ABSTRACT
Background: Gastric carcinoma (GC) is a common malignant tumor. Recently, it has been found that long non-coding RNAs (lncRNAs) play important role in cancer. In this paper, we investigated the effects and mechanism of lncRNA GASL1 in GC cells.
Methods: GASL1 level in GC cells was up-regulated via cell transfection. Cell proliferation, migration, invasion were detected by CCK-8, BrdU, Transwell assays and western blot. In addition, the regulation of GASL1 on microRNA (miR)-106a level was detected using RT-qPCR and the binding between GASL1 and miR-106a was confirmed by bioinformatic prediction and luciferase reporter assay. The effects of overexpressing miR-106a on GASL1-regulated GC cell behaviors were further explored. Moreover, western blot also was used to detect the pathway-related proteins.
Results: Overexpression of GASL1 decreased the viability and BrdU levels. Meanwhile, CyclinD1 level was decreased while p53 and p21 levels were strengthened by overexpression of GASL1. On cell metastasis, up-regulation of GASL1 decreased cell migration, invasion and related proteins matrix metalloproteinase (MMP)-9 and Vimentin levels. Meanwhile, silencing GASL1 exerted opposite effects on GC cells. Moreover, GASL1 negatively regulated and targeted miR-106a. Up-regulation of miR-106a weakened the functions of GASL1 in cell proliferation and metastasis. Besides, GASL1 decreased the relate-protein levels of PI3K/AKT and ras/raf/MEK/ERK pathways while miR-106a weakened these changes.
ConclusionGASL1 restrained GC cell proliferation and metastasis and blocked PI3K/AKT and ras/raf/MEK/ERK pathways by sponging miR-106a.
KEYWORDS: Gastric carcinoma, proliferation, metastasis, lncRNA, GASL1, miR-106a
Introduction
Gastric carcinoma (GC) is the most common malignant tumor derived from digestive tract [1]. Due to differences in dietary culture, the incidence GC in China is much higher than that in European countries [2]. Due to the lack of diagnostic indicators for specificity and sensitivity, it is difficult to diagnosis in the early phase of GC [3]. Therefore, patients with GC are diagnosed and confirmed at the middle and late stages, that increase the difficulty of treatment [4]. With the development of genetic engineering, numerous studies have confirmed that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) played important part on the development of GC [5]. Therefore, strengthening the research on the molecular mechanism of GC is of great significance for early diagnosis and treatment of GC [6].
LncRNAs are a class of non-coding RNAs with little or no protein coding capacity, whose length is more than 200 nt [7]. With the in-depth research on lncRNAs, the important role of lncRNAs in malignant carcinomas has attracted extensive attention of scientists. Besides, the potential as carcinoma diagnostic or therapeutic marker is becoming more and more obvious [8]. LncRNA GASL1 is a newly discovered lncRNA in recent years, which presented low expression in lung carcinoma and exerted anti-tumor effects [9]. Further research discovered GASL1 was also down-regulated in GC [10] and it was closely related to prognosis of GC [11]. Although studies had confirmed the GASL1 expression in GC, the functions and mechanism of GASL1 in GC was not completely clear. Based on the previous literature, we deeply explored the functions and mechanism of GASL1 in GC process.
MicroRNAs (miRNAs) are a short endogenous non-coding RNA whose length is about 22 nt [12]. It negatively binds to the 3' untranslated region of mRNA and negatively regulates gene expression at the post-transcriptional level [13]. Moreover, it can exert effects in cell proliferation, differentiation and apoptosis [14]. MiRNAs are characteristically expressed in different types of tumor tissues, hint the potential for tumor diagnosis [15]. MiR-106a, a 23 nt miRNA [16], is located on chromosome Xq26.2 [17]. MiR-106a was overexpressed in GC and played a tumorigenic role [18]. There is enough evidence to prove that miR-106a function in carcinoma and can be used as a diagnostic marker for carcinomas [19]. Therefore, we boldly put forward a hypothesis, does GASL1 connected with miR-106a?
We examined the role of GASL1 and miR-106a in cell proliferation and migration of GC cell line MKN45 and SGC-7901. In addition, we briefly explored the relationship between GASL1 and miR-106a and the associated signal pathways. The whole article is a further development on GC-related molecules GASL1 and miR-106a, providing a new idea for clinical detection and treatment of GC.
Materials and methods
Cell culture and treatment
The GC cells cell line MKN45 and SGC-7901 (laboratory retention) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, 12100, Solarbio, Beijing, China), which were supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, US). Cells was cultured at sterility, 37 °C and 5% CO2. The DMEM were replaced every 2 days.
Viability assay
CCK8 kit (CA1210, Solarbio) was used in this experiment. Briefly, after lncRNA and miRNA were transfected, a total of 5 × 103 cells per well was plated in a 96-well cell plate for 24 h incubation. The 10 µL CCK-8 was added into each well in 100 μL DMEM, and the cell plate was incubated at 37°C for 1 h (in a dark place). After that, the absorbance was measured at 450 nm in microplate reader (Bio-Rad, Sunnyvale, CA, US). Cellular viability rate was calculated as the percentage of absorbance under 450 nm.
BrdU assay
Fluorescein isothiocyanate (FITC) BrdU Flow kit (557891, FITC, BD Bioscience, San Jose, CA, US) was used in this experiment. The cell inoculation density was 1 × 105 cells/well. After lncRNA and miRNA were transfected, Bromodeoxyuridine (BrdU, 5 μL) was added into cells (24 h). Then, we fixed and denatured cells. Next, Fluorochrome-conjugated anti-BrdU antibody (100 μL, from FITC BrdU Flow Kit) were added in cells. At last, FACS can (BD) analyzed the results.
Migration and invasion assay
Migration and invasion assay were detected by transwell (24 well and 8 μm pore size, Millipore Corp, Bedford, MA, US). GC cells (1 × 105/mL, 200 μL) were plated in upper chambers of 24-well transwell plate, cultured in FBS-free DMEM and pre-treated with 1 μg/mL mitomycin C for 1 h to exclude the effect of cell proliferation. The lower chambers were added with 600 μL DMEM contained 10% FBS. Then, cells were cultured at 37°C for 24 h. Finally, the cells in the upper membrane were wiped with a cotton swab. Cells at under chamber were fixed and stained. The number of migrated cells was counted under inverted microscope (Olympus Japan Co., Tokyo, Japan). At cell invasion assay, the only difference was that the upper membrane (8 μm) had a layer of matrigel (356232, Corning incorporated, Corning, NY, US).
Plasmids and transfection
A shRNA directed against GASL1 (5ʹ-GAC GTG TCA GGA CCT TCG T-3ʹ) was cloned into the retroviral vector, pRETRO-SUPER. The fragment of GASL1 (NR_149020) was cloned into pEFIRES-P vector to over-express GASL1. Besides, GASL1 were transfected using polyjet transfection reagent (SignaGen, Gaithersburg, MD, USA). MiR-106a mimic (sense: 5-‘AAA AGU GCU UAC AGU GCA GGU AG-3ʹ, antisense: 5ʹ-ACC UGC ACU GUA AGC ACU UUU UU-3ʹ) and NC mimic (sense: 5ʹ-UGG CAG UGU CUU AGC UGG UUG U-3ʹ, antisense: 5ʹ-ACA ACC AGC UAA GAC ACU GCC A − 3ʹ) were synthesized (GenePharma, Shanghai, China) and transfected utilizing Lipofectamine 3000 (Life Technologies, Carlsbad, CA, US). Concretely speaking, 24 h before performing cell transfection, cells were plated in cell plates or cell culture flask containing DMEM with 10% FBS. The plasmids and lipofectamine reagent were diluted in Opti-MEM™ medium (Life Technologies) and hatched together for 20 min. Next, cells were hatched with plasmids-lipofectamine complex for 6 h. The medium was changed to DMEM containing 10% FBS. After 48 h incubation, total RNA was isolated from the transfected cells to check transfection efficiency using RT-qPCR.
Reverse transcription-quantitative PCR (RT-qPCR)
Trizol was used to extract RNA. After quantitating, RNA (10 ng) and primers were used to detect reverse transcription of miR-106a. The reverse transcription was 42°C, 5 min and 95°C for 10 s. The PCR progress was 95°C and 5 s, 60°C and 30 s. The cycles were 40 times. The GASL1 levels were detected by PrimeScript RT Master Mix (RR036A, TaKaRa). The reverse transcription conditions were 25°C, 10 min at and 48°C for 30 min, final step was 95°C, 5 min. The cycles also were 40 times. Primers used were presented as follows: miR-106a forward primer, 5ʹ-CGC AAA AGT GCT TAC AGT GCA-3ʹ and reverse, 5ʹ-GTG CAG GGT CCG AGG T-3ʹ; U6 primer forward, 5ʹ-GCT TCG GCA GCA CAT ATA CTA AAA T-3ʹand reverse, 5ʹ-CGC TTC ACG AAT TTG CGT GTC AT-3ʹ; GASL1 forward primer: 5ʹ-CTG AGG CCA AAG TTT CCA AC-3ʹ and reverse, 5ʹ-CAG CCT GAC TTT CCC TCT TCT-3ʹ; GAPDH forward primer: 5ʹ-CAT GTT CCA ATA TGA TTC CAC C-3ʹ and reverse, 5ʹ-GAT GGG ATT TCC ATT GAT GAC-3ʹ. The experiments were repeated three times. Data were calculated according to 2−ΔΔCt.
Luciferase reporter gene assay
The wild type (WT) sequence of GASL1 was inserted into psiCHECK2 luciferase reporter plasmid (Promega, Madison, WI, USA) and the recombinant plasmid was named as GASL1-WT. In addition, the mutant (MUT) sequence was inserted into psiCHECK2 and the recombinant one was named as GASL1-MUT. HEK-293 cells (CRL-1573, American Type Culture Collection, Manassas, VA, USA) cultured in MEM (Solarbio) were transfected with luciferase reporter plasmid expressing WT or MUT of GASL1 combined with miR-106a mimic (or NC mimic) and renilla luciferase plasmid (Promega) using Lipfectamine 3000. Following a 24-h incubation, cells were lysed and analyzed for dual luciferase activity using Dual-Luciferase® Reporter Assay System (Promega). The renilla luciferase activity was utilized to normalize firefly luciferase.
Western bolt
RIPA buffer, protease and phosphatase (Sigma, St. Louis, US) were used to obtain proteins from cells. Protein quantification was tested by BCA kit (P0012, Beyotime, Jiangsu, China). The protein samples were parted by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Then, protein samples were transferred to polyvinylidene fluoride (PVDF) membrane (Millipore). The membrane was blocked in 5% bovine serum albumin (BSA) for 2 h and covered with primary antibodies (Abcam, Cambridge, MA, US): cyclinD1 (ab226977), p53 (ab131442), p21 (ab227443), matrix metalloproteinase (MMP)-9 (ab73734), Vimentin (ab137321), β-actin (ab119716), t-PI3K (ab191606), p-PI3K (ab182651), t-AKT (ab126811), p-AKT (ab131443), ras (ab69747), raf (ab200653), t-MEK (ab178876), p-MEK (ab194754), t-ERK (ab109282) and p-ERK (ab131438) at 4°C and overnight. Then, it was incubated by HRP marked Goat Anti-Rabbit IgG H&L (ab205718, Abcam). Signals were detected by ECL Detection System (Thermo Scientific, Rockford, IL, USA) and β-actin was an internal control.
Statistical analysis
All experiments were run at least three times. Data were expressed as mean ± standard deviation (SD). Statistical analysis was performed by using Graphpad 6.0 (Graph Pad Software, CA, US). The P-values were calculated using analysis of variance (ANOVA). P < 0.05 was considered statistically significant.
Results
GASL1 was silenced and overexpressed in GC cells
GASL1 levels were detected by RT-qPCR in MKN45 and (P < 0.001, Figure 1a) SGC-7901 cells (P < 0.001, Figure 1b). The data revealed that overexpression of GASL1 meaningfully strengthened GASL1 levels and silencing GASL1 declined GASL1 levels, implying that we successfully transfected the plasmids into GC cells.
Figure 1.
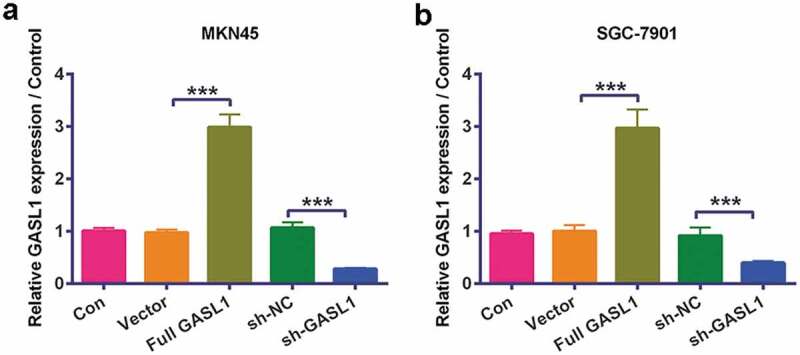
GASL1 was silenced and overexpressed in MKN45 and SGC-7901 cells
Overexpression of GASL1 meaningfully strengthened GASL1 levels and silencing GASL1 declined GASL1 levels in (a) MKN45 and (b) SGC-7901 cells. Data were expressed as mean ± SD and compared by one-way ANOVA followed by Tukey’s multiple comparison test (n = 3 per group).*** P < 0.001.
GASL1 restrained cell proliferation
Based on Figure 1 results, the functions of GASL1 in cell proliferation were detected. The results discovered when transfected full GASL1 into MKN45 and SGC-7901 cells, cell viability (P < 0.05, Figure 2a) and BrdU (P < 0.05, Figure 2b) levels were meaningfully declined. On the molecular level, the cyclinD1 levels were meaningfully declined while p53 and p21 levels were strengthened (P < 0.05, P < 0.01, P < 0.001, Figure 2c,e) by regulation of GASL1. Silencing GASL1 played opposite effects on cell proliferation. All results implied that GASL1 restrained MKN45 and SGC-7901 cell proliferation.
Figure 2.
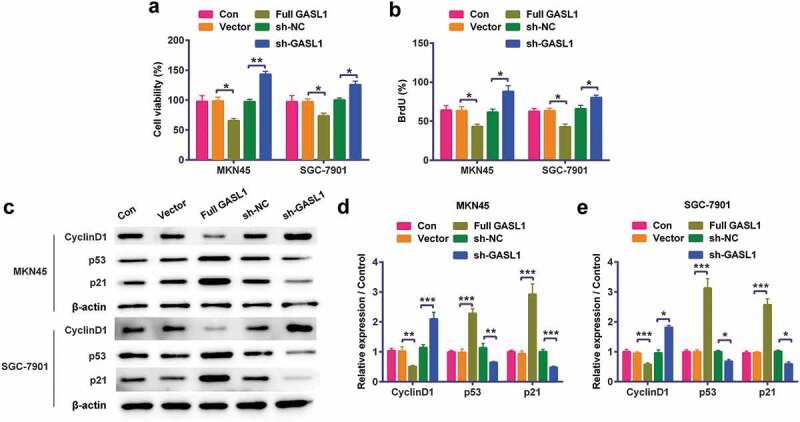
GASL1 GASL1 restrained MKN45 and SGC-7901 cell proliferation
Overexpression of GASL1 meaningfully declined (a) cell viability, (b) BrdU and (c-e) cyclinD1 levels but strengthened p53 and p21 levels in cells. At the same time, silencing GASL1 meaningfully strengthened cell viability, BrdU and cyclinD1 levels but declined p53 and p21 levels in cells. Data were expressed as mean ± SD and compared by two-way ANOVA followed by Tukey’s multiple comparison test (n = 3 per group).* P < 0.05; ** P < 0.01; *** P < 0.001.
GASL1 restrained cell metastasis
Cell metastases were further detected in MKN45 and SGC-7901 cells. The results discovered when transfected full GASL1 into MKN45 and SGC-7901 cells, cell migration (P < 0.05 or P < 0.001, Figure 3a) and invasion (P < 0.01 or P < 0.001, Figure 3b) were meaningfully declined. On the molecular level, the levels of MMP-9 and Vimentin were also meaningfully declined (P < 0.01 or P < 0.001, Figure 3c,e). Silencing GASL1 played opposite effects on cell metastasis. All results implied GASL1 restrained MKN45 and SGC-7901 cell metastasis.
Figure 3.
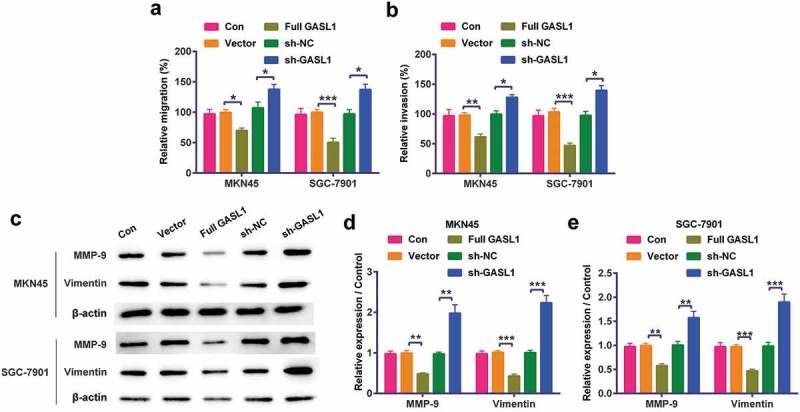
GASL1 restrained MKN45 and SGC-7901 cell metastasis
Overexpression of GASL1 meaningfully declined (a) cell migration, (b) invasion, (c-e) matrix metalloproteinase (MMP)-9 and Vimentin levels in cells. At the same time, silencing GASL1 played opposite effects on cell metastasis. Data were expressed as mean ± SD and compared by two-way ANOVA followed by Tukey’s multiple comparison test (n = 3 per group).* P < 0.05; ** P < 0.01; *** P < 0.001.
GASL1 negatively regulated miR-106a
The level of miR-106a was detected by RT-qPCR in MKN45 (Figure 4a) and SGC-7901 (Figure 4b) cells. The data revealed that over-expression of GASL1 meaningfully declined miR-106a levels (P < 0.001) and silencing GASL1 strengthened miR-106a levels (P < 0.01 or P < 0.001), implying GASL1 negatively regulated miR-106a. Moreover, binding of GASL1 to its target miRNA was further explored. Similar in concept to bioinformatics prediction from Starbase (http://starbase.sysu.edu.cn) (Figure 4c), dual luciferase reporter assay was utilized to test the specificity of GASL1 for miR-106a. HEK-293 cells were transfected with GASL1-WT or GASL1-MUT combined with miR-106a mimic of NC mimic, followed by detection of luciferase activity. GASL1 targeted to miR-106a as evident by reduction of relative luciferase activity that was presented in GASL1-WT along with miR-106a mimic group (P < 0.05, Figure 4d), but absent in GASL1-MUT and miR-106a mimic-transfected group. The data indicated the direct binding of GASL1 to miR-106a. Furthermore, in order to further support that the regulation between GASL1 and miR-106a, the expression level of GASL1 was detected after over-expression of miR-106a. Result in Figure 4e revealed that up-regulation of miR-106a inhibited GASL1 level in both MKN45and SGC-7901 (P < 0.05, P < 0.01), hinting that GASL1 and miR-106a were negatively regulated by each other.
Figure 4.
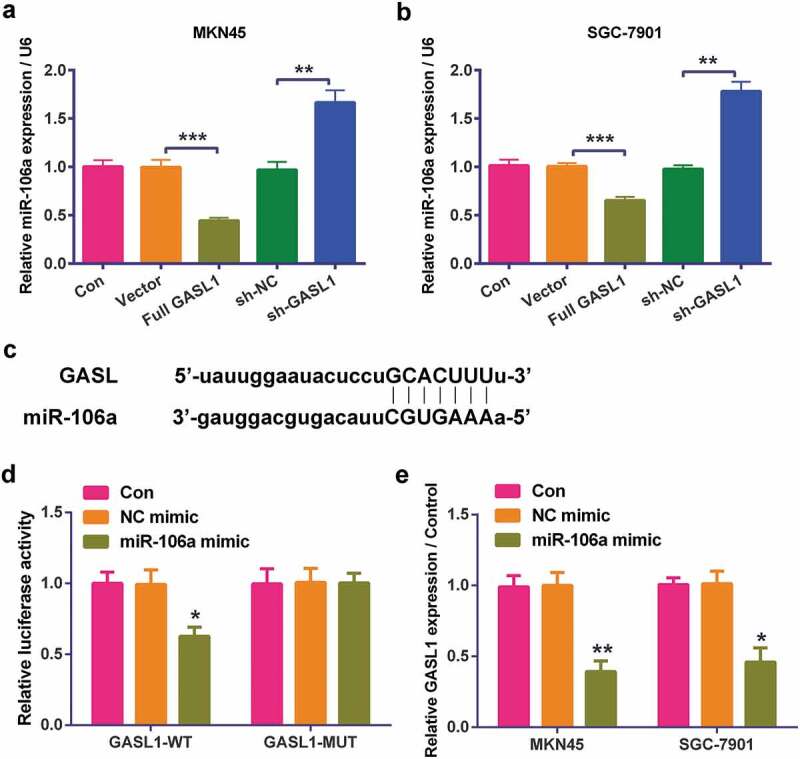
GASL1 negatively regulated miR-106a in MKN45 and SGC-7901 cells
Overexpression of GASL1 meaningfully declined miR-106a levels and sh-GASL1 strengthened miR-106a levels in (a) MKN45 and (b) SGC-7901 cells. (c) Binding between GASL1 and miR-106a. (d) GASL1 targeted miR-106a. Data were expressed as mean ± SD and compared by one-way ANOVA (for a-b) and two-way ANOVA (for D-E) followed by Tukey’s multiple comparison test (n = 3 per group).** P < 0.01; *** P < 0.001.
Full GASL1 restrained cell proliferation and metastasis by sponging miR-106a
In order to detect the functions of miR-106a, we overexpressed miR-106a in MKN45 and SGC-7901 cells (P < 0.001, Figure 5a). Based on this, the results discovered that miR-106a mimic statistically strengthened full GASL1-induced decline in cell viability (Figure 5b) and BrdU (Figure 5c levels of MKN45 and SGC-7901 cells (P < 0.05 or P < 0.01). Meanwhile, Figure 5d–f proved that when miR-106a mimic and full GASL1 were co-transfected into cells, Cyclin D1 level (P < 0.01 or P < 0.001) was conspicuously strengthened but p53 and p21 levels (P < 0.01 or P < 0.001) were conspicuously declined. On the metastasis, miR-106a mimic statistically strengthened full GASL1-induced decline in cell migration (P < 0.05, Figure 5i–5h) and invasion (P < 0.05, Figure 5i). Moreover, the levels of MMP-9 and Vimentin (Figure 5j–l–Figure 5i–k) were strengthened when miR-106a mimic and full GASL1 were co-transfected into cells (P < 0.01 or P < 0.001). In short, overexpression of GASL1 restrained cell proliferation and metastasis by sponging miR-106a.
Figure 5.
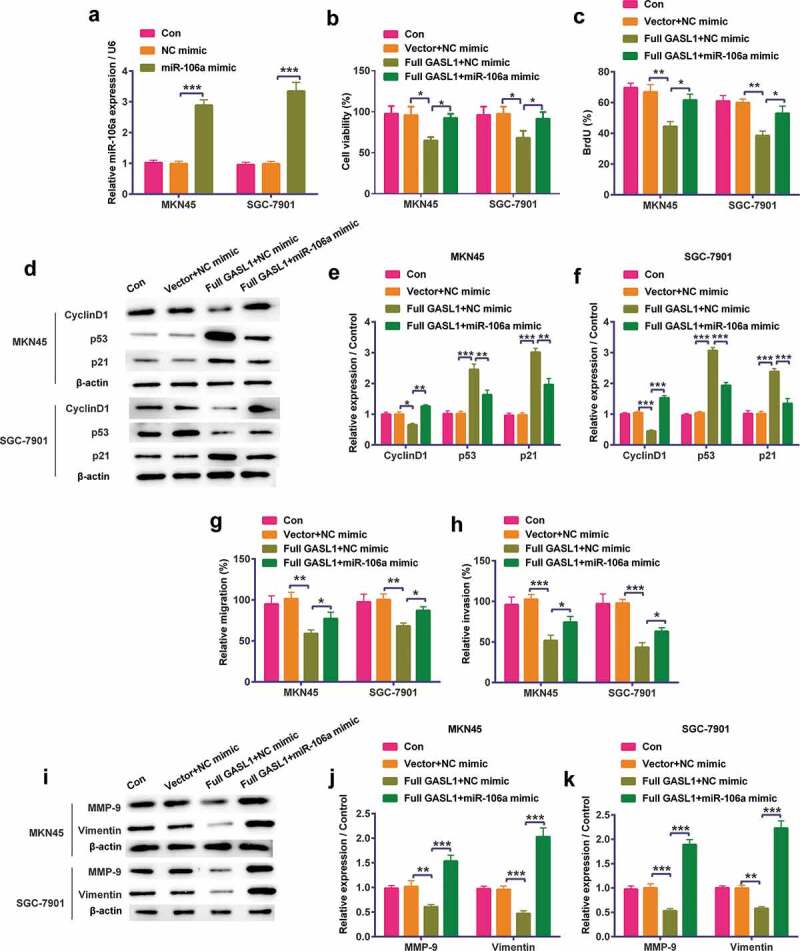
Full GASL1 restrained cell proliferation and metastasis by sponging miR-106a in MKN45 and SGC-7901 cells
(a) miR-106a mimic was transfected into cells. When miR-106a mimic and full GASL1 were co-transfected into cells, (b) cell viability, (c) BrdU and (d-g) Cyclin D1 levels were strengthened and p53 and p21 levels were declined. Meanwhile, (h) migration, (i) invasion and (j-l) matrix metalloproteinase (MMP)-9 and Vimentin levels were also strengthened. Data were expressed as mean ± SD and compared by two-way ANOVA followed by Tukey’s multiple comparison test (n = 3 per group). * P < 0.05; ** P < 0.01; *** P < 0.001.
GASL1 restrained the PI3K/AKT and ras/raf/MEK/ERK signaling pathways by sponging miR-106a
Western blot was used to detect the GASL1 and miR-106a functions on PI3K/AKT and ras/raf/MEK/ERK pathways. Overexpression of GASL1 declined the phosphorylation levels of PI3K (P < 0.01, Figure 6a,b), AKT (P < 0.001, Figure 6a,b), ras, raf, MEK and ERK (P < 0.05 or P < 0.01 or P < 0.001, Figure 6c,d) in MKN45 and SGC-7901 cells. This change was strengthened when miR-106a mimic was transfected. All results implied that GASL1 restrained the PI3K/AKT and ras/raf/MEK/ERK pathways via sponging miR-106a.
Figure 6.
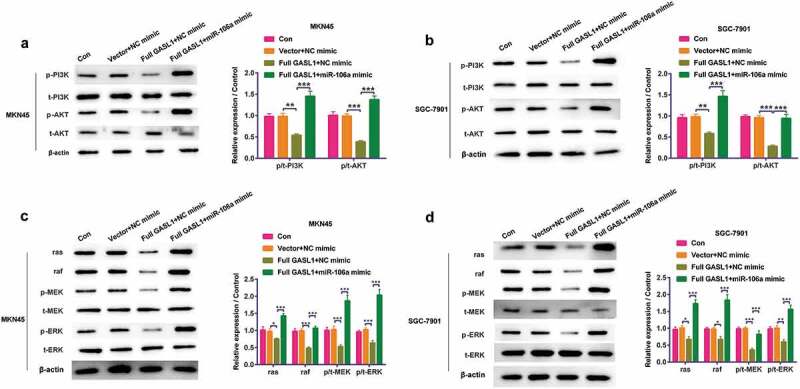
GASL1 restrained the PI3K/AKT and ras/raf/MEK/ERK signaling pathways via sponging miR-106a. (a and c are MKN45 cells when b and d are SGC-7901 cells)
Overexpression of GASL1 declined the phosphorylation levels of (a and b) PI3K, AKT, (c and d), MEK and ERK and ras/raf levels in MKN45 and SGC-7901 cells while miR-106a mimic strengthened the expression. Data were expressed as mean ± SD and compared by two-way ANOVA followed by Tukey’s multiple comparison test (n = 3 per group). * P < 0.05; ** P < 0.01; *** P < 0.001.
Discussion
GC is a digestive cancer with high mortality and poor prognosis. Through the above experiments, we found that GASL1 inhibited tumor proliferation and metastasis and negatively regulated miR-106a level. MiR-106a can partially restore GASL1 inhibition in MKN45 and SGC-7901 cells. Finally, we discovered GASL1 blocked PI3K/AKT and ras/raf/MEK/ERK pathways via sponging miR-106a.
With the deepening of research, researchers discovered that lncRNAs could participate in cell metabolism, development and other processes [20]. On the other hand, studies display that numerous lncRNAs are abnormally expressed in GC and play anti-tumor or pro-tumor roles. For instance, Wang et al. proved that lncRNA SNHG7 was high expressed and exerted a promoting tumor effects in GC [21]. Chen et al. proved that lncRNA VPS9D1-AS1 was low expressed in GC and related to tumor size [22]. Zhou et al. proved that lncRNA AF147447 was low expressed and exerted an inhibiting tumor effects in GC [23]. GASL1 was a newly discovered lncRNA, which played vital part on cancer. Studies have discovered GASL1 could regulate the cell cycle of human osteosarcoma cell [24] and it had low expression in prostate carcinoma and associated with poor prognosis [25]. Further studies have found that GASL1 was also lowly expressed in GC [9,11]. In this paper, we tested the proliferation and metastasis of GC cells by CCK8, BrdU, migration, invasion and western blot experiments. The results displayed that GASL1 meaningfully declined cell viability, BrdU, and cell migration, invasion, cyclinD1, MMP-9 and Vimentin levels but strengthened p53 and p21 levels in cells. This indicated that GASL1 exerted an anti-carcinoma effect in GC cells in vitro, which was consistent with previous research.
As the research progressed, scientists discovered that lncRNA usually worked in GC by targeting miRNAs. For example, Yan et al. proved that lncRNA SNHG6 prompted GC cells EMT by sponging miR-101-3p [26]. YiRen et al. proved that lncRNA MALAT1 regulated GC cell autophagy by sponging miR-23b-3p [27]. Moreover, Yang et al. proved that LINC01133 restrained GC progress by sponging miR-106a-3p [28]. Therefore, we further explored the relationship between GASL1 and miR-106a. Figure 4 displays GASL1 meaningfully declined miR-106a levels and silencing GASL1 strengthened miR-106a levels in MKN45 and SGC-7901 cell, indicating GASL1 negatively regulated miR-106a.
MiR-106a belongs to the miR-17 family [29]. Studies had shown that miR-106a was highly expressed and exerted promoting role in GC [30], prostate carcinoma [31] and other carcinomas while it was down-regulated and exerted inhibiting role in glioma [32], renal cell carcinoma [33] and other carcinomas. In addition, miR-106a also participated in carcinoma diagnosis and prognosis assessment. For instance, Shirmohammadi et al. proved that miR-106a could be an early diagnosis in GC [34]. Song et al. proved that the serum miR-106a could be used to assess prognosis of chemotherapy [35]. In this paper, we displayed when miR-106a mimic and full GASL1was co-transfected into cells, cell viability, BrdU and Cyclin D1 levels were strengthened and p53 and p21 levels were declined. Meanwhile, migration, invasion and MMP-9 and Vimentin levels were also strengthened. This also confirmed the carcinoma-promoting effect of miR-106a in GC.
PI3K/AKT and ras/raf/MEK/ERK pathways were involved in the development of GC [36,37]. Further evidence suggested that lncRNAs and miRNAs also worked through these two pathways. For instance, studies had proved lncRNA MALAT1 worked in carcinoma via PI3K/AKT [38] and ras/raf/MEK/ERK [39] pathways while miR-145 worked in injury via these two pathways [40]. In our research, we also discovered that GASL1 declined the phosphorylation levels of PI3K, AKT, MEK and ERK and ras/raf levels in MKN45 and SGC-7901 cells while miR-106a mimic strengthened the expression.
In conclusion, GASL1 restrained GC cell proliferation and metastasis and blocked PI3K/AKT and ras/raf/MEK/ERK pathways by sponging miR-106a. This article developed the functions of GASL1 in GC and provided a new idea for clinical diagnosis of GC.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure statement
The authors declare that they have no conflict of interest.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nie Y, Wu K, Yu J, et al. A global burden of gastric cancer: the major impact of China. Expert Rev Gastroenterol Hepatol. 2017;11:651–661. [DOI] [PubMed] [Google Scholar]
- [3].Sung JK. Diagnosis and management of gastric dysplasia. Korean J Intern Med. 2016;31:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wu J, Li G, Wang Z, et al. Circulating microRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers. 2015;2015:435656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhou X, Ye F, Yin C, et al. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem. 2015;36:1440–1452. [DOI] [PubMed] [Google Scholar]
- [6].Fu DG. Epigenetic alterations in gastric cancer (Review). Mol Med Rep. 2015;12:3223–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. [DOI] [PubMed] [Google Scholar]
- [8].Li Z, Dou P, Liu T, et al. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. [DOI] [PubMed] [Google Scholar]
- [9].Su WZ, Yuan X. LncRNA GASL1 inhibits tumor growth of non-small cell lung cancer by inactivating TGF-beta pathway. Eur Rev Med Pharmacol Sci. 2018;22:7282–7288. [DOI] [PubMed] [Google Scholar]
- [10].Peng C, Li X, Yu Y, et al. LncRNA GASL1 inhibits tumor growth in gastric carcinoma by inactivating the Wnt/beta-catenin signaling pathway. Exp Ther Med. 2019;17:4039–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang B, Chen H, Zhang Y. Involvement of GASL1 in postoperative distant recurrence of gastric adenocarcinoma after gastrectomy distal resection and the possible mechanism. J Cell Biochem. 2019;120:11454–11461. [DOI] [PubMed] [Google Scholar]
- [12].Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. [DOI] [PubMed] [Google Scholar]
- [14].Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. [DOI] [PubMed] [Google Scholar]
- [15].Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. [DOI] [PubMed] [Google Scholar]
- [16].Zeiner GM, Boothroyd JC. Use of two novel approaches to discriminate between closely related host microRNAs that are manipulated by Toxoplasma gondii during infection. RNA. 2010;16:1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang G, Zhang R, Chen X, et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J Mol Med. 2011;89:1037. [DOI] [PubMed] [Google Scholar]
- [18].Zhu M, Zhang N, He S, et al. MicroRNA-106a functions as an oncogene in human gastric cancer and contributes to proliferation and metastasis in vitro and in vivo. Clin Exp Metastasis. 2016;33:509–519. [DOI] [PubMed] [Google Scholar]
- [19].Pan YJ, Zhuang Y, Zheng JN, et al. MiR-106a: promising biomarker for cancer. Bioorg Med Chem Lett. 2016;26:5373–5377. [DOI] [PubMed] [Google Scholar]
- [20].Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–4622. [PubMed] [Google Scholar]
- [22].Chen M, Wu X, Ma W, et al. Decreased expression of lncRNA VPS9D1-AS1 in gastric cancer and its clinical significance. Cancer Biomarkers. 2017;21:23–28. [DOI] [PubMed] [Google Scholar]
- [23].Zhou X, Chen H, Zhu L, et al. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget. 2016;7:82770–82782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gasri-Plotnitsky L, Ovadia A, Shamalov K, et al. A novel lncRNA, GASL1, inhibits cell proliferation and restricts E2F1 activity. Oncotarget. 2017;8:23775–23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li Z, Liu H, Ju W, et al. LncRNA GASL1 inhibits growth and promotes expression of apoptosis-associated proteins in prostate carcinoma cells through GLUT-1. Oncol Lett. 2019;17:5327–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yan K, Tian J, Shi W, et al. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. [DOI] [PubMed] [Google Scholar]
- [27].YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Z, Liu M, Zhu H, et al. miR‐106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol Carcinog. 2013;52:634–646. [DOI] [PubMed] [Google Scholar]
- [30].Zhu M, Zhang N, He S, et al. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588:600–607. [DOI] [PubMed] [Google Scholar]
- [31].Luo B, Kang N, Chen Y, et al. Oncogene miR-106a promotes proliferation and metastasis of prostate cancer cells by directly targeting PTEN in vivo and in vitro. Minerva Med. 2018;109:24–30. [DOI] [PubMed] [Google Scholar]
- [32].Yang G, Zhang R, Chen X, et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J Mol Med (Berl). 2011;89:1037–1050. [DOI] [PubMed] [Google Scholar]
- [33].Pan YJ, Wei LL, Wu XJ, et al. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8:e3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shirmohammadi K, Sohrabi S, Jafarzadeh Samani Z, et al. Evaluation of altered expression of miR-9 and miR-106a as an early diagnostic approach in gastric cancer. J Gastrointest Oncol. 2018;9:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Song J, Yin J, Bai Z, et al. the profile of serum microRNAs predicts prognosis for resected gastric cancer patients receiving platinum-based chemotherapy. Dig Dis Sci. 2017;62:1223–1234. [DOI] [PubMed] [Google Scholar]
- [36].Gonzalez-Hormazabal P, Musleh M. Polymorphisms in RAS/RAF/MEK/ERK pathway are associated with gastric cancer. Genes (Basel). 201. 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Riquelme I, Tapia O, Espinoza JA, et al. The gene expression status of the PI3K/AKT/mTOR pathway in gastric cancer tissues and cell lines. Pathol Oncol Res. 2016;22:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin Y, Feng SJ, Qiu S, et al. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- [39].Dong P, Xiong Y, Yue J, et al. Exploring lncRNA-mediated regulatory networks in endometrial cancer cells and the tumor microenvironment: advances and challenges. Cancers (Basel). 2019;11:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang P, Yuan Y. LncRNA-ROR alleviates hypoxia-triggered damages by downregulating miR-145 in rat cardiomyocytes H9c2 cells. J Cell Physiol. 2019;234:23695–23704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


