Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is an emerging health concern due to its high mortality rate of 35%. At present, no vaccine is available to protect against MERS-CoV infections. Therefore, an in silico search for potential antigenic epitopes in the non-redundant proteome of MERS-CoV was performed herein. First, a subtractive proteome-based approach was employed to look for the surface exposed and host non-homologous proteins. Following, immunoinformatics analysis was performed to predict antigenic B and T cell epitopes that were used in the design of a multi-epitopes peptide. Molecular docking study was carried out to predict vaccine construct affinity of binding to Toll-like receptor 3 (TLR3) and understand its binding conformation to extract ideas about its processing by the host immune system. We identified membrane protein, envelope small membrane protein, non-structural protein ORF3, non-structural protein ORF5, and spike glycoprotein as potential candidates for subunit vaccine designing. The designed multi-epitope peptide then linked to β-defensin adjuvant is showing high antigenicity. Further, the sequence of the designed vaccine construct is optimized for maximum expression in the Escherichia coli expression system. A rich pattern of hydrogen and hydrophobic interactions of the construct was observed with the TLR3 allowing stable binding of the construct at the docked site as predicted by the molecular dynamics simulation and MM-PBSA binding energies. We expect that the panel of subunit vaccine candidates and the designed vaccine construct could be highly effective in immunizing populations from infections caused by MERS-CoV and could possible applied on the current pandemic COVID-19.
Keywords: Middle East respiratory syndrome coronavirus, Middle East respiratory syndrome, Multi-epitope peptide, Toll-like receptor 3
1. Introduction
Middle East respiratory syndrome caused by a novel coronavirus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), is a highly fatal viral respiratory tract infection in humans and is a global public health threat [1,2]. MERS-CoV was first described on June 13, 2012 in a patient from Dr. Soliman Fakeeh Hospital in Jeddah, Kingdom of Saudi Arabia [3,4]. The patient died 11 days after the virus detection. Since then, the virus was being reported frequently from across the Arabian Peninsula [4,5]. Furthermore, the spread of MERS-CoV to the countries of Middle East and other continents has brought international attention to the severe implications of the MERS pandemic [1]. Till to date, a total of 2279 laboratory-confirmed cases of MERS are reported worldwide according to the World Health Organization (WHO). This include 806 deaths with fatality rate of 35.3%. The bulk of these case occurred in Saudi Arabia: 1901 cases with death troll climbed to 732 at fatality rate of 38.5% (https://www.who.int/emergencies/mers-cov/en/). Dromedary camels (Camelus dromedaries) are implicated as a primary reservoir for the spillover of infection to the humans [3]. Person-to-person transmission of the virus have been shown to occur frequently especially in the hospital settings [6]. Direct contact with the camel or consumption of its products appear to be the main route for camel-to-human transmission, while prolonged contact among human hosts leads to person-to-person transmission [7]. Both camel-to-human and person-to-person transmission have been well documented in Saudi Arabia [5]. In South Korea, transmission of the diseases is entirely human-to-human contact [8]. In between 2012 and 2016, small outbreaks and sporadic episodes of the diseases have been reported across the Arabian Peninsula and were detected in humans from Bahrain, Kuwait, Oman, Qatar, the most notorious outbreak was seen in 2014 with more than 500 confirmed cases in the province of Riyadh and Makkah [9]. Till to date, cases of MERS-CoV are continuously reported from Saudi Arabia every month and that's why this country is known as critical global epicenter for MERS-CoV [10].
A range of therapeutics are investigated for MERS-CoV but still no approved treatment is available [11]. Development of a vaccine is a cost effective prophylactic measure and remains the most scalable [12]. In this regard, several candidates have been evaluated using variety of different approaches: however, still vaccine for MERS-CoV is not available [13]. Initially, vaccine development studies were hampered due to the absence of an appropriate small animal model of the MERS-CoV disease [14]. Barriers like these have been overcome in the recent past by developing in vivo approaches that allow and facilitate the testing vaccine candidate for MERS-CoV in small animal models [14]. Subunit vaccines that contains one or more pathogen immunogenic component are immunologically focused and have gained popularity because of reduced risk in vivo and relative ease of production [15]. The virus proteome comprises several vital vaccine targets that contribute in infection and pathogenesis of MERS-CoV [16]. The receptor-binding domain of spike (S) glycoprotein in particular mediate virus interaction with the host cell [15]. The envelope E protein plays a significant role in host cell recognition [17]. The interferon (IFN)-antagonizing properties of membrane protein aid in reducing IFN levels in infected cells [18]. Similarly, proteins from the open reading frame (ORF) group like ORF3, ORF5 and ORF4a are important from viral infection and pathogenesis point of view and mutational studies revealed that mutation in these genes leads to abrupt inflammation, host cell processes disruption and dysregulation IFN pathway activation [6]. The use of these single subunit vaccine in vaccine formulation is also not an appropriate choice to have as they harbor several different antigenic determinants, all of which are not desired and some may have detrimental effects on protective immunity induction [19]. This rationale has led to create an interest in designing a peptide vaccine for MERS-CoV that comprise of epitopes required for desired cellular and humoral immune response [20]. Peptide vaccines are considered sufficient enough to induce appropriate T and B-cell immunity, while at the same time excluding the risk of reactogenic and allergenic responses [21]. Moreover, peptide vaccine can be implicated for induction of broad-spectrum immunity against multiple variant of the pathogen [22]. On the contrary, because of the small size peptide vaccine is weakly immunogenic and require a carrier molecule for adjuvanting and chemical stability [19]. Several peptide vaccines are under process of development for different viral diseases, such as hepatitis C virus (HCV) [23], human papilloma virus (HPV) [24], human immunodeficiency virus (HIV) [25], and influenza [26] etc. A multi-epitope vaccine contains a series of overlapping epitopes that can elicit effective cytotoxic T cells, T helper cells and B cells responses against viral pathogen [27]. Unlike, subunit or single epitope-based vaccines, multi-epitope vaccines have a distinctive concept design with additional properties. These properties include: (i) the chances of adverse effects or pathological responses are less because of reduced unwanted components, the introduction of adjuvant capacity enhances chances of long-lasting and immunogenicity responses, (iii) as the final formation has multiple epitope the spectra of targeted viruses is expanded, (iv) they are capable of producing strong cellular and humoral immunity simultaneously, (v) the epitopes in multi-epitope peptide can be recognized by T cell receptors (TCRs) of various T-cell subsets [19].
Keeping in view the broad applicability of multi-epitope vaccine, in this work we explored the complete proteome of MERS-CoV exoproteome and secretome for cytotoxic T lymphocyte (CTL) and helper T lymphocyte (HTL) epitopes containing B-cell epitopes. Additionally, molecular docking, molecular dynamics simulation and MM-PBSA binding free energies were performed to underpin multi-epitope peptide binding pose and interactions with TLR3 receptor proteins.
2. Materials and methods
2.1. Retrieval of MERS-CoV proteome and subtractive proteomics
The complete reference proteome of MERS-CoV (Proteome ID: UP000139997) was retrieved from Swiss-Prot reviewed universal protein knowledgebase database (Uniprot) [28] and subjected to subtractive proteomics pipeline for identification of potential vaccine candidates [29]. First in the process, proteins subcellular localization was predicted using an online tools: CELLO2GO [30] tool with E-value set at 0.001 and Virus-PLoc server (http://www.csbio.sjtu.edu.cn/bioinf/virus/). Proteins having localization in extracellular matrix and membrane were considered and analyzed further along the framework. All the screened proteins were evaluated for redundancy in Cluster Database at High Identity with Tolerance (CD-Hit) with sequence identity threshold set at 50%. [31] Next, comparative homology analysis of the filtered proteins with the human proteome (H. sapiens, taxid: 9606) was performed with parameters of E-value: 0.005, bit score: ≥100 and sequence identity: ≤30% in online BlASTp tool of the National Center for Biotechnological Information (NCBI) [32,33]. To avoid any homology of the protein sequence with the beneficial bacteria of human gut, a subsequent BlASTp was run against Lactobacillus rhamnosus (taxid: 47715) taking the identity sequence cut-off ≤ 30%, bit score of 100 and E-value of 0.005 [34,35].
2.2. Prediction of epitopes
The CTL epitopes for the proteins were predicted using NetCTL 1.2 server (http://www.cbs.dtu.dk/services/NetCTL/). NetCTL 1.2 server combines prediction of C-terminal cleavage, binding affinity for reference set of MHC class I which is restricted to 12 MHC class I super type, and TAP transport efficiency. The prediction of MHC peptide binding, proteasome cleavage event, TAP transport efficiency is done using neural networks, NetChop neural networks, and weight matrix based methods, respectively. Predicted CTL epitopes with overall combine score of 1 were selected. CTL epitopes from the pathogen proteins will elicit cell mediated immunity for inhibiting the growth and development of the pathogen and produce memory cells for remembering the pathogen in future encounters [36,37]. The predicted CTL epitopes were then subjected to VaxiJen to evaluate antigenicity of the epitopes [38]. Those with values greater >0.4 were considered antigenic and considered only. The response from Helper T-lymphocyte (HTL) epitopes constitute the major part of cellular immunity and aid in producing various cytokines and immune cell to clear the pathogen [39]. The HTL epitopes induce both CTL and HTL immune responses by secreting different lymphokines and could be crucial part of prophylactic and immunotherapeutic vaccine [40]. HTL epitopes interacting with a reference set of HTL alleles were predicted using IEDB MHC-II epitope prediction module keeping the available parameters as default [41]. All predicted epitopes further subjected to allergenicity, toxicity and IFN-gamma inducing peptides prediction by using online AllergenFP v.1.0 [42], ToxinPred [43], and IFNepitope server [44], respectively. The predicted epitopes were ranked based percentile rank score: lower score describe higher affinity binding of the epitope for HTL binding. The predicted HTL epitopes for its affinity to activate T1 type immune response were cross-validated using IFN epitope server [44].
2.3. Multi-epitope peptide vaccine construction
The shortlisted conserved epitopes were then linked to each other through a flexible linker AAY. β-defensin was used as an adjuvant to recruit naive T-cells and immature dendritic cells to the infection site [45]. The CTL-epitopes peptide is linked to β-defensin at the N-terminal using EAAAK linker while the CTL and HTL epitopes were linked to each other AAY and GPGPG linker, respectively. The structure of the construct was predicted using Raptor X [46]and ab ignition I-tasser [47]. The physicochemical parameters of the vaccine construct were evaluated using ProtParam server [48]. The generated structures of the multi-epitope peptide were then utilized in PDBsum to create Ramachandran plot for the structures [49].The antigenicity and allergenicity were predicted by employing VaxiJen [38] and AlgPred servers [50], respectively.
2.4. B-cell epitopes assessment
Identification of B-cell epitopes is vital in vaccine designing considering the fact that it leads to production of B lymphocytes that differentiate into antibody producing plasma cells and memory cells [51]. B-cell epitopes of 20 amino acids long in the input protein sequences were identified using BCPREDS [52]. The epitope specificity threshold was allowed as default by 75%.
2.5. Complementary DNA analysis for cloning and expression
Reverse translational analysis was carried out using Codon Usage Wrangler Tool (https://www.mrc-lmb.cam.ac.uk/ms/methods/codon.html) to generate optimized cDNA. The cDNA sequence was then used in GeneScript to compute GC content and codon adaptation index (CAI) score. Ideally, the GC content of a sequence should be in between 30% to 70%, while the CAI score of 1 is considered good.
2.6. Molecular docking with TLR3
Multi-epitope peptide vaccine sequence was then used in molecular docking studies in PatchDock [53]. TLR3 (PDB ID, 1ZIW) was used as receptor proteins in the process. The best predate complexes were subsequently subjected to FireDock for refinement [54]. The affinity of the vaccine construct for TLR3 was also validated by ClusPro 2.0 docking software [55].
2.7. In silico immune simulation of the vaccine construct
The vaccine construct immunogenic potential was elucidated using C-immune server [56]. The simulation parameters were treated as: 10 number of antigen dose, random seed (12345), volume of simulation 100, number of simulation steps,1000, HLA alleles used (MHC class I A0101 allele, B MHC class I B0702, DR MHC class II DRB1_0101 allele), and time step of injection employed is 1.
2.8. Molecular dynamics simulation
MD simulation was run for 120 ns on the most suitable docked solution of TLR3-MEPVC. AMBER16 was used to execute this MD simulation [57]. The ff14SB force field was used to parameterize the complex [58]. The complex was solvated into a TIP3P water box. Na + ions were added to the system to counterbalance its charge. Hydrogen atoms of the system were minimized for 500 steps, solvation box and carbon alpha atoms for 1000 steps, and all non-heavy atoms for 300 steps. Later on, in an attempt to manage the system temperature, it was heated to 300 K (NVT) for 20 ps through langevin dynamics [59] where a 2 femtosecond time step long restraint of 5 kcal/mol-A2 on carbon alpha atoms was permitted. System was then unrestrained for 100 ps while being equilibrated. NPT ensemble was put in practice for 50 ps to manage the pressure on the system. Finally, simulation run of 120 ns was achieved at a time step of 2 femtoseconds. AMBER CPPTRAJ [60] was then used to examine the trajectories for to evaluate complex dynamics and stability.
2.9. Calculation of binding free energies
MMPBSA [55].py package was used to predict binding free energies of the system [61]. A total of 100 frames were derived from simulation trajectories to be employed in MMGBSA (Molecular Mechanics Generalized Born Surface Area) and MMPBSA (Molecular Mechanics Poisson–Boltzmann Surface Area) [62]. The examination of the free energy difference between solvated phase and gas phase of the system is the ultimate goal of this assay. The determination of net binding free energy was carried out as,
The binding free energy calculation was done on 100 frames extracted from complete length of MD simulation as well as from last 20-ns to thoroughly validate intermolecular affinity and stability. Entropy of the system was estimated using a bash script following the protocol provided by Duan et al., 2016 [63].
3. Results and discussion
3.1. Subtractive proteomics
The complete MERS-CoV proteome contains 13 proteins in total, namely: Replicase polyprotein 1ab, Replicas polyprotein 1a, Membrane protein, Envelope small membrane protein, Non-structural protein ORF4b, Non-structural protein ORF3, Non-structural protein ORF5, Non-structural protein ORF4a, Nucleoprotein, and Spike glycoprotein. These proteins were first investigated in subcellular localization of subtractive proteomics to screen only those candidates that are exposed for direct interaction with the host cells. Such proteins identification represent an important goal as they are involved in virulence and pathogenesis and their vital importance from stimulating immune responses [64]. Only proteins that are localized to either extracellular (exoproteome) or outer membrane (secretome) were opted in subcellular localization filter. Five proteins: Membrane protein, Envelope small membrane protein, Non-structural protein ORF3, Non-structural protein ORF5, Spike glycoprotein were shortlisted in this analysis as highlighted in bold in Table 1 . Next in the framework, these proteins were subjected non-redundancy check. In this, non-redundant dataset was only considered. All the five proteins mentioned above were non-redundant which means that each protein represented in the dataset is only singly repeated [35]. The comparative homology check with the humans revealed no hit, indicating highly non-similarity between these virus proteins and the human proteome. This is quite vital in avoiding the auto-immune responses and designing a safe vaccine [65,66]. Lastly, a comparative homology analysis between the MERS-CoV shortlisted proteins and the beneficial gut bacterium, Lactobacillus rhamnosus, was performed to eliminate the risk of accidental inhibition of good bacteria in the host. Bacteria of such type play profound role in improving functions of the host immune system, aid in digestion of food for extracting vital nutirents, and prevent the growth of hostile infection causing bacteria etc. [34]. All the five proteins were noticed to show no homology to the said representative gut bacterium proteome allowing the safe use of the filtered proteins epitopes.
Table 1.
Proteins that constitute the proteome of the MERS-CoV. Bold highlighted proteins are the one selected in subcellular localization analysis.
| Protein ID | Protein | CELLO2GO (Prediction score) | Virus-PLoc |
|---|---|---|---|
| sp|K9N7C7|R1AB_CVEMC | Replicase polyprotein 1ab | Plasma membrane (2.35) | Plasma membrane |
| >sp.|K9N638|R1A_CVEMC | Replicase polyprotein 1a | Plasma membrane (2.54) | Nucleus |
| >sp|K9N7A1|VME1_CVEMC | Membrane protein | Plasma membrane (4.95) | Plasma membrane |
| >sp|K9N5R3|VEMP_CVEMC | Envelope small membrane protein | Plasma membrane (3.33) | Plasma membrane |
| >sp.|K9N643|ORF4B_CVEMC | Non-structural protein ORF4b | Mitochondrial (1.63) | Plasma membrane |
| >sp|K9N796|ORF3_CVEMC | Non-structural protein ORF3 | Extracellular (3.62) | Plasma membrane |
| >sp|K9N7D2|ORF5_CVEMC | Non-structural protein ORF5 | Plasma membrane (4.94) | Plasma membrane |
| >sp.|K9N4V0|ORF4A_CVEMC | Non-structural protein ORF4a | Extracellular (2.21) | Cytoplasm |
| >sp.|K9N4V7|NCAP_CVEMC | Nucleoprotein | Nucleus (3.07) | Nucleus |
| >sp|K9N5Q8|SPIKE_CVEMC | Spike glycoprotein | Plasma membrane (3.31) | Plasma membrane |
3.2. Prediction of cytotoxic T-lymphocyte epitopes
Cytotoxic T cell lymphocytes (also known as Killer T cells) are important component of immune system for tumor surveillance and against intracellular pathogens including bacteria, protozoan and viruses [36]. When these cells recognized its antigen, it become activated and follow three step mechanisms to kill the infected or malignant cells [67]. First, it secretes cytokines primarily IFN-γ and TNF-α which have antiviral and antitumor effects [68]. Secondly, it produces and release cytotoxic granules. These granules have perforin that make the target cell membrane porous and granzymes which being serine proteases hydrolyze intracellular proteins, block viral proteins synthesis and eventually leads to apoptosis [67]. All the five proteins shortlisted in the previous step was analyzed for 9mer cytotoxic T-cell receptor epitopes. Briefly, 5 CTL epitopes were obtained for membrane protein, 3 for envelop small membrane protein, 3 for non-structural protein ORF3, 4 for non-structural protein ORF7 and 38 for the spike protein. Among these predicted epitopes, we found only 4 antigenic epitopes for membrane protein, 0 for envelop small membrane protein, 1 for non-structural protein ORF3 and ORF5, and 17 for spike protein. Only antigenic epitopes were considered while non-antigenic were discarded. The selected epitopes are also non-allergic, non-toxic and are IFN-gamma inducing peptide. Predicted CTL epitopes for the screened 5 subunit vaccine proteins are shown in Table 2 .
Table 2.
CTL epitopes for shortlisted 5 proteins with antigenicity scores.
| Protein | Peptide | VaxiJen Score | Result | Allerganicity | Toxicity |
|---|---|---|---|---|---|
| Membrane protein | LLITIVLQY | 0.8719 | Probable ANTIGEN | Probable non-allergen | −1.09(Non-Toxin) |
| ITIVLQYGY | 1.2147 | Probable ANTIGEN | Probable-non-allergen | −0.65(Non-Toxin) | |
| ALSIFSAVY | 0.1615 | Probable NON-ANTIGEN | Probable non-allergen | −1.37(Non-Toxin) | |
| VSAMMWISY | 0.6059 | Probable ANTIGEN | Probable non-allergen | −0.79(Non-Toxin) | |
| GTNSGVAIY | 0.4620 | Probable ANTIGEN | Probable non-allergen | −1.16(Non-Toxin) | |
| Envelope small membrane protein | MTGFNTLLV | 0.0681 | Probable NON-ANTIGEN | Probable non-allergen | −1.41(Non-Toxin) |
| TLLVQPALY | 0.2772 | Probable NON-ANTIGEN | Probable allergen | −1.23(Non-Toxin) | |
| LVQPALYLY | 0.0512 | Probable NON-ANTIGEN | Probable non-allergen | −0.87(Non-Toxin) | |
| Non-structural protein ORF3 | VTAFSKPLY | 0.2377 | Probable NON-ANTIGEN | Probable non-allergen | −1.12(Non-Toxin) |
| AQADTAGLY | 0.1360 | Probable NON-ANTIGEN | Probable non-allergen | −0.66(Non-Toxin) | |
| SVNLFDVGY | 1.2493 | Probable ANTIGEN | Probable non-allergen | −1.01(Non-Toxin) | |
| Non-structural protein ORF5 | STDSIVFTY | 0.3229 | Probable NON-ANTIGEN | Probable non-allergen | −0.91(Non-Toxin) |
| FTYIPASGY | 0.1568 | Probable NON-ANTIGEN | Probable allergen | −0.89(Non-Toxin) | |
| MVLYFLVLY | 0.1428 | Probable NON-ANTIGEN | Probable allergen | −0.81(Non-Toxin) | |
| SSTYIECTY | 0.8016 | Probable ANTIGEN | Probable non-allergen | −0.30(Non-Toxin) | |
| Spike glycoprotein | FLLTPTESY | 0.2812 | Probable NON-ANTIGEN | Probable allergen | −1.38(Non-Toxin) |
| EVDIQQTFF | 0.8992 | Probable ANTIGEN | Probable non-allergen | −1.29(Non-Toxin) | |
| VSKADGIIY | 0.1659 | Probable NON-ANTIGEN | Probable allergen | −1.19(Non-Toxin) | |
| TYSNITITY | 1.5697 | Probable ANTIGEN | Probable non-allergen | −1.10(Non-Toxin) | |
| ITYQGLFPY | 0.9146 | Probable ANTIGEN | Probable non-allergen | −0.89(Non-Toxin) | |
| YQGDHGDMY | 0.2627 | Probable NON-ANTIGEN | Probable allergen | −0.73(Non-Toxin) | |
| TSATIRKIY | 0.1760 | Probable NON-ANTIGEN | Probable allergen | −1.32(Non-Toxin) | |
| FSDGKMGRF | 1.0567 | Probable ANTIGEN | Probable non-allergen | −0.24(Non-Toxin) | |
| NSYTSFATY | 0.2171 | Probable NON-ANTIGEN | Probable non-allergen | −0.96(Non-Toxin) | |
| ATDCSDGNY | 0.7838 | Probable ANTIGEN | Probable non-allergen | −0.19(Non-Toxin) | |
| ASLNSFKEY | −0.155 | Probable NON-ANTIGEN | Probable non-allergen | −0.50(Non-Toxin) | |
| ITEDEILEW | 0.1074 | Probable NON-ANTIGEN | Probable allergen | −1.21(Non-Toxin) | |
| QTAQGVHLF | 0.3715 | Probable NON-ANTIGEN | Probable allergen | −0.98(Non-Toxin) | |
| GVHLFSSRY | 0.1530 | Probable NON-ANTIGEN | Probable non-allergen | −0.54(Non-Toxin) | |
| FSSRYVDLY | 1.0488 | Probable ANTIGEN | Probable non-allergen | −0.44(Non-Toxin) | |
| YVDLYGGNM | 0.2930 | Probable NON-ANTIGEN | Probable allergen | −0.81(Non-Toxin) | |
| QSDRKAWAA | 0.2840 | Probable NON-ANTIGEN | Probable allergen | −0.87(Non-Toxin) | |
| LLDFSVDGY | 0.0436 | Probable NON-ANTIGEN | Probable allergen | −0.95(Non-Toxin) | |
| DLSQLHCSY | 0.9444 | Probable ANTIGEN | Probable non-allergen | −0.83(Non-Toxin) | |
| SFDVESGVY | 0.9858 | Probable ANTIGEN | Probable non-allergen | −1.33(Non-Toxin) | |
| LSGTPPQVY | 0.2591 | Probable NON-ANTIGEN | Probable non-allergen | −0.96(Non-Toxin) | |
| PAAIASNCY | 0.5141 | Probable ANTIGEN | Probable non-allergen | −0.61(Non-Toxin) | |
| CYSSLILDY | 0.4038 | Probable ANTIGEN | Probable non-allergen | −0.65(Non-Toxin) | |
| YSSLILDYF | 0.0327 | Probable NON-ANTIGEN | Probable allergen | −1.03(Non-Toxin) | |
| TTITKPLKY | 0.1377 | Probable NON-ANTIGEN | Probable allergen | −1.48(Non-Toxin) | |
| ITKPLKYSY | 0.6911 | Probable ANTIGEN | Probable non-allergen | −1.77(Non-Toxin) | |
| STVWEDGDY | −0.0613 | Probable NON-ANTIGEN | Probable non-allergen | −0.73(Non-Toxin) | |
| MTEQLQMGF | 1.2508 | Probable ANTIGEN | Probable non-allergen | −1.40(Non-Toxin) | |
| YYSDDGNYY | −0.0986 | Probable NON-ANTIGEN | Probable allergen | −0.68(Non-Toxin) | |
| YSDDGNYYC | 0.1491 | Probable NON-ANTIGEN | Probable allergen | −0.52(Non-Toxin) | |
| VSVPVSVIY | 0.0758 | Probable NON-ANTIGEN | Probable allergen | −1.15(Non-Toxin) | |
| HISSTMSQY | 0.8394 | Probable ANTIGEN | Probable non-allergen | −0.92(Non-Toxin) | |
| QVDQLNSSY | 0.4655 | Probable ANTIGEN | Probable non-allergen | −0.68(Non-Toxin) | |
| FSFGVTQEY | 1.6924 | Probable ANTIGEN | Probable non-allergen | −1.30(Non-Toxin) | |
| RSAIEDLLF | 0.1712 | Probable NON-ANTIGEN | Probable allergen | −1.01(Non-Toxin) | |
| VVNAPNGLY | 0.0211 | Probable NON-ANTIGEN | Probable allergen | −0.68(Non-Toxin) | |
| WSYTGSSFY | 1.0625 | Probable ANTIGEN | Probable non-allergen | −1.27(Non-Toxin) | |
| NTTLLDLTY | 1.5316 | Probable ANTIGEN | Probable non-allergen | −1.40(Non-Toxin) |
3.3. Prediction of helper T-lymphocyte epitopes
T helper lymphocytes or CD4+ cells are important player of the immune system by instigating and shaping adaptive immune system. T helper lymphocytes recognize peptides displayed on the MHC class II molecules. They release cytokines that aid in activating several other immune cells. They are essential for B cell antibody class switching, enhancing bactericidal activity of macrophages, and growth and activation of cytotoxic T cells [51,69]. The five proteins were subjected to MHC-II epitope prediction of 15mer length as shown in Table 3 . Only epitopes of lowest percentile score were selected and as such epitopes with percentile score < 3 were opted only. Using this criterion, we predicted 3 HTL epitopes for membrane protein, 9 for envelope small membrane protein and non-structural protein ORF3, 25 for non-structural protein ORF5 and 31 for the spike protein. Antigenicity evaluation revealed 0 epitopes for membrane protein, envelope small membrane protein and non-structural protein ORF5 4 and 3 respectively, 6 for non-structural protein ORF3 and 17 for spike proteins. The selected epitopes besides being antigenic are also non-allergic, non-toxic and are IFN-gamma inducing peptide.
Table 3.
HTL epitopes for shortlisted 5 proteins with antigenicity scores.
| Protein | Start amino acid | End amino acid | Peptide | Percentile score | Vaxijen antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|
| Membrane protein | 16 | 30 | SQIISGIVAAVSAMM | 3.02 | 0.1060 (Probable non-antigen) | Probable allergen | −1.13 (Non-Toxin) |
| 17 | 31 | QIISGIVAAVSAMMW | 3.02 | 0.1324 (Probable non-antigen). | Probable non-allergen | −1.07(Non-Toxin) | |
| 18 | 32 | IISGIVAAVSAMMWI | 4.57 | 0.1871 (Probable non-antigen). | Probable non-allergen | −0.73(Non-Toxin) | |
| Envelope small membrane protein | 44 | 58 | MTGFNTLLVQPALYL | 0.32 | 0.2107 (Probable non-antigen) | Probable non-allergen | −1.52(Non-Toxin) |
| 43 | 57 | CMTGFNTLLVQPALY | 0.71 | 0.1828 (Probable non-antigen). | Probable non-allergen | −1.20(Non-Toxin) | |
| 45 | 59 | TGFNTLLVQPALYLY | 0.88 | 0.1509 (Probable non-antigen) | Probable non-allergen | −1.15(Non-Toxin) | |
| 42 | 56 | QCMTGFNTLLVQPAL | 1.24 | 0.0771 (Probable non-antigen). | Probable non-allergen | −1.28(Non-Toxin) | |
| 46 | 60 | GFNTLLVQPALYLYN | 1.71 | 0.1781 (Probable non-antigen). | Probable non-allergen | −1.26(Non-Toxin) | |
| 27 | 41 | LLVCMAFLTATRLCV | 2.27 | 0.6923 (Probable antigen). | Probable non-allergen | −0.67(Non-Toxin) | |
| 28 | 42 | LVCMAFLTATRLCVQ | 2.51 | 1.0320 (Probable antigen). | Probable non-allergen | −0.56(Non-Toxin) | |
| 26 | 40 | TLLVCMAFLTATRLC | 2.74 | 0.6878 (Probable antigen). | Probable non-allergen | −1.03(Non-Toxin) | |
| 25 | 39 | ITLLVCMAFLTATRL | 2.91 | 0.6608 (Probable antigen). | Probable non-allergen | −1.30(Non-Toxin) | |
| Non-structural protein ORF3 | 6 | 20 | PPTLLLVFSLSLLVT | 0.96 | 0.5922 (Probable antigen). | Probable non-allergen | −1.25 Non-Toxin |
| 7 | 21 | PTLLLVFSLSLLVTA | 0.96 | 0.5642 (Probable antigen). | Probable non-allergen | −1.44(Non-Toxin) | |
| 5 | 19 | RPPTLLLVFSLSLLV | 1.36 | 0.4001 (Probable antigen). | Probable non-allergen | −1.12(Non-Toxin) | |
| 9 | 23 | LLLVFSLSLLVTAFS | 1.58 | 0.5310 (Probable antigen). | Probable non-allergen | −1.36(Non-Toxin) | |
| 4 | 18 | QRPPTLLLVFSLSLL | 1.81 | 0.5021 (Probable antigen). | Probable non-allergen | −1.13(Non-Toxin) | |
| 8 | 22 | TLLLVFSLSLLVTAF | 1.81 | 0.4190 (Probable antigen). | Probable non-allergen | −1.45(Non-Toxin) | |
| 3 | 17 | VQRPPTLLLVFSLSL | 2.05 | 0.3279 (Probable non-antigen). | Probable non-allergen | −1.17(Non-Toxin) | |
| 14 | 28 | SLSLLVTAFSKPLYV | 2.18 | 0.2561 (Probable non-antigen). | Probable non-allergen | −1.48(Non-Toxin) | |
| 13 | 27 | FSLSLLVTAFSKPLY | 4.05 | 0.2759 (Probable non-antigen). | Probable non-allergen | −1.45(Non-Toxin) | |
| Non-structural protein ORF5 | 11 | 25 | LYFLVLYNFLLAIVL | 0.04 | 0.4775 (Probable antigen). | Probable non-allergen | −1.11(Non-Toxin) |
| 10 | 24 | VLYFLVLYNFLLAIV | 0.14 | 0.3188 (Probable non-antigen). | Probable non-allergen | −1.11(Non-Toxin) | |
| 12 | 26 | YFLVLYNFLLAIVLV | 0.14 | 0.5150 (Probable antigen). | Probable non-allergen | −1.32(Non-Toxin) | |
| 13 | 27 | FLVLYNFLLAIVLVN | 0.14 | 0.5732 (Probable antigen). | Probable non-allergen | −1.40(Non-Toxin) | |
| 7 | 21 | RTMVLYFLVLYNFLL | 0.28 | 0.0324 (Probable non-antigen). | Probable allergen | −0.87(Non-Toxin) | |
| 8 | 22 | TMVLYFLVLYNFLLA | 0.28 | 0.2192 (Probable non-antigen). | Probable non-allergen | −0.93(Non-Toxin) | |
| 9 | 23 | MVLYFLVLYNFLLAI | 0.28 | 0.2900 (Probable non-antigen). | Probable non-allergen | −1.18(Non-Toxin) | |
| 35 | 49 | PASGYVAALAVNVCL | 0.77 | 0.0202 (Probable non-antigen). | Probable non-allergen | −0.88(Non-Toxin) | |
| 36 | 50 | ASGYVAALAVNVCLI | 0.77 | 0.1282 (Probable non-antigen). | Probable non-allergen | −0.85(Non-Toxin) | |
| 37 | 51 | SGYVAALAVNVCLIP | 1.06 | 0.3421 (Probable non-antigen). | Probable non-allergen | −0.85(Non-Toxin) | |
| 33 | 47 | YIPASGYVAALAVNV | 1.15 | 0.1243 (Probable non-antigen). | Probable non-allergen | −1.19(Non-Toxin) | |
| 34 | 48 | IPASGYVAALAVNVC | 1.33 | −0.1842 (Probable non-antigen). | Probable allergen | −1.18() | |
| 30 | 44 | DSVPLHIIAPSSLIV | 1.81 | 0.2266 (Probable non-antigen). | Probable non-allergen | −0.90(Non-Toxin) | |
| 28 | 42 | SIVFTYIPASGYVAA | 2.05 | 0.3680 (Probable non-antigen). | Probable non-allergen | −1.12(Non-Toxin) | |
| 29 | 43 | IVFTYIPASGYVAAL | 2.05 | 0.2499 (Probable non-antigen). | Probable non-allergen | −0.84(Non-Toxin) | |
| 27 | 41 | DSIVFTYIPASGYVA | 2.27 | 0.1703 (Probable non-antigen). | Probable allergen | −1.06(Non-Toxin) | |
| 9 | 23 | RSHFIRVSTVSSHGM | 2.51 | 0.1042 (Probable non-antigen). | Probable non-allergen | −1.08(Non-Toxin) | |
| 10 | 24 | SHFIRVSTVSSHGMV | 2.51 | 0.1973 (Probable non-antigen). | Probable non-allergen | −1.11(Non-Toxin) | |
| 8 | 22 | MRSHFIRVSTVSSHG | 2.74 | 0.0550 (Probable non-antigen). | Probable non-allergen | −1.43(Non-Toxin) | |
| 7 | 21 | DMRSHFIRVSTVSSH | 3.36 | 0.3701 (Probable non-antigen). | Probable non-allergen | −1.26(Non-Toxin) | |
| 31 | 45 | FTYIPASGYVAALAV | 3.49 | 0.2843 (Probable non-antigen). | Probable non-allergen | −1.16(Non-Toxin) | |
| 29 | 43 | RDSVPLHIIAPSSLI | 3.88 | 0.2191 (Probable non-antigen). | Probable non-allergen | −1.07(Non-Toxin) | |
| 30 | 44 | VFTYIPASGYVAALA | 3.95 | −0.0681 (Probable non-antigen). | Probable non-allergen | −1.13(Non-Toxin) | |
| 26 | 40 | TDSIVFTYIPASGYV | 4.16 | 0.0533 (Probable non-antigen). | Probable allergen | −0.97(Non-Toxin) | |
| 25 | 39 | STDSIVFTYIPASGY | 4.72 | 0.0749 (Probable non-antigen). | Probable non-allergen | −0.89(Non-Toxin) | |
| Spike protein | 14 | 28 | QSIFYRLNGVGITQQ | 0.39 | 0.7573 (Probable antigen). | Probable non-allergen | −2.15(Non-Toxin) |
| 15 | 29 | SIFYRLNGVGITQQV | 0.62 | 0.7288 (Probable antigen). | Probable non-allergen | −2.30(Non-Toxin) | |
| 4 | 18 | QQLVRSESAALSAQL | 0.62 | 0.3902 (Probable non-antigen). | Probable non-allergen | −1.16(Non-Toxin) | |
| 43 | 57 | IVSFVVNAPNGLYFM | 0.71 | 0.1671 (Probable non-antigen). | Probable non-allergen | −0.39(Non-Toxin) | |
| 38 | 52 | YIWLGFIAGLVALAL | 0.77 | 0.7583 (Probable antigen). | Probable non-allergen | −0.79(Non-Toxin) | |
| 40 | 54 | WLGFIAGLVALALCV | 0.77 | 0.4240 (Probable antigen). | Probable non-allergen | −0.74(Non-Toxin) | |
| 41 | 55 | LGFIAGLVALALCVF | 0.77 | 0.4118 (Probable antigen). | Probable non-allergen | −0.84(Non-Toxin) | |
| 13 | 27 | AQSIFYRLNGVGITQ | 0.79 | 0.6283 (Probable antigen). | Probable non-allergen | −1.97(Non-Toxin) | |
| 46 | 60 | VYDTIKYYSIIPHSI | 0.88 | 0.2157 (Probable non-antigen). | Probable non-allergen | −1.09(Non-Toxin) | |
| 12 | 26 | FAQSIFYRLNGVGIT | 0.96 | 0.4940 (Probable antigen). | Probable non-allergen | −1.77(Non-Toxin) | |
| 39 | 53 | IWLGFIAGLVALALC | 0.96 | 0.4564 (Probable antigen). | Probable non-allergen | −0.77(Non-Toxin) | |
| 16 | 30 | IFYRLNGVGITQQVL | 1.15 | 0.6444 (Probable antigen). | Probable non-allergen | −2.29(Non-Toxin) | |
| 42 | 56 | GFIAGLVALALCVFF | 1.15 | 0.2054 (Probable non-antigen). | Probable allergen | −0.59(Non-Toxin) | |
| 5 | 19 | QLVRSESAALSAQLA | 1.33 | 0.5139 (Probable antigen). | Probable non-allergen | −1.22(Non-Toxin) | |
| 11 | 25 | PFAQSIFYRLNGVGI | 1.36 | 0.4443 (Probable antigen) | Probable non-allergen | −1.66(Non-Toxin) | |
| 44 | 58 | VSFVVNAPNGLYFMH | 1.71 | 0.4899 (Probable antigen). | Probable allergen | −0.43(Non-Toxin) | |
| 3 | 17 | AQQLVRSESAALSAQ | 1.81 | 0.4991 (Probable antigen). | Probable non-allergen | −0.91(Non-Toxin) | |
| 6 | 20 | LVRSESAALSAQLAK | 1.9 | 0.4473 (Probable antigen). | Probable non-allergen | −1.02(Non-Toxin) | |
| 42 | 56 | HIVSFVVNAPNGLYF | 2.18 | 0.1797 (Probable non-antigen). | Probable non-allergen | −0.67(Non-Toxin) | |
| 37 | 51 | WYIWLGFIAGLVALA | 2.27 | 0.6569 (Probable antigen). | Probable non-allergen | −0.72(Non-Toxin) | |
| 17 | 31 | FYRLNGVGITQQVLS | 2.55 | 0.6337 (Probable antigen). | Probable non-allergen | −2.25(Non-Toxin) | |
| 28 | 42 | DFNLTLLEPVSISTG | 2.91 | 1.5583 (Probable antigen). | Probable non-allergen | −1.73(Non-Toxin) | |
| 2 | 16 | VAQQLVRSESAALSA | 3.09 | 0.3358 (Probable non-antigen). | Probable non-allergen | −1.11(Non-Toxin) | |
| 21 | 35 | HGDMYVYSAGHATGT | 3.49 | −0.1383 (Probable non-antigen). | Probable non-allergen | −1.27(Non-Toxin) | |
| 22 | 36 | GDMYVYSAGHATGTT | 3.49 | 0.0241 (Probable non-antigen). | Probable allergen | −0.99(Non-Toxin) | |
| 45 | 59 | SFVVNAPNGLYFMHV | 3.63 | 0.3855 (Probable non-antigen). | Probable non-allergen | −0.48(Non-Toxin) | |
| 23 | 37 | DMYVYSAGHATGTTP | 3.72 | 0.1080 (Probable non-antigen). | Probable non-allergen | −1.03(Non-Toxin) | |
| 9 | 23 | EGGGWLVASGSTVAM | 3.95 | 0.1129 (Probable non-antigen). | Probable non-allergen | −1.03(Non-Toxin) | |
| 10 | 24 | GGGWLVASGSTVAMT | 3.95 | 0.2673 (Probable non-antigen). | Probable non-allergen | −1.12(Non-Toxin) | |
| 43 | 57 | FIAGLVALALCVFFI | 4.16 | 0.2672 (Probable non-antigen). | Probable non-allergen | −0.60(Non-Toxin) | |
| 39 | 53 | NKFNQALGAMQTGFT | 4.77 | 0.3893 (Probable non-antigen). | Probable allergen | −0.79(Non-Toxin) |
3.4. Multi-epitope peptide construction
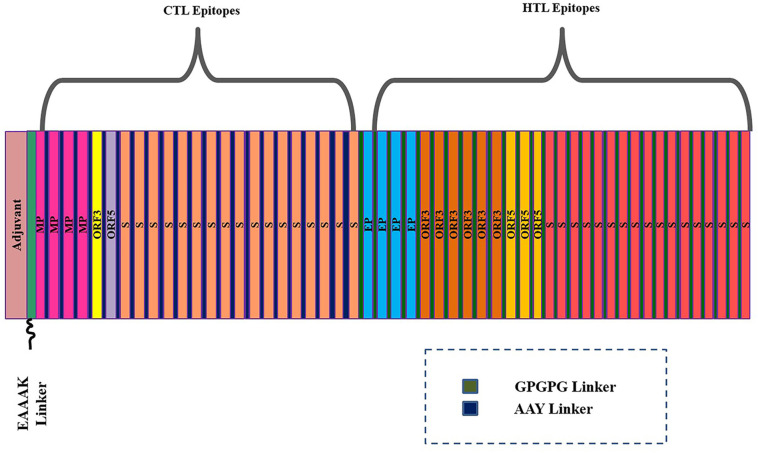
A multi-epitope peptide of 874 residues containing sequence conserved epitopes of both T cell immunity (CTL and HTL) and B cell immunity was designed. This design was based on the fact that individual peptide epitope is weakly immunogenic, therefore, fusing the screened epitopes to make a multi-epitope peptide could potentially enhance their immunogenicity capacity. Further, linking this peptide to an adjuvant can further increase the chance of good immune presentation [19]. In total, 53 epitopes were predicted for cytotoxic T cell immunity among which 19 epitopes were found antigenic and only considered for multi-epitope vaccine construct designing. Similarly, 77 HTL epitopes were predicted among which 30 were antigenic and considered. The CTL epitopes were joined together using AAY linkers. These linkers possess the cleavage sites for proteasome in the mammalian cells thus aiding in the production of natural epitopes and stopping the generation of junctional epitopes and by this way enhancing the presentation of the epitopes presentation to the host immune system [70]. The HTL epitopes were interconnected to each other through GPGPG linker for adequate presentation and stimulation of T cell responses. The EAAAK linker was employed to link the peptide of CTL epitopes to β-defensin adjuvant at the N-terminal and HTL epitopes at the C-terminal. β-defensin is a potent immune adjuvant and functions by stimulating the production of lymphokines which subsequently cellular immunity and production of antigen specific immunoglobulin [45]. The multi-epitope vaccine construct is schematically presented in Fig. 1 .
Fig. 1.
Schematic presentation of the multi-epitope vaccine construct design.
The full sequence of the construct contains large portion of B cell epitopes indicating the construct high potential of activating both arms of the host immunity. The predicted B cell epitope region in the vaccine construct is shown in S-Fig. 1. The average value for the construct is 0.602 with maximum value of 0.713 and minimum value of 0.177. Humoral immunity is a key component of the host immune system in clearing the microbial pathogen in extracellular fluid and block the spread of intracellular infection.
3.5. Allergenicity and antigenicity prediction of multi-epitope peptide construct
To ensure the non-allergic responses of the vaccine construct upon administration, the allergenicity of the construct was evaluated [71]. The prediction was based on SVM that securitized the amino acids composition of the input sequence for allergic sequences. The construct sequence was declared as non-allergen with score of −0.80559987 (cut-off = −0.4). The percent of positive predictive value deduced was 0% in contrast to negative predictive value that was equal to 0%. Similarly, the antigenicity of the construct was re-evaluated to certain its immune system evoking potential. The overall antigen prediction score for the vaccine construct is 0.4744 that indicate the probable antigenic behavior of the designed chimeric peptide.
3.6. Physiochemical properties of the multi-epitope peptide vaccine construct
The physiochemical properties of the designed construct were predicted to guide the experimentalists in synthesis and understanding the formulation of the vaccine. The molecular weight estimated for the construct is 140 kDa that explain two things: (i) first the medium size of the construct will allow its convenient purification and secondly, small and medium size chimeric proteins harbor appropriate number of epitopes for specific and targeted immune responses against the pathogen thus limiting the chances of allergic and adverse reactions [72]. The vaccines composed of whole organism or a single large protein have large number of antigenic determinants all of which are probably not required as they could led to non-specific immunity [19]. The theoretical pI computed for the vaccine construct is 8.05 indicating its basic nature. The instability index of the protein is estimated as 27.07 and classify it as stable. This instability index tells an estimate of protein stability in a test tube and is based on statistical analysis of 32 stable and 12 unstable proteins [73]. It has been determined that stable nature of proteins is due to the occurrence of certain dipeptides that differ significantly from non-stable. The protein instability index calculated by Protparam assigned a weightage of instability to different 400 dipeptides. Proteins with instability index of higher than 40 are categorized as unstable. The aliphatic index represents relative volume occupied by a protein aliphatic side chains involving amino acid residues: leucine, isoleucine, alanine and valine [74]. It is regarded as a key factor conferring thermostability to globular proteins. The aliphatic index for the query vaccine peptide is 106.10 presenting the high thermostability of the construct. The estimated half-life of the construct in mammalian reticulocytes (in-vitro), yeast (in vivo) and Escherichia coli (in vivo) is 5.5 h, 3 and 2 min, respectively. This half-life tells the predicted time for the protein to be disappeared after its synthesis. The grand average of hydropathicity (GRAVY) is 0.664. Lower the value implies greater hydrophilicity of the protein and has more probability of interacting with water residues. GRAVY is the cumulative hydrophilicity score of all the amino acids divided by total number of protein sequence.
3.7. Prediction of multi-epitope peptide secondary and tertiary structure
The secondary structure of the designed multi-epitope vaccine construct consists of 23 (2.7%), 338 (39.8%), 6 3–10 helix (0.7%), and 482 (56.8%) among other. More specifically, the remaining are composed of 3 β-sheets, 3 β-hairpins, 2 beta bulges, 6 strands, 20 helices, 22 helix-helix interactions, 51 β-turns, and 46 γ-turns. All the 874 residues of the input construct were modelled. The best structure was modelled using the A chain of the 4KNH template by Raptor-X with p-value of 3.18e-04. The 3D model of the predicted structure is shown in Fig. 2 .
Fig. 2.
Predicted 3D structure of in silico designed multi-epitope peptide vaccine construct.
3.8. Ramachandran plot analysis of the multi-epitope peptide construct
Ramachandran diagram or Ramachandran plot is a method of visualizing Phi or φ (the bonds between nitrogen and carbon alpha) and Psi or ψ (the bond between carbon and carbon alpha) of a given polypeptide [75]. The Phi and Psi are also known as Ramachandran or dihederal angles and hold control local structural importance vital for protein folding. Ramachandran plot is a vital assessment factor for evaluating three dimensional structure of a protein. This plot can be divided into four Quadrants (from I to IV) that give important information about secondary structure of a protein. Quadrant I represent conformations that are left handed alpha helices. Quadrant II mostly accommodates beta strands and is the region for most favorable conformations and biggest among all of the regions. Quadrant III is the following biggest region after II and is the place for right handed alpha helices. Lastly, Quadrant IV is the disfavored region and is the area for steric clash conformations. The Ramachandran plot statistics are given in Table 4 . It can be observed that majority of the protein residues i.e. 527 that make 80.5% of the total protein residues are in the most favored regions. This means that most of the residues are not in correct and usual geometry. In count to these regions, additionally allowed and generously allowed regions covers 122 residues (18.6%) and 4 (0.6%), respectively. Only 2 residues (0.3%) were found in the disallowed regions having conformation φ around 0–180 degrees and ψ around −180 to 0 degrees. The number of glycine and proline residues are 123 and 170, respectively. Proline is frequently found to be present in the outlier region of the Ramachandran plot and there is a reason to explain this. The cyclic side chain of proline face constrains upon rotation by its inclusion in pyrrolidine ring thus Phi values are confined around −60. Glycine, on the contrast, lacks beta carbon atom and therefore least steric compared to other amino acids. This support the large area distribution of glycine on the Ramachandran plot and allows conformation forbidden to other amino acids: allowing their occurrence in protein turn regions where others would be sterically hindered. The different parameters of G-factor of the vaccine construct are also provided in the Table 4. G-factor provides information about geometry of proteins that how unusual it is. Values below −0.5 are considered unusual, while those having values less −1.0 are highly unusual. The structure was immediately submitted to GalaxyRefine server to rebuilds the side chains and run side chains repacking followed by structure relaxation [76]. The Ramachandran for the refined vaccine construct is provided in S-Fig. 2. It is quite clear that the structure quality is improved by increasing the percentage of residues in the most favored region. Also, the G-factor value of the structure is improved to 0.00.
Table 4.
Structure assessment of multi-epitope peptide vaccine construct.
| Ramachandran plot statistics | Parameters | Number of residues unrefined/refined | Percentage unrefined/refined |
|---|---|---|---|
| ProCheck analysis | Most favored regions (A,B,L) | 527/587 | 80.5%/ 90.3% |
| Additional allowed regions [a,b,l,p] | 122/64 | 18.6%/ 9.8% | |
| Generously allowed regions [~a,~b,~l,~p] | 4/ 3 | 0.6%/0.5% | |
| Disallowed regions [XX] | 2/1 | 0.3%/0.1% | |
| Non-glycine and non-proline residues | 655/655 | 100.0%/100.0% | |
| End residues (Excluding glycine and proline) | 1/1 | ||
| Glycine residues | 123/123 | ||
| Proline residues | 70/70 | ||
| Total number of residues | 849/849 | ||
| Score | Average score | ||
| G-factors | Phi-psi distribution | −0.43/ | −0.64/0.23 |
| Chi1-chi2 distribution | −0.32/−0.10 | ||
| Chi1 only | 0.01/0.28 | ||
| Chi3 & chi4 | 0.90/0.47 | ||
| Omega | 1.32/0.98 | ||
| Main-chain bond lengths | 0.47/−0.11 | ||
| Main-chain bond angles | −0.13/−0.55 | 0.12/−0.37 | |
| Overall average | −0.31/0.00 | ||
3.9. Disulphide engineering
Disulphide engineering is a key approach in biotechnology to increase thermal stability of protein under study and assist in understanding its dynamics [77]. The introduction of disulphide bonds in the protein is believed to lower the conformational entropy and at the same time increases the free energy of the denatured state [78]. Thus both these actions lead to overall stability of protein fold conformation. Total of 70 pair of residues were found that could be manipulated for the purpose of increasing protein stability. However, only pair of residues with combined energy of >2 kcal/mol were opted. The complete set of residues with potential to be mutated is listed in Table 5 . The wild and mutated multi-epitope vaccine construct are provided in Fig. 3 .
Table 5.
Residue pairs shortlisted for disulfide engineering.
| Res1 chain | Res1 seq # | Res1 AA | Res2 chain | Res2 seq # | Res2 AA | Chi3 | Energy | Sum B-factors |
|---|---|---|---|---|---|---|---|---|
| A | 97 | PHE | A | 102 | MET | −86.29 | 1.23 | 0 |
| A | 110 | THR | A | 114 | ASP | 109.74 | 5.39 | 0 |
| A | 158 | TYR | A | 190 | ALA | −108.6 | 5.63 | 0 |
| A | 229 | TRP | A | 234 | SER | 126.5 | 7.66 | 0 |
| A | 229 | TRP | A | 235 | SER | −100.22 | 3.63 | 0 |
| A | 247 | LEU | A | 251 | ALA | 86.04 | 3.56 | 0 |
| A | 260 | ALA | A | 282 | THR | 118.89 | 2.5 | 0 |
| A | 261 | PHE | A | 302 | PHE | 105.89 | 5.82 | 0 |
| A | 263 | THR | A | 280 | PHE | −106.33 | 2.62 | 0 |
| A | 267 | LEU | A | 278 | MET | −83.19 | 3.18 | 0 |
| A | 268 | CYS | A | 305 | ALA | −112.99 | 6.14 | 0 |
| A | 270 | GLY | A | 275 | LEU | 124.91 | 6.54 | 0 |
| A | 274 | GLY | A | 295 | THR | −107.75 | 4.37 | 0 |
| A | 277 | CYS | A | 305 | ALA | −78.11 | 1.24 | 0 |
| A | 279 | ALA | A | 299 | CYS | −76.8 | 5.24 | 0 |
| A | 285 | ARG | A | 300 | MET | −76.39 | 4.49 | 0 |
| A | 293 | PRO | A | 311 | PRO | 95.37 | 4.87 | 0 |
| A | 320 | CYS | A | 342 | PHE | 114.39 | 2.37 | 0 |
| A | 321 | MET | A | 324 | LEU | 123.71 | 3.02 | 0 |
| A | 323 | PHE | A | 340 | LEU | −87.62 | 2.83 | 0 |
| A | 336 | PRO | A | 369 | ALA | 99.99 | 2.84 | 0 |
| A | 348 | VAL | A | 389 | VAL | 125.53 | 5.9 | 0 |
| A | 350 | GLY | A | 354 | GLY | 102.65 | 5.14 | 0 |
| A | 363 | LEU | A | 384 | SER | 87.87 | 1.22 | 0 |
| A | 372 | GLY | A | 375 | ARG | 105.76 | 4.29 | 0 |
| A | 377 | PRO | A | 408 | PHE | 112.75 | 3.23 | 0 |
| A | 380 | LEU | A | 403 | LEU | 80.23 | 5.29 | 0 |
| A | 387 | LEU | A | 396 | LEU | 111.66 | 4.04 | 0 |
| A | 390 | GLY | A | 395 | LEU | −62.98 | 5.65 | 0 |
| A | 400 | SER | A | 441 | SER | −75.24 | 3.78 | 0 |
| A | 448 | ALA | A | 467 | ILE | −91.3 | 5.12 | 0 |
| A | 469 | LEU | A | 492 | GLY | −85.2 | 2.81 | 0 |
| A | 472 | GLY | A | 491 | PRO | 74.27 | 4.2 | 0 |
| A | 477 | LEU | A | 497 | VAL | 124.74 | 5.55 | 0 |
| A | 499 | TYR | A | 562 | ALA | 84 | 1.2 | 0 |
| A | 512 | GLY | A | 533 | PRO | 83.4 | 3.1 | 0 |
| A | 514 | GLY | A | 531 | PRO | −97.54 | 5.75 | 0 |
| A | 516 | SER | A | 537 | PHE | 82.22 | 2.65 | 0 |
| A | 518 | PHE | A | 526 | ILE | 110.01 | 2.55 | 0 |
| A | 522 | ASN | A | 540 | LEU | 97.31 | 3.12 | 0 |
| A | 523 | GLY | A | 794 | GLY | −94.11 | 2.48 | 0 |
| A | 523 | GLY | A | 798 | TRP | −90.23 | 2.84 | 0 |
| A | 542 | GLY | A | 790 | GLY | −113.82 | 5.53 | 0 |
| A | 556 | ILE | A | 590 | GLY | −91.62 | 1.66 | 0 |
| A | 559 | GLY | A | 583 | VAL | 79.53 | 6.6 | 0 |
| A | 562 | ALA | A | 582 | LEU | 101.05 | 2.72 | 0 |
| A | 577 | GLY | A | 808 | LEU | 89.37 | 2.67 | 0 |
| A | 599 | ALA | A | 621 | ARG | −102.21 | 3.64 | 0 |
| A | 614 | GLY | A | 657 | LEU | −80.8 | 2.37 | 0 |
| A | 614 | GLY | A | 661 | ALA | −72.29 | 4.59 | 0 |
| A | 632 | GLY | A | 635 | PHE | 121.17 | 4.49 | 0 |
| A | 642 | ARG | A | 662 | GLY | −82.84 | 3.57 | 0 |
| A | 650 | GLY | A | 655 | ILE | 88.7 | 3.96 | 0 |
| A | 670 | GLY | A | 726 | GLY | −116.69 | 5.07 | 0 |
| A | 671 | PRO | A | 726 | GLY | −66.97 | 5.27 | 0 |
| A | 672 | GLY | A | 726 | GLY | −65.36 | 5.59 | 0 |
| A | 699 | SER | A | 702 | ALA | −92.32 | 2.2 | 0 |
| A | 708 | LEU | A | 717 | ALA | 106.12 | 3.41 | 0 |
| A | 723 | ARG | A | 748 | MET | −95.54 | 3.87 | 0 |
| A | 737 | PHE | A | 749 | HIS | 103.9 | 3.82 | 0 |
| A | 740 | ASN | A | 807 | ALA | 99.13 | 5.01 | 0 |
| A | 741 | ALA | A | 745 | LEU | 124.88 | 2.64 | 0 |
| A | 752 | GLY | A | 755 | ALA | −103.23 | 3 | 0 |
| A | 764 | ALA | A | 826 | GLN | 84.38 | 4.6 | 0 |
| A | 765 | ALA | A | 777 | ARG | 75.84 | 1.42 | 0 |
| A | 765 | ALA | A | 782 | ALA | −83.1 | 3.88 | 0 |
| A | 799 | LEU | A | 841 | LEU | −90.24 | 5.33 | 0 |
| A | 799 | LEU | A | 842 | GLU | 67.85 | 5.1 | 0 |
| A | 822 | GLY | A | 846 | ILE | −77.27 | 4.69 | 0 |
| A | 832 | GLY | A | 835 | ASP | 120.01 | 2.47 | 0 |
Fig. 3.
Disulfide engineering of the multi-epitope vaccine construct.
3.10. Adaptation of codon usage to Escherichia coli K 12 strain
The codon adaptation tool is a simple way to adjust the codon usage of the input sequence to the selected prokaryotic and eukaryotic system [79,80]. The difference in codon usage of the host from that organism from which the desired sequence stems from plays a major role in minor expression of that sequence. Codon adaptation of the vaccine construct primary sequence to E. coli K12 expression system is vital in ensuring its maximum expression. The codon adaptive index and GC content of the improve sequence are 0.86 and 57.75, respectively. The adaptive score of the vaccine and it's in silico restriction cloning are provided in S-Fig. 3 and Fig. 4 .
Fig. 4.
Cloned vaccine construct shown in red into the expression vector.
3.11. Molecular docking with TLR3
Molecular docking of the designed vaccine construct with TLR3 as a receptor was accomplished using PatchDock, which generated more than 20 complexes of TLR3 and vaccine construct conformations. The top 10 best predicted solutions were subjected to the fast interaction refinement in molecular docking (firedock) to refine the predicted solutions based the global energy. Lower the global energy of a solution implies the stable formation of complex and vice versa. As such solution 3 with global energy of −24.02 kcal/mol, attractive van der Waals energy of −38.57 kcal/mol, repulsive van der Waals energy of 28.88 kcal/mol, atomic contact energy of 11.49 kcal/mol and hydrogen bonding energy of −4.00 kcal/mol was considered the best predicted complex (Table 6 ). The docked conformation of the TLR3 and construct with respect to each is shown in Fig. 5 . The top complex conformation of the TLR3-MEVC was also understand via ClusPro that complement good binding of the molecules with lowest energy of −1268.5 kcal/mol.
Table 6.
Refined complexes of FireDock.
| Rank | Solution number | Global energy (kcal/mol) | Attractive Van der Waals | Repulsive Van dar Waals | Atomic contact energy (kcal/mol) | Hydrogen bonding |
|---|---|---|---|---|---|---|
| 1 | 3 | −24.02 | −38.57 | 28.88 | 11.49 | −4.00 |
| 2 | 7 | −18.39 | −25.30 | 19.27 | −2.78 | −1.87 |
| 3 | 10 | −8.94 | −21.98 | 11.85 | 4.64 | −1.22 |
| 4 | 4 | −8.93 | −15.55 | 6.26 | 1.15 | −1.13 |
| 5 | 9 | −6.05 | −6.53 | 1.51 | −2.06 | 0.00 |
| 6 | 6 | −3.57 | −17.34 | 10.10 | 0.14 | −0.98 |
| 7 | 5 | 5.95 | −11.60 | 14.02 | 3.33 | 0.00 |
| 8 | 8 | 13.59 | −4.79 | 1.11 | 1.08 | −1.43 |
| 9 | 1 | 17.90 | −8.84 | 28.80 | 1.84 | −1.58 |
| 10 | 2 | 2825.46 | −44.83 | 3622.84 | −0.61 | −5.55 |
Fig. 5.
Docked complex of the designed multi-epitope peptide vaccine construct (shown in red) and chain A of TLR3 (shown in yellow).
3.12. Immune simulation
The vaccine construct appears to produce robust IgM antibody response even in the absence of recurrent antigen exposure. The IgM antibody titer seen reach close to 700,000 scales (Fig. 6 ). Among the cytokines and interleukins, high level interferon gamma was reported to the antigen compared to others (Fig. 6). Additionally, appropriate and decent cellular activity with their respective memory from the host immune system were revealed along with proliferating dendritic cells and enhanced activity from macrophages (S-Fig. 4). Different humoral immunity isotypes can be easily noticed, an indicator of isotype switching and memory cell formation (S-Fig. 5). This immune profile leads to a conclusion of a good agreement on natural development of immunity against the virus and protection against the virus.
Fig. 6.
The vaccine construct antigen, and antibodies and immune complexes response (left). Cytokines and interleukins concentration in response to the vaccine construct antigen (right).
3.13. Molecular dynamics simulations
The multi-epitope peptide vaccine construct complex was seen stably docked with the TLR3 receptor and the epitopes are exposed throughout the simulation time for efficient and easy processing by the host immune system. The dynamics of the complex was evaluated through two structural parameters. First, root mean square deviation (rmsd) was investigated that measures the distance between the carbon alpha atoms of superimposed proteins [81]. The average rmsd estimated for the system is 5.2 Å. The system is reported with some major rmsd peaks that after visual inspection found because of the moving loops of the system. This may be an approach towards complex stability as both the interacting molecules are stably docked with each other's. Secondly, we evaluted root mean square fluctuations (rmsf) for the system. RMSF sheds light on the fluctuating residues from there mean position [82]. The average rmsf for the system is 4.6 Å. As mentioned earlier, these variation are because of fluctuating loops of the system which is not altering intermolecular conformations and does not affect overall stability. The rmsd and rmsf plots of the system can be seen in Fig. 7 . Additionally, we performed hydrogen bond analysis for the complex to decipher the strength of intermolecular interactions during simulation time. As can be seen in Fig. 8 , the molecules are involved in good number of hydrogen bonds throughout the simulation time, enhancing stability of the complex.
Fig. 7.
RMSD (left) and RMSF (right) for the simulated TLR3-MEPVC complex.
Fig. 8.

Hydrogen bonds analysis for TLR3-MEPVC complex.
3.14. MM-PBSA binding energies calculation
The docking prediction for the vaccine construct conformation with respect to the TLR3 receptor was validated by performing computationally cost effective and sufficiently reliable MMMGBSA and MMPBSA approaches. Both these approaches were run over trajectories of the MD simulations. It is evident from the Table 7 that robust energies are noticed for the system and the values are quite low indicating a good agreement on the docking findings. The intermolecular ineractions are dominated by both van der Waals and electrostatic energies with energy value of −188.44 kcal/mol and −473.80 kcal/mol, respectively. Summing, both these mentioned values are contributing factor to the significantly low gas phase energy of −626.25 kcal/mol. Opposed to this, non-favorable energy of solvation was noticed for the system (564.3822 kcal/mol in MM-GBSA and 527.0717 kcal/mol in MM-PBSA). The net binding energy of the system in MM-GBSA is −61.8682 kcal/mol whereas in MM-PBSA this energy is −99.1787 kcal/mol. Both these net values support highly stable system and excellent intermolecular binding affinity between the vaccine construct and TLR3 receptor. The net entropy concluded for the system is 57.76 kcal/mol which again suggesting stable nature of the vaccine at the docked site. Additionally, to evaluate intermolecular affinity towards end of the simulation time, MMPB/GBSA of last 20-ns was performed (S-Table 1). The net MMGBSA and MMPBSA in this time period are −77.4 kcal/mol and − 107.4 kcal/mol, respectively.
Table 7.
Binding free energies for the TLR3-MEPVC complex.
| Energy component | Average | Std. dev. | Std. err. of mean |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −188.4473 | 7.4753 | 0.7475 |
| EEL | −437.8031 | 15.8762 | 1.5876 |
| EGB | 587.7871 | 15.3673 | 1.5367 |
| ESURF | −23.4049 | 0.9328 | 0.0933 |
| DELTA G gas | −626.2504 | 16.5671 | 1.6567 |
| DELTA G solv | 564.3822 | 15.1083 | 1.5108 |
| DELTA TOTAL | −61.8682 | 4.7152 | 0.4715 |
| MM-PBSA | |||
| VDWAALS | −188.4473 | 7.4753 | 0.7475 |
| EEL | −437.8031 | 15.8762 | 1.5876 |
| EPB | 547.1448 | 15.0112 | 1.5011 |
| ENPOLAR | −20.0731 | 0.4982 | 0.0498 |
| EDISPER | 0.0000 | 0.0000 | 0.0000 |
| DELTA G gas | −626.2504 | 16.5671 | 1.6567 |
| DELTA G solv | 527.0717 | 14.8625 | 1.4863 |
| DELTA TOTAL | −99.1787 | 7.2585 | 0.7258 |
4. Conclusions
This study shows that membrane protein, envelope small membrane protein, non-structural protein ORF3, non-structural protein ORF5 and spike glycoprotein could act as potential subunit vaccine candidates for designing a subunit-based vaccine against MERS-CoV. Similarly, these proteins were mapped for novel set of antigenic B and T cell epitopes that could be used in the design of a multi-epitope peptide vaccine construct to circumvent the limitations of subunit vaccines. Further, we have shown that the designed multi-epitope vaccine construct docking affinity for TLR3 immune receptor, thus maximizing its presentation to the human immune system and ensuring the evoking potential of both humoral and cellular immune responses. The prediction of docking was validated by molecular dynamics simulation and binding free energies that validated complex high stability and intermolecular affinity. Lastly, the use of the subunit vaccine candidates and multi-epitope vaccine construct in experimental in vivo model is highly recommended to underpin their immune protective potential against MERS-CoV and the current pandemic COVID-19.
The following are the supplementary data related to this article.
S-Fig. 1.
Predicted B cell epitope region in the vaccine construct.
S-Fig. 2.

Ramachandran plot for the 3D structure of multi-epitope peptide vaccine construct.
S-Fig. 3.
Codon optimization score of the vaccine construct.
S-Fig. 4.
T cell immunity profile against the vaccine construct antigen.
S-Fig. 5.
B cell immunity profile against the vaccine construct antigen.
Binding free energies for the TLR3-MEPVC complex.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- 1.Sikkema R.S., Farag E., Islam M., Atta M., Reusken C., Al-Hajri M.M., Koopmans M.P.G. Global status of Middle East respiratory syndrome coronavirus in dromedary camels: a systematic review. Epidemiol. Infect. 2019;147 doi: 10.1017/S095026881800345X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fehr A.R., Channappanavar R., Perlman S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.-J., Jonges M., Farag E., Diab A., et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall A.J., Tokars J.I., Badreddine S.A., Bin Saad Z., Furukawa E., Al Masri M., Haynes L.M., Gerber S.I., Kuhar D.T., Miao C., et al. Health care worker contact with MERS patient, Saudi Arabia. Emerg. Infect. Dis. 2014;20:2148. doi: 10.3201/eid2012.141211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alshukairi A.N., Zheng J., Zhao J., Nehdi A., Baharoon S.A., Layqah L., Bokhari A., Al Johani S.M., Samman N., Boudjelal M., et al. High prevalence of MERS-CoV infection in camel workers in Saudi Arabia. MBio. 2018;9:e01985–e02018. doi: 10.1128/mBio.01985-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F.W., Lau S.K.P., K.K.W. To, Cheng V.C.C., Woo P.C.Y., Yuen K.-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gytis D., Carvalho L.M., Rambaut A., Bedford T. MERS-CoV spillover at the camel-human interface. Elife. 2018;7 doi: 10.7554/eLife.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang K., Ki M., Lee E.G., Lee S.Y., Yoo B., Choi J.H. MERS epidemiological investigation to detect potential mode of transmission in the 178th MERS confirmed case in Pyeongtaek, Korea. Epidemiol. Health. 2015;37 doi: 10.4178/epih/e2015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossner C., Danielson N., Gervelmeyer A., Berthe F., Faye B., Kaasik Aaslav K., Adlhoch C., Zeller H., Penttinen P., Coulombier D. Human--dromedary camel interactions and the risk of acquiring zoonotic Middle East respiratory syndrome coronavirus infection. Zoonoses Public Health. 2016;63:1–9. doi: 10.1111/zph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkhamis M.A., Fernández-Fontelo A., VanderWaal K., Abuhadida S., Puig P., Alba-Casals A. Temporal dynamics of Middle East respiratory syndrome coronavirus in the Arabian Peninsula, 2012–2017. Epidemiol. Infect. 2019;147 doi: 10.1017/S0950268818002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modjarrad K. Treatment strategies for Middle East respiratory syndrome coronavirus. J. Virus Erad. 2016;2:1. doi: 10.1016/S2055-6640(20)30696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindewolf C., Menachery V.D. Middle East respiratory syndrome vaccine candidates: cautious optimism. Viruses. 2019;11:74. doi: 10.3390/v11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly-Cirino C., Mazzola L.T., Chua A., Oxenford C.J., Van Kerkhove M.D. An updated roadmap for MERS-CoV research and product development: focus on diagnostics. BMJ Glob. Heal. 2019;4 doi: 10.1136/bmjgh-2018-001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergara-Alert J., Vidal E., Bensaid A., Segalés J. Searching for animal models and potential target species for emerging pathogens: experience gained from Middle East respiratory syndrome (MERS) coronavirus. One Heal. 2017;3:34–40. doi: 10.1016/j.onehlt.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccin. 2018;17:677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goo J., Jeong Y., Park Y.-S., Yang E., Jung D.-I., Rho S., Park U., Sung H., Park P.-G., Choi J., et al. Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Res. 2020;251:22–29. doi: 10.1016/j.virusres.2020.197863. 197863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surya W., Li Y., Verdià-Bàguena C., Aguilella V.M., Torres J. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 2015;201:61–66. doi: 10.1016/j.virusres.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lui P.-Y., Wong L.-Y.R., Fung C.-L., Siu K.-L., Yeung M.-L., Yuen K.-S., Chan C.-P., Woo P.C.-Y., Yuen K.-Y., Jin D.-Y. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microb. Infect. 2016;5:1–9. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W., Joshi M., Singhania S., Ramsey K., Murthy A. Peptide vaccine: progress and challenges. Vaccines. 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skwarczynski M., Toth I. Peptide-based synthetic vaccines. Chem. Sci. 2016;7:842–854. doi: 10.1039/c5sc03892h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bijker M.S., Melief C.J.M., Offringa R., Van Der Burg S.H. Design and development of synthetic peptide vaccines: past, present and future. Expert Rev. Vaccin. 2007;6:591–603. doi: 10.1586/14760584.6.4.591. [DOI] [PubMed] [Google Scholar]
- 22.Faisal A.-R.M., Imtiaz S.H., Zerin T., Rahman T., Shekhar H.U. Computer aided epitope design as a peptide vaccine component against Lassa virus. Bioinformation. 2017;13:417. doi: 10.6026/97320630013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolesanova E.F., Sanzhakov M.A., Kharybin O.N. Development of the schedule for multiple parallel “difficult” peptide synthesis on pins. Int. J. Pept. 2013;2013 doi: 10.1155/2013/197317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solares A.M., Baladron I., Ramos T., Valenzuela C., Borbon Z., Fanjull S., Gonzalez L., Castillo D., Esmir J., Granadillo M., et al. Safety and immunogenicity of a human papillomavirus peptide vaccine (CIGB-228) in women with high-grade cervical intraepithelial neoplasia: first-in-human, proof-of-concept trial. ISRN Obstet. Gynecol. 2011;2011 doi: 10.5402/2011/292951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., McNevin J., Zhao H., Tebit D.M., Troyer R.M., McSweyn M., Ghosh A.K., Shriner D., Arts E.J., McElrath M.J., et al. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J. Virol. 2007;81:12179–12188. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanekova Z., Kiraly J., Stropkovska A., Mikušková T., Mucha V., Kostolansky F., Varečková E. Heterosubtypic protective immunity against influenza A virus induced by fusion peptide of the hemagglutinin in comparison to ectodomain of M2 protein. Acta Virol. 2011;55:61–67. doi: 10.4149/av_2011_01_61. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell. Mol. Immunol. 2018;15:182. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium U. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta R., Pradhan D., Jain A.K., Rai C.S. TiD: standalone software for mining putative drug targets from bacterial proteome. Genomics. 2017;109:51–57. doi: 10.1016/j.ygeno.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Yu C.-S., Cheng C.-W., Su W.-C., Chang K.-C., Huang S.-W., Hwang J.-K., Lu C.-H. CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blast N. Basic local alignment search tool. Natl. Libr. Med. Natl. Cent. Biotechnol. Inf. 2015 [Google Scholar]
- 33.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45:D12. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bermúdez-Humarán L.G., Salinas E., Ortiz G.G., Ramirez-Jirano L.J., Morales J.A., Bitzer-Quintero O.K. From probiotics to psychobiotics: live beneficial bacteria which act on the brain-gut axis. Nutrients. 2019;11:890. doi: 10.3390/nu11040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadood A., Jamal A., Riaz M., Khan A., Uddin R., Jelani M., Azam S.S. Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb. Pathog. 2018;115:194–198. doi: 10.1016/j.micpath.2017.12.063. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamoto T., Yamamoto H., Matano T. CD8+ cytotoxic-T-lymphocyte breadth could facilitate early immune detection of immunodeficiency virus-derived epitopes with limited expression levels. MSphere. 2019;4:e00381–e00418. doi: 10.1128/mSphere.00381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luckheeram R.V., Zhou R., Verma A.D., Xia B. CD4+ T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012 doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saadi M., Karkhah A., Nouri H.R. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect. Genet. Evol. 2017;51:227–234. doi: 10.1016/j.meegid.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2018;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P.S., Consortium O.S.D.D., et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhanda S.K., Vir P., Raghava G.P.S. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugiarto H., Yu P.-L. Avian antimicrobial peptides: the defense role of β-defensins. Biochem. Biophys. Res. Commun. 2004;323:721–727. doi: 10.1016/j.bbrc.2004.08.162. [DOI] [PubMed] [Google Scholar]
- 46.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ProtParam E. 2017. ExPASy-ProtParam Tool. [Google Scholar]
- 49.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha S., Raghava G.P.S. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T-and B-cell epitope prediction. J Immunol Res. 2017;2017 doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrusier N., Nussinov R., Wolfson H.J. FireDock: fast interaction refinement in molecular docking. Proteins Struct. Funct. Bioinforma. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 55.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein—protein docking. Nat. Protoc. 2017;12:255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Case D.A., Cerutti D.S., Cheateham T.E., Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Greene D., Homeyer N., et al. Univ. California; San Fr: 2016. AMBER16 Package. [Google Scholar]
- 58.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., et al. The FF14SB force field. Amber. 2014;14:29–31. [Google Scholar]
- 59.Izaguirre J.A., Catarello D.P., Wozniak J.M., Skeel R.D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001;114:2090–2098. [Google Scholar]
- 60.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 61.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 62.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duan L., Liu X., Zhang J.Z.H. Interaction entropy: a new paradigm for highly efficient and reliable computation of protein—ligand binding free energy. J. Am. Chem. Soc. 2016;138:5722–5728. doi: 10.1021/jacs.6b02682. [DOI] [PubMed] [Google Scholar]
- 64.Barh D., Barve N., Gupta K., Chandra S., Jain N., Tiwari S., Leon-Sicairos N., Canizalez-Roman A., dos Santos A.R., Hassan S.S., et al. Exoproteome and secretome derived broad spectrum novel drug and vaccine candidates in Vibrio cholerae targeted by Piper betel derived compounds. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ali A., Naz A., Soares S.C., Bakhtiar M., Tiwari S., Hassan S.S., Hanan F., Ramos R., Pereira U., Barh D., et al. Pan-genome analysis of human gastric pathogen H. pylori: comparative genomics and pathogenomics approaches to identify regions associated with pathogenicity and prediction of potential core therapeutic targets. Biomed Res. Int. 2015;2015 doi: 10.1155/2015/139580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naz A., Awan F.M., Obaid A., Muhammad S.A., Paracha R.Z., Ahmad J., Ali A. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect. Genet. Evol. 2015;32:280–291. doi: 10.1016/j.meegid.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt M.E., Varga S.M. The CD8 T cell response to respiratory virus infections. Front. Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian Y., da Silva Antunes R., Sidney J., Arlehamn C.S.L., Grifoni A., Dhanda S.K., Paul S., Peters B., Weiskopf D., Sette A. A review on T cell epitopes identified using prediction and cell-mediated immune models for Mycobacterium tuberculosis and Bordetella pertussis. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Mol. Biol. Cell. 4th ed. Garland Science; 2002. Helper T cells and lymphocyte activation. [Google Scholar]
- 70.Shamriz S., Ofoghi H. Design, structure prediction and molecular dynamics simulation of a fusion construct containing malaria pre-erythrocytic vaccine candidate, PfCelTOS, and human interleukin 2 as adjuvant. BMC Bioinformatics. 2016;17:1–15. doi: 10.1186/s12859-016-0918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung E.H. Vaccine allergies. Clin. Exp. Vaccine Res. 2014;3:50–57. doi: 10.7774/cevr.2014.3.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hassan A., Naz A., Obaid A., Paracha R.Z., Naz K., Awan F.M., Muhmmad S.A., Janjua H.A., Ahmad J., Ali A. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genomics. 2016;17:732. doi: 10.1186/s12864-016-2951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guruprasad K., Reddy B.V.B., Pandit M.W. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 74.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 75.Hooft R.W.W., Sander C., Vriend G. Objectively judging the quality of a protein structure from a Ramachandran plot. Bioinformatics. 1997;13:425–430. doi: 10.1093/bioinformatics/13.4.425. [DOI] [PubMed] [Google Scholar]
- 76.Heo L., Park H., Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dombkowski A.A., Sultana K.Z., Craig D.B. Protein disulfide engineering. FEBS Lett. 2014;588:206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 78.Craig D.B., Dombkowski A.A. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics. 2013;14:346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Angov E. Codon usage: nature’s roadmap to expression and folding of proteins. Biotechnol. J. 2011;6:650–659. doi: 10.1002/biot.201000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maiorov V.N., Crippen G.M. 1994. Significance of Root-mean-Square Deviation in Comparing Three-dimensional Structures of Globular Proteins. [DOI] [PubMed] [Google Scholar]
- 82.Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010;98:861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Binding free energies for the TLR3-MEPVC complex.