Abstract
To understand SARS-CoV-2 microevolution, this study explored the genome-wide frequency, gene-wise distribution, and molecular nature of all point-mutations detected across its 71,703 RNA-genomes deposited in GISAID till 21 August 2020. Globally, nsp1/nsp2 and orf7a/orf3a were the most mutation-ridden non-structural and structural genes respectively. Phylogeny of 4618 spatiotemporally-representative genomes revealed that entities belonging to the early lineages are mostly spread over Asian countries, including India, whereas the recently-derived lineages are more globally distributed. Of the total 20,163 instances of polymorphism detected across global genomes, 12,594 and 7569 involved transitions and transversions, predominated by cytidine-to-uridine and guanosine-to-uridine conversions, respectively. Positive selection of nonsynonymous mutations (dN/dS >1) in most of the structural, but not the non-structural, genes indicated that SARS-CoV-2 has already harmonized its replication/transcription machineries with the host metabolism, while it is still redefining virulence/transmissibility strategies at the molecular level. Mechanistic bases and evolutionary/pathogenicity-related implications are discussed for the predominant mutation-types.
Keywords: SARS-CoV-2, Genome-wide mutations, Transition, Transversion, Nonsynonymous and synonymous mutations, Microevolution
Highlights
-
•
SARS-CoV-2 microevolution was studied in 71,703 whole-genomes sequenced worldwide until 21.08.2020.
-
•
Globally, nsp1/nsp2 and orf7a/orf3a were the most mutation-ridden non-structural and structural genes respectively.
-
•
Whole-genome phylogeny revealed that entities belonging to the early lineages are mostly spread over Asian countries.
-
•
A transition:transversion ratio of 2.66 characterized SARS-CoV-2, with C→U and G→U being the predominant mutation-types.
-
•
Missense mutations are positively selected mostly in structural genes; missense C→Us tend to usher hydrophobic amino acids.
1. Introduction
On 30 Dec 2019, Li Wenliang in Wuhan, China, first recognized and communicated about the outbreak of a contagious illness resembling severe acute respiratory syndrome (SARS), which subsequently got identified as the 2019 novel coronavirus disease (COVID-19; causative agent: SARS coronavirus 2, abbreviated as SARS-CoV-2 [1]). Since then COVID-19 has spread to hundreds of countries and infected tens of millions of people, killing more than a million. The first whole-genome sequence of SARS-CoV-2 was deposited in GenBank (NC_045512.2) on January 5 [2]. The positive-sense, single-stranded, 29,903 nucleotide long RNA genome contained 16 and 9 non-structural and structural genes respectively, plus two untranslated segments of 254 and 229 nucleotides at the 5′ and 3′ ends respectively [2]. High gene-arrangement similitudes of SARS-CoV-2 with coronaviruses found in bats (Rhinolophus sinicus) [3,4] and Sunda Pangolin (Manis javanica) [5] indicated COVID-19 to be a zoonotic disease [6], even though human to human transmission of SARS-CoV-2 is now very well established.
At the same time as the scientific community is racing to develop vaccines and therapeutics against COVID-19 [7], the virus on its part is busy accumulating mutations across its pan-genome, some of which may well help it evade clinical interventions [[8], [9], [10], [11]]. In this microevolutionary context, the present study analyzes 71,703 global whole-genome sequences of this novel coronavirus to reconstruct the phylogeny and reveal the trends of point-mutation accumulation. Besides identifying the genome-wide frequency, gene-wise distribution, and molecular characteristics of all point-mutations detected across these genomes, the ratio between the rates of nonsynonymous (missense) and synonymous mutations (dN/dS) was determined to understand the selection pressures on the different genes. Mechanistic basis and evolutionary implications have been discussed for the preponderance of some specific types of mutations in SARS-CoV-2.
2. Methods and algorithms
2.1. Comparative genomics
Of the 83,475 SARS-CoV-2 whole-genome sequences available in the repository of Global Initiative on Sharing All Influenza Data (GISAID) on 21 August 2020, 42.22% were from UK, while the rest were from 107 other countries. All these sequences were downloaded together with their metadata, and the dataset was filtered using the Augur tool kit [12] to eliminate undesired sequences. 11,723 entries were removed based on the minimum 29,000 nucleotide length cut-off that was set with reference to the genome size of the Wuhan strain (NC_045512.2); another 49 were removed because they originated from non-human sources. In this way, 71,703 GISAID entries remained in the final dataset used for further study. For all downstream analyses, again, the 29,903 nucleotide long complete whole-genome of the earliest-sequenced SARS-CoV-2 strain from Wuhan (NC_045512.2) was used as the reference sequence. The software package called MicroGMT or Microbial Genomics Mutation Tracker [13] was used to identify modifications in the genome sequences analyzed. This package essentially uses Minimap2 [14] and Bcftools [15] to map individual genomes against the reference and store the results in a Variant Call Format (VCF) table. It further utilizes the SnpEff tool [16] to characterize all the detected mutations at the level of the nucleotide as well as the amino acid in the translated sequence. Although MicroGMT also reports instances of insertion and deletion in the sequences compared, the current study focused only on the point-mutation data, which were further verified as follows. The software MAFFT [17] was used with default options to align all the whole-genome sequences included in the dataset. Polymorphisms (base substitutions) were identified in the individual genomes using the software SNP-sites [18], which specifically identifies single nucleotide polymorphisms (SNPs) from aligned multi-fasta sequence files. Subsequently, the VCF file generated from the SNP-site analysis was processed using the software VCFtools [19] to enumerate all transition and transversion events within the dataset of aligned whole-genome sequences. Frequency of point mutations (M f) in the SARS-CoV-2 pan-genome, or a given segment (locus) of the pan-genome, was calculated as P i / (L n × N s), where P i is the number of instances of polymorphism detected within the genome/locus, L n is the nucleotide length of the genome/locus, and N s is the number of sequenced entities present in the dataset. dN/dS (also known as ω or Ka/Ks), which is the ratio between the rates of nonsynonymous (dN) and synonymous (dS) mutations, was determined for all the individual genes of SARS-CoV-2, based on likelihood analysis using the software package HyPhy [20]. Sequence similarities between SARS-CoV-2 genome pairs were computed using the software FastANI, which uses a high throughput method for average nucleotide identity analysis [21].
2.2. Phylogenomic analyses
Evolutionary relationship between the existing SARS-CoV-2 lineages was inferred from a phylogenetic tree constructed based on a subset of the 71,703 whole-genome sequences used for studying mutation accumulation trends. Sub-sampling was necessary because it is not possible to meaningfully display 71,703 sequences in a single phylogenetic tree. This sub-dataset, comprising 4618 complete whole-genome sequences, was created using the software package Augur [12], and by means of including (in an unbiased way) 150 genomes per geographical region (continent) per month since the first Wuhan strain was sequenced (NC_045512). Multiple sequence alignment was also created using the Augur tool kit of the Nextstrain package. Further alignment was carried out using the software IQ-TREE 2 which uses the maximum likelihood method for tree construction [22]; Generalised Time Reversible (GTR) model was followed to construct the phylogenetic tree, which was finally visualized in the software Auspice (https://auspice.us). For the labeling of clades in the phylogenetic tree, type-defining marker mutations were downloaded from the Nextstrain github repository which comes as a package within the Nextstrain tool (https://github.com/nextstrain/ncov). Rules of clade-labeling followed were those mentioned in the website located at https://nextstrain.github.io/ncov/naming_clades.html. Thus, clades were labeled based on the geographical origin of the sequences, plus three different concepts of clade nomenclature that are in use for COVID-19, namely (i) the dynamic clade nomenclature system PANGOLIN [23] (ii) Year-Letter nomenclature system proposed by Hodcroft et al. (https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming), and (iii) the system proposed by Tang et al. [24], and followed by GISAID, which names major clades based on nine distinct marker mutations spread over 95% of the known SARS-Cov-2 diversity.
In order to elucidate the biogeography and microevolution of SARS-CoV-2 in India, the latest hotspot of the COVID-19 pandemic, we reconstructed the phylogeny using a separate sub-dataset (derived from the same 71,703 GISAID sequences) that included a large number of sequences from Indian strains, alongside representative sequences from all other geographical areas to enable understanding of the whole dynamics from a global perspective. This sub-dataset building involved ‘focal’ sampling for India and ‘selective’ sampling for other geographical areas, both following custom rules laid down in Nextstrain: for the ‘focal’ country (India), up to 300 sequences, or whatever maximum number (<300) is available, per month for each year under consideration; for contextual sampling, 50 such whole-genome sequences per month per country that are genetically associated to the ‘focal’ samples based on the priority call criterion called ‘Proximity’. This approach short-listed 5778 whole-genome sequences, of which 1148 belonged to the ‘focal’ country India. These 5778 sequences were analyzed using the same methodology as the one described above for the global phylogentic tree, following which the Indian sequences were mapped as per their clade affiliation and indicated using the GISAID and Year-Letter clade nomenclature systems.
3. Results and discussions
3.1. Small but phylogenetically significant divergences in global SARS-CoV-2 genomes
Average nucleotide identity (ANI, for a Kmer size of 16, over a fragment size of 1000 nucleotides) and sequence length coverage for all the pairwise alignments possible between the 11,189 complete whole-genome sequences available simultaneously in GISAID and NCBI SARS-CoV-2 database (https://www.ncbi.nlm.nih.gov/sars-cov-2/) on 21 August 2020 showed that in all the cases both identity and coverage were within 99 and 100% (notably, ANI calculation was not possible for all the 71,703 GISAID genomes retrieved on 21 August 2020). Whilst individual SARS-CoV-2 genomes differed only by a few nucleotides, the small sequence divergences across geographies indicated that within the short time span of the current pandemic, the pan-genome has diversified, and the quasispecies reservoir has expanded, rapidly for this novel coronavirus. This holds major implications for the adaptation of the virus within human hosts, and in doing so have serious consequences on the resultant pathogenesis, disease complications, and control [25].
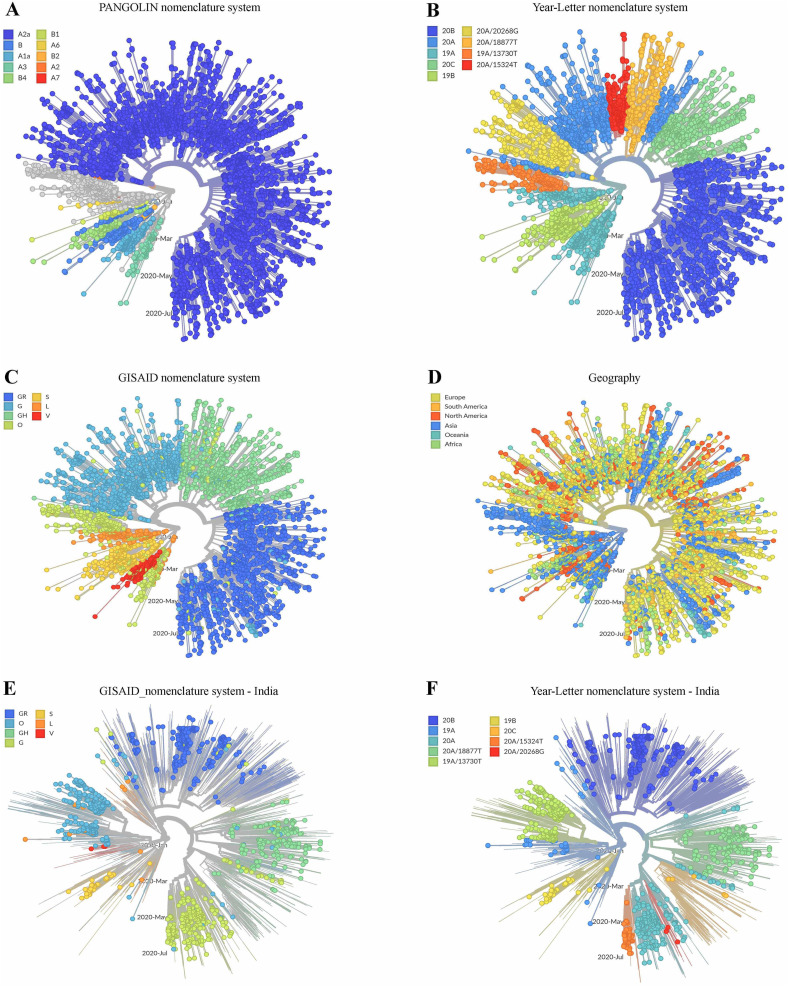
The overall evolutionary paths traced thus far by SARS-CoV-2 was delineated by labeling the 4618 global (GISAID) sequences on the phylogenetic tree using three different concepts of clade nomenclature defined in the web-based resoure https://nextstrain.github.io/ncov/ (Figs. 1A-1C). Information regarding the geographical origin of the sequences analyzed was also used to label the tree (Fig. 1D). Fig. 1A, where the tree topology was labeled according to the dynamic clade nomenclature system [23] called Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN), reflected the global preponderance of the ancestral SARS-CoV-2 lineage identified as Clade A. Notably, this ancestral clade [23] is epitomized by the 29,872 nucleotide long genome LR757995, which was isolated from Wuhan on 26 December 2019, sequenced, and submitted to GenBank on 30 January 2020. The PANGOLIN nomenclatural approach also illustrated the clear divergence of Clade A from the other SARS-CoV-2 major-clade named B, the typical representative (NC_045512.2) of which was also isolated from Wuhan on 26 December 2019, but submitted to GenBank on 12 January 2020. Albeit the genome sequence NC_045512.2 was deposited at an earlier date, the clade it represents (B) has apparently diverged at a later stage of evolution from Clade A alongside the other A-derived linages A1a and A7.
Fig. 1.
Radial trees representing the phylogenetic relationships among the different SARS-COV-2 genomes sequenced till 21 August 2020. (A-D) shows the phylogeny reconstructed based on 4618 global sequences extracted from the universal dataset of 71,703 complete whole-genomes. (A) identifies and labels the clades based on the dynamic clade nomenclature system PANGOLIN [23]. This convention currently defines 62 evolved lineages based on shared mutations, of which 10 initially-described lineages (old Nextstrian Clades) have been shown. (B) identifies and labels the clades based on Year-Letter naming as per the nomenclature system proposed by Hodcroft et al. (https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming). (C) identifies and labels the clades based on the nomenclature system proposed by Tang et al. (https://academic.oup.com/nsr/article/7/6/1012/5775463) and which is also followed by GISAID. (D) labels the entities analyzed based on the geographical region (continent) from the sequences were obtained. (E-F) shows phylogeny based on 1148 Indian and 4630 global sequences extracted from the universal dataset of 71,703 complete whole-genomes. (E) shows only the Indian sequences, and identifies and labels the clades based on Year-Letter nomenclature system. (F) also shows only the Indian sequences, and identifies and labels the clades based on GISAID nomenclature system.
On the other hand, Fig. 1B, where branches of the phylogenetic tree have been labeled according to the Year-Letter nomenclature system (i.e. with the year of identification followed by an alphabet) of Hodcroft et al., 2020 (https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming), showed that the largest lineage A2 identified by PANGOLIN clade-nomenclature system, emerged in the year 2020 and evolved further into a number of sub-lineages characterized by mutations in specific nucleotide positions (these have been designated in Fig. 1B as branches 20A, 20B, 20C, etc.). This system, which names new major clades only when the frequency of a clade exceeds 20% in a representative global sample and that clade differs in at least two nucleotide positions from its parent clade, not only corroborated the early (i.e. 2019) advent of the ancestral lineages of the PANGOLIN clade A but also identified their derivatives which formed PANGOLIN Clade B.
Consistent with the above phylogenetic interpretations, labeling of the tree with the third clade-nomenclature convention, which was proposed by Tang et al. [24] and is also followed by GISAID, indicated that the two original lineages, named as S and L (essentially equivalent to 19A and 19B of the Year-Letter nomenclature system), has diversified and thus far given rise to a total of seven clades, based on nine distinct marker mutations spread over 95% of the known SARS-Cov-2 diversity (Fig. 1C). As per the data available till 21 August 2020, Clade L is apparently more populous than Clade S, and has diversified further into V and G, with G splitting further into G, GH and GR (essentially equivalent to the old A2a clade of PANGOLIN, or the 20A, 20C and 20B of Year-Letter, nomenclature systems).
Labeling of the phylogenetic tree on the basis of the geographical origin of the sequences showed that members of the original and early-diverged clades (S and L, and V, respectively) are still mostly spread over Asian countries, whereas the recently derived clades (G, GH and GR) are distributed across the globe, especially in Europe and North America (Fig. 1D). India being the latest hotspot of the COVID-19 pandemic, recording >50,000 cases of infection and > 700 cases of fatality daily between July-end and October-middle 2020 (https://www.worldometers.info/coronavirus/country/india/), the phylogeny and biogeography of Indian SARS-CoV-2 isolates was analyzed using the specialized (GISAID-derived) dataset encompassing 1148 and 4630 genome sequences of Indian and global origins respectively. The phylogenetic tree topology obtained with this India-focused dataset (Fig. 1E and F) was essentially congruent with that obtained for the global dataset of 4618 GISAID sequences (Figs. 1A-1D). Mapping of the Indian sequences on this tree topology using the GISAID (Fig. 1E) and Year-Letter (Fig. 1F) clade nomenclature systems showed that all the mutation-types which epitomize the major clades of global SARS-CoV-2 evolution are also present in India, albeit at potentially different frequencies of distribution within the country's viral population. For instance, the relatively lower number of sequences populating the two emerging lineages 20A/20268G and 20A/15324 T can be clearly seen in Fig. 1F which, in turn, corroborated the hypothesis that in the Asian countries the ancestral lineages are still more prevalent than the recently-derived mutational groups.
3.2. Gene-wise mapping of the substitution mutations recruited in global SARS-CoV-2 genomes
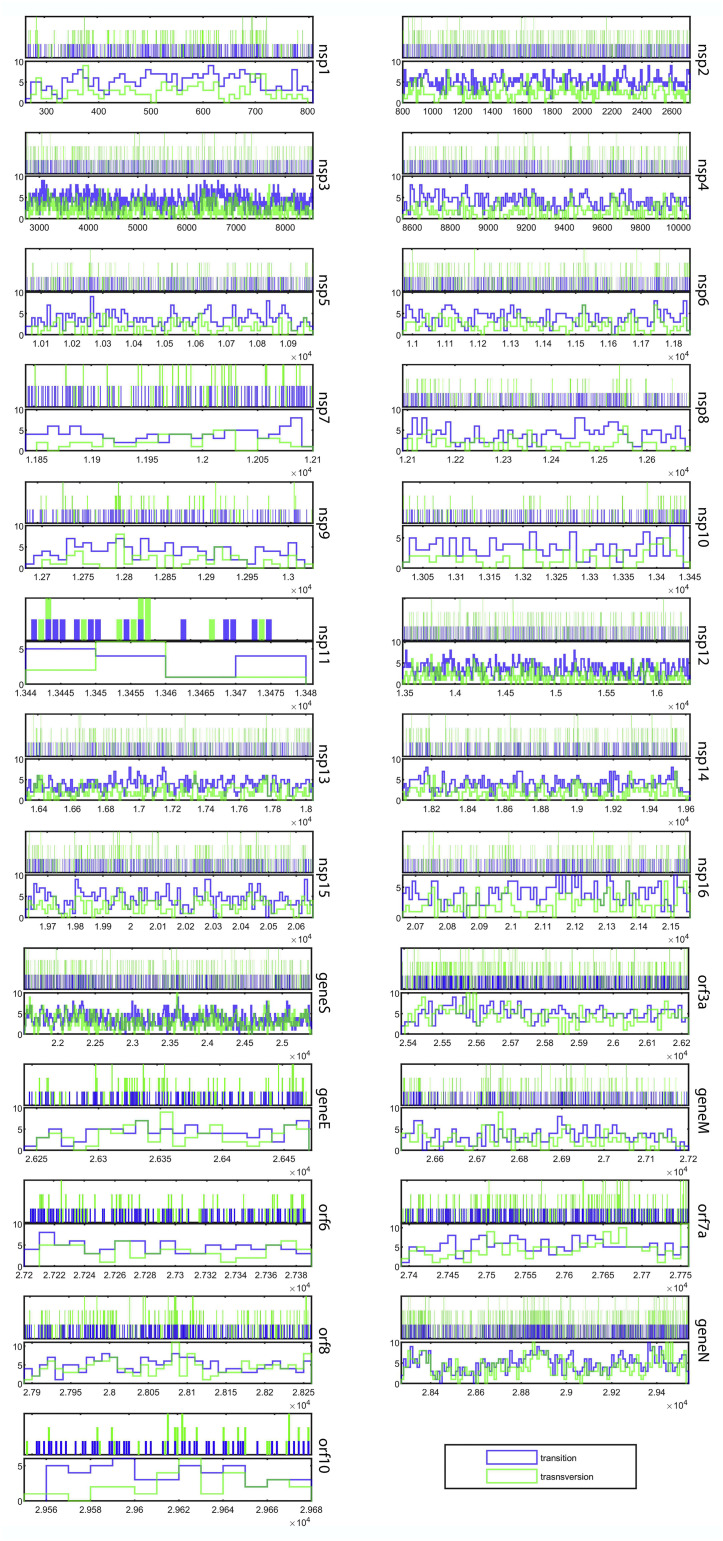
Multiple alignment of the 71,703 SARS-CoV-2 whole-genome sequences investigated in this study (29,903 completely aligned nucleotide positions, with reference to the 5′ to 3′ sequence of NC_045512.2, the earliest-sequenced genome from Wuhan), revealed 20,163 instances of single nucleotide substitution (polymorphism) across the genomes participating in the alignment (Supplementary File 1, Table S1). Overall, these point mutations have taken place at a frequency (M f) of 9.4 × 10−6, i.e. [20,163 / (29,903 × 71,703)]. On the other hand, frequency of point mutations (M f) in the 21,290 nucleotide long genomic locus encoding non-structural proteins or Nsps (Fig. 2 ) was found to be 8.78 × 10−6, i.e. [13,417 / (21,290 × 71,703)]. M f for the 8112 nucleotide long genomic locus encoding structural proteins (Fig. 2) was higher, i.e. 1.07 × 10−5 = [6199 / (8112 × 71,703)], while that for the 493 nucleotide long total untranscribed region (UTR) was highest, i.e. 1.54 × 10−5 = [547 / (493 × 71,703)]. Genes-wise, nsp1 and orf7a were the most mutation-prone non-structural and structural gene, as their M f values were 1.12 × 10−5 = [433 / (541 × 71,703)] and 1.37 × 10−5 = [359 / (366 × 71,703)] respectively; M f was also comparably high for nsp2 (1.08 × 10−5) and orf3a (1.35 × 10−5). The 20,163 instances of point mutation (polymorphisms / single nucleotide substitutions) detected across 71,703 SARS-CoV-2 genomes corresponded to only 16,002 nucleotide positions of the global alignment. This has happened in such a way that 12,203 positions each involved one specific substitution in one particular strain; 3437 positions each involved two different substitutions in two different strains; and 362 positions each involved three different substitutions in three different strains (Supplementary File 1, Table S1). This distribution showed that 53.5% (i.e. 16,002 / 29,903) of the SARS-CoV-2 pan-genome has developed polymorphism via generation of small but definite mutations across the plethora of strains disseminated globally since the COVID outbreak in December 2019.
Fig. 2.
Gene-wise localization of all the transitions and transversions detected in the 71,703 SARS-CoV-2 genomes analyzed (the graphics are based on the data given in Supplementary File 1, Table S1). Probability density plots (showing the distributions of the mutation-types) are given for all the individual genes in their respective lower panels. Nucleotide positions (with reference to the 5′ to 3′ sequence of NC_045512.2, the earliest-sequenced SARS-CoV-2 genome) covered by each gene is plotted in the X axis. Multiple mutation-types, when detected at a single nucleotide-position, are indicated as multi-color (stacked) vertical bars.
3.3. High rate of missense (nonsynonymous) mutations in the structural protein-coding genes
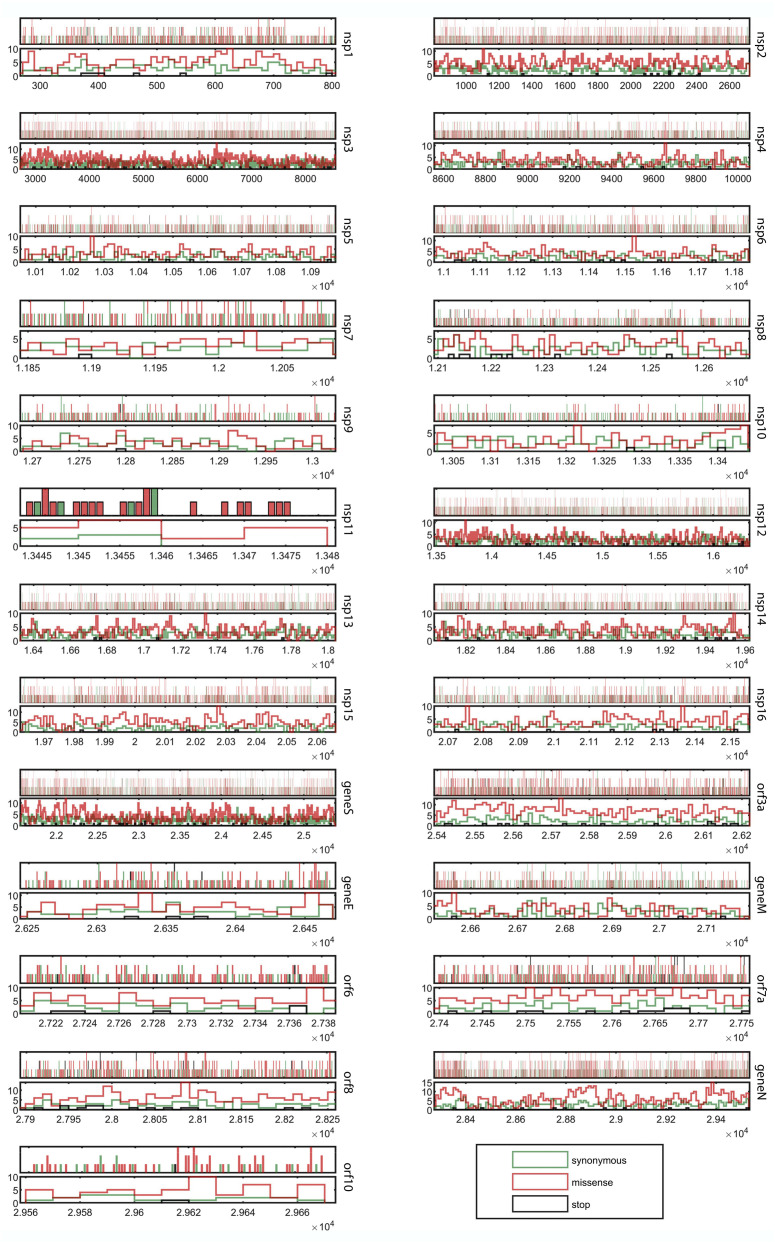
SARS-CoV-2 has experienced strong selection pressure over a short period of time. For animal viruses, in general, forces of selection (fitness constraints) emanate from host immunogenic responses, and also during replication and transmission between hosts. Evolutionarily fit (selected) strains develop tropism, and infect different cell-types or tissues of the host, reproduce within them, and in turn give rise to a variety of new strains having diverse chronic to acute infectious characteristics [26,27]. Genomic data can reveal where, when, and (sometimes) how viral pathogens have responded to various forces of natural selection. In the context of codon models, natural selection of any genetic locus is typically measured using the parameter dN/dS, which represents the ratio between the global rates of nonsynonymous (dN) and synonymous (dS) mutation accumulation in that locus. Fig. 3 graphically depicts the synonymous, missense, or stop-codon-generating nature of all point mutations detected in the 71,703 SARS-CoV-2 genomes, while their molecular details are all given in Supplementary File 1, Table S1. Based on these data a likelihood-based analysis was carried out to determine dN/dS values for all the individual genes of SARS-CoV-2. For any genetic locus, trends of positive Darwinian selection yield dN/dS >1, whereas tendencies of negative selection, or selective removal of alleles that are deleterious, result in dN/dS <1 [28]. In our analysis, all the SARS-CoV-2 genomic loci encoding Nsps, except nsp11, were found to have dN/dS values <1; among the structural genes, the same was true for S and M (genes for the structural proteins Orf3a, E, Orf6, Orf7a, Orf8, N, and Orf10, however, had dN/dS >1; Table 1 ). These numbers indicated that in the Nsp-coding genes of SARS-CoV-2 (except nsp11) missense point mutations are under purifying selection; in contrast, for the structural protein-coding genes (except S and M), missense point mutations tend to result in positive selection, thereby becoming potent drivers of evolution of this virus. Interestingly, most of the structural protein-coding genes that are under positive selection (i.e. the ones having dN/dS >1) confer abilities to infect host cells via evading the immune system (specifically, the innate immune system), and eventually induce apoptotic pathways [[29], [30], [31], [32], [33], [34], [35]]. Consequently, brisk amino acid changes in these protein sequences may well be instrumental in allowing the virus innovate newer techniques to fulfil its pathogenic objectives. From a holistic evolutionary perspective based on the above considerations, SARS-CoV-2 seems to have already succeeded in stably synchronizing its replication and transcription machineries with the host's metabolic environment (as its non-structural genes are clearly recruiting less missense mutations). The virus, however, by means of actively recruiting more missense mutations in its structural genes, is still testing newer biophysical options to increase the efficiency of its molecular contrivances for virulence and transmissibility (pathogenicity).
Fig. 3.
Gene-wise representation of the synonymous, missense, or stop-codon-generating nature of all point mutations detected in the 71,703 SARS-CoV-2 genomes analyzed (graphics based on the data given in Supplementary File 1, Table S1). Probability density plots (showing the distributions of the mutation-types) are given for all the individual genes in their respective lower panels. Nucleotide positions (with reference to the 5′ to 3′ sequence of NC_045512.2, the earliest-sequenced SARS-CoV-2 genome) covered by each gene is plotted in the X axis. Multiple mutation-types, when detected at a single nucleotide-position, are indicated as multi-color (stacked) vertical bars.
Table 1.
Locus-wise distribution of the total 20,163 instances of polymorphism detected in the SARS-CoV-2 pan-genome based on 71,703 complete whole-genomes sequenced globally until 21 August 2020.
| Locus (length in bp) | Number of transitions detected (Ts) |
ΣTs | Number of transversions detected (Tv) |
ΣTv | Σ (Ts + Tv) | Point mutation frequency (Mf) | No. of missense mutations | No. of synonymous mutations | dN/dS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A➔G | G➔A | C➔U | U➔C | A➔U | U➔A | C➔A | A➔C | C➔G | G➔C | G➔U | U➔G | ||||||||
| 5’ UTR (265) | 31 | 33 | 47 | 35 | 146 | 21 | 17 | 15 | 11 | 8 | 10 | 30 | 10 | 122 | 268 | 1.41 × 10−5 | NA | NA | NA |
| nsp1 (541) | 57 | 79 | 81 | 68 | 285 | 20 | 25 | 14 | 7 | 4 | 9 | 54 | 15 | 148 | 433 | 1.12 × 10−5 | 271 | 155 | 0.7398 |
| nsp2 (1914) | 253 | 219 | 294 | 215 | 981 | 52 | 67 | 75 | 82 | 7 | 12 | 156 | 52 | 503 | 1484 | 1.08 × 10−5 | 996 | 477 | 0.9479 |
| nsp3 (5836) | 707 | 477 | 718 | 632 | 2534 | 154 | 169 | 175 | 200 | 34 | 53 | 388 | 124 | 1297 | 3831 | 9.15 × 10−6 | 2448 | 1351 | 0.5803 |
| nsp4 (1500) | 146 | 107 | 191 | 178 | 622 | 36 | 45 | 40 | 19 | 4 | 16 | 69 | 37 | 266 | 888 | 8.25 × 10−6 | 521 | 360 | 0.5126 |
| nsp5 (918) | 86 | 52 | 112 | 97 | 347 | 16 | 24 | 21 | 23 | 1 | 6 | 50 | 17 | 158 | 505 | 7.67 × 10−6 | 310 | 190 | 0.6417 |
| nsp6 (870) | 83 | 67 | 103 | 104 | 357 | 23 | 28 | 24 | 13 | 7 | 15 | 65 | 24 | 199 | 556 | 8.91 × 10−6 | 337 | 210 | 0.7000 |
| nsp7 (249) | 29 | 16 | 35 | 23 | 103 | 4 | 8 | 8 | 7 | 3 | 2 | 14 | 8 | 54 | 157 | 8.79 × 10−6 | 86 | 70 | 0.4999 |
| nsp8 (594) | 58 | 45 | 71 | 59 | 233 | 12 | 12 | 8 | 15 | 2 | 4 | 33 | 8 | 94 | 327 | 7.67 × 10−6 | 187 | 132 | 0.4892 |
| nsp9 (339) | 34 | 30 | 48 | 26 | 138 | 5 | 7 | 13 | 5 | 0 | 2 | 17 | 10 | 59 | 197 | 8.10 × 10−6 | 108 | 88 | 0.4933 |
| nsp10 (417) | 31 | 19 | 51 | 45 | 146 | 8 | 7 | 12 | 8 | 3 | 5 | 18 | 8 | 69 | 215 | 7.19 × 10−6 | 119 | 94 | 0.4187 |
| nsp11 (39) | 2 | 4 | 5 | 3 | 14 | 1 | 2 | 0 | 1 | 2 | 1 | 3 | 0 | 10 | 24 | 8.58 × 10−6 | 19 | 5 | 1.132 |
| nsp12 (2847) | 259 | 175 | 319 | 285 | 1038 | 55 | 65 | 54 | 53 | 13 | 20 | 219 | 44 | 523 | 1561 | 7.64 × 10−6 | 906 | 637 | 0.6057 |
| nsp13 (1713) | 181 | 96 | 200 | 173 | 650 | 37 | 39 | 53 | 33 | 6 | 12 | 133 | 29 | 342 | 992 | 8.08 × 10−6 | 583 | 405 | 0.4500 |
| nsp14 (1581) | 140 | 107 | 198 | 172 | 617 | 24 | 38 | 32 | 49 | 10 | 15 | 133 | 33 | 334 | 951 | 8.39 × 10−6 | 568 | 373 | 0.4024 |
| nsp15 (1038) | 149 | 96 | 112 | 110 | 467 | 38 | 30 | 32 | 39 | 4 | 21 | 94 | 21 | 279 | 746 | 1.00 × 10−5 | 514 | 228 | 0.3937 |
| nsp16 (894) | 88 | 68 | 90 | 106 | 352 | 29 | 19 | 23 | 20 | 7 | 7 | 70 | 23 | 198 | 550 | 8.58 × 10−6 | 342 | 200 | 0.4554 |
| geneS (3822) | 346 | 246 | 428 | 417 | 1437 | 173 | 107 | 141 | 117 | 47 | 122 | 309 | 103 | 1119 | 2556 | 9.32 × 10−6 | 1615 | 906 | 0.6193 |
| orf3a (828) | 89 | 86 | 137 | 114 | 426 | 42 | 38 | 55 | 40 | 15 | 36 | 117 | 35 | 378 | 804 | 1.35 × 10−5 | 588 | 195 | 1.5013 |
| gap | 2 | 1 | 2 | 1 | 6 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 8 | ND | NA | NA | NA |
| geneE (228) | 15 | 19 | 30 | 32 | 96 | 8 | 10 | 12 | 6 | 8 | 6 | 24 | 6 | 80 | 176 | 1.08 × 10−5 | 110 | 63 | 1.0206 |
| gap | 4 | 1 | 4 | 9 | 18 | 3 | 1 | 0 | 0 | 2 | 1 | 5 | 1 | 13 | 31 | ND | NA | NA | NA |
| geneM (669) | 50 | 40 | 82 | 60 | 232 | 22 | 14 | 21 | 11 | 7 | 17 | 55 | 18 | 165 | 397 | 8.28 × 10−6 | 209 | 183 | 0.6548 |
| gap | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 5 | ND | NA | NA | NA |
| orf6 (186) | 22 | 11 | 19 | 35 | 87 | 15 | 7 | 8 | 4 | 1 | 5 | 19 | 5 | 64 | 151 | 1.13 × 10−5 | 99 | 46 | 1.3944 |
| gap | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 5 | ND | NA | NA | NA |
| orf7a (366) | 44 | 30 | 65 | 54 | 193 | 23 | 22 | 20 | 17 | 12 | 13 | 47 | 12 | 166 | 359 | 1.37 × 10−5 | 241 | 95 | 1.1946 |
| gap | 15 | 11 | 22 | 25 | 73 | 6 | 10 | 1 | 4 | 0 | 6 | 15 | 6 | 48 | 121 | ND | NA | NA | NA |
| orf8 (366) | 40 | 32 | 46 | 63 | 181 | 22 | 14 | 22 | 10 | 8 | 19 | 51 | 13 | 159 | 340 | 1.30 × 10−5 | 228 | 92 | 1.4522 |
| gap | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 4 | 7 | ND | NA | NA | NA |
| geneN (1260) | 160 | 142 | 215 | 102 | 619 | 92 | 33 | 65 | 52 | 30 | 58 | 169 | 24 | 523 | 1142 | 1.26 × 10−5 | 763 | 366 | 1.2633 |
| gap | 1 | 3 | 6 | 0 | 10 | 1 | 0 | 4 | 0 | 0 | 1 | 4 | 0 | 10 | 20 | ND | NA | NA | NA |
| orf10 (117) | 10 | 8 | 16 | 14 | 48 | 6 | 3 | 1 | 3 | 2 | 3 | 9 | 2 | 29 | 77 | 9.18 × 10−6 | 53 | 19 | 1.2981 |
| 3’ UTR (229) | 37 | 29 | 34 | 30 | 130 | 19 | 15 | 17 | 12 | 9 | 21 | 43 | 13 | 149 | 279 | 1.70 × 10−5 | NA | NA | NA |
| Pan-genome (29903) | 3174 | 2350 | 3783 | 3287 | 12,594 | 969 | 877 | 967 | 865 | 256 | 520 | 2414 | 701 | 7569 | 20,163 | 9.4 × 10−6 | 12,221 | 6940 | NA |
ND = not determined.
NA = not applicable.
dN = rate of missense (non-synonymous) mutation accumulation (ratio between the number of non-synonymous mutations and non-synonymous sites).
dS = rate of synonymous mutation accumulation (ratio between the number of synonymous mutations and synonymous sites).
3.4. High frequency of C➔U and G➔U mutations across global SARS-CoV-2 genomes
Of the 20,163 instances of polymorphism identified across global SARS-CoV-2 genomes, 12,594 and 7569 involved transition and transversion mutations respectively. In this way, a transition:transversion ratio of 1.66 characterized the nucleotide substitution bias of SARS-CoV-2. Notably, the prevalence of transition mutations in SARS-CoV-2 is higher than what is expected if transition and transversion events took place randomly. Individually also, all the SARS-CoV-2 genes had transition:transversion ratios >1.
Of the total 12,594 transition mutations encountered, maximum, i.e. 3783, featured C➔U conversion, which was 30% of the total transition count (Table 1). Individually, again, most of the SARS-CoV-2 genes were found to have C➔U conversion as the predominant transition-type across the global genomes analyzed; only in nsp16, geneE, orf6 and orf8 was U➔C most prevalent (Table1). Of the 7569 transversions detected across global SARS-CoV-2 genomes, an overwhelming 2414 (31.9%) featured G➔U conversion (Table 1). Individually, all the SARS-CoV-2 genes had G➔U conversion as the most predominant transversions-type.
3.5. Significance of copious mutations in non-structural genes 1 and 2, and most structural genes, especially orf3a and orf7a
Since RNA viruses encode their own genome replication machineries (and do not depend on the hosts' replication systems as the DNA viruses do), they can optimize their mutation rates to achieve evolutionary fitness. This leads to an unrelenting generation of genomic variants for any RNA virus, alongside a rivalry among the extant variants, including the more advanced ones that are added to the viro-diversity over time [36]. Consequently, all active genomic variants maintained within global/local RNA virus populations (quasispecies) come to possess equal abilities to replicate and complete the infection cycle [36]. In this context, the divergence of several lineages and sub-lineages of SARS-CoV-2 since the December-2019 outbreak (via generation of small mutations across its world-wide strains) - alongside the more or less efficient circulation of its two original major-lineages (clades indicated as S and L in Fig. 1) across distinct geographies - reflects the equivalent pathological and evolutionary fitness of all its extant quasispecies. This rich stock of genotypic, and therefore potentially phenotypic, variants is likely to hold major implications for potential multifaceted adaptations of this novel coronavirus within human hosts, and in doing so have serious consequences on the resultant pathogenesis, disease complications and control [25].
Viruses that have evolved to survive via changing their hosts are extremely skilled molecular manipulators; the key to their ecological fitness is attributed to their ability to subvert host defense systems to ensure survival, replication and proliferation [37]. Coronavirus-encoded accessory proteins, in general, play critical roles in virus-host interactions and modulation of host-immune responses, thereby contributing to their pathogenicity [38,39]. nsp1 and nsp2 are the most mutation-prone non-structural genes of SARS-CoV-2, as they have the highest M f values among all such genes (Table 1). Nsp1 is known to inhibit translation by binding to the host's 40S ribosome, and also inhibit IFN signaling, while Nsp2 inhibits the two host proteins proinhibitin1 and proinhibitin2 to disrupt the cellular environment [33]. Copious mutations in these two genes, therefore, can help the virus innovate novel molecular routes to evade host immunogenic response.
With regard to the 16 non-structural genes of SARS-CoV-2 it is remarkable that only nsp11 has a dN/dS value >1 (Table 1). The exact function of Nsp11 is not known. However, in Arterivirus, this protein has been characterized as a Nidoviral uridylate-specific endoribonuclease (NendoU) that is associated with RNA processing [29]. So, a dN/dS vaue >1 for nsp11 could be indicative of an intrinsic versatility of this gene in contriving newer ways of shielding the genetic material from the host's innate-immune system.
There is a clearcut distinction in the cell-death related consequences of different viral infections. While Herpesviruses, Poxviruses, Adenoviruses, and Baculoviruses bring about reduction of cell death, SARS-CoV, Ebola, Poliovirus, West Nile virus and Hepatitis B virus are capable of increasing cell death [40]. Earlier studies had reported that the accessory protein Orf3a of SARS-CoVs has pro-apoptotic activity [41]; very recent studies further implicated this protein of SARS-CoV-2 in inducing extrinsic apoptotic pathway through a unique membrane-anchoring strategy [34]. In view of these key roles of Orf3a in SARS-CoV-2 pathogenicity, and thereby transmissibility, the global existence of 804 instances of point mutations (426 transitions with 137 C➔U substitutions and 378 transversions with 117 G➔U substitutions) in the orf3a locus (Table 1; Supplementary File 1, Table S1; Figure2; Supplementary File 2, Fig. S1), including several nonsynonymous ones (Fig. 3), appears to be a part the insidious strategies of the virus towards completion of its life cycle and killing of host cells. The intrinsic molecular plasticity of orf3a activity is underscored by the fact that these copious mutations have not hampered the pathogenic aptitude of the virus. Furthermore, in this context it is noteworthy that orf3a is not only one of the most mutation-prone structural genes (M f second highest among all such genes), its dN/dS value is also >1 (Table 1).
Furthermore, in the context of the structural genes of SARS-CoV-2 it is noteworthy that orf7a is the most mutation-prone (M f = 1.37 × 10−5), and also has a dN/dS value of 1.2 (Table 1). Globally, the gene encompasses 359 instances of point mutations, of which 193 are transitions with 65C➔U substitutions, and 166 are transversions with 47 G➔U substitutions (Table 1; Supplementary File 1, Table S1; Figure2; Supplementary File 2, Fig. S1). In all SARS-CoVs, the type I membrane protein encoded by this gene is known to interact with bone marrow stromal antigen-2 (BST-2) and may play a role in viral assembly or budding events unique to SARS-CoVs [33]. Budding events are central to the transmissibility of SARS-CoV-2, so recruitment of copious mutations, especially nonsynonymous ones, in this structural gene (Fig. 3) affords novel molecular options to increase the efficiency of virulence (pathogenicity) of the virus.
3.6. Physicochemical underpinnings of the preponderance of C➔U and G➔U substitutions
In view of the overwhelming preponderance of C➔U and G➔U transitions in the global mutation spectrum of SARS-CoV-2 (as compared to all other transition and transversion mutations respectively) it seems likely that in the ecological context of this novel coronavirus some physicochemical and/or biochemical mutagen is more instrumental in bringing about this selective change, over and above the general replication error-induced mechanism of mutagenesis. Cytosine can convert to uracil through processes akin to hydrolytic deamination under the action of ultra-violet (UV) irradiation, which is well established in the context of DNA [42]. C➔U conversion is also possible chemically under the mediation of bisulfite reagents [43] that are frequently used as disinfectants, antioxidants and preservative agents. Incidentally, several control techniques involving heating, sterilization, ultraviolet germicidal irradiation (UVGI) [44] and/or chemical disinfectants [45] are being used currently to reduce the risk of viral infection from contaminated surfaces. Of these, intense UV-C irradiation is at the forefront of our fight against COVID-19, so indiscriminate use of the same may well accelerate the incidence of C➔U mutations in global SARS-CoV-2 genomes. Furthermore, UV's specificity for targeting two adjacent pyrimidine nucleotides is long known [46], while in the context of DNA, UV-induced signature mutations collated from existing data on cells exposed to UVC, UVB, UVA or solar simulator light, have been confirmed as C➔T in ≥60% dipyrimidine sites, of which again ≥5% is CC➔TT [47]. In consideration of the above facts, it seems likely that UV irradiation is the potential cause of not only the global preponderance of C➔U point mutations across SARS-CoV-2 genomes, but also the low abundance of two consecutive cytidines in all lineages of this novel coronavirus. For instance, the 29,903 nucleotide RNA genome (NC_045512.2) of the SARS-CoV-2 reference strain from Wuhan (China) has 22.28% of its genome in the form of two consecutive pyrimidine nucleotides (YY), with the most predominant being UU (8.15%) followed by CU (6.85%), UC (4.70%), and lastly CC (2.57%).
Errors resulting from replication as well as translation may be instrumental in rendering the G➔U mutations prevalent across global SARS-CoV-2 genomes. RNA viruses mutate vastly as a result of their RNA-dependent RNA polymerases (RdRPs) being error prone. From the host's view point, a propensity for incorrect protein synthesis is ushered when cells are stressed due to viral infection, and under such circumstances the viral RNA itself becomes prone to mistranslation [48]. It is therefore conceivable that SARS-CoV-2, in addition to classical mutations acquired from error-prone replication at the genomic level, uses the mistranslated replication-cum-transcription complex for the development of diverged genomic lineages [49,50]. In other words, when the viral infection discharges its positively-sensed RNA-genome into the host cell, errors in the RdRP crops up via mistranslation [51,52]; the consequent blend of wild-type and changed RdRP enzymes through its replication activities give rise to a range of viral genome-variants or quasispecies, even within a single transmission event [50]. Those variants which have the best viral fitness, eventually, endure and become predominant in the population. In this context, it is further noteworthy that both tautomeric and anionic Watson-Crick(W—C)-like mismatches can increase the recruitment of replication and translation errors [53,54]. A sequence-dependent kinetic network system connects G•T/U wobbles with three particular W—C mismatches comprising of two quickly exchanging tautomeric species (Genol•T/U⇌G•Tenol/Uenol, population < 0.4%) and one anionic species (G•T−/U−, population ≈0.001% at unbiased pH) [55].
3.7. Interpreting S gene mutations at the amino acid level
The array of highly glycosylated spike (S) proteins present on the surface of SARS-CoV-2 bind to the host cell receptor called angiotensin-converting enzyme 2 (hACE2), and upon activation by a Type II transmembrane serine protease located on the host cell membrane, facilitate viral entry into the cell [56]. Owing to its crucial role in SARS-CoV-2 infection the spike constitutes a key target for vaccine and drug development against COVID-19 [[57], [58], [59]], and for the same reason it is imperative to interpret the mutations accumulating globally in the S gene at the amino acid level and evaluate their molecular biological significance. Furthermore, in this context, it is noteworthy that some/many of the 2556 point mutations (1437 transitions with 428C➔U substitutions and 1119 transversions with 309 G➔U substitutions) detected across global S gene homologs (Table 1, Table 2 ) may put serious question marks on the eventual effectiveness of S-targeting vaccines/drugs.
Table 2.
Structural domain-wise distribution of the major mutation-types detected across spike protein-encoding genes in 71,703 complete SARS-CoV-2 whole-genomes sequenced globally until 21 August 2020.
| Domains of SARS-CoV-2 spike protein (span in amino acid positions) | No. of mutations detected | No. of missense mutations | No. of synonymous mutations | No. of stops generated due to mutations | No. of C➔U transitions (NCU) | NCU resulting in non-synomymous mutations | No. of G➔U transversions (NGU) | NGU resulting in missense mutations |
|---|---|---|---|---|---|---|---|---|
| Upstream region | 3a | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Signal peptide (1−13) | 38 | 24 | 14 | 0 | 6 | 4 | 4 | 4 |
| N-terminal Domain or NTD (14–305) | 696 | 468 | 221 | 7 | 101 | 58 | 85 | 77 |
| Peptide linking NTD with Receptor Binding Domain or RBD (306–318) | 31 | 21 | 10 | 0 | 5 | 2 | 1 | 1 |
| RBD (319–541) | 375 | 208 | 157 | 10 | 61 | 24 | 33 | 29 |
| Peptide linking RBD with Fusion Peptide or FP (542–787) | 464 | 295 | 165 | 4 | 98 | 63 | 46 | 41 |
| FP (788–806) | 37 | 26 | 11 | 0 | 4 | 3 | 2 | 2 |
| Peptide linking FP with Heptapeptide Repeat Sequence or HR1 (807–911) | 227 | 140 | 85 | 2 | 41 | 23 | 28 | 25 |
| HR1 (912–984) | 130 | 78 | 51 | 1 | 24 | 11 | 17 | 17 |
| Peptide linking HR1 with Heptapeptide Repeat Sequence or HR2 (985–1162) | 315 | 192 | 118 | 5 | 51 | 32 | 46 | 41 |
| HR2 (1163–1213) | 101 | 72 | 29 | 0 | 14 | 6 | 14 | 14 |
| Transmembrane domain (1213–1237) | 50 | 32 | 18 | 0 | 5 | 1 | 15 | 14 |
| Cytoplasmic domain (1237–1273) | 87 | 59 | 27 | 1 | 17 | 7 | 18 | 17 |
| Splice region | 2b | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 2556 | 1615 | 906 | 30 | 428 | 234 | 309 | 282 |
These three mutations have non-coding effect.
One of these two mutations involved stop loss and the other a stop retention.
2551 out of the 2556 instances of single nucleotide polymorphism detected were found to be distributed over the 12 different domains of the S protein; only 3 and 2 are in the upstream and splice regions respectively (Table 2). Of the 2551 mutations, again, 1615 are missense and 906 synonymous, while 30 generate stop codons within the reading frame (Table 2 and Fig. 3). Domain-wise, and across the 71,703 S protein homologs analyzed, mutation count per amino acid of the domain-length is maximum for the signal peptide (2.92) and lowest for the receptor binding domain (RBD, 1.68).
Like most other SARS-CoV-2 genes, C➔U and G➔U were also found to be the most dominant transition and transversion types across global S gene homologs (Table 1). Out of the 428 C➔U transitions detected across S homologs, 234 (54.7%) constituted missense mutations, of which again 102 have resulted in the conversion of hydrophilic amino acids to hydrophobic ones (58 Thr➔Ile, 1 Thr➔Met, 25 Ser➔Phe, and 18 Ser➔Leu; Supplementary File 2, Table S2). On the other hand, out of the 309 G➔U transversions detected across global S gene homologs, 282 (91.3%) constitute missense mutations, of which however only 41 have resulted in the conversion of hydrophilic amino acids to hydrophobic ones (16 Cys➔Phe, 12 Ser➔Ile, 6 Arg➔Ile, 5 Arg➔Leu, 2 Arg➔Met; Supplementary File 1, Table S1). Remarkably, in all the other genes of SARS-CoV-2, nonsynonymous amino acid replacements resulting from C➔U mutations have similar propensities for changing threonine and serine to isoleucine, leucine, methionine or phenylalanine (Supplementary File 2, Table S2). Corroboratively, a significant positive correlation (R = 0.99; P = 0.00001) was observed between the number of missense C➔U mutations in a gene and the number of hydrophilic to hydrophobic amino acid substitutions in the corresponding translated sequence.
Hydrophobic interactions play critical roles in protein folding and structure determination, so replacement of amino acids by those having distinct hydrophobicities can contribute significantly to the evolution of proteins. Mutation trends presently revealed in global SARS-CoV-2 genomes corroborated a previous theory that most protein architectures are designed in such a way that in the early phases of evolution they have apparently unique functional specializations but in the course of evolution they can manipulate the effects of C➔U mutations in such a way as to expand the probability of hidden, optional plans being extricated [[60], [61], [62]]. C➔U changes continually push amino acid contents of proteins towards more hydrophobic states, thereby unleashing the auxiliary plans of proteins on a fairly ordinary premise; in case suitable sections are present such optional plans can emerge as the new normals.
C➔U mutations in the spike have also resulted in quite a few events of proline replacement (proline codons involve no guanosine, so expectedly none of the G➔U transversions affected any proline residue). Of the 234 global C➔U transitions yielding missense mutations in the spike, 50 have resulted in the substitution of proline residues (27 Pro➔Ser, 23 Pro➔Leu; Supplementary File 2, Table S2). In all the other structural and non-structural genes as well, nonsynonymous amino acid substitution-yielding C➔U mutations exhibited global propensity for replacing proline to serine or leucine residues (Supplementary File 2, Table S2). Furthermore, a significant positive correlation (R = 0.97; P = 0.00001) was observed between the number of missense C➔U mutations accumulating in a gene and the number of proline replacements occurring in the corresponding translated sequence. In a protein sequence, when proline is replaced by serine or leucine a strong helix breaker is removed and replaced by a residue indifferent to helix formation [63]. In the process conformational freedom of the protein is increased, which in the context of the spike can implicate versatile infectivity.
The core of SARS-CoV-2 RBD consists of five antiparallel β sheets (β1, β2, β3, β4 and β7) connected by petite helices and loops. The receptor-binding motif (RBM), which mediates the contact with ACE2, lies between the β4 and β7 strands of the RBD core [64]. There are nine cysteine residues in RBD, of which eight are involved in the formation of Cys-Cys pairs: the three pairs Cys336-Cys361, Cys379-Cys432, and Cys391-Cys525 stabilize the β sheet structure, while the fourth one, Cys480-Cys488, bridges the loops present in the distal end of RBM [64]. The codons for Cys480, Cys488 and Cys525 are globally unaffected by any mutation; Cys361 and Cys391 involve instances of only synonymous mutations, while the remaining three cysteine residues have undergone substitution mutations, namely Cys336Arg, Cys379Phe and Cys432Phe (Supplementary File 1, Table S1). The fact that the cysteine pair Cys480-Cys488, through the entire evolutionary path of SARS-CoV-2, has remained unaffected by mutations reflects the indispensible (evolutionarily chosen/selected) status of the relevant disulfide bonding in the spike protein's structure and function. Corroborative to the apparent pivotal role of Cys480-Cys488 in spike architecture, this cysteine pair is also closest to the spike-hACE2 receptor interface as compared to the other cysteine pairs [65]. Likewise, from the trends of mutation revealed, structural indispensability was also apparent for Cys525 which has thus far remained unaffected by any mutation, and Cys391 where only one synonymous mutation has been selected across the 71,703 homologs analyzed. The globally-conserved status of the Cys391-Cys525 pair reinforces a previous hypothesis that this disulfide linkage which, in solution, is easily accessible to solvent molecules, could be a potent target for thiol group-containing therapeutic biochemicals towards structural dismantling of the spike protein [66]. Out of 39 amino acid residues of the spike that are reportedly responsible for binding with hACE2 [64,67], four residues (Phe486, Asn487, Cys488, and Gln498) are globally unaffected by any mutation; ten (Asp405, Tyr421, Leu455, Tyr473, Gln474, Tyr489, Leu492, Tyr495, Thr500, and Gly502) involve instances of only synonymous mutations, while the remaining 25 have undergone one or more instances of nonsynonymous substitutions (Supplementary File 2, Table S3). The likelihood of mutations being fixed in the genome depends on various factors, such as fitness of the phenotype or the position of the residues in the three-dimensional structure. Renewed studies of structural biology are required to reveal how these 25 substitutions individually alter the existing paradigm of RBD-hACE2 interaction.
The unique amino acid residue Lys417, albeit lying outside the RBM, forms salt-bridge with the Asp30 residue of hACE2 [64]. In two separate instances of polymorphism across spike homologs, this Lys417 has been substituted by arginine (A➔G: Lys417Arg, a conservative replacement) and asparagine (G➔U: Lys417Asn, a radical replacement) (Supplementary File 2, Table S3). Lysine and arginine have similar size and charge, so their interchange may cause minimal secondary structure rearrangement [68]; but how such changes eventually influence the salt-bridge interaction with Asp30 of hACE2 is still unclear. Likewise, how the Lys417Asn substitution alters the spike-hACE2 interaction paradigm as a whole is also completely unknown. Whereas the jury is still out on the biophysical significances of the global array of missense mutations in the spike, they surely pose matters of concern for drug designers and vaccine developers worldwide.
4. Conclusion
The current investigation of 71,703 complete whole-genome sequences of SARS-CoV-2 isolates from across the world brought to the fore a number of remarkable aspects of microevolution of this novel coronavirus. Phylogenomic analysis illustrated that the two major-lineages of the virus have thus far contributed almost equivalently to the pandemic, even as members of the early lineages are still mostly spread over Asian countries and those of the relatively recent lineages have undergone more global distribution. In the coming days it would be worth exploring whether this viro-geography has any bearing on the differential death rates of COVID-19 in Asian and European/American countries (https://www.worldometers.info/coronavirus/). An overwhelming preponderance of transition mutations, and far less frequency of transversions, was observed in the pan-genome of the virus, irrespective of whether the genetic locus encoded a non-structural or structural protein. In this context it is noteworthy that the 29,903 nucleotide long SARS-CoV-2 pan-genome was found to have maintained a substantive 4965 transversion mutations, notwithstanding the fact that natural selection disfavors transversion mutations because they are often missense, so less likely to conserve the structural biological properties of the original amino acids. Likewise, positive selection of missense mutations (reflected in dN/dS values >1) in most of the structural genes of SARS-CoV-2 is indicative of vigorous molecular maneuvering by the virus to augment its virulence potentials, escape human immunity, and ensure enhanced global transmissibility. Among all transitions and transversion events in SARS-CoV-2, a molecular bias was observed for C➔U and G➔U substitutions respectively. Furthermore, in all the genes, nonsynonymous amino acid replacement-yeilding C➔U mutations were found to have a remarkable propensity for changing hydrophilic residues to hydrophobic ones. More comprehensive and multi-faceted surveillance of the microevolution of SARS-CoV-2 is needed to gain constant insights into the pathogenic dynamism of the virus, and improvise control and therapeutic strategies accordingly.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
RC conceived and designed the study. RC and WG analyzed the data, interpreted the results and wrote the paper. CR and TM brought in methodologies, performed the experiments and analyses, and contributed to the manuscript. SMa, SKM and SMu participated in data analysis. All authors read and vetted the manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgement
The authors are grateful to all the researchers worldwide who sequenced and shared SARS-CoV-2 whole-genome sequence data via the NCBI (https://www.ncbi.nlm.nih.gov/sars-cov-2/) and GISAID (https://www.gisaid.org/) respositories. The research has benefited from the valuable data available at the websites https://nextstrain.org/ncov/ and https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2020.11.003.
Appendix A. Supplementary data
Nucleotide positions of the SARS-CoV-2 pan-genome over which the 20,163 instances of global polymorphism are distributed [positions are numbered with reference to the 5′ to 3′ sequence (NC_045512.2) of the earliest-sequenced strain from Wuhan, China; molecular characteristics of the mutations are also given].
Supplementary Table S2: Gene-wise distribution of the missense mutation-yielding CàU transitions (particularly those involving hydrophilic to hydrophobic amino acid replacement and proline replacement) detected across 71703 complete SARS-CoV-2 whole-genomes sequenced globally until 21 August 2020; Supplementary Table S3: Spectrum of mutations detected globally till 21 August 2020 in the hACE-2-interacting amino acid residues of the Receptor Binding Domain (RBD) of SARS-CoV-2 spike protein; Supplementary Fig. S1: Gene-wise localization of all the C→U and G→U mutations detected in the 71703 SARS-CoV-2 genomes analyzed.
References
- 1.Green A., Wenliang Li. The Lancet. 2020;395(10225):682. doi: 10.1016/S0140-6736(20)30382-2. [DOI] [Google Scholar]
- 2.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yxl P., Zhou X.-G., Wang B., Hu Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 doi: 10.1101/2020.01.22.914952. Epub Jan 23. [DOI] [Google Scholar]
- 4.Paraskevis D., Kostaki E.G., Magiorkinis G. Full genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P., Chen W., Chen J.P. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) Viruses. 2019;11:979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. M. Folegatti, K. J. Ewer, P. K. Aley, et al., Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial, Lancet doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed]
- 8.Benvenuto D., Angeletti S., Giovanetti M. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J. Inf. Secur. 2020;S0163-4453(20):30186–30189. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korber B., Fischer W.M., Gnanakaran S. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054. 04.29.069054. [DOI] [Google Scholar]
- 10.P. Saha, R. Majumder, S. Chakraborty, et al., Mutations in spike protein of SARS-CoV-2 modulate receptor binding, membrane fusion and immunogenicity: an Insight into viral tropism and pathogenesis of COVID-19, ChemRxiv. doi:10.26434/chemrxiv.12320567.v1.
- 11.Wang Q., Zhang Y., Wu L. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing Y., Li X., Gao X., Dong Q. MicroGMT: a mutation tracker for SARS-CoV-2 and other microbial genome sequences. Front. Microbiol. 2020;11:1502. doi: 10.3389/fmicb.2020.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani P., Adrian P., Le Lily W. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page A.J., Taylor B., Delaney A.J. SNP-sites: rapid efficient of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2 doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danecek P., Auton A., Abecasis G. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pond S.L.K., Frost S.D.W., Muse S.V. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 21.Jain C., Rodriguez R.L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Comm. 2018;9:1–8. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minh B.Q., Schmidt H.A., Chernomor O. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut A., Holmes E.C., O’Toole A. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X., Wu C., Li X., Song Y. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domingo E., Holland J.J. Complications of RNA heterogeneity for the engineering of virus vaccines and antiviral agents. In: Setlow K.K., editor. Genetic engineering, principles and methods. Vol. 14. Plenum press; New York, NY: 1992. pp. 13–32. [DOI] [PubMed] [Google Scholar]
- 26.Aiamkitsumrit B., Sullivan N.T., Nonnemacher M.R., Pirrone V., Wigdahl B. Human immunodeficiency virus type 1 cellular entry and exit in the T lymphocytic and monocytic compartments: mechanisms and target opportunities during viral disease. Adv. Virus Res. 2015;93:257–311. doi: 10.1016/bs.aivir.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Spielman S.J., Weaver S., Shank S.D., Magalis B.R., Li M., Pond S.L.K. Evolution of viral genomes: interplay between selection, recombination, and other forces. Methods Mol. Biol. 2019;910:427–468. doi: 10.1007/978-1-4939-9074-0_14. [DOI] [PubMed] [Google Scholar]
- 28.Kryazhimskiy S., Plotkin J.B. The population genetics of dN/dS. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M., Li X., Deng Z., Chen Z., Liu Y., Gao Y., Wu W., Chen Z. Structural biology of the arterivirus nsp11 endoribonucleases. J. Virol. 2016;91:e01309–e01316. doi: 10.1128/JVI.01309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., Ding C. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimoto F.K. The proteins of severe acute respiratory syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eigen M., McCaskill J., Schuster P. Molecular quasispecies. J. Phys. Chem. 1988;92:6881–6891. [Google Scholar]
- 37.Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signaling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prete M., Favoino E., Catacchio G. SARS-CoV-2 Inflammatory syndrome. clinical features and rationale for immunological treatment. Int. J. Mol. Sci. 2020;21:3377. doi: 10.3390/ijms21093377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke P., Tyler K.L. Apoptosis in animal models of virus-induced disease. Nat. Rev. Microbiol. 2009;7:144–155. doi: 10.1038/nrmicro2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padhan K., Minakshi R., Mohammad M.A.B., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89 doi: 10.1099/vir.0.83665-0. 1960–196. [DOI] [PubMed] [Google Scholar]
- 42.Peng W., Shaw B.R. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC→TT transitions. Biochemistry. 1996;35:10172–10181. doi: 10.1021/bi960001x. [DOI] [PubMed] [Google Scholar]
- 43.Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis- a personal account. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2008;84:321–330. doi: 10.2183/pjab/84.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng C.C., Li C.-S. Inactivation of viruses on surfaces by ultraviolet germicidal irradiation. J. Occup. Environ. Hyg. 2007;4:400–405. doi: 10.1080/15459620701329012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matallana-Surget S., Meador J.A., Joux F., Douki T. Effect of the GC content of DNA on the distribution of UVB-induced bipyrimidine photoproducts. Photochem. Photobiol. Sci. 2008;7:794–801. doi: 10.1039/b719929e. [DOI] [PubMed] [Google Scholar]
- 46.Miller J.H. Mutagenic specificity of ultraviolet light. J. Mol. Biol. 1985;182:45–68. doi: 10.1016/0022-2836(85)90026-9. [DOI] [PubMed] [Google Scholar]
- 47.Brash D.E. UV signature mutations. Photochem. Photobiol. 2015;91:15–26. doi: 10.1111/php.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohler K., Ibba M. Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2017;2:17117. doi: 10.1038/nmicrobiol.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribas de Pouplana L., Santos M.A., Zhu J.H., Farabaugh P.J., Javid B. Protein mistranslation: friend or foe? Trends Biochem. Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Ou X., Cao J., Cheng A. Errors in translational decoding: tRNA wobbling or misincorporation? PLoS Genet. 2019;15(3) doi: 10.1371/journal.pgen.1008017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma N.J., Hemez C.F., Barber K.W. Organisms with alternative genetic codes resolve unassigned codons via mistranslation and ribosomal rescue. eLife. 2018;7:1–23. doi: 10.7554/eLife.34878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nibert M.L. Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology. 2019;507:96–100. doi: 10.1016/j.virol.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koag M.C., Nam K., Lee S. The spontaneous replication error and the mismatch discrimination mechanisms of human DNA polymerase β. Nucleic Acids Res. 2014;42:11233–11245. doi: 10.1093/nar/gku789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G. New structural insights into translational miscoding. Trends. Biochem. Sci. 2016;41:798–814. doi: 10.1016/j.tibs.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Kimsey I.J., Szymanski E.S., Zahurancik W.J. Dynamic basis for dG•dT misincorporation via tautomerization and ionization. Nature. 2018;554:195–201. doi: 10.1038/nature25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARSCoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pang J., Wang M.X., Ang I.Y.H. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9:E623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salvatori G., Luberto L., Maffei M. SARS-CoV-2 spike protein: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poole A., Penny D., Sjöberg B.M. Confounded cytosine! Tinkering and the evolution of DNA. Nat. Rev. Mol. Cell Biol. 2001;2:147–151. doi: 10.1038/35052091. [DOI] [PubMed] [Google Scholar]
- 61.Studer R.A., Dessailly B.H., Ornego C.A. Residue mutations and their mpact on protein structure and function: detecting beneficial and pathogenic changes. Biochem. J. 2013;449:581–594. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- 62.Matyasek R., Koverik A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased toward C>U transitions, indicating rapid evolution in their hosts. Genes. 2020;11:761. doi: 10.3390/genes11070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S.C., Goto N.K., Williams K.A., Deber C.M. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 65.Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Debnath U., Dewaker V., Prabhakar Y.S., Bhattacharyya P., Mandal A. Conformational perturbation of SARS-CoV-2 spike protein using N-acetyl cysteine, a molecular scissor: a probable strategy to combat COVID-19. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12687923.v1. Preprint. [DOI] [Google Scholar]
- 67.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Betts M.J., Russell R.B. Amino acid properties and consequences of substitutions. In: Barnes M.R., Gray I.C., editors. Bioinformatics for Geneticists. John Wiley & Sons, Inc; 2003. pp. 289–316. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide positions of the SARS-CoV-2 pan-genome over which the 20,163 instances of global polymorphism are distributed [positions are numbered with reference to the 5′ to 3′ sequence (NC_045512.2) of the earliest-sequenced strain from Wuhan, China; molecular characteristics of the mutations are also given].
Supplementary Table S2: Gene-wise distribution of the missense mutation-yielding CàU transitions (particularly those involving hydrophilic to hydrophobic amino acid replacement and proline replacement) detected across 71703 complete SARS-CoV-2 whole-genomes sequenced globally until 21 August 2020; Supplementary Table S3: Spectrum of mutations detected globally till 21 August 2020 in the hACE-2-interacting amino acid residues of the Receptor Binding Domain (RBD) of SARS-CoV-2 spike protein; Supplementary Fig. S1: Gene-wise localization of all the C→U and G→U mutations detected in the 71703 SARS-CoV-2 genomes analyzed.