimensional Gas Chromatography Time-of-Flight Mass Spectrometry (GC×GC-TOFMS) Data Using a Null Distribution Approach. Anal. Chem. 2015, 87, 3812–3819. 10.1021/ac504472s. [DOI] [PubMed] [Google Scholar]
- Zhao B.; Bryant L.; Wilde M.; Cordell R.; Salman D.; Ruszkiewicz D.; Ibrahim W.; Singapuri A.; Coats T.; Gaillard E.. LabPipe: an extensible informatics platform to streamline management of metabolomics data and metadata. 2019, arXiv:1910.13246. arXiv preprint. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.