ABSTRACT
Breast cancer is one of the dominant cancers of women-related death universal. This inquiry aims to disclose the probable role of circABCC4 in breast cancer. The level of circABCC4 was discovered through qRT-PCR. The reactions of circABCC4 and miR-154-5p on the cell viability, apoptosis, migration as well as invasion were, respectively, inspected by CCK-8, flow cytometry, and transwell assays. The association betwixt circABCC4 and miR-154-5p was investigated. The accumulation of NF-κB and Wnt/β-catenin pathway proteins was discovered through Western blot. The expression of circABCC4 was far great in tumor tissues than in normal tissues. Knockdown of circABCC4 could subdue cell viability, migration, invasion, and enhance apoptosis in breast cancer cell lines. CircABCC4 negatively regulated the manifestation of miR-154-5p and shared binding sites with the latter. Suppression of miR-154-5p expression partially conversed the repressive effect of circABCC4 knockdown on breast cancer cell viability, migration, invasion, and NF-κB and Wnt/β-catenin pathways. CircABCC4 knockdown repressed breast cancer cells viability, migration, and invasion by up-regulating miR-154-5p via inhibiting NF-κB and Wnt/β-catenin signal pathways.
KEYWORDS: CircABCC4, miR-154-5p, breast cancer, proliferation, migration
Introduction
Breast cancer is the greatest familiar malicious tumor in females and the occurrence rises progressively worldwide. According to statistics, 20.8 million new cases were diagnosed in 2018, resulting in 626,679 deaths, higher than colorectal cancer and lung cancer [1]. While significant development has been made in the latest years in patients with early diagnosis and systemic treatment, breast cancer recurrence and distant metastasis have become obstacles to successful treatment. The molecular pathogenesis of breast cancer is still mostly restricted [2]. Consequently, there is no doubt that we should comprehend molecular mechanisms to promote the advance of operative beneficial strategies and find extra accurately predictive markers for the prevention of breast cancer.
Circular RNAs (circRNAs) are initially thought to be splicing errors [3]. However, with the development of whole transcriptome sequencing technology, a large number of circRNAs have recently been identified. CircRNAs are found to be a class of endogenous non-protein coding RNAs shorn of 5ʹ cap or 3ʹ poly A tail [4]. A growing number of studies have found that circRNAs contribute to biological developments through a variety of mechanisms, including acting as a ceRNA of microRNAs (miRNAs), thereby affecting translation and splicing [5–7]. Cancer-related circRNAs are constantly discovered [8,9], but the character in breast cancer progress has been less studied. CircABCC4 (circBase ID: hsa_circ_0030586; chr13:95,813,442–95,840,796) is a novel circRNA that is derived from ABCC4 mRNA back-splicing and located in chromosome 13q32.1 [10]. Previous literature reported the character of circABCC4 in prostate cancer, but it has not been described in breast cancer. Therefore, we explored the character of circABCC4 in breast cancer.
miRNAs are well-preserved non-coding RNAs (18–25 nucleotides) and show a regulatory role in gene expression through post-transcriptional results [11]. A previous report showed that multiple miRNAs were significantly dysregulated in breast cancer [12]. miR-154-5p is closely related to various cancers, and research findings are closely related to prostate cancer [13] as well as colorectal cancer [14], but there are few studies on miR-154-5p in breast cancer. Therefore, we inspected the effect of miR-154-5p on breast cancer and explored whether the influences of circABCC4 knockdown on the biotic function of breast cancer cells were mediated by miR-154-5p.
This study was performed in human MCF-7 and MDA-MB-231 cells, which aimed to investigate the character of circABCC4 in cell proliferation, migration as well as invasion. In this examination, the influences of circABCC4 and miR-154-5p on the NF-κB and Wnt/β-catenin signal pathways were studied for further explaining the tumor-promoting role of circABCC4. This study may offer an experimental basis for the treatment of breast cancer.
Materials and methods
Clinical samples
The human breast tissue and normal breast tissue were gained from Anhui Chest Hospital (Hefei, China) (n = 25). No patients received any management previous to surgery. Acquainted consensus was gained for each patient and the investigation was approved by the Medical Ethics Committee of Anhui Chest Hospital. All patients are female. Besides, among them 8 people <50 years of age, 17 people ≥50 years of age; TNM stage I/II: 18 people, TNM stage III: 7 people; lymph nodes metastasis: 6 people, no metastasis: 19 people; ER positive: 13 people, ER negative people: 12; PR positives: 17 people, PR negatives: 8 people; HER-2 positive: 16 people, HER-29 negative: 9 people; differentiation G1/G2: 20 people, G3: 5 people; Size <2.5 cm 17 people, ≥2.5 cm: 8 people.
Cell cultivation
The human breast cancer cell lines, which incorporated MDA-MB-231 (ATCC number: HTB-26) and MCF-7 (ATCC number: HTB-22, Manassas, VA, USA) were used as the main study material. These cells were cultivated in 10% fetal calf serum (FBS, Sigma-Aldrich) and 10 μg/ml bovine insulin in Leibovitz L-15 medium (Sigma-Aldrich). A damp environment comprising 5% CO2 also 95% air, including 37°C for cell cultivation. It is also necessary to study whether further miRNAs play a character in circABCC4-regulated tumorigenesis in the future.
Cell transfection
The specific circABCC4 small-interfering RNA, si-NC, miR-154-5p inhibitor and NC inhibitor (Dojindo Molecular Technologies, Gaithersburg, MD) were synthesized also transfected into breast cancer cell lines. Since the uppermost transfection efficacy happened at 48 hours, it was considered to be the yield time in the following experiments. The transfection sequences included si-circABCC4: 5‘-AAAUCCAAUAGGCAUCA GAGA-3‘ and miR-154-5p inhibitor: 5‘-CGAAGGCAACACGGAUAACCUA-3‘.
Cell viability assessment
The CCK-8 (Dojindo Molecular Technologies, Gaithersburg, MD) was consumed to detect cell viability in the current study. First of all, different group cell lines were preparedinto 96-well plates based on 1 × 103 cells/well, and cultivated in a CO2 incubator for 1 h. Secondly, referring to the instruction, the CCK-8 was joined in and mixed with the cells for 1 h, after the 24 h conventional culture. Finally, absorbance measurement was tested employing a Microplate Reader (ThermoFisher, Massachusetts, USA).
Apoptosis assay
Apoptosis analysis was implemented via propidium iodide (PI, BBI Solution, Crumlin, UK) and fluorescein isothiocyanate (FITC, BBI Solution, Crumlin, UK)-conjugated Annexin V staining. Briefly, the breast cancer cell lines were rinsed with pre-PBS twice in buffer and fixed in ethanol. The fixed cells were washed with pre-cold PBS twice. Then, the breast cancer cell lines were resuspended in buffer and fixed in 70% ethanol. Then, fixed cells were dealt following the manufacturer’s protocol and examined with a flow cytometer (ThermoFisher, Massachusetts, USA).
Western blot
The protein in breast cancer cell lines was taken out by RIPA lysis buffer (Sangon Biotech, Shanghai, China) permitting the manufacturer’s protocol. After quantified with the Pierce BCA Protein Assay Kit (ThermoFisher, Massachusetts, USA), the proteins were separated by Bio-Rad Bis-Tris Gel system. The membrane was blocked in (Cambridge, UK) 5% blocking buffer and incubated at 4°C overnight. The information of catalog was as following: CyclinD1 (ab226977), cyclin-dependent kinases (CDK) 6 (ab54576), Cleaved-caspase 3 (ab2302), Bcl-2 (ab692), Bax (ab32503), t-p65 (ab16502), p-p65 (ab86299), Wnt3a (ab28472), β-catenin (ab32572) and β-actin (ab8226). Then, it was washed with horseradish peroxidase-labeled secondary antibody and incubated for 1 h. Finally, the membranes were rinsed with HRP Substrate-marked secondary antibody for 1 h at 25°C (Millipore, MA, USA), and the expression levels of proteins were evaluated with Image Lab™ Software (Bio-Rad, Shanghai, China).
Migration and invasion assay
The chamber migration (8.0 μm Millipore, Bedford, Massachusetts, USA) was conducted to test migration and invasion. Briefly, we added 200 μl of the cell suspension to the upper chamber and co-cultured with 600 μl of medium containing 5% FBS in the lower chamber for 20–24 h. The chamber is then detached and the unmigrated cells on the membrane are wiped off. Breast cancer cell lines were fixed by using 10% paraformaldehyde and stained with crystal violet, then placed under an inverted microscope and finally counted. In addition, the upper chamber was coated with BD Matrigel (BD Biosciences, USA) for the transwell invasion assay to evaluate cell invasion capability.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was taken out from transfected cell lines utilizing Trizol reagent (Takara, Japan). Reverse transcription was manufactured using a One Step PrimeScript miRNA cDNA Synthesis Kit (Takara). qRT-PCR was accomplished with the SYBR Geen Realtime PCR Master Mix (Takara), and the data were synthesized on an ABI 7300 system (ABI). Besides, U6 as well as GAPDH were used as interior controls. The primer sequences used in our PCR assay were as following:
circABCC4: Forward: 5ʹ-TGAGTCAATTCTGAAAGCTCCG-3ʹ, Reverse: 5ʹ-GG CCGACCACAGCTAACAAT-3ʹ; GAPDH: Forward: 5ʹ-GGGAAACTGTGGCGT GAT-3ʹ; Reverse: 5ʹ-GGGTGTCGCTGTTGAAGT-3ʹ; miR-154-5p: Forward: 5ʹ-ACACTCCAGCTGGGTAGGTTATCCGTGTTG-3ʹ, Reverse: 5ʹ-TGGTGTCGT GGAGTCG-3ʹ; U6, Forward, 5ʹ-CTCGCTT CGGCAGCACA-3ʹ, Reverse, 5ʹ-AA CGCTTCACGAATTTGCGT-3ʹ.
Statistical analysis
The statistics represented by means of mean ± SD from three independent experiments. The SPSS 19.0 software was the mean tool for analyses (SPSS Inc., Chicago, IL, USA). ANOVA remained responsible for P-values, which P < 0.05 was adopted as statistical differences.
Results
CircABCC4 was enhanced in breast cancer patient tissues
We first detected the level of circRNA in the human normal tissues and breast cancer patient tumor tissues to measure the expression of circABCC4 in breast cancer. We discovered that circABCC4 level was upper in tumor tissues through qRT-PCR (P < 0.001, Figure 1) than the control tissues. The results proposed that circABCC4 upregulation might be interrelated with the occurrence and evolution of breast cancer.
Figure 1.
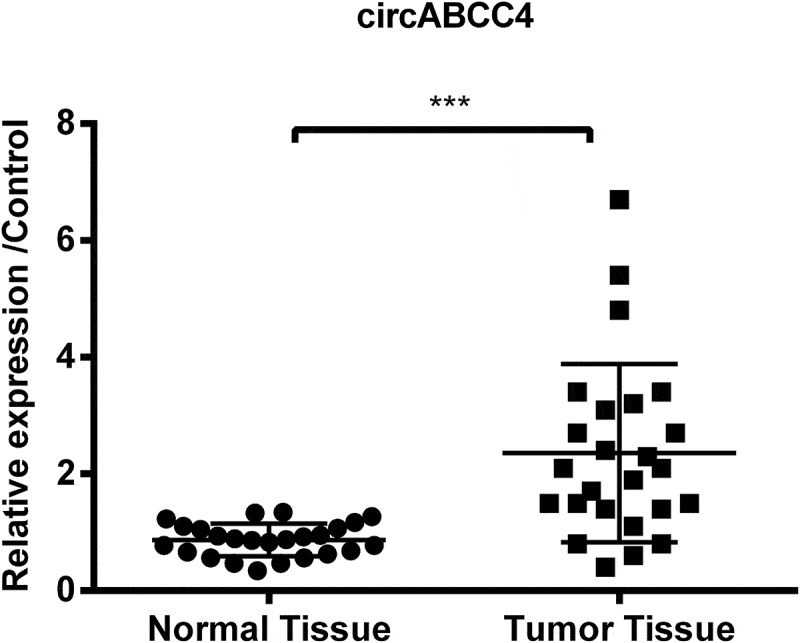
CircABCC4 level was more high in breast tumor tissues than in normal breast tissues. The level of circABCC4 was discovered by qRT-PCR. qRT-PCR, Quantitative real-time polymerase chain reaction; (n = 25). ANOVA remained responsible for P-values, ***P < 0.001 was considered as significant results
CircABCC4 knockdown constrained breast cancer cells viability and promoted apoptosis
We transfected circABCC4-siRNA into breast cancer cells to silence circABCC4, and inspected the purpose of circABCC4. We found that following si-circABCC4 transfection, circABCC4 expression was meaningfully reduced in both breast cancer cell lines (P < 0.01, Figure 2(a)). We detected the cell viability by CCK-8 assays and found declined cell viability (P < 0.01, Figure 2(b)), and the down-regulated protein levels of CyclinD1 also CDK6 (P < 0.01, Figure 2(d-f)) in circABCC4 silencing cells. Then, we used the flow cytometry to detect apoptosis rates of cells transfected with circABCC4-siRNA or else NC-siRNA. The data suggested that circABCC4 down-regulation significantly promoted apoptosis (P < 0.001, Figure 2(c)). Meanwhile, we tested the level of apoptosis-linked proteins and found that circABCC4 knockdown significantly down-regulated Bcl-2, but up-regulated Bax also Cleaved/Caspase-3 (P < 0.01 or P < 0.001, Figure 2(g-i)). Our consequences proved that circABCC4 knockdown might suppress cell viability as well as promote apoptosis of breast cancer cells.
Figure 2.
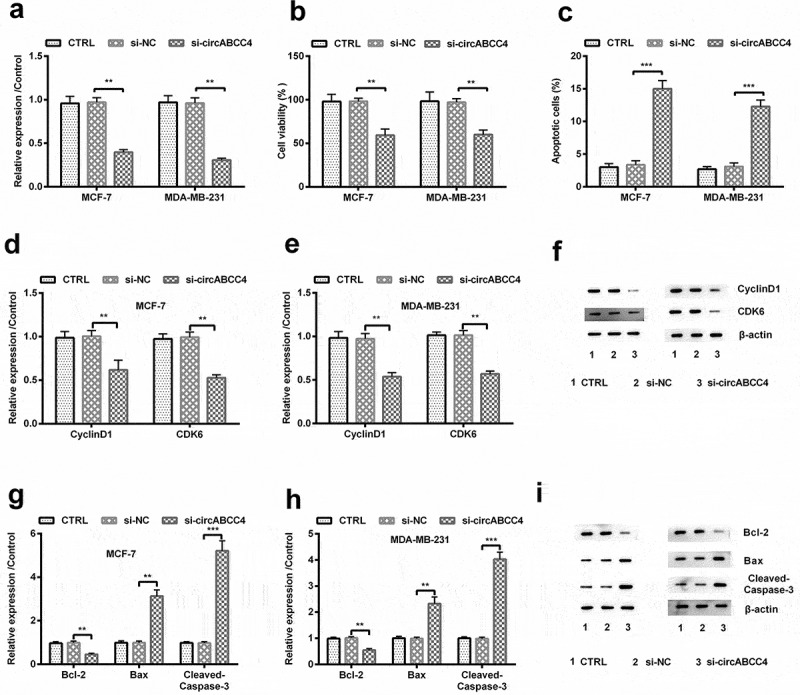
CircABCC4 knockdown repressed breast cancer cells viability and encouraged apoptosis. (a) qRT-PCR analysis for detection of circABCC4 after breast cancer cell lines were transfected with si-NC, si-circABCC4 or corresponding controls. (b) The cell viability was detected by CCK-8. (c) The apoptosis of breast cancer cell lines were identified by flow cytometry. (df) The expression of CyclinD1 as well as CDK6 was investigated by western blot. (gi) The expression of cells apoptosis-correlated proteins (Bcl-2, Bax, Cleaved/Caspase-3) were examined by western blot. CTRL, control; si, small interfering; NC, Negative control; CCK-8, Cell Counting Kit-8; qRT-PCR, Quantitative real-time polymerase chain reaction (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 and *** P < 0.001 were considered as significant results
CircABCC4 knockdown inhibited breast cancer cells migration and invasion
Previous studies have proved that migration and invasion are the main causes of breast cancer-associated deaths [15,16]. The effects of circABCC4 knockdown on migration and invasion of cells were examined. We found that circABCC4 knockdown decreased migration and invasion ability compared with si-NC in cells (P < 0.01, Figure 3(a-b)). These data revealed that circABCC4 knockdown could constrain the metastatic ability of breast cancer cells.
Figure 3.
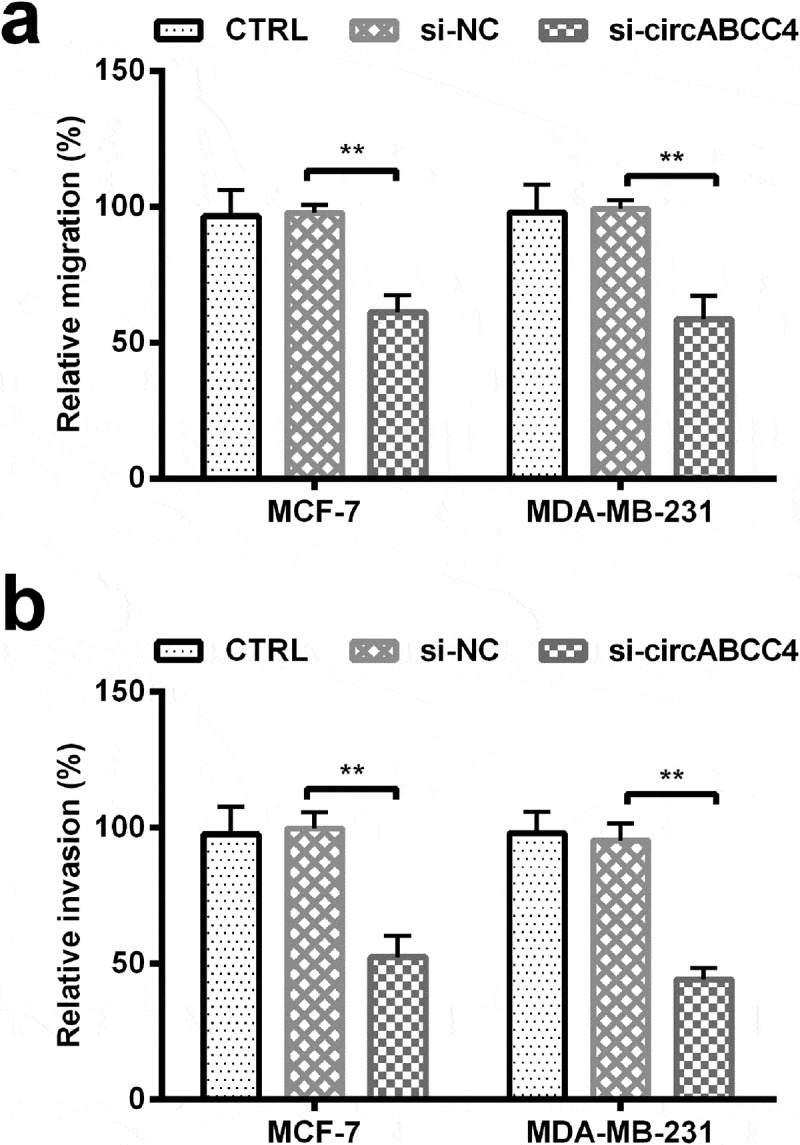
CircABCC4 knockdown inhibited breast cancer cells migration as well as invasion. (ab) The cell migration as well as cell invasion of breast cancer cell lines were detected by Transwell chamber assay. CTRL, control; si, small interfering; NC, Negative control (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 was considered as significant results
miR-154-5p was negatively regulated by circABCC4
We explored the relationship between circABCC4 and miR-154-5p for exploring the indepth mechanisms of circABCC4 involving in breast cancer progression. Firstly, we explored the link between circABCC4 and miR-154-5p based on bioinformatics analysis, and found that there were complementary sequences between them (Figure 4(a)). As a further confirmation of this result, qRT-PCR revealed that circABCC4 knockdown might negatively regulate the level of miR-154-5p compared with si-NC (P < 0.01, Figure 4(b)) in circABCC4-siRNA-transfected breast cancer cell lines. In conclusion, these above results showed that circABCC4 negatively regulated the level of miR-154-5p.
Figure 4.
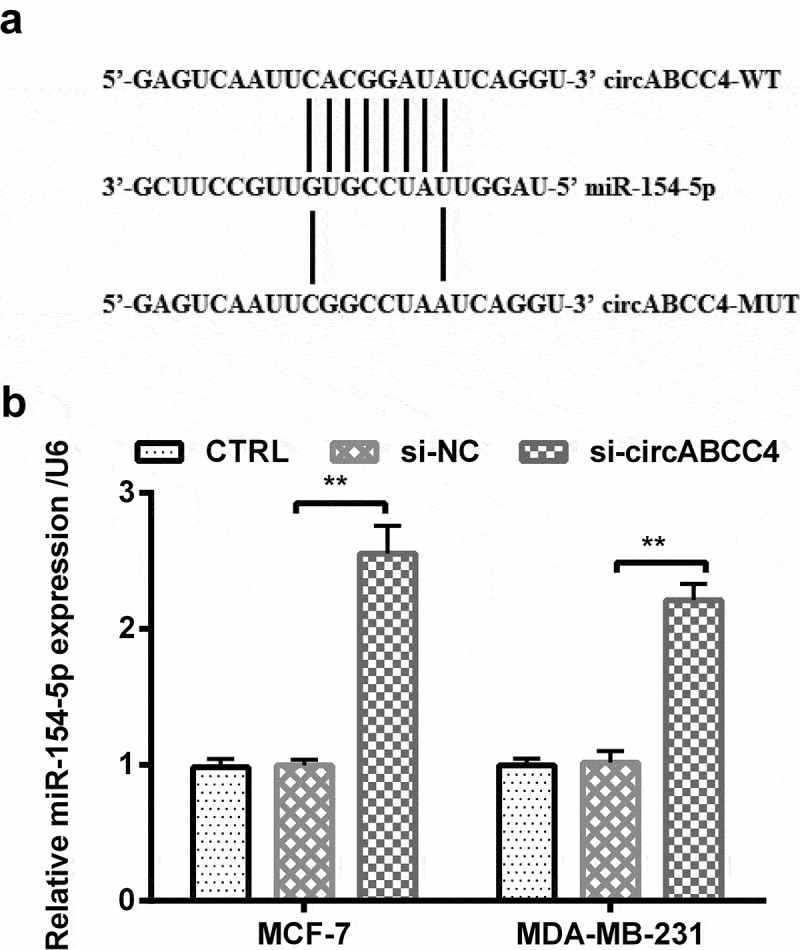
CircABCC4 depressingly controlled the level of miR-154-5p. (a) The complementary binding sites were explored via bioinformatics analysis, and (b) level of miR-154-5p in breast cancer cell lines was measured by RT-qPCR after transfection of si-NC, si-circABCC4 or the corresponding control. TRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 was considered as significant results
CircABCC4 knockdown inhibited cell viability and improved cell apoptosis by promoting miR-154-5p
We then explored the functions and mechanisms of miR-154-5p of cell viability as well as apoptosis. We transfected miR-154-5p inhibitor into cells for knocking down miR-154-5p. The level of miR-154-5p was meaningfully decreased after knocking down circABCC4 (P < 0.01, Figure 5(a)). Then, our results proved that knockdown of circABCC4 and miR-154-5p inhibition increased cell viability compared with si-NC + NC inhibitor (P < 0.05 or P < 0.01, Figure 5(b)), besides significantly down-regulated the protein levels of CyclinD1 also CDK6 (P < 0.05 or P < 0.01, Figure 5(d-f)). In addition, these results displayed that knockdown of circABCC4 and miR-154-5p inhibitor inhibited apoptosis compared with si-NC + NC inhibitor (P < 0.05 or P < 0.01, Figure 5(c)). Meanwhile, these above results displayed that knockdown of circABCC4 and miR-154-5p inhibitor significantly raised the level cells, but downgraded the expression of Bax as well as Cleaved/Caspase-3 (P < 0.05, P < 0.01 or P < 0.001, Figure 5(g-i)). In general, our results demonstrated circABCC4 knockdown might suppress cell viability and encourage apoptosis of breast cancer cells by promoting miR-154-5p.
Figure 5.
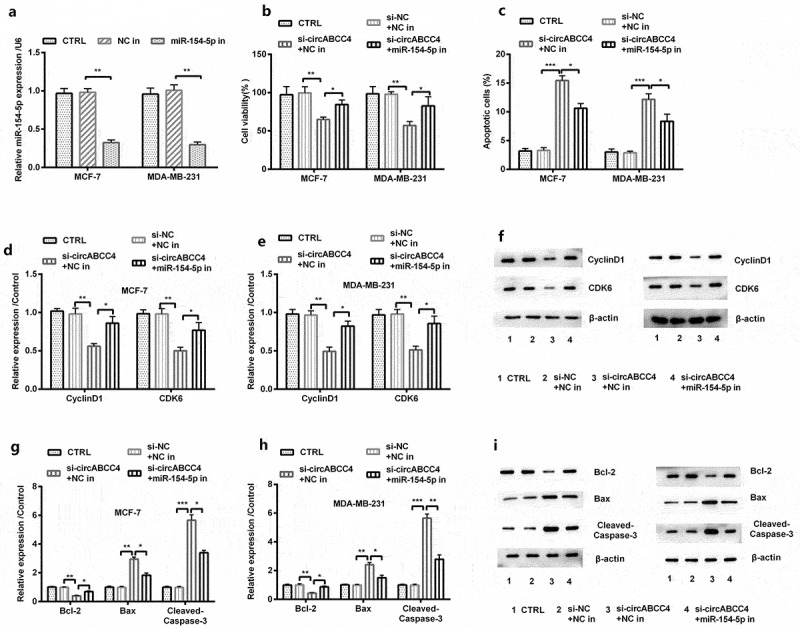
CircABCC4 knockdown subdued cell viability and promoted apoptosis of breast cancer cells by promoting miR-154-5p. (a) NC inhibitor and inhibitory miR-154-5p were transfected into breast cancer cell lines as well as the level of miR-154-5p was detected by qRT-PCR. (b) The cell viability was detected by CCK-8. (c) The apoptosis of breast cancer cell lines were detected by flow cytometry. (df) The level of CyclinD1 also CDK6 was analyzed through western blot. (gi) The expression of breast cancer cell lines apoptosis-correlated proteins (Bcl-2, Bax, Cleaved/Caspase-3) were examined through western blot. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA; CCK-8, Cell Counting Kit-8; qRT-PCR, Quantitative real-time polymerase chain reaction; CDK, cyclin-dependent kinases (n = 25). ANOVA remained responsible for P-values, * P < 0.05, ** P < 0.01 and *** P < 0.001 were considered as significant results
CircABCC4 knockdown inhibited cell migration also invasion of breast cancer cells by promoting miR-154-5p
Other than this, we examined the meanings of miR-154-5p on migration as well as invasion of cells. We found that miR-154-5p inhibitor increased cell migration and invasion ability compared with si-circABCC4 + NC inhibitor (P < 0.05 or P < 0.01, Figure 6(a,b)). These above results uncovered circABCC4 knockdown inhibited cell migration, invasion of breast cancer cells via promoting miR-154-5p.
Figure 6.
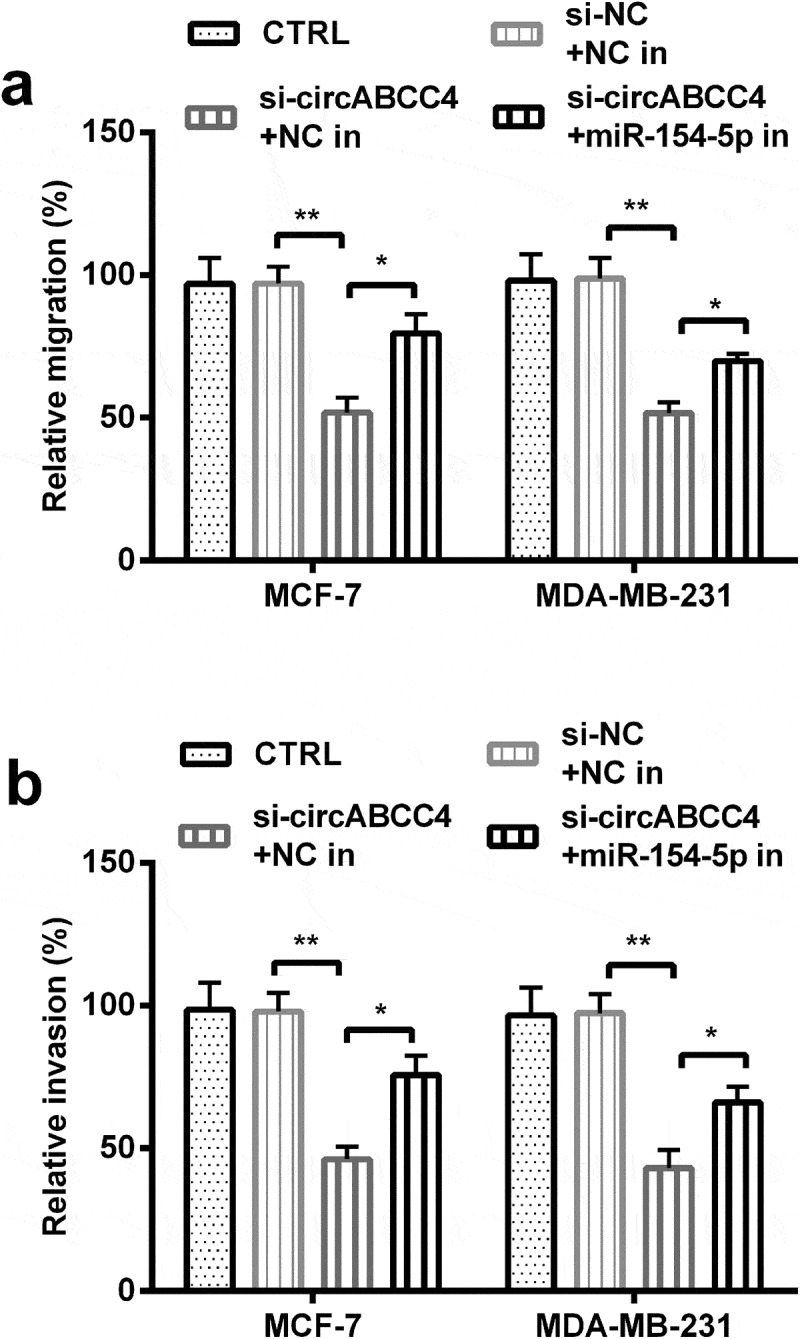
CircABCC4 knockdown inhibited cell migration besides invasion of breast cancer cells by promoting miR-154-5p. (ab) The cell migration also cell invasion of breast cancer cell lines were detected by Transwell chamber assay. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, * P < 0.05 and ** P < 0.01 were considered as significant results
CircABCC4 knockdown inhibited NF-κB and Wnt/β-catenin signal pathways by raising miR-154-5p
The NF-κB and Wnt/β-catenin signal pathways are vital pathways involving in breast cancer [17,18]. We examined whether the circABCC4-miR-154-5p axis could adjust the activation of both pathways. We observed the protein accumulation of several well-known proteins in these pathways by Western blot. As shown in the Figures, circABCC4 knockdown and miR-154-5p inhibitor raised the expression of p/t-NF-κB and Wnt3a compared with si-circABCC4 + NC inhibitor (P < 0.05 or P < 0.01, Figure 7(a-f)). These above results displayed that circABCC4 knockdown restrained NF-κB and Wnt/β-catenin signal pathways by raising miR-154-5p.
Figure 7.
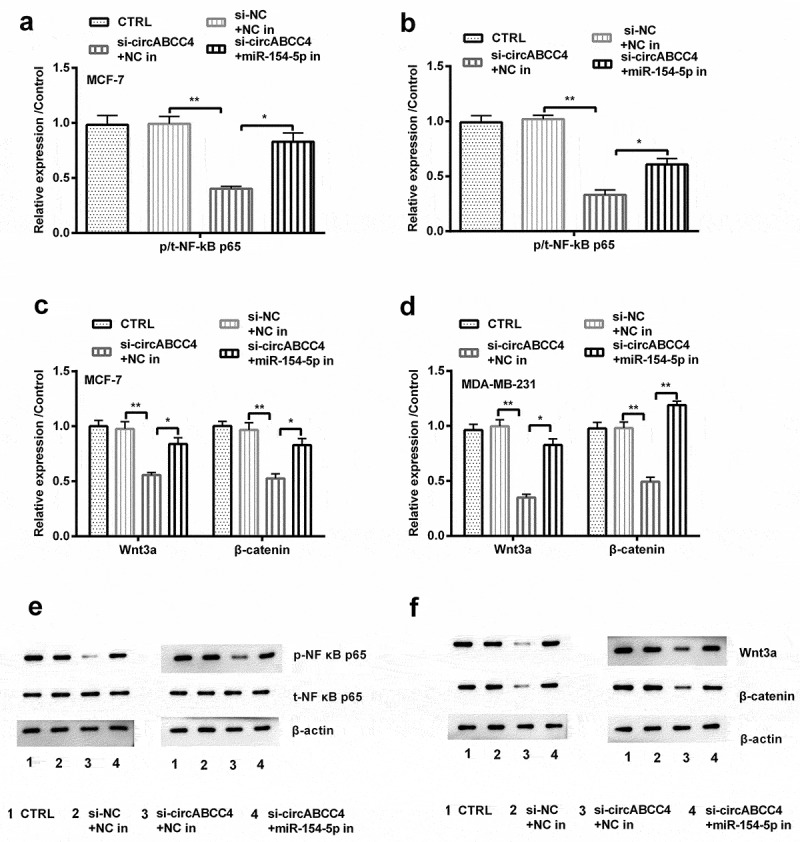
CircABCC4 knockdown inhibited NF-κB and Wnt/β-catenin signal pathway via raising miR-154-5p. (ab) After transfection with si-circABCC4, miR-154-5p inhibitor or their corresponding controls, the protein of the NF-κB and Wnt/β-catenin signal pathways were analyzed by Western blot. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, * P < 0.05 and ** P < 0.01 were considered as significant results
Discussion
In our study, we revealed that the level of circABCC4 was greatly raised in breast cancer tissues. Besides, circABCC4 knockdown dramatically inhibited breast cancer cell lines viability, invasion, migration also promote apoptosis. Besides, we found that circABCC4 promoted breast cancer progression by modulating miR-154-5p. In addition, circABCC4 knockdown could inhibit NF-κB and Wnt/β-catenin pathways, suppressing cell viability, migration and invasion, promoting apoptosis by raising the level of miR-154-5p.
Accumulating evidence showed that circRNAs could act as an important regulator in cancer pathogenesis [19–21]. According to previous reports, one study found that circGFRA1 was up-regulated in breast cancer [22]. As proved by Tang et al., hsa_circ_0001982 was overexpressed in all breast cancer tissues as well as cell lines, hsa_circ_0001982 knockdown inhibited breast cancer cell proliferation, invasion in addition to induced apoptosis [23]. CircABCC4 is a novel circRNA that is derived from ABCC4 mRNA back-splicing and located in chromosome 13q32.1 [10]. A previous in vitro study designated that circABCC4 was significantly raised in prostate cancer, and the study also found circABCC4 knockdown meaningfully inhibited prostate cancer cell survival ability, migration as well as invasion [10]. However, we know very little about the part of circABCC4 in a cancer that women are prone to. Therefore, we aimed to investigate the potential regulatory role of circABCC4. We found that the expression of circABCC4 was significantly upregulated in breast cancer tissues. Moreover, knockdown of circABCC4 was correlated with poorer cell viability, migration, and invasion in breast cancer cell lines. CircABCC4 knockdown significantly promoted cells apoptosis. Our experimental results were consistent with the results of prostate cancer studies [10].
Studies have found that the circRNA-miRNA-mRNA axis is associated with cancer progression [24,25]. CircRNAs can be used as ceRNAs to counteract miRNA mediated complex post-transcriptional regulatory networks [26–28]. It was first reported that a circRNA named CDR1as acted as a sponge of miR-7 [29]. Another circRNA called Sry regulates the expression of miR-138 [30]. A previous study has demonstrated that circABCC4 sponged miR-1182 to stimulate FOXP4 level, and circABCC4 knockdown significantly inhibited prostate cancer cell ability, migration, as well as invasion [10]. Therefore, we detected the possible character of circABCC4 in miRNAs in breast cancer. miR-154-5p plays a tumor-inhibiting role in a variety of cancers. For instance, small nucleolar RNA host gene (SNHG) 1 acted as a sponge for miR-154-5p in colon cancer, repressing Cyclin D2 (CCND2) expression [14]. miR-154-5p may suppress prostate cancer cell ability, migration by targeting E2F5 [13]. In breast cancer, the SNHG5 level was obviously and negatively associated with miR-154-5p expression level, and SNHG5 sponged miR-154-5p to repress proliferating cell nuclear antigen (PCNA) [31]. Therefore, we speculated that circABCC4 might also play a vital role in breast cancer by adaptable miR-154-5p. To prove this hypothesis, we made a bioinformatic predication and identified there were complementary sequences between circABCC4 and miR-154-5p. Besides, we implemented qRT-PCR to further verify the link between circABCC4 and miR-154-5p and found that circABCC4 negatively regulated the level of miR-154-5p. The effects of circABCC4 knockdown on cell viability, apoptosis, migration, as well as invasion could be partially attenuated by miR-154-5p expression, which indicated that circABCC4 might be a ceRNA of miR-154-5p in breast cancer cells.
The NF-κB and Wnt/β-catenin signal pathways are closely associated with breast cancer [18,32]. For instance, Deng et al. reported that β-catenin might play a vital role in tumorigenesis through the cross-regulation of NF-κB [33]. Chandra P et al. showed that abnormal motivation of the Wnt/β-catenin signal pathway was involved in the growth of breast cancer and the promotion of β-catenin-drive downstream marks, c-Myc and cyclin D1, and was also connected with breast cancer progress [34]. In addition, Jia et al. indicated that the small molecule carbamelin, which inhibited both β-catenin and NF-κB, inhibited the improvement and advancement of breast cancer [35]. One reach found that miR-154-5p overexpression reduced the activation of the pathway, which was non-canonical Wnt/PCP (RhoA-ROCK) [36]. Another study found that miR-154 encouraged myocardial fibrosis via β-catenin [37], but less research on cancer. In this research, we found that inhibition of circABCC4 knockdown deactivated the NF-κB as well as Wnt/β-catenin pathways, whereas miR-154-5p displayed an opposing effect on those signaling pathways to circABCC4 silencing. These above results revealed that motivation of the NF-κB, as well as Wnt/β-catenin signal pathways, was achieved by miR-154-5p up-regulation. Our experimental results were similar to the results of the above studies.
Conclusion
In summary, our study indicates that circABCC4 is an innovative potential oncogene in breast cancer. Knockdown of circABCC4 inhibits cell viability, migration also invasion by inhibiting NF-κB as well as Wnt/β-catenin pathway via targeting miR-154-5p. CircABCC4 may serve as a cancerous-gene in breast cancer progression, and further studies are still necessary to demonstrate the roles of miRNAs in circABCC4-regulated tumorigenesis.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure statement
The authors declare that there are no conflicts of interest.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].F Alonso D, V Ripoll G, Garona JB, et al. Metastasis: recent discoveries and novel perioperative treatment strategies with particular interest in the hemostatic compound desmopressin. Curr Pharm Biotechnol. 2011;12:1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nigro J, Cho K, Fearon E, et al. Scrambled exons. Cell. 1991;64:607–613. [DOI] [PubMed] [Google Scholar]
- [4].Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333. [DOI] [PubMed] [Google Scholar]
- [6].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- [7].Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(9–21):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kristensen L, Hansen T, Venø M, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang C, Deng H, Wang Y, et al. RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23:6112–6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ha M, Kim VN.. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- [12].Iorio MV, Ferracin M, Liu C-G, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. [DOI] [PubMed] [Google Scholar]
- [13].Zheng Y, Zhu C, Ma L, et al. miRNA-154-5p inhibits proliferation, migration and invasion by targeting E2F5 in prostate cancer cell lines. Urol Int. 2017;98:102–110. [DOI] [PubMed] [Google Scholar]
- [14].Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jin X, Mu P. Targeting breast cancer metastasis. Breast Cancer. 2015;9(BCBCR):S25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang J, Liu D, Feng Z, et al. MicroRNA-138 modulates metastasis and EMT in breast cancer cells by targeting vimentin. Biomed Pharmacother. 2016;77:135–141. [DOI] [PubMed] [Google Scholar]
- [17].Hseu Y-C, Lin Y-C, Rajendran P, et al. Antrodia salmonea suppresses invasion and metastasis in triple-negative breast cancer cells by reversing EMT through the NF-κB and Wnt/β-catenin signaling pathway. Food Chem Toxicol. 2019;124:219–230. [DOI] [PubMed] [Google Scholar]
- [18].Mu J, Zhu D, Shen Z, et al. The repressive effect of miR-148a on Wnt/β-catenin signaling involved in Glabridin-induced anti-angiogenesis in human breast cancer cells. BMC Cancer. 2017;17:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. [DOI] [PubMed] [Google Scholar]
- [20].Okholm TLH, Nielsen MM, Hamilton MP, et al. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2017;2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo J-N, Li J, Zhu C-L, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tang -Y-Y, Zhao P, Zou T-N, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. [DOI] [PubMed] [Google Scholar]
- [24].Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yin W-B, Yan M-G, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. [DOI] [PubMed] [Google Scholar]
- [26].Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [30].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [31].Chi J-R, Yu Z-H, Liu B-W, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. 2019;17:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu L, Ahn KS, Shanmugam MK, et al. Oleuropein induces apoptosis via abrogating NF‐κB activation cascade in estrogen receptor–negative breast cancer cells. J Cell Biochem. 2019;120:4504–4513. [DOI] [PubMed] [Google Scholar]
- [33].Deng J, Miller SA, Wang H-Y, et al. β-catenin interacts with and inhibits NF-κB in human colon and breast cancer. Cancer Cell. 2002;2:323–334. [DOI] [PubMed] [Google Scholar]
- [34].Prasad CP, Rath G, Mathur S, et al. Potent growth suppressive activity of curcumin in human breast cancer cells: modulation of Wnt/β-catenin signaling. Chem Biol Interact. 2009;181:263–271. [DOI] [PubMed] [Google Scholar]
- [35].Jia D, Yang W, Li L, et al. β-Catenin and NF-κB co-activation triggered by TLR3 stimulation facilitates stem cell-like phenotypes in breast cancer. Cell Death Differ. 2015;22:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li J, Hu C, Han L, et al. MiR-154-5p regulates osteogenic differentiation of adipose-derived mesenchymal stem cells under tensile stress through the Wnt/PCP pathway by targeting Wnt11. Bone. 2015;78:130–141. [DOI] [PubMed] [Google Scholar]
- [37].Dong P, Liu W, Wang Z. MiR-154 promotes myocardial fibrosis through β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:2052–2060. [DOI] [PubMed] [Google Scholar]


