Summary
Advances in whole-genome sequencing (WGS) technologies have documented genetic diversity and epidemiology of the major foodborne pathogen Listeria monocytogenes (Lm) in Europe and North America, but data concerning South America are scarce. Here, we examined the population structure and genetic diversity of this major foodborne pathogen collected in Brazil. Based on core genome multilocus sequence typing (cgMLST), isolates from lineages I (n = 22; 63%) and II (n = 13; 37%) were distributed into 10 different sublineages (SLs) and represented 31 new cgMLST types (CTs). The most prevalent SLs were SL9 (n = 9; 26%), SL3 (n = 6; 17%) and SL2 and SL218 (n = 5; 14%). Isolates belonging to CTs L2-SL9-ST9-CT4420 and L1-SL315-ST520-CT4429 were collected 3 and 9 years apart, respectively, revealing long-term persistence of Lm in Brazil. Genetic elements associated with stress survival were present in 60% of isolates (57% SSI-1 and 3% SSI-2). Pathogenic islands were present in 100% (LIPI-1), 43% (LIPI-3) and 6% (LIPI-4) of the isolates. Mutations leading to premature stop codons were detected in the prfA and inlA virulence genes. This study is an important contribution to understanding the genomic diversity and epidemiology of Lm in South America. In addition, the results highlight the importance of using WGS to reveal Lm long-term persistence.
Introduction
The gram-positive bacterium Listeria monocytogenes (Lm) is an important foodborne pathogen that is able to survive in stressful environmental conditions and grow in refrigerated foods. The consumption of contaminated food has been linked with both epidemic and sporadic listeriosis. Despite the low overall incidence of listeriosis, the disease is associated with high hospitalization and mortality rates (Orsi et al., 2011; de Noordhout et al., 2014; Lomonaco et al., 2015; Charlier et al., 2017). Invasive forms of the disease are particularly dangerous for immunocompromised individuals and pregnant woman, leading to sepsis, meningitis, encephalitis, or abortion (Silk et al., 2012; de Noordhout et al., 2014; Charlier et al., 2017).
Lm population structure is relatively clonal, divided into four phylogenetic lineages (I, II, III and IV; Orsi et al., 2011) and multiple clonal complexes (CCs, defined on the basis of multilocus sequence typing [MLST]; Ragon et al., 2008) or sublineages (SLs, defined on the basis of core genome MLST; Moura et al., 2016). Major CCs and SLs are distributed globally (Chenal-Francisque et al., 2011; Haase et al., 2014; Moura et al., 2016) and can be heterogeneous in terms of virulence (Maury et al., 2016; Maury et al., 2017). Although with limited discriminatory power, Lm can also be distinguished into 13 different serotypes on the basis of the flagellar and somatic antigenic differences, which can be grouped into six PCR-based genoserogroups (IIa, IIb, IIc, IVb, IVb-v1, L; Doumith et al., 2004; Leclercq et al., 2011). Together, serogroups IVb and IIb (lineage I) and IIa (lineage II) are responsible for a large part of confirmed cases of listeriosis (Lomonaco et al., 2015).
Listeriosis is a notifiable disease in North America and in many European countries (Camargo et al., 2017), but not in Brazil. Existing national epidemiological surveillance programs actively detect and investigate clusters of cases, identify the origin of food contamination and implement control measures. Whole-genome sequencing (WGS) has recently emerged as a powerful tool for national and international outbreak investigations (Schmid et al., 2014; Ruppitsch et al., 2015; Moura et al., 2017; Nielsen et al., 2017; Van Walle et al., 2018). WGS allows an unprecedented subtyping resolution by analysis of the core genome, as well as the accessory genome. Questions related to potential virulence heterogeneity and antimicrobial resistance genes can also be answered, and outbreak investigations have become more precise by correlating epidemiological data with genetic characteristics of the isolates involved (Nielsen et al., 2017; Lüth et al., 2018; Schürch et al., 2018).
Considering the importance of listeriosis, the widespread distribution of Lm, and the absence of data from Brazil, we used WGS to subtype and characterize Lm isolates recovered in the last decades from beef, food production environment (PE) and clinical samples from eight Brazilian states. This information was then compared with genomes from other countries in South America, North America and Europe.
Results and discussion
In Brazil, few reports of listeriosis outbreaks or sporadic cases have been described and often no epidemiological link has been established (Camargo et al., 2017). In addition, isolates from outbreaks have only been superficially characterized. To better understand the population structure and molecular diversity of Lm in Brazil, we sequenced 35 genomes representative of isolation dates, geographic extent and sample origin. The quality metrics of the sequence data and draft assemblies are available in Supporting Information Table S1. All draft genomes met the quality standards necessary for core genome multilocus sequence typing (cgMLST) analysis (Moura et al., 2016).
Lm population structure
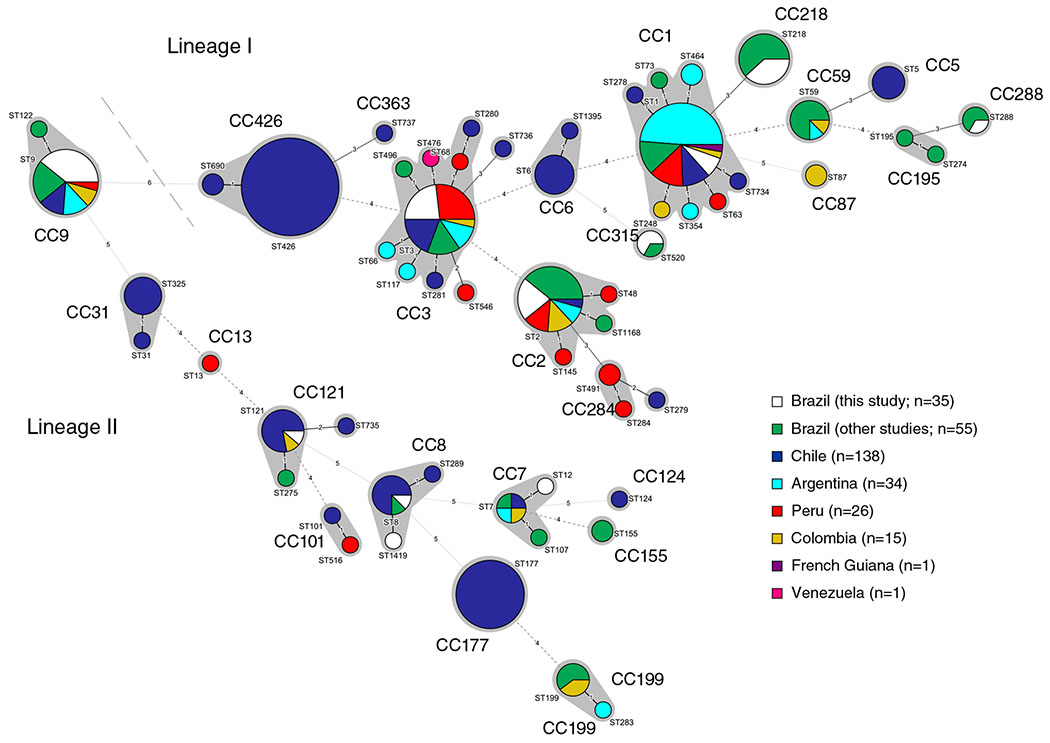
The Brazilian isolates were grouped by lineage I (n = 22; 63%) and lineage II (n = 13; 37%) and by genoserogroups IVb (n = 10; 28.6%), IIc (n = 9; 25.7%), IIb (n = 7; 20%), IVb-v1 (n = 5; 14.3%) and IIa (n = 4; 11.4%). On the basis of 7-loci MLST, isolates were distributed among 11 different sequence types (STs; Table 1) and 10 CCs, most of them previously reported in South America (Fig. 1 and Supporting Information Table S2). ST1, ST2 and ST520 (serogroup IVb), ST218 (IVb-v1), ST3, ST288 (IIb), ST9 (IIc) and ST8 (IIa) had been previously reported in dairy industries and retail products in Brazil (Chenal-Francisque et al., 2011; Haase et al., 2014).
Table 1.
Genomic typing of Listeria monocytogenes isolates recovered in Brazil from 1978 to 2013.
| Isolate | Sourcea | Stateb | Year | Lineage | Serotype | Serogroupc | CC (MLST)d | ST (MLST)d | SL (cgMLST)e | CT (cgMLST)e | Accession numbers |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CLIST 2140 | Human – Placenta | PE | 1978 | II | 1/2a | IIa | CC7 | ST12 | SL7 | CT720 | ERR3199887 |
| CLIST 732 | Raw beef | MT | 2009 | II | 1/2a | IIa | CC8 | ST8 | SL8 | CT4418 | ERR3199900 |
| CLIST 3727 | Raw beef | SP | 2003 | II | 1/2a | IIa | CC8 | ST1419 | SL8 | CT4419 | ERR3199902 |
| CLIST 3864 | Raw beef | RS | 2002 | II | 1/2a | IIa | CC121 | ST121 | SL121 | CT4428 | ERR3199903 |
| 19 | PE | MG | 2009 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4420 | ERR3199909 |
| 45 | Raw beef | MG | 2010 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4424 | ERR3199911 |
| 227 | PE | RS | 2010 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4422 | ERR3199910 |
| 508 | Raw beef | MG | 2012 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4420 | ERR3199912 |
| 581 | Raw beef | MG | 2012 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4423 | ERR3199913 |
| CLIST 2137 | Human – CSF | PR | 1983 | II | 1/2a | IIc | CC9 | ST9 | SL9 | CT4425 | ERR3199886 |
| CLIST 3186 | Raw beef | SP | 2006 | II | 1/2a | IIc | CC9 | ST9 | SL9 | CT4426 | ERR3199901 |
| CLIST 3726 | Raw beef | SP | 2003 | II | 1/2c | IIc | CC9 | ST9 | SL9 | CT4421 | ERR3199914 |
| CLIST 3732 | Human – CSF | DF | 1989 | II | 1/2a | IIc | CC9 | ST9 | SL9 | CT4427 | ERR3199888 |
| 7 | Raw beef | MG | 2009 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4448 | ERR3199904 |
| CLIST 441 | Raw beef | MT | 2010 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4447 | ERR3199905 |
| CLIST 2083 | Human – Blood | RJ | 2008 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4444 | ERR3199889 |
| CLIST 2138 | Human – CSF | SP | 1982 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4445 | ERR3199890 |
| CLIST 3865 | Raw beef | SP | 2002 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4446 | ERR3199906 |
| CLIST 3869 | Raw beef | RJ | 2004 | I | 1/2b | IIb | CC3 | ST3 | SL3 | CT4449 | ERR3199907 |
| CLIST 3870 | Raw beef | MT | 2004 | I | 1/2b | IIb | CC288 | ST288 | SL288 | CT4439 | ERR3199908 |
| CLIST 1011 | Human – Blood | SP | 1985 | I | 4b | IVb | CC1 | ST1 | SL1 | CT4437 | ERR3199891 |
| CLIST 1015 | Human – Blood | SP | 1985 | I | 4b | IVb | CC1 | ST1 | SL1 | CT4438 | ERR3199892 |
| CLIST 3735 | Human – Blood | SP | 1985 | I | 4b | IVb | CC1 | ST1 | SL1 | CT4436 | ERR3199898 |
| 233 | PE | MG | 2012 | I | 4b | IVb | CC2 | ST2 | SL2 | CT4431 | ERR3199915 |
| CLIST 3723 | Human – CSF | RJ | 1990 | I | 4b | IVb | CC2 | ST2 | SL2 | CT4433 | ERR3199896 |
| CLIST 3724 | Raw beef | RJ | 2003 | I | 4b | IVb | CC2 | ST2 | SL2 | CT4434 | ERR3199920 |
| CLIST 3729 | PE-animal | RS | 1991 | I | 4b | IVb | CC2 | ST2 | SL2 | CT4430 | ERR3199917 |
| CLIST 3739 | Human – CSF | PR | 2000 | I | 4b | IVb | CC2 | ST2 | SL2 | CT4432 | ERR3199899 |
| CLIST 3734 | Human – CSF | PE | 1989 | I | 4b | IVb-v1 | CC218 | ST218 | SL218 | CT4015 | ERR3199897 |
| CLIST 2930 | Human – Blood | RJ | 2004 | I | 4b | IVb-v1 | CC218 | ST218 | SL218 | CT4440 | ERR3199895 |
| CLIST 2168 | Human – CSF | RJ | 2007 | I | 4b | IVb-v1 | CC218 | ST218 | SL218 | CT4441 | ERR3199894 |
| 1282 | PE | SP | 2013 | I | 4b | IVb-v1 | CC218 | ST218 | SL218 | CT4442 | ERR3199916 |
| 74 | Raw beef | MG | 2010 | I | 4b | IVb-v1 | CC218 | ST218 | SL218 | CT4443 | ERR3199918 |
| CLIST 2167 | Human – CSF | SP | 1997 | I | 4b | IVb | CC315 | ST520 | SL315 | CT4429 | ERR3199893 |
| CLIST 3192 | Raw beef | SP | 2006 | I | 4b | IVb | CC315 | ST520 | SL315 | CT4429 | ERR3199919 |
PE, production environment; CSF, cerebrospinal fluid.
Brazilian state: PE, Pernambuco; MT, Mato Grosso; SP, São Paulo; RS, Rio Grande do Sul; MG, Minas Gerais; PR, Paraná; DF, Distrito Federal; RJ, Rio de Janeiro.
According to Doumith et al. (2004) and Leclercq et al. (2011).
Clonal complex (CC) and sequence type (ST) defined according to Ragon et al. (2008).
Sublineage (SL) and cgMLST type (CT) defined according to Moura et al. (2016).
Fig. 1.
Minimum spanning tree based on the 7-loci MLST profiles of the 35 isolates included in this study together with the 270 profiles from South America publicly available at BIGSdb-Lm (listed in Supporting Information Table S2). STs are represented by coloured circles where size is proportional to the number of isolates. Grey zones denote MLST CCs. The number of allelic differences between profiles is indicated on the branches. [Color figure can be viewed at wileyonlinelibrary.com]
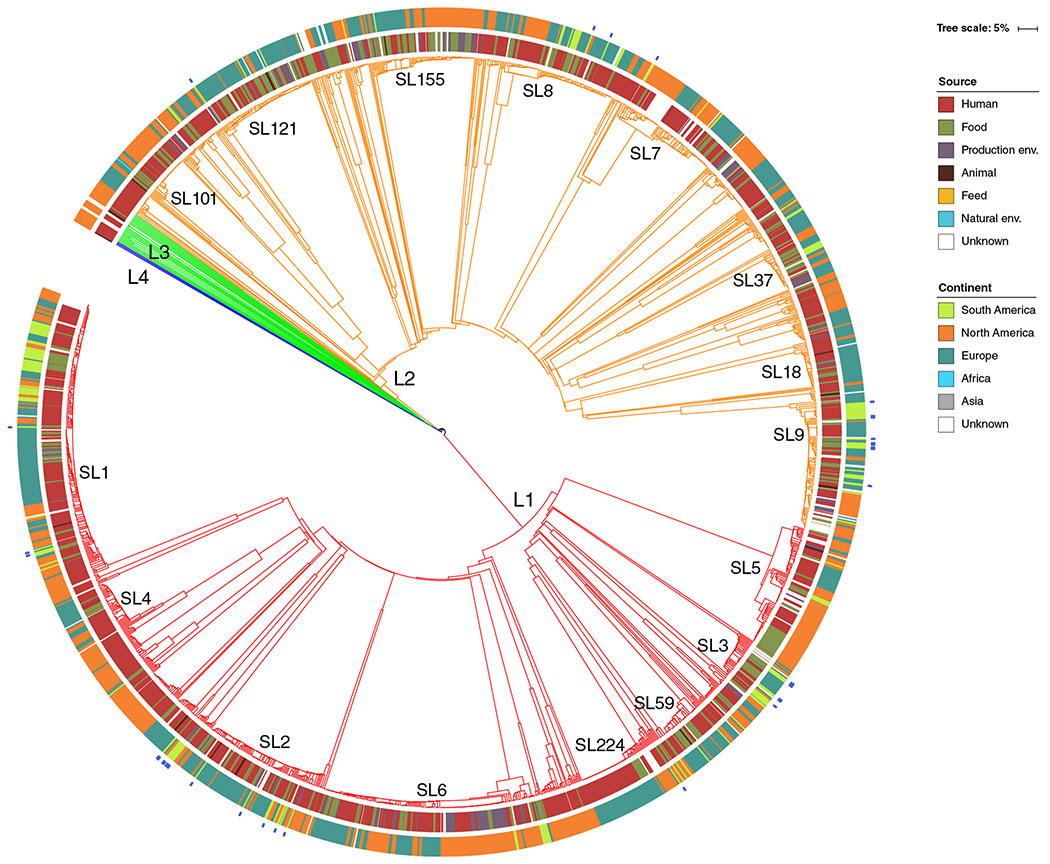
At the level of core genome, isolates were distributed among 10 SLs and represented 33 different cgMLST types (CTs; Fig. 2A). For a better understanding of the broader phylogenetic context, the 35 Brazilian Lm isolates were included in a comparison of global data from 1696 genomes (Moura et al., 2016) and 113 additional genomes from South America (Supporting Information Table S3) from Chile (n = 95), Peru (n = 13), Colombia (n = 2), Brazil (n = 1), Venezuela (n = 1) and Argentina (n = 1). Consistent with MLST, the Brazilian isolates were mostly represented by SLs that have been identified worldwide (Fig. 3). Out of 33 CTs identified in this study, 31 were new in BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/; Moura et al., 2016) and were assigned to CT4418-CT4434 and CT4436-CT4449 (Fig. 2A). Only L1-SL218-ST218-CT4015 and L2-SL7-ST12-CT720 types have been previous reported in Europe (Charlier et al., 2017; Maury et al., 2017) and in the United States (SRA accession no. SRR1664806), suggesting a global intercontinental spread of these strains. The high proportion of new CTs (94%) found in this study highlights the current scarcity of genomes from South America in public databases (currently mostly populated by genomes from North America, Europe and Australia) and the importance of more genomic studies to fully understand Lm biodiversity.
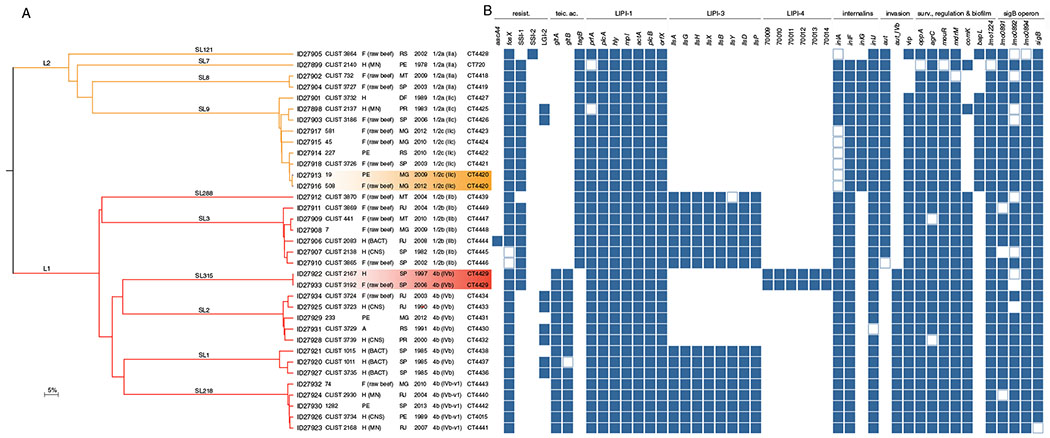
Fig. 2.
Virulence and resistance gene profiles of the 35 isolates sequenced in this study.
A. Single-linkage clustering based on the cgMLST profiles. Branches are coloured by phylogenetic lineage (L1, red; L2, orange) and labelled by SL. Information on the BIGSdb identifier, isolate’s name, origin (H, human; F, food; PE, food production environment), state (PE, Pernambuco; MT, Mato Grosso; SP, São Paulo; RS, Rio Grande do Sul; MG, Minas Gerais; PR, Paraná; DF, Distrito Federal; and RJ, Rio de Janeiro), year of isolation, serotype (serogroup) and CT are provided. B. Resistance and virulence genes patterns of either presence, absence or truncation. Colour-filled boxes represent the presence of the different genetic traits. Empty boxes represent genes with truncations leading to premature stop codons. Absent genes are marked in white. [Color figure can be viewed at wileyonlinelibrary.com]
Fig. 3.
Phylogenetic placement of the 35 isolates included in this study together with the worldwide collection of 1696 genomes (Moura et al., 2016) and 113 additional genomes from South America retrieved from NCBI database (last accessed 01-10-2018). The dendrogram was obtained using the single-linkage method on the cgMLST allelic profiles of the 1844 isolates. Branches are coloured by phylogenetic lineage (L1, red; L2, orange; L3, green; L4, blue). SLs with more than 30 isolates are labelled in the tree. Inner rings show the isolate’s source type and geographic location, according to the colour codes shown on the left. Isolates from this study are marked in blue in the most external ring. [Color figure can be viewed at wileyonlinelibrary.com]
Long-term strain persistence in Brazil
Interestingly, two CTs (highlighted in Fig. 2A) included more than one isolate (cgMLST similarity ≥99.6%), suggesting a possible epidemiological link (Moura et al., 2016). To confirm the close phylogenetic relationships observed using cgMLST (which covers 1748 genes, corresponding to ~1.6 Mb of Lm genome), isolates belonging to the same CT were also compared at the level of whole genome (~3.0 Mb, comprising all coding and non-coding regions) by single nucleotide polymorphisms analysis (wgSNP).
Isolates CLIST 2167 (collected in 1997, human source) and CLIST 3192 (collected in 2007 from raw beef), assigned as L1-SL315-ST520-CT4429, were collected a decade apart in Sao Paulo (SP) state and showed no allelic differences on the basis of cgMLST and wgSNP analyses.
The two isolates assigned to L2-SL9-ST9-CT4420 (isolate 19, collected in 2009 from meat handles and isolate 508, collected in 2012 from raw beef) were recovered 4 years apart from the same food processing plant in Minas Gerais (MG) state. These isolates differed by only 5 cgMLST alleles and 108 wgSNPs, the latter located in 12 genes (85 synonymous and 19 non-synonymous SNPs) and 4 non-coding regions. The high number of SNPs detected within only 12 genes suggests the occurrence of genetic recombination events. Whether these genetic modifications contributed to the adaptation or environmental persistence of these isolates remains unclear.
Despite the lack of trace back and/or forward investigations to confirm the epidemiological association between these isolate pairs, their high genetic relatedness highlights a possible long-term Lm contamination in these food production plants.
Antibiotic and stress resistance traits
None of the Lm isolates harboured any of the screened resistance genes towards disinfectants (Fig. 2B and Supporting Information Table S4). Intrinsic antibiotic resistance genes (lmo0919, norB, lmo0441, sul, fosX) were present in all isolates. Two isolates (CTs L1-SL3-ST3-CT4445 and L1-SL3-ST3-CT4446; Fig. 2B) exhibited a C(187)T mutation in the fosX gene (resistance to fosfomycin). This new mutation results in a premature stop codon (PMSC) at residue 63, likely resulting in complete susceptibility to fosfomycin (Scortti et al., 2018). Acquired antibiotic resistance traits were not detected, with the exception of aacA4 (resistance to aminoglycosides) present in one isolate (isolate CLIST 2083, L1-SL3-ST3-CT4444).
The Listeria genomic island (LGI-2), which was previously identified in lineage I isolates and that is associated with arsenic resistance (Lee et al., 2017) was detected in 23% (8/35) of isolates from both lineages I and II (Fig. 2B). Nearly all lineage II isolates carried SSI-1 (Fig. 2B), whereas SSI-2 was only present in one isolate (CLIST 3864, L2-SL121-ST121-CT4428). These stress survival islets contribute to Lm survival and grow in adverse environmental conditions, such as low pH and high salt concentrations (Ryan et al., 2010) or alkaline and oxidative stress conditions (Harter et al., 2017), facilitating the adaptation and persistence in production environments. A similar SSI prevalence was previously reported in Chile (Toledo et al., 2018; P = 0.13; Chi-square test), suggesting these mechanisms are typically present in L. monocytogenes, particularly in lineage II. Further studies will allow a better understanding as to whether these genes confer a selective advantage to the isolates described here.
Virulence genetic traits
As expected, the pathogenic island LIPI-1 was present in all isolates (Fig. 2B). Despite its high degree of conservation, two clinical isolates CLIST 2137 (L2-SL9-ST9-CT4425) and CLIST 2140 (L2-SL7-ST7-CT720) carried frameshift and mutations in the prfA gene leading to PMSCs [L(221) and G(16)Stop respectively] (Fig. 2B). The prfA gene regulates the transcription of several Lm genes expressed during infection (Chakraborty et al., 1992; Milohanic et al., 2003; de las Heras et al., 2011) and truncations in this gene can impact haemolysis and impair Lm virulence (Rupp et al., 2015; Maury et al., 2017). Results are in agreement with the previous reports that showed that the CT L2-SL7-ST7-CT720 is non-haemolytic due to a prfA PMSC (Maury et al., 2017).
Consistent with Lm phylogeny, isolates from lineage I also carried LIPI-3 and LIPI-4 islands. LIPI-3, which encodes listeriolysin S (Cotter et al., 2008; Quereda et al., 2016), was present in all lineage I isolates except those from SLs SL2 and SL315. LIPI-4, which has been shown to increase Lm neuro- and placental tropism (Maury et al., 2016) was present only in two isolates (L1-SL315-ST520-CT4429; Fig. 2), consistent with previous reports of its presence in SL315 (Moura et al., 2016).
Alleles encoding InlA-truncated variants, impaired in epithelial invasion (Lecuit et al., 1997; Lecuit et al., 2001), were identified in all SL121 and SL9 isolates, obtained from food or associated environments (Fig. 2B). The detected PMSCs included the types 6 (Q(492)Stop), 11 (W(685)Stop) and 19 (E(326)Stop), previously described (Olier et al., 2003; Rousseaux et al., 2004; Gelbíčová et al., 2015; Moura et al., 2016; Toledo et al., 2018). Interestingly, both CTs involved in the persistent contaminations described above carried either deletions or mutations in the inlA gene. Isolates belonging to L1-SL315-ST520-CT4429 harboured a 3 amino acid deletion at residue 737 (pre-anchor domain) in InlA internalin, whereas in L2-SL9-ST9-CT4420, isolates carried the PMSC at residue 326 (leucine-rich repeat motifs). Whether these favour strain, persistence remains to be clarified.
Other genes important for Lm virulence and stress response also carried mutations or frameshifts leading to PMSCs (e.g., aut, oppA, agrC, mouR) and/or were evenly distributed across the Lm phylogeny (e.g., inlG, bapL; Fig. 2B). Further studies will clarify the impact of these mutations on Lm virulence phenotypes.
Conclusions
Our results constitute a first contribution to understanding the genomic diversity and epidemiology of Lm in Brazil and South America. This study also highlights the importance of implementing WGS-based epidemiological surveillance programs to detect transmission chains and uncover long-term persistence of Lm in PEs. Future work will clarify the impact of the genetic traits and mutations found in this study on Lm virulence and persistence.
Experimental procedures
Lm isolates selection
A collection of Brazilian isolates, most of them kindly provided by FioCruz (CLIST collection), were previously characterized (Camargo et al., 2015; Camargo et al., 2016). Based on this work, a total of 35 representative Lm isolates were selected for WGS based on their year of isolation (from 1978 to 2013), serotypes (1/2a, n = 7; 1/2b, n = 7; 1/2c, n = 6; 4b, n = 15), sources (PE, n = 5; beef, n = 16; and clinical, n = 14) and geographic distribution in the country (from eight states). The major serotypes from lineages I and II were included due to their epidemiological relevance (Hofer et al., 2006; Vallim et al., 2015). Pure cultures were maintained at −80°C in brain heart infusion broth (BHI, Oxoid, Basingstoke, England) in the presence of 20% (v/v) glycerol (Merck, Whitehouse Station, NJ, USA), and recovered by growth on BHI agar prior use.
Genome sequencing and assembly
The genome sequence data were obtained after DNA extraction using the DNeasy Blood and Tissue kit (QIAGEN, Germany) following the protocol for purification of total DNA from gram-positive bacteria, library preparation with Ovation Ultralow Library System V2 (Nugen, San Carlos, CA, USA) and sequencing by Illumina MiSeq platform. Paired-ends reads of 300 bp (Illumina, San Diego, CA, USA) included an average genome coverage of 169× (listed in Supporting Information Table S1). Reads were trimmed using Trimmomatic version 0.35 (Bolger et al., 2014) and assemblies were obtained using SPAdes version 3.10 (Anton et al., 2012).
In silico molecular typing
PCR-serogroups (Doumith et al., 2004), MLST (Ragon et al., 2008) and cgMLST (Moura et al., 2016) profiles were extracted from draft assemblies as previously described (Moura et al., 2016). MLST profiles were compared against the 270 other public profiles from South America available at the Institute Pasteur MLST database (BIGSdb-Lm, http://bigsdb.pasteur.fr/listeria/), using the minimum spanning tree clustering method implemented in BioNumerics v7.6 (Applied-Maths).
SLs and CTs were assigned in BIGSdb-Lm database based on the cgMLST profiles and the allelic cutoffs previously established (150 allelic differences for SLs and 7 allelic differences from CTs; Moura et al. (2016)). New CTs were assigned to profiles not previously registered in BIGSdb-Lm.
Genomes were compared against the public collection of 1696 Lm genomes collected mostly in Europe and North America (Moura et al., 2016) and 113 additional genomes from South America (listed in Supporting Information Table S3) retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/; accessed on 01 Oct 2018). Dendrograms were built based on cgMLST profiles similarities in BioNumerics v.7.6 (http://www.applied-maths.com) using categorical differences and single-linkage clustering method. Trees were visualized by using iTol v.4.2 (Letunic and Bork, 2016).
Isolates belonging to the same CT were further analysed at the level of wgSNP by mapping their sequence reads against a representative reference assembly for each corresponding CT (isolates 19 and CLIST 2167, for CT4420 and CT4429 respectively) using the snippy pipeline (https://github.com/tseemann/snippy).
Virulence and resistance genotyping
The presence of 248 genes (listed in Supporting Information Table S4) previously identified as involved in stress and antibiotic resistance, biofilm formation and virulence (Chen et al., 2005; Kuenne et al., 2013; Gahan and Hill, 2014; Wattam et al., 2014; Maury et al., 2016) was accessed in silico in BIGSdb-Lm using the BLASTN algorithm (Jolley and Maiden, 2010) as previously described (Moura et al., 2016). Individual gene alignments were performed using MUSCLE (Edgar, 2004).
Data availability
The genomes obtained in this study were deposited in NCBI/EMBL/DDBJ databases under the project accession number PRJEB31124.
Supplementary Material
Table S1. Quality metrics and accession numbers of the 35 Listeria monocytogenes assembled genomes.
Table S2. South American isolates and profiles (n = 305) included in MLST analysis (Institut Pasteur MLST scheme of 7 loci; Ragon et al., 2008).
Table S4. Listeria monocytogenes genes distribution according to their functional categories.
Table S3. South American isolates and profiles (n = 148) included in cgMLST analyses (Institut Pasteur cgMLST scheme of 1748 loci; Moura et al., 2008).
Acknowledgements
The authors express their gratitude to Deyse Vallim and Ernesto Hofer from the Laboratory of Bacterial Zoonoses (FioCruz, Brazil) for provision of bacterial strains from CLIST collection. The present material is based upon work supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenção de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Anton B, Sergey N, Dmitry A, Alexey AG, Mikhail D, Alexander SK, et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo AC, Castilho NPA, Silva DAL, Vallim DC, Hofer E, and Nero LA (2015) Antibiotic resistance of Listeria monocytogenes isolated from meat-processing environments, beef products, and clinical cases in Brazil. Microb Drug Resist 21: 458–462. [DOI] [PubMed] [Google Scholar]
- Camargo AC, Vallim DC, Hofer E, and Nero LA (2016) Molecular serogrouping of Listeria monocytogenes from Brazil using PCR. J Food Prot 79: 144–147. [DOI] [PubMed] [Google Scholar]
- Camargo AC, Woodward JJ, Call DR, and Nero LA (2017) Listeria monocytogenes in food-processing facilities, food contamination, and human listeriosis: the Brazilian scenario. Foodborne Pathog Dis 14: 623–636. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Leimeister-Wächter M, Domann E, Hartl M, Goebel W, Nichterlein T, and Notermans S (1992) Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol 174: 568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Perrodeau É, Leclercq A, Cazenave B, Pilmis B, Henry B, et al. (2017) Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 17: 510–519. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, and Jin Q (2005) VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33: D325–D328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, et al. (2011) Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis 17: 1110–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Draper LA, Lawton EM, Daly KM, Groeger DS, Casey PG, et al. (2008) Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria monocytogenes. PLoS Pathog 4: e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M, Buchrieser C, Glaser P, Jacquet C, and Martin P (2004) Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42: 3819–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan C, and Hill C (2014) Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 4: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbíčová T, Koláčková I, Pantůček R, and Karpíšková R (2015) A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol 38: 293–296. [PubMed] [Google Scholar]
- Haase JK, Didelot X, Lecuit M, Korkeala H, and Achtman M (2014) The ubiquitous nature of Listeria monocytogenes clones: a large-scale multilocus sequence typing study. Environ Microbiol 16: 405–416. [DOI] [PubMed] [Google Scholar]
- Harter E, Wagner EM, Zaiser A, Halecker S, Wagner M, and Rychli K (2017) The novel stress survival islet 2 (SSI-2), predominantly present in Listeria monocytogenes strains of ST121, is involved in alkaline and oxidative stress response. Appl Environ Microbiol 83: e00827–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E, Reis C.M.F.d., and Hofer CB (2006) Sorovares de Listeria monocytogenes e espécies relacionadas, isoladas de material clínico humano. Rev Soc Bras Med Trop 39: 32–37. [DOI] [PubMed] [Google Scholar]
- Jolley KA, and Maiden MC (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, et al. (2013) Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Heras A, Cain RJ, Bielecka MK, and Vázquez-Boland JA (2011) Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14: 118–127. [DOI] [PubMed] [Google Scholar]
- Leclercq A, Chenal-Francisque V, Dieye H, Cantinelli T, Drali R, Brisse S, and Lecuit M (2011) Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int J Food Microbiol 147: 74–77. [DOI] [PubMed] [Google Scholar]
- Lecuit M, Ohayon H, Braun L, Mengaud J, and Cossart P (1997) Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun 65: 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, et al. (2001) A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292: 1722–1725. [DOI] [PubMed] [Google Scholar]
- Lee S, Ward TJ, Jima DD, Parsons C, and Kathariou S (2017) The arsenic resistance Listeria genomic island LGI2 exhibits sequence and integration site diversity and propensity for three Listeria monocytogene clones with enhanced virulence. Appl Environ Microbiol 83: e01189–e01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonaco S, Nucera D, and Filipello V (2015) The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect Genet Evol 35: 172–183. [DOI] [PubMed] [Google Scholar]
- Lüth S, Kleta S, and Al Dahouk S (2018) Whole genome sequencing as a typing tool for foodborne pathogens like Listeria monocytogenes – the way towards global harmonisation and data exchange. Trends Food Sci Technol 73: 67–75. [Google Scholar]
- Maury MM, Tsai Y-H, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, et al. (2016) Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48: 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury MM, Chenal-Francisque V, Bracq-Dieye H, Han L, Leclercq A, Vales G, et al. (2017) Spontaneous loss of virulence in natural populations of Listeria monocytogenes. Infect Immun 85: e00541–e00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppée J-Y, Frangeul L, Vega Y, Vázquez-Boland JA, et al. (2003) Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47: 1613–1625. [DOI] [PubMed] [Google Scholar]
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, et al. (2016) Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2: 16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Tourdjman M, Leclercq A, Hamelin E, Laurent E, Fredriksen N, et al. (2017) Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg Infect Dis 23: 1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen EM, Björkman JT, Kiil K, Grant K, Dallman T, Painset A, et al. (2017) Closing gaps for performing a risk assessment on Listeria monocytogenes in ready-to-eat (RTE) foods: activity 3, the comparison of isolates from different compartments along the food chain, and from humans using whole genome sequencing (WGS) analysis. EFSA Support. Publ 14: 1151E. [Google Scholar]
- de Noordhout CM, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, et al. (2014) The global burden of listeriosis: a systematic review and metaanalysis. Lancet Infect Dis 14: 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olier M, Pierre F, Rousseaux S, Lemaître J-P, Rousset A, Piveteau P, and Guzzo J (2003) Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect Immun 71: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, Bakker H.C.d., and Wiedmann M (2011) Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301: 79–96. [DOI] [PubMed] [Google Scholar]
- Quereda JJ, Dussurget O, Nahori M-A, Ghozlane A, Volant S, Dillies M-A, et al. (2016) Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci 113: 5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, and Brisse S (2008) A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4: e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S, Olier M, Lemaître JP, Piveteau P, and Guzzo J (2004) Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl Environ Microbiol 70: 2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Aguilar-Bultet L, Jagannathan V, Guldimann C, Drögemüller C, Pfarrer C, et al. (2015) A naturally occurring prfA truncation in a Listeria monocytogenes field strain contributes to reduced replication and cell-to-cell spread. Vet Microbiol 179: 91–101. [DOI] [PubMed] [Google Scholar]
- Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. (2015) Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53: 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S, Begley M, Hill C, and Gahan CGM (2010) A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J Appl Microbiol 109: 984–995. [DOI] [PubMed] [Google Scholar]
- Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, et al. (2014) Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch AC, Arredondo-Alonso S, Willems RJL, and Goering RV (2018) Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin Microbiol Infect 24: 350–354. [DOI] [PubMed] [Google Scholar]
- Scortti M, Han L, Alvarez S, Leclercq A, Moura A, Lecuit M, and Vazquez-Boland J (2018) Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet 14: e1007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk BJ, Date KA, Jackson KA, Pouillot R, Holt KG, Graves LM, et al. (2012) Invasive listeriosis in the foodborne diseases active surveillance network (foodNet), 2004–2009: further targeted prevention needed for higher-risk groups. Clin Infect Dis 54: S396–S404. [DOI] [PubMed] [Google Scholar]
- Toledo V, den Bakker HC, Hormazábal JC, González-Rocha G, Bello-Toledo H, Toro M, and Moreno-Switt AI (2018) Genomic diversity of Listeria monocytogenesis isolated from clinical and non-clinical samples in Chile. Genes 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim DC, Barroso Hofer C, Lisba R.d.C., Victor Barbosa A, Alves Rusak L, Reis C.M.F.d., and Hofer E (2015) Twenty years of Listeria in Brazil: occurrence of Listeria species and Listeria monocytogenes serovars in food samples in Brazil between 1990 and 2012. Biomed Res Int 2015: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Walle I, Björkman JT, Cormican M, Dallman T, Mossong J, Moura A, et al. (2018) Retrospective validation of whole genome sequencing-enhanced surveillance of listeriosis in Europe, 2010 to 2015. Eurosurveillance 23: 1700798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42: D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Quality metrics and accession numbers of the 35 Listeria monocytogenes assembled genomes.
Table S2. South American isolates and profiles (n = 305) included in MLST analysis (Institut Pasteur MLST scheme of 7 loci; Ragon et al., 2008).
Table S4. Listeria monocytogenes genes distribution according to their functional categories.
Table S3. South American isolates and profiles (n = 148) included in cgMLST analyses (Institut Pasteur cgMLST scheme of 1748 loci; Moura et al., 2008).
Data Availability Statement
The genomes obtained in this study were deposited in NCBI/EMBL/DDBJ databases under the project accession number PRJEB31124.