Abstract
NF-YA transcription factors function in modulating tolerance to abiotic stresses that are serious threats to crop yields. In this study, GmNFYA13, an NF-YA gene in soybean, was strongly induced by salt, drought, ABA, and H2O2, and suppressed by tungstate, an ABA synthesis inhibitor. The GmNFYA13 transcripts were detected in different tissues in seedling and flowering stages, and the expression levels in roots were highest. GmNFYA13 is a nuclear localization protein with self-activating activity. Transgenic Arabidopsis plants overexpressing GmNFYA13 with higher transcript levels of stress-related genes showed ABA hypersensitivity and enhanced tolerance to salt and drought stresses compared with WT plants. Moreover, overexpression of GmNFYA13 resulted in higher salt and drought tolerance in OE soybean plants, while suppressing it produced the opposite results. In addition, GmNFYA13 could bind to the promoters of GmSALT3, GmMYB84, GmNCED3, and GmRbohB to regulate their expression abundance in vivo. The data in this study suggested that GmNFYA13 enhanced salt and drought tolerance in soybean plants.
Keywords: nuclear factor YA, salt and drought tolerance, ABA hypersensitivity, soybean, crop yield
Introduction
As types of osmotic stress, salt and drought stresses are major challenges to growth and development of plants, resulting in severe yield losses (Zhu, 2002; Golldack et al., 2014). Many changes related to metabolism and physiology occur when plants are subjected to osmotic stresses (Golldack et al., 2011). These stresses are negatively affecting the available arable land, which is paralleled by the pressing need for food all over the world (Golldack et al., 2011). The signaling pathways of salt and drought stresses have been uncovered in previous researches (Zhu, 2002; Golldack et al., 2011; Lawlor, 2013). Abscisic acid (ABA) is produced in plants under abiotic stresses, especially salt and drought stresses, and the accumulation level of ABA is positively related to salt and drought tolerance of plants (Nakashima and Yamaguchi-Shinozaki, 2013). ABA signaling is complex, including receptors PYR/PYL/RCARs, PP2Cs, SnRK2s, AREB/ABFs, and ABA-responsive genes (Ma et al., 2009; Park et al., 2009; Vlad et al., 2009). Moreover, NCED3 was certified as a key ABA synthetase (Barrero et al., 2006; Hao et al., 2009; Sussmilch et al., 2017; Zhang et al., 2020). Many transcription factors (TFs) were identified to play vital roles in regulating osmotic stress via an ABA-dependent pathway, such as NAC, WRKY, MYB, ERF, and bZIP (Hu et al., 2006; Zhou et al., 2008; Lippold et al., 2009; Peng et al., 2009; Zhang et al., 2009; Amir Hossain et al., 2010; Hsieh et al., 2010; Ren et al., 2010).
NF-Y TFs function in affecting plant growth and development (Kwong et al., 2003; Miyoshi et al., 2003; Xu et al., 2005; Wenkel et al., 2006; Cai et al., 2007; Mu et al., 2008; Wei et al., 2010; Ballif et al., 2011; Stephenson et al., 2011; Hackenberg et al., 2012; Hou et al., 2014; Myers et al., 2016) and are essential in abiotic stress responses. Overexpression of NFYA5 (Li et al., 2008), GmNFYA3 (Ni et al., 2013),GmNFYB1 (Li et al., 2017), and NF-YB2 and NF-YB3 (Sato et al., 2019) conferred drought tolerance to transgenic Arabidopsis plants. Transgenic rice plants overexpressing OsNFYA7 (Lee et al., 2015), OsHAP2E (Alam et al., 2015), and Cdt-NFYC1 (Chen et al., 2015) showed enhanced tolerance to osmotic stresses.
Enhanced tolerance to osmotic stresses is associated with increased transcript levels of stress-related genes. For example, overexpression of SOS1, SOS2, SOS3 (Qiu et al., 2002), GmSALT3 (Guan et al., 2014), GmSOS1, and GmNHX1 (Zhang W. et al., 2019) resulted in enhanced salt tolerance in plants. In addition, DREB2A (Nakashima et al., 2000), ABI3 (Mittal et al., 2014), RD29A (Bihmidine et al., 2013), GmMYB84 (Wang N. et al., 2017), GmDREB2 (Chen et al., 2007), and GmWRKY46 (Luo et al., 2013) were vital genes that positively regulated drought tolerance in plants. During osmotic stresses, reactive oxygen species (ROS) are produced and result in oxidative damage to cells, especially cytomembranes (Uchida et al., 2002; Zhou et al., 2018; Cui et al., 2019). A respirator burst oxidase homolog B (RbohB) gene in soybean, GmRbohB, was determined to function in ROS scavenging (Wang N. et al., 2017). Therefore, these genes are indispensable for tolerance to salt and drought stresses in plants.
Soybean is an important oil-bearing crop all over the world, and its production is constrained by salt and drought stresses. In this study, overexpression of an NF-Y gene in soybean, GmNFYA13, conferred enhanced salt and drought tolerance to transgenic soybean plants, while its abolition resulted in opposite results. Considering these results, GmNFYA13 should be a candidate gene in molecular breeding of soybean.
Materials and Methods
Plant Growth and Treatments
Seeds of the soybean (Glycine max L. Merr.) cultivar Williams 82 were grown in plastic pots (14 cm diameter, 15 cm depth) containing nutrient soil and grown for 20 days in a tissue culture room at 28/18°C day/night with a 14-h photoperiod and 70% relative humidity as described previously (Ma et al., 2020), which are considered as normal conditions for soybean plants. The seedlings were used to assess gene expression patterns under several treatments. For H2O and salt treatment, the seedlings were subjected to H2O and 200 mM NaCl solution for 12 h; for drought treatment, the seedlings were pulled out from soil and placed on a plastic plate for 12 h; for ABA and H2O2 treatment, the roots were immersed in 100 μM ABA and 10 mM H2O2 for 12 h. The leaves under various treatments were harvested at 0, 1, 2, 4, 8, and 12 h and frozen at −80°C for isolation of RNA. To test whether an ABA synthesis inhibitor (tungstate) could affect salt-induced GmNFYA13 transcripts, two detached leaves of soybean plants were placed into ddH2O to eliminate the effect of wound stress for 1 h, and then transferred into ddH2O and 2 mM tungstate for 3 h, followed by being subjected to 200 mM NaCl solution for 1 h. To determine whether tungstate could suppress the drought-induced expression level of GmNFYA13, the “drought + tungstate” treatment was performed as described previously (Ma et al., 2020) and 1 mM naproxen was replaced with 2 mM tungstate. The leaves immersed in ddH2O were used as the control. Samples were collected and frozen at −80°C.
To evaluate the transcription levels of GmNFYA13 in different tissues, the tissues of 20-day-old and 50-day-old soybean plants were collected and stored as described previously (Ma et al., 2020). All of the experiments were performed in a tissue culture room at 28/18°C day/night with a 14-h photoperiod and 70% relative humidity.
Cloning of GmNFYA13
Williams 82 soybean plants were used for RNA isolation, followed by reverse transcription to cDNA. The full-length coding sequence (CDS) of GmNFYA13 was isolated using PrimerSTAR Max DNA polymerase (Takara, Japan) and primer set 5′-GGTAGTCAATAGTCATCACTT-3′ and 5′-CAAGTTGGATGAGATAAAGCC-3′. The PCR products were cloned into the vector pEASY-Blunt (TransGen, China) and sequenced; the correct clone was stored at −80°C for isolation of the CDS of GmNFYA13.
Generation of Transgenic Arabidopsis Lines
The CDS without the termination codon was fused in pCAMBIA1302 vector and ligated with NcoI site to generate the pCAMBIA1302:GmNFYA13 fusion construct, which is driven by the promoter of cauliflower mosaic virus 35S (CaMV35S). The primer set used in this assay was 5′-GGGACTCTTGACCATGATGCCAGGGAAACCT-3′ and 5′-TCAGATCTACCATGGCTTTGAAATCCCCATT-3′. The pCAMBIA1302:GmNFYA13 fusion constructs were transformed into Agrobacterium tumefaciens strain GV3101, followed by infection of Arabidopsis flowers. Hygromycin (Roche, Germany) was used to screen the positive transgenic lines to obtain the T3 homozygotes. The 35S:GmNFYA13-2, 35S:GmNFYA13-4, and 35S:GmNFYA13-5 transgenic Arabidopsis lines were used to explore the function of GmNFYA13.
Subcellular Localization
Five-day-old 35S:GmNFYA13 and WT Arabidopsis plants on 1/2 MS medium were used to determine the localization of GmNFYA13 in cells. Roots of 35S:GmNFYA13 plants could express GFP-GmNFYA13 fusion protein. Roots of 35S:GmNFYA13 and WT Arabidopsis plants were detached and observed using a confocal laser scanning microscope (Zeiss, LSIM700) as described previously (Liu et al., 2013; Ma et al., 2020).
Promoter:GUS Analysis
The 2000-bp DNA fragment upstream of cDNA was amplified to obtain the GmNFYA13 promoter using PrimerSTAR Max DNA polymerase (Takara, Japan) with the primer set 5′-GGACACGTGTCTCGATCTTCG-3′ and 5′-CTTAGCACAACTTTTCCCTAT-3′. The products were cloned into a modified pCAMBIA1305 vector to generate the pCAMBIA1305:PGmNFYA13 fusion construct with the forward primer 5′-CCATGATTACGAATTCGGACACGTGTCTCGATCTTCG-3′ and reverse primer 5′-CTCAGATCTACCATGGCTTAGCACAA CTTTTCCCTAT-3′ as previously described (Ma et al., 2020). The A. tumefaciens strain GV3101 harboring pCAMBIA1305:PGmNFYA13 was used to infect Arabidopsis flowers. The positive seeds were screened with hygromycin to obtain the T3 homozygotes. T3-generation transgenic Arabidopsis plants were used to carry out further experiments. The PGmNFYA13:GUS analysis was performed as described previously (Jefferson et al., 1987).
To measure the transcripts of GUS in PGmNFYA13:GUS transgenic Arabidopsis plants under normal condition, the rosette leaves, cauline leaves, roots, flowers, and siliques were sampled and used for RNA isolation. The special primer set of GUS was listed in Supplementary Table 1.
Transactivation Activity Assay in Yeast Strains
The full-length CDS of GmNFYA13 was ligated into the NdeI restriction site of the pGBKT7 vector to generate the BD:GmNFYA13 construct with the forward primer 5′-AGGAGGACCTGCATATGATGCCAGGGAAACCT-3′ and reverse primer 5′-GCCTCCATGGCCATATGTTATTTGAAATC CCC-3′. The construct was transformed into yeast strain AH109. The yeast strains were grown on SD -Trp plates at 30°C for 3 days and then transferred to SD -Trp/-His/-Ade and SD -Trp/-His/-Ade with 4 mg/ml α-Gal. The yeast strains harboring the empty pGBKT7 construct were used as the negative control.
Germination and Root Growth of Arabidopsis Plants
35S:GmNFYA13 seeds were sterilized as described previously (Ma et al., 2020). For the germination assay, the sterilized seeds were sown on 1/2 MS medium containing 50–80 mM NaCl, 0.5–0.8 μM ABA, and 8–10% PEG 6000 (PEG), followed by stratification for 2 days at 4°C, and then the mediums were transferred in a greenhouse at 22°C with a 16-h photoperiod and 70% relative humidity, which are considered normal conditions for Arabidopsis plants. The seeds sown on 1/2 MS medium were set as the control.
For the root growth assay, sterilized seeds were sown on 1/2 MS medium and stratified for 2 days, and then the mediums were transferred into a greenhouse under normal conditions. The 3-day-old seedlings were grown on 1/2 MS medium containing 100–120 mM NaCl, 0.5–0.8 μM ABA, and 10–12% PEG for 7 days, respectively, and then the root lengths were measured using a straightedge.
Salt and Drought Treatment of Arabidopsis Lines
The stratified seeds were grown for 7 days under normal conditions, and then 7-day-old seedlings were transferred into plastic pots (4.5 cm in length and width, 5 cm in depth) for 2 weeks. For salt treatment, the 3-week-old seedlings were irrigated with 350 mM NaCl solution; for drought treatment, water supply was shut off until significant difference was detected between 35S:GmNFYA13 and WT Arabidopsis plants, and then the seedlings were watered and allowed to recover for a week.
To assess the transcription levels of salt-responsive genes in the 35S:GmNFYA13 transgenic Arabidopsis plants, the leaves of 3-week-old seedlings were collected at 1 h after salt treatment. To assess the transcription levels of drought-related genes in 35S:GmNFYA13 lines, the leaves of 3-week-old seedlings were sampled at 2 h after drought treatment. The collected leaves were immediately frozen with liquid nitrogen for RNA isolation. The leaves of 3-week-old seedlings under normal conditions were set as the control.
Generation of Transgenic Soybean Plants
To generate transgenic soybean plants overexpressing GmNFYA13, the CDS of GmNFYA13 without the termination codon was introduced into the vector pCAMBIA3301 to generate the pCAMBIA3301:GmNFYA13 construct driven by the CaMV35S promoter and ligated with NcoI and BstEII sites with the primer set 5′-GGACTCTTGACCATGATGCCAGGGAAACCTGAC-3′ and 5′-ATTCGAGCTGGTCACCTTTGAAATCCCCATTAGG-3′. To generate transgenic soybean plants suppressing GmNFYA13, a 705-bp RNA interference fragment (Supplementary Figure 1) was artificially synthesized and inserted into the same vector to generate the pCAMBIA3301:RNAi-GmNFYA13 construct. The pCAMBIA3301:GmNFYA13 (OE), pCAMBIA3301:RNAi-GmNFYA13 (RNAi), and pCAMBIA3301 empty vector (EV) were transformed into A. rhizogenes K599 and stored at −80°C.
Soybean seeds were sown into nutrient soil in plastic pots (14 cm diameter, 15 cm depth) and the hypocotyls of 5-day-old seedlings were infected by A. rhizogenes K599 harboring OE, RNAi, and EV construct with a syringe tip, and then covered by plastic cups for 24 h without light. The soybean plants were uncovered, and the infected hypocotyls were surrounded by peat and vermiculite mixture (1:1, v/v). They were irrigated with enough water to create a warm and humid environment for 2 weeks, and then hairy roots were induced. The principal roots were cut off, and soybean plants with positive hairy roots screened by QuickStix kit (EnviroLogix, America) and qRT-PCR were used to perform subsequent experiments; they were, respectively, named OE, RNAi, and EV transgenic soybean plants.
Salt and Drought Treatments of Transgenic Soybean Plants
The transgenic soybean plants with positive hairy roots were planted in plastic pots (14 cm diameter, 15 cm depth) containing nutrient soil for 7 days to establish normal growth. It is difficult to complete the entire growth period using plants with only a few positive hairy roots under salt and drought stresses. However, the transgenic soybean plants are sufficient for drought tolerance at the seedling stage.
For salt treatment, the soybean plants were irrigated with 500 mM NaCl solution; for drought treatment, the water supply to soybean plants was shut off until significant difference was observed among the three transgenic soybean lines.
To evaluate the transcription level of salt/drought-related genes in transgenic soybean plants, for salt and drought treatments, the positive hairy roots were sampled at 1 h after salt stress and 2 h after drought stress. The control was the non-treated positive hairy roots.
Quantitative Real-Time PCR (qRT-PCR) Analysis
The qRT-PCR analysis was performed using an Applied Biosystems 7500 real-time PCR system and the data were analyzed with the 2–ΔΔCT method based on CT values (Ni et al., 2013). The specific primers for qRT-PCR in this study are in Supplementary Table 1.
Measurement of Water Loss, Soil Water Potential (SWP), and Proline Content
In 35S:GmNFYA13 Arabidopsis and three transgenic soybean lines, water loss and SWP were measured as described previously (Li et al., 2008; Ma et al., 2020). The proline content was measured as described previously (Cui et al., 2019) after drought treatment for 4 days and 5 days with respect to 35S:GmNFYA13 Arabidopsis and transgenic soybean plants, respectively.
Measurement of Relative Water Content (RWC), Malondialdehyde (MDA), Chlorophyll, and Ion Leakage
In 35S:GmNFYA13 Arabidopsis and three transgenic soybean lines, RWC, MDA, chlorophyll, and ion leakage were measured after drought and salt treatment as described previously (Ni et al., 2013; Ma et al., 2020).
Measurement of ABA Concentration and Stomatal Aperture
Abscisic acid concentration was measured when 35S:GmNFYA13 transgenic Arabidopsis plants were subjected to salt and drought treatment for 4 days as described previously (Wang et al., 2018). Leaves of 3-week-old 35S:GmNFYA13 plants were detached and immersed into stomatal opening buffer, which was prepared for 3 h as previously reported (Tian et al., 2015); leaves were then transferred to 0, 5, and 10 μM ABA solution for 2 h. Stomatal apertures were detected using a confocal laser scanning microscope (Zeiss, LSIM700) and measured with the ruler tool in Adobe Photoshop CS5.
Transcriptional Activation Assay in Tobacco Plants
Tobacco seeds were sterilized with 1% sodium hypochlorite for 15 min, washed three times using sterile water, and then sown on 1/2 MS medium. Seven-day-old seedlings were transferred into nutrient soil in a greenhouse at 25/22°C (day/night) with a 16-h photoperiod and 50–60% relative humidity as described previously (Zhang et al., 2017). Tobacco plants with four leaves were infected.
The promotors of target genes were inserted into pGreen II 0800 as previously described (Ma et al., 2020). The primer set for GmSALT3 was 5′-GTCGACGGTATCGATAAGCTTCGATGATACTACATAAGT-3′ and 5′-CAGGAATTCGATATCAAGCTTGGCCAAAGACTC AGTGCT-3′, with 5′-GTCGACGGTATCGATAAGCTTCCTGG TCCACTCTAGTAG-3′ and 5′-CAGGAATTCGATATCAAGCT TGGTGAATGGTCGGTTGAA-3′ for GmNCED3. Primers for GmMYB84 were 5′-GTCGACGGTATCGATAAGCTTCTCAG CTTCAATTACAAT-3′ and 5′-CAGGAATTCGATATCAAGCT TGGCTATTCTCACTCACTA-3′; primers for GmRbohB were 5′-GTCGACGGTATCGATAAGCTTACGACACAGTCTGAGAAT-3′ and 5′-CAGGAATTCGATATCAAGCTTCTCCTCGTCGTCC TTGAA-3′. The fusion constructs pGreen II 0800:PGmSALT3/GmNCED3/GmMYB84/GmRbohB were transformed into A. tumefaciens strain GV3101. GV3101 harboring the pCAMBIA1302 empty vector and pCAMBIA1302:GmNFYA13 fusion construct were co-transformed into tobacco leaves with GV3101 harboring pGreen II 0800:PGmSALT3/GmNCED3/GmMYB84/GmRbohB. The GUS fluorescence was observed using LB985 NightSHADE (Berthold, Germany) following the manufacturer’s instructions.
To quantify the LUC activity in the tobacco leaves of equal weight, the protein was extracted. The extraction buffer consisted of 50 mM Tris, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, and 5 mM DTT, and then pH was adjusted to 8.0. The LUC expression levels of the protein solution were evaluated with GloMax Multi Jr (Promega, America) according to the manufacturer’s instructions.
Statistical Analysis
The data in this research were subjected to Duncan’s tests using SPSS Statistics 22 (IBM, United States). The data of germination rate, water loss, and SWP were assessed with Student’s t-test. Three biological replications were performed for each experiment in this study.
Results
Expression Pattern of NF-YA Genes in Soybean
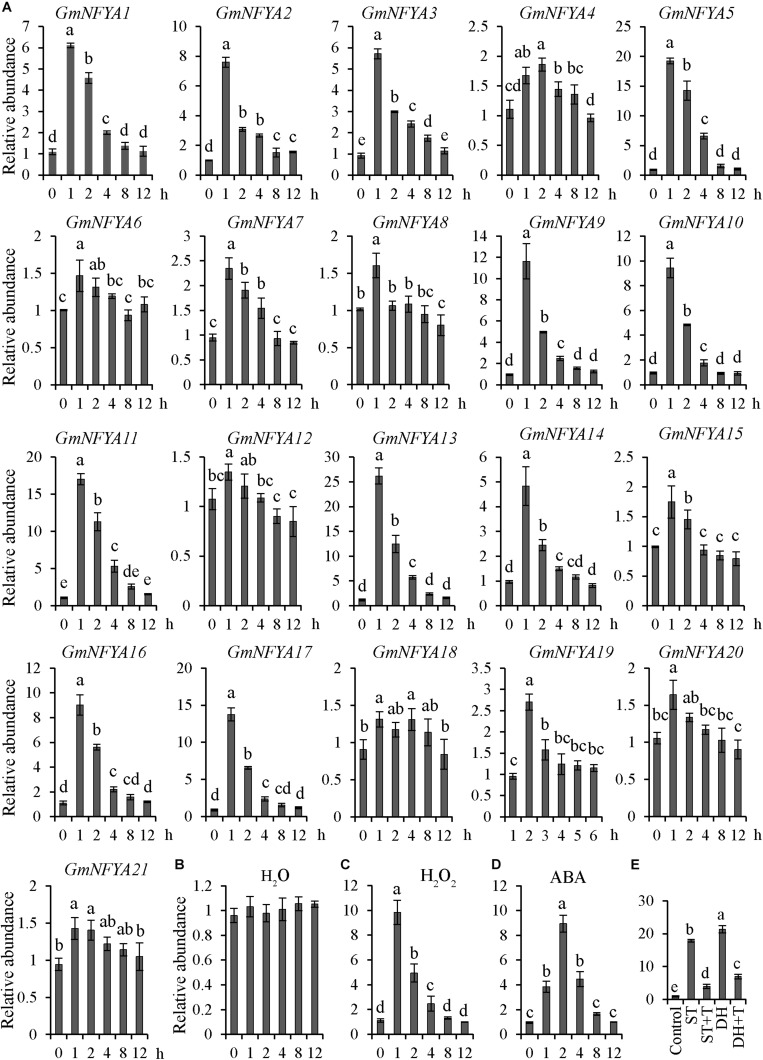
As described previously, the transcription level of GmNFYA5 was the highest among all 21 GmNFYAs under drought conditions (Ma et al., 2020). To determine whether GmNFYAs were induced by salt treatment, the transcription levels of 21 GmNFYAs were analyzed by qRT-PCR. The results showed that 12 members were highly responsive to salt stress and the transcript level of GmNFYA13 was the highest (Figure 1A). In addition, the GmNFYA13 transcript was higher than other GmNFYAs except GmNFYA5 and GmNFYA11 under drought treatment (Ma et al., 2020). Moreover, the transcript level of GmNFYA13 was induced by H2O2 and ABA treatment (Figures 1C,D) but not by H2O (Figure 1B). To further explore whether ABA was involved in salt- and drought-induced GmNFYA13 transcription, detached leaves were subjected to tungstate solution, which is an ABA biosynthesis inhibitor, followed by salt treatment for 1 h, or drought treatment for 2 h. Compared with the control condition, the GmNFYA13 transcript under salt/drought treatment was similar to the above expression pattern (Figure 1E). However, pretreatment with tungstate suppressed the induction of GmNFYA13 expression (Figure 1E). These results showed that ABA played an essential role in salt- and drought-induced GmNFYA13 transcription.
FIGURE 1.
GmNFYA13 transcripts were induced by treatment of salt, H2O2, and ABA. (A) The expression levels of 21 NF-YA members in Glycine max were evaluated with qRT-PCR under salt treatment. (B–D) H2O, H2O2, and ABA induced GmNFYA13 transcripts. (E) Tungstate suppressed salt- and drought-induced GmNFYA13 transcripts. Control, salt, salt + tungstate, drought, and drought + tungstate are indicated by CTR, ST, ST + T, DH, and DH + T, respectively. GmCYP2 was used as the internal control and transcripts of all genes used in the assay were normalized to the non-treated expression level, which was set as 1.0. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05.
The coding sequence of GmNFYA13 (Glyma12g36540) is 912 bp in length, encoding a 34.06-KD polypeptide with an isoelectric point (pI) of 9.50. The conserved core regions of GmNFYA13, which consists of an NF-YB/C binding subdomain and CCAAT binding sequences, are similar to those of Arabidopsis (Supplementary Figure 2A). Phylogenetic analysis of conserved amino acid sequences showed that GmNFYA13 clustered with AtNFYA2 and AtNFYA10 in Arabidopsis (Supplementary Figure 2B).
Tissue-Specific Expression Analysis and Characterization of GmNFYA13
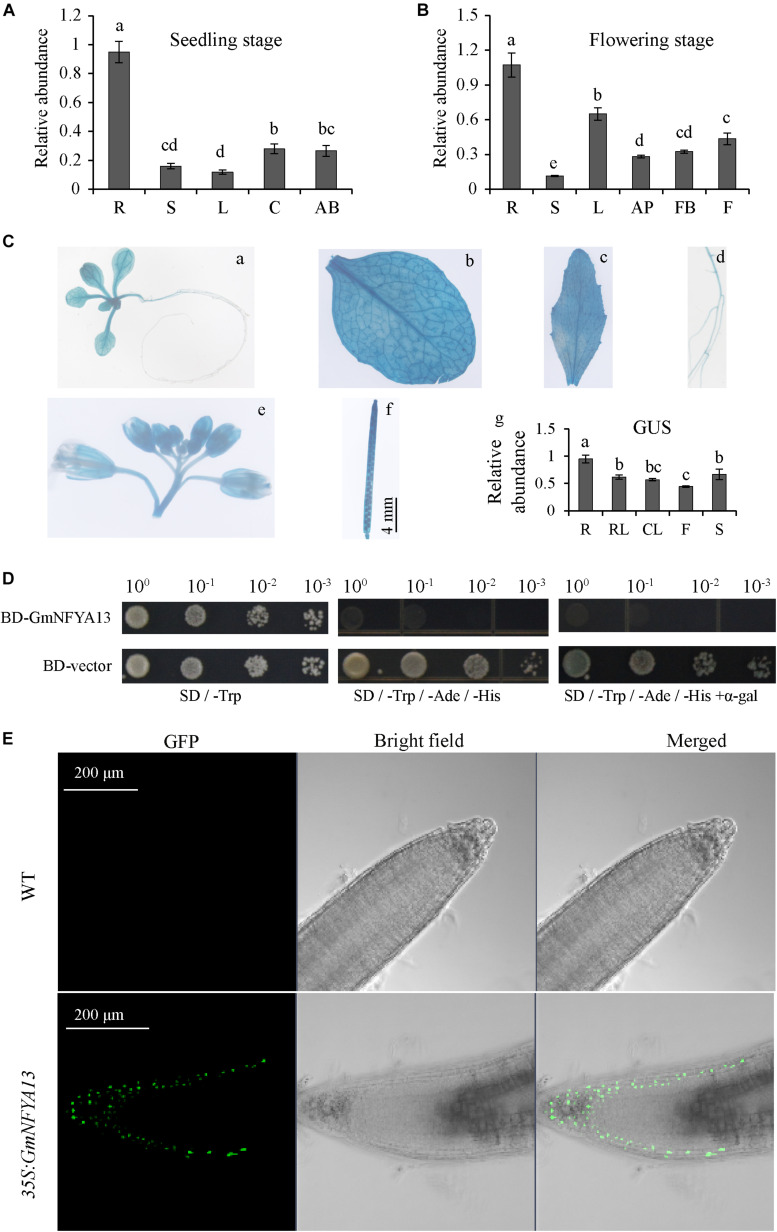
The transcription of GmNFYA13 was detected in various tissues at seedling and flowering growth phases based on qRT-PCR (Figures 2A,B). The GmNFYA13 transcript was highest in roots in both stages and significantly improved in leaves of the former period compared with that of the latter (Figures 2A,B). To better evaluate the tissue-specific expression of GmNFYA13, T3-generation transgenic Arabidopsis plants overexpressing PGmNFYA13:GUS were generated. GUS staining was observed in Arabidopsis seedling, rosette leaf, cauline leaf, root, flower, and silique (Figure 2C). The GUS expression level in leaves was lower than that in roots (Figure 2C), consistent with the data of qRT-PCR in soybean plants.
FIGURE 2.
Analysis of tissue-specific expression, self-activating activity, and subcellular localization of GmNFYA13. (A,B) GmNFYA13 transcripts were assessed in various tissues at two growth periods. The expression level of GmNFYA13 in roots was set as 1.0, and GmCYP2 was the internal control. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05. (C) GUS staining in each tissue in PGmNFYA13:GUS transgenic Arabidopsis plants. a, 5-day-old seedling; b, rosette leaf (RL); c, cauline leaf (CL); d, root (R); e, flower (F); f, silique (S); g, transcripts of GUS were measured in various tissues of transgenic Arabidopsis by qRT-PCR. Scale bar = 4 mm. The transcripts of GUS in roots were set as 1.0, and AtTub8 was the internal control. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05. (D) Self-activating activity analysis of GmNFYA13. (E) GFP fluorescence was detected in roots of WT and 35S:GmNFYA13 Arabidopsis plants. Scale bar = 200 μm. Each experiment had three biological replicates.
To test whether GmNFYA13 independently regulated downstream genes, the self-activating activity was evaluated in yeast strains. The yeast strains harboring BD:GmNFYA13 fusion construct could grow on SD -Trp/-His/-Ade and SD -Trp/-His/-Ade with α-Gal, where no yeast strains harboring BD empty vector could grow. The results implied that GmNFYA13 had self-activating activity (Figure 2D).
To observe the localization of GmNFYA13 in plant cells, roots of WT and 35S:GmNFYA13 Arabidopsis plants were detached. The GFP fluorescence in roots of 35S:GmNFYA13 plants was exclusively detected in the nuclei with no fluorescence observed in roots of WT plants (Figure 2E), which showed that GmNFYA13 was a nuclear localization protein.
Sensitivity of 35S:GmNFYA13 Arabidopsis Plants to ABA, NaCl, and PEG
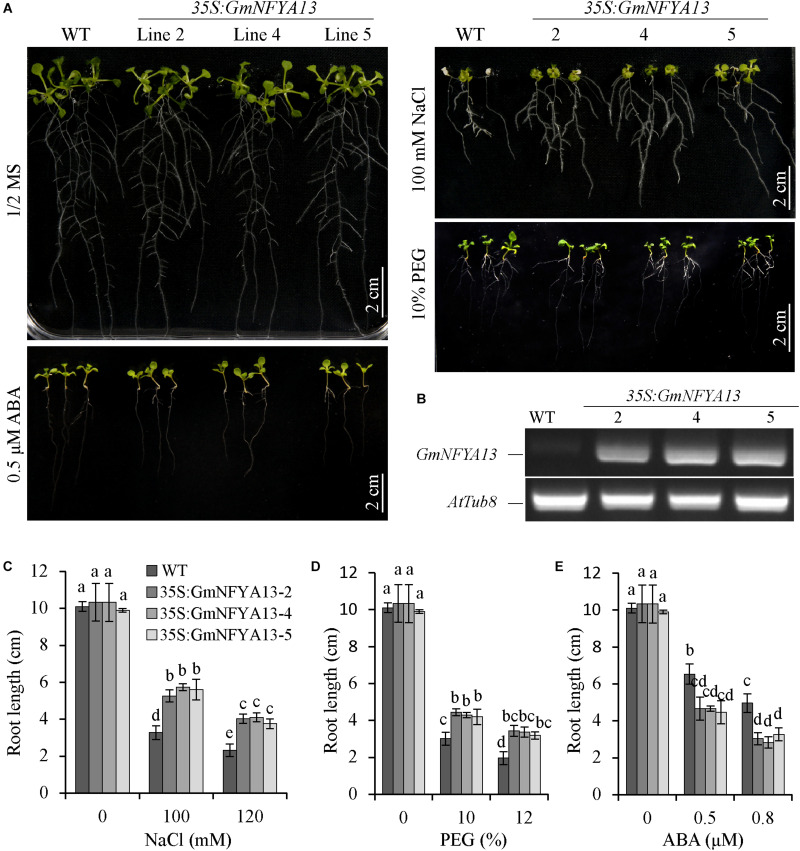
Three 35S:GmNFYA13 Arabidopsis lines (35S:GmNFYA13-2, 35S:GmNFYA13-4, and 35S:GmNFYA13-5) were used to carry out further assays based on RT-PCR analysis (Figure 3B and Supplementary Figure 5). In the germination assay, there was no significant difference between 35S:GmNFYA13 and WT Arabidopsis seeds with respect to germination rates under control conditions (Supplementary Figure 3A). However, in comparison to WT Arabidopsis seeds, the germination rate of 35S:GmNFYA13 seeds was higher when subjected to different concentrations of NaCl and PEG, but lower under treatment of ABA (Supplementary Figures 3B–G).
FIGURE 3.
Root growth of WT and 35S:GmNFYA13 transgenic Arabidopsis seedlings under treatments of salt, drought, and ABA. (A) The root elongation of WT and 35S:GmNFYA13 with/without treatments. Scale bar = 2 cm. (B) The transcript levels of GmNFYA13 in WT and 35S:GmNFYA13 lines were evaluated with RT-PCR. AtTub8 was the internal control. (C–E) The root lengths of WT and 35S:GmNFYA13 lines subjected to various concentrations of NaCl, PEG, and ABA. Data indicate three biological replicates (n = 18) ± SD. Different letters above the columns represent significant differences at P < 0.05. Three biological replicates were used in each experiment.
In the root elongation assay, there was no significant difference between root lengths of 35S:GmNFYA13 and WT seedlings prior to these three treatments (Figure 3A). Consistent with the germination assay results, under treatments of salt and PEG, the root elongation of WT seedlings was more severely suppressed in comparison to that of 35S:GmNFYA13 lines (Figures 3A,C,D). Under ABA treatments, the root length of WT seedlings was significantly longer than that of 35S:GmNFYA13 seedlings (Figures 3A,E).
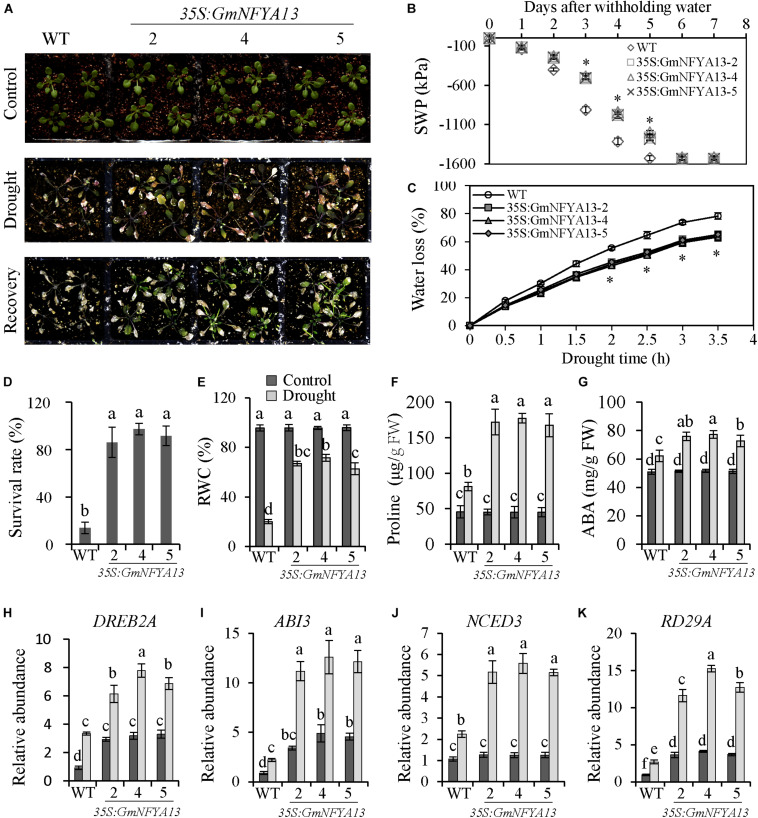
Tolerance of 35S:GmNFYA13 Arabidopsis Lines to Salt Treatment
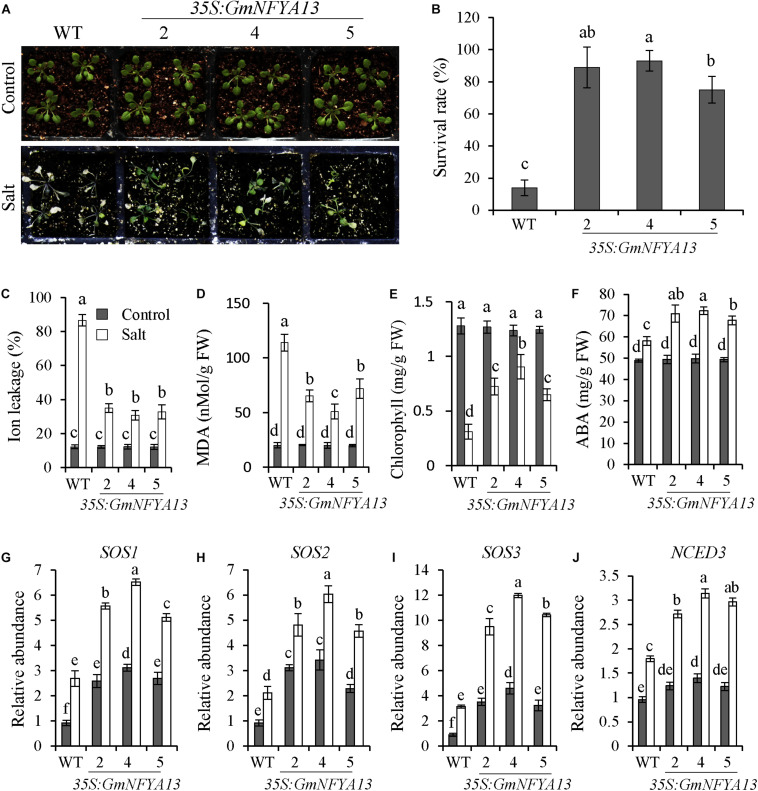
To elucidate the function of GmNFYA13 in regulating salt tolerance of Arabidopsis plants, 3-week-old 35S:GmNFYA13 and WT plants were subjected to 350 mM NaCl solution for 7 days. Leaves of WT plants were bleached, whereas most leaves from 35S:GmNFYA13 plants remained green with no significant difference observed between the former and latter sets of plants under control conditions (Figure 4A). Consistently, compared with 35S:GmNFYA13 plants, the survival rate and chlorophyll content were lower, and the ion leakage and MDA contents in WT plants were apparently higher (Figures 4B–E). No significant difference was noted between 35S:GmNFYA13 and WT Arabidopsis lines with respect to physiological indices under control conditions (Figures 4B–E).
FIGURE 4.
35S:GmNFYA13 Arabidopsis seedlings showed enhanced salt tolerance compared to WT lines. (A) Assessment of salt tolerance in WT and 35S:GmNFYA13 transgenic Arabidopsis plants. Three-week-old seedlings were treated with 350 mM NaCl solution for 7 days. (B–E) The survival rate, ion leakage, MDA, and chlorophyll content of WT and 35S:GmNFYA13 lines were measured after salt treatment. (F) The ABA concentration of WT and 35S:GmNFYA13 lines were measured following salt treatment for 4 days. (G–J) The expression levels of SOS1, SOS2, SOS3, and NCED3 in WT and 35S:GmNFYA13 lines were evaluated under control and salt treatments. Transcript levels of these genes were normalized to those in WT plants under control conditions, and AtTub8 was used as the internal control. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05. Each experiment had three biological replicates.
To determine how GmNFYA13 regulated salt tolerance of 35S:GmNFYA13 Arabidopsis plants, salt-related genes were analyzed in 35S:GmNFYA13 and WT lines by qRT-PCR under control and salt treatments. Under control conditions, transcript abundance of SOS1, SOS2, and SOS3 in 35S:GmNFYA13 lines were higher than those in WT lines with no significant difference detected for the NCED3 transcript. However, under salt conditions, transcript levels of SOS1, SOS2, SOS3, and NCED3 in 35S:GmNFYA13 plants were markedly higher compared with WT plants (Figures 4G–J).
Tolerance of 35S:GmNFYA5 Arabidopsis Lines to Drought Treatment
To evaluate the role of GmNFYA13 in drought tolerance, water supply was shut off for 9 days. More extreme wilting was observed in WT seedlings compared with 35S:GmNFYA13 seedlings with no difference detected under control conditions (Figure 5A). During drought treatment, the SWP was measured. Compared with WT lines, the SWP of 35S:GmNFYA13 lines fell more slowly, and the minimum level was reached at 5 and 6 days after drought treatment with respect to the former and latter sets of seedlings (Figure 5B). Consistently, in comparison to WT lines, detached leaves in 35S:GmNFYA13 lines lost water more slowly (Figure 5C). The survival rate and RWC of WT lines was lower compared with that of 35S:GmNFYA13 lines (Figures 5D,E). The proline and ABA contents were measured at 4 days after drought stress, and proline content was higher in 35S:GmNFYA13 lines compared with WT lines (Figure 5F).
FIGURE 5.
35S:GmNFYA13 Arabidopsis seedlings showed enhanced drought tolerance compared to WT lines at the seedling stage. (A) Evaluation of drought tolerance in WT and 35S:GmNFYA13 transgenic Arabidopsis plants. Three-week-old seedlings were subjected to drought treatment for 9 days and recovered for 7 days. (B) The SWP of the soil where WT and 35S:GmNFYA13 were grown. (C) The water loss of detached leaves from WT and 35S:GmNFYA13 Arabidopsis seedlings. (D) The survival rate of WT and 35S:GmNFYA13 Arabidopsis seedlings after recovery from drought treatment. (E,F) The relative water content and proline content of WT and 35S:GmNFYA13 Arabidopsis seedlings after drought treatment. (G) The ABA concentration of WT and 35S:GmNFYA13 Arabidopsis seedlings following drought treatment for 4 days. (H–K) Transcript levels of DREB2A, ABI3, NCED3, and RD29A under control and drought treatment. Transcript levels of these genes were normalized to those in WT plants under control conditions, AtTub8 was used as the internal control. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05. Three biological replicates were used in each experiment.
In addition, ABA was produced in Arabidopsis plants treated with drought and salt stresses and ABA accumulation in 35S:GmNFYA13 lines was higher than WT lines (Figures 4F, 5G). ABA triggers stomatal pore closure in leaves (Nakashima and Yamaguchi-Shinozaki, 2013), so the stomatal apertures were observed and measured. When treated with different concentrations of ABA, the stomatal apertures of detached leaves in WT seedlings were significantly bigger than those in 35S:GmNFYA13 seedlings with no difference noted under control conditions (Supplementary Figures 4A,B).
To further understand how GmNFYA13 functioned in regulating drought tolerance of 35S:GmNFYA13 Arabidopsis plants, the drought-responsive genes were evaluated by qRT-PCR. Under control and drought conditions, expression levels of DREB2A, ABI3, RD29A, and NCED3 in 35S:GmNFYA13 Arabidopsis plants were higher than those in WT plants with no significant difference detected for the NCED3 transcript under control conditions (Figures 5H–K).
Tolerance of Transgenic Soybean Plants to Salt Treatment
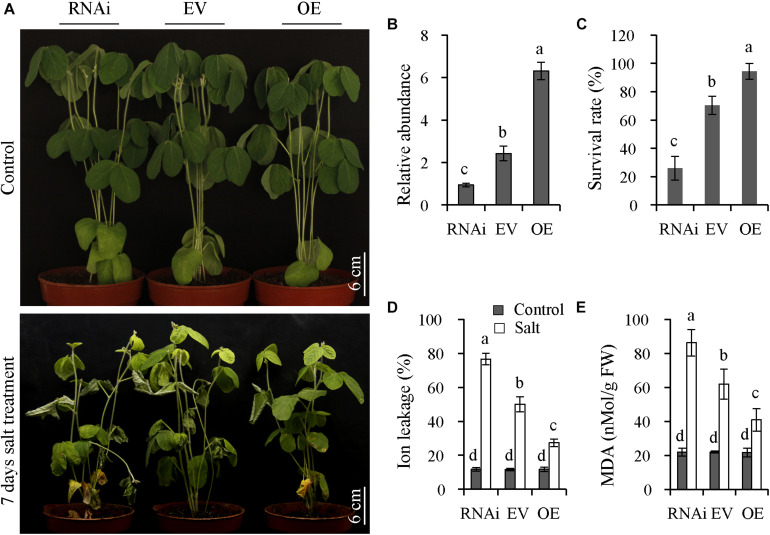
To explore the functions of GmNFYA13, OE, RNAi, and EV transgenic soybean lines with positive hairy roots were generated. Compared with EV lines, the expression levels of GmNFYA13 in OE and RNAi lines were significantly higher and lower, respectively (Figure 6B). The transgenic soybean lines were subjected to 500 mM NaCl solution for 7 days. Under salt treatment, the RNAi plants showed chlorosis, and the EV plants displayed less damage than RNAi lines with turgor maintained in OE plants (Figure 6A); the survival rate of OE plants overexpressing GmNFYA13 was significantly higher than EV plants, while its abolition resulted in the opposite effect (Figure 6C); compared with EV plants, the ion leakage and MDA content in OE and RNAi plants were significantly lower and higher, respectively (Figures 6D,E). However, no significant difference was detected among these indices in OE, EV, and RNAi plants under control conditions (Figures 6C–E).
FIGURE 6.
OE and RNAi transgenic soybean plants, respectively, displayed improved and decreased salt tolerance compared with EV plants. (A) Assessment of salt tolerance in RNAi, EV, and OE transgenic soybean plants. Soybean plants with positive hairy roots were subjected to 500 mM NaCl solution for 7 days. Scale bar = 6 cm. (B) GmNFYA13 transcript was evaluated in RNAi, EV and OE transgenic soybean plants. GmCYP2 was used as the internal control and transcripts of GmNFYA13 were normalized to the expression level in RNAi transgenic soybean plants, which was set as 1.0. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05. (C–E) The survival rate, ion leakage, and MDA content in RNAi, EV, and OE transgenic soybean plants. Data indicate three biological replicates (n = 18) ± SD. Different letters above the columns represent significant differences at P < 0.05.
Tolerance of Transgenic Soybean Plants to Drought Treatment
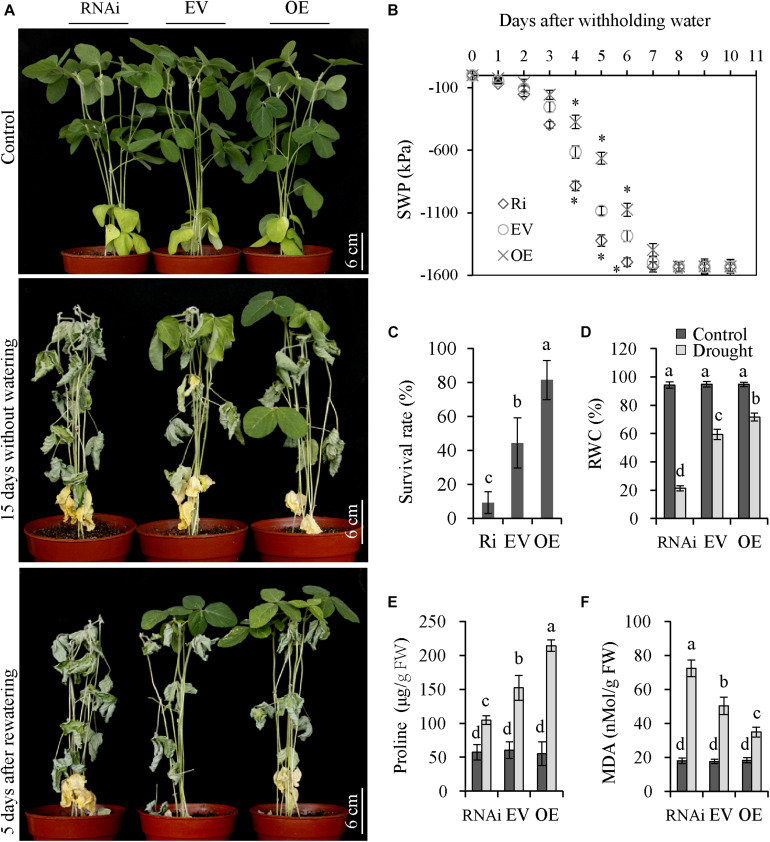
To assess the role of GmNFYA13 in drought tolerance, OE, RNAi, and EV transgenic soybean lines were deprived of water for 15 days. The OE plants displayed less wilting compared with EV plants, while RNAi plants showed the opposite results (Figure 7A). During drought treatment, the SWP of the soil was measured. After initiation of drought stress, SWP of OE, EV, and RNAi lines declined to the minimum levels at 8, 7, and 6 days, respectively (Figure 7B).
FIGURE 7.
OE and RNAi transgenic soybean plants, respectively, displayed increased and decreased tolerance to drought stress compared with EV plants. (A) Drought tolerances were evaluated in RNAi, EV, and OE transgenic soybean plants. Soybean plants were subjected to drought treatment for 15 days and recovery for 5 days. Scale bar = 6 cm. (B) The SWP of the soil where RNAi, EV, and OE transgenic plants grew were assessed. Asterisks represent significant differences at P < 0.05 in comparison to the corresponding controls. (C) The survival rate was measured in RNAi, EV, and OE transgenic plants after recovery from drought treatment. (D–F) The relative water content, proline, and MDA content were measured in RNAi, EV, and OE transgenic plants after drought treatment. Data indicate three biological replicates (n = 18) ± SD. Different letters above the columns represent significant differences at P < 0.05. Each experiment had three biological replicates.
Compared with EV plants, the survival rate, RWC, and proline content of OE and RNAi plants were significantly higher and lower, respectively (Figures 7C–E). The OE plants overexpressing GmNFYA13 produced significantly lower MDA content, while RNAi plants where it was abolished showed the opposite results (Figure 7F). No significant difference was noted among physiological indices of OE, RNAi, and EV plants under control conditions (Figures 7C–F).
Transcript Levels of Salt/Drought-Responsive Genes
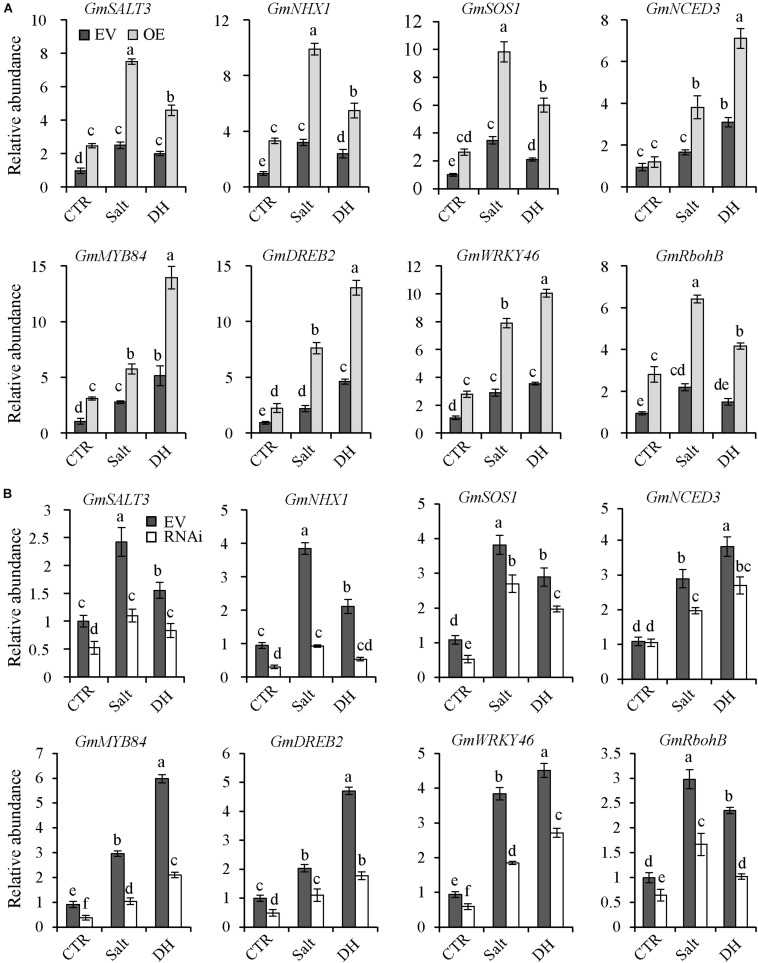
To investigate how GmNFYA13 functioned in salt/drought tolerance in transgenic soybean plants, three salt-related genes (GmSALT3, GmSOS1, and GmNHX1) (Guan et al., 2014; Zhang W. et al., 2019), one key gene involved in ABA synthesis (GmNCED3) (Zhang X. Z. et al., 2019), three drought-responsive genes (GmDREB1, GmWRKY46 and GmMYB84) (Chen et al., 2007; Luo et al., 2013; Wang N. et al., 2017; Ma et al., 2020), and one gene, GmRbohB, related to oxidative stress (Wang N. et al., 2017), were selected for further analysis.
Under control conditions, no significant difference was detectable for transcript abundance of GmNCED3 among OE, EV, and RNAi soybean plants (Figures 8A,B); transcript levels of GmSALT3, GmNHX1, GmSOS1, GmDREB1, GmWRKY46, GmMYB84, and GmRbohB in OE plants were higher compared with EV plants (Figure 8A); the opposite results were detected in RNAi plants (Figure 8B). Under salt and drought treatments, compared with WT plants, expression levels of all genes in OE plants were significantly higher and those in RNAi plants were significantly lower (Figures 8A,B).
FIGURE 8.
The expression levels of GmSALT3, GmNHX1, GmSOS1, GmNCED3, GmMYB84, GmDREB2, GmWRKY46, and GmRbohB in RNAi, EV, and OE transgenic soybean plants under control, salt, and drought conditions. (A) Transcripts of GmSALT3, GmNHX1, GmSOS1, GmNCED3, GmMYB84, GmDREB2, GmWRKY46, and GmRbohB in EV and OE transgenic lines under control, salt, and drought conditions. (B) Transcript levels of these genes in RNAi and EV transgenic lines under control, salt, and drought conditions. GmCYP2 was used as the internal control and transcripts of GmNFYA13 were normalized to the expression level in EV transgenic soybean plants under control conditions, which was set as 1.0. CTR represents the control condition, and salt and DH represent salt and drought treatment, respectively. For salt treatment, roots were subjected to 500 mM NaCl solution for 1 h; for drought treatment, roots were placed on a plastic plate for 2 h; roots under normal condition were used as the control sample. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05.
Transcriptional Activation Assays
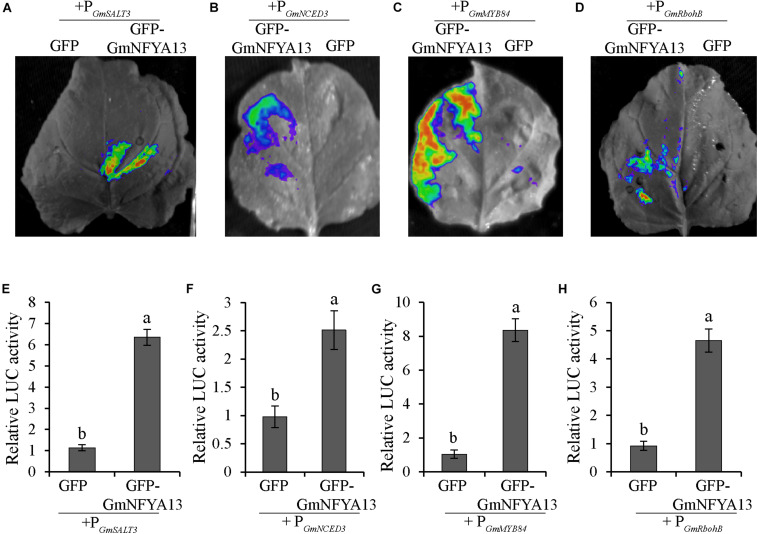
To test whether GmNFYA13 could directly regulate the downstream genes in vivo, LUC activity in the tobacco leaves harboring recombinant pCAMBIA1302:GmNFYA13 or pCAMBIA1302 empty vector in consort with pGreen II 0800:PGmSALT3/GmNCED3/GmMYB84/GmRbohB were assessed. Consistent with our expectation, the LUC expression levels in the former sets of leaves were significantly higher than those in the latter sets, which showed that GmNFYA13 could positively modulate the expression levels of GmSALT3, GmMYB84, GmNCED3, and GmRbohB by binding to their promotors (Figures 9A–H).
FIGURE 9.
LUC activity was enhanced due to the regulation of GmNFYA13 on promoters of GmSALT3, GmNCED3, GmMYB84, and GmRbohB. (A–D) The LUC activity was evaluated in tobacco leaves with LB985 NightSHADE. (E–H) The LUC expression levels of the protein solution extracted from the leaves, indicated by ratio of LUC/REN, were measured with GloMax Multi Jr. The ratio of LUC/REN of GFP-GmNFYA13 + PGmSALT3/GmNCED3/GmMYB84/GmRbohB was normalized to that of GFP + PGmSALT3/GmNCED3/GmMYB84/GmRbohB, which was set as 1.0. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05.
Discussion
Compared with Arabidopsis and rice plants, the soybean genome is larger, containing 115 NF-Y genes and 21 NF-YA members (Thao and Tran, 2012). However, only GmNFYA3 (Ni et al., 2013) and GmNFYA5 (Ma et al., 2020) had been functionally characterized. In addition, overexpression of GmNFYA3 and GmNFYA5 could only confer drought tolerance to transgenic plants (Ni et al., 2013; Ma et al., 2020). In this study, transgenic Arabidopsis and soybean plants carrying extra copies of GmNFYA13 displayed improved salt and drought tolerance.
The GmNFYA13 transcript was highest among 21 NF-YA genes in soybean following salt treatment (Figure 1A) and higher than other NF-YA genes except GmNFYA5 and GmNFYA11 when subjected to drought treatment (Ma et al., 2020). In addition, the expression level of GmNFYA13 was highest in roots compared with other tissues (Figure 2A). Likewise, GmNFYA3, AtNFYA5, and Cdt-NFYC1 (Li et al., 2008; Ni et al., 2013; Chen et al., 2015), which were induced by osmotic stresses with high levels in roots, functioned in modulating salt or drought tolerance of plants. These results implied that GmNFYA13 could play important roles in regulating salt and drought tolerance in plants.
In addition, GmNFYA13 was induced by ABA treatment (Figure 1D), and pretreatment with tungstate suppressed the salt- and drought-induced GmNFYA13 transcript (Figure 1E), showing that ABA was involved in response to osmotic stresses of GmNFYA13. ABA is a hormonal molecule produced during drought and salt stresses (Zhu, 2002; Barrero et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2013). Under salt and drought treatments, higher NCED3 transcripts in 35S:GmNFYA13 plants (Figures 4J, 5J) resulted in increased ABA biosynthesis and accumulation (Figures 4F, 5G), causing smaller stomatal apertures (Supplementary Figure 4) and less water loss in leaves (Figure 5C) to enhance tolerance to osmotic stresses. Moreover, 35S:GmNFYA13 Arabidopsis seeds and seedlings displayed higher sensitivity to ABA compared with WT lines (Figure 3A and Supplementary Figure 3). Enhanced drought and salt tolerance was accompanied by ABA hypersensitivity (Ni et al., 2013; Chen et al., 2015; Ma et al., 2020). These results indicated that overexpression of GmNFYA13 enhanced tolerance to osmotic stresses in an ABA-dependent manner. In addition, compared with corresponding lines with the normal GmNFYA13 transcript, the higher expression levels of DREB2A and GmDREB2 in 35S:GmNFYA13 Arabidopsis and OE soybean lines (Figures 5H, 8A) showed that overexpression of GmNFYA13 also enhanced osmotic stresses through another mechanism, an ABA-independent pathway.
The functional studies of GmNFYA3 and GmNFYB1 in transgenic Arabidopsis plants were professional (Ni et al., 2013; Li et al., 2017). However, the functions of these genes were determined in transgenic Arabidopsis plants. Introduction of SWP in this study (Figures 5B, 6B) during drought treatment was not performed in some other reports (Li et al., 2008; Chen et al., 2015; Li et al., 2017; Du et al., 2018; Li et al., 2019). Moreover, the genes in soybean should be identified in transgenic soybean plants.
Consistently, in comparison to EV soybean lines, the OE lines overexpressing GmNFYA13 and RNAi lines suppressing it, respectively, displayed increased and decreased tolerance to osmotic stresses. The analysis of qRT-PCR (Figure 8) and LUC activity (Figure 9) indicated that GmNFYA13 could positively and directly regulate the transcript levels of downstream genes by binding to the promoters in vivo. GmRbohB was determined to be a key gene scavenging ROS to improve tolerance to abiotic stresses in soybean plants (Wang N. et al., 2017). Consistent with this result, transcript levels of GmNFYA13 were induced by H2O2 solution (Figure 1C). In contrast to EV plants, the lower content of MDA (Figures 6E, 7F), which reflected the degree of the damage of ROS on membranes (Imahori et al., 2016; Wang X. Y. et al., 2017), was associated with the lower level of oxidative stress in OE transgenic soybean lines under salt and drought treatments. These results showed that overexpression of GmNFYA13 enhanced tolerance to osmotic and oxidative stresses in OE transgenic soybean plants.
In conclusion, physiological indices and expression levels of stress-related genes shed light on the essential roles of multifunctional GmNFYA13 and give further insights into the modulation of responses to osmotic stresses in soybean plants. Accordingly, GmNFYA13 should be a positive regulator of tolerance to osmotic stresses in soybean and has application value to improve salt and drought tolerance in other plants.
Data Availability Statement
Soybean gene sequences in this study were downloaded from the plant genomics resource (https://phytozome.jgi.doe.gov/pz/portal.html). Arabidopsis gene sequences in this study were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). The gene accessions are listed in Supplementary Table 1.
Author Contributions
Z-SX coordinated the project, conceived and designedexperiments, and edited the manuscript. X-JM performed experiments and wrote the first draft. J-DF and Y-MT conducted the bioinformatic work and performed experiments. T-FY, Z-GY, JC, Y-BZ, MC, and Y-ZM provided analytical tools and managed reagents, coordinated the project, and contributed valuable discussions. All authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Bin Liu (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the soybean seeds.
Footnotes
Funding. This research was financially supported by the National Natural Science Foundation of China (31871624) for designing and performing experiments, the Fundamental Research Funds for Central Non-Profit of Institute of Crop Sciences, the Chinese Academy of Agricultural Sciences (Y2020PT12 and S2020ZC02), and the China Postdoctoral Science Foundation (2019M660886) for data analysis, writing, and revising the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.587244/full#supplementary-material
The synthetic fragment RNAi-GmNFYA13. The hairpin structure contained three parts: the positive sequence in blue, the reverse complementary sequence in green and the intron of GmNFYA13 in purple.
The sequence alignment and phylogenetic analysis of GmNFYA13 and ten NF-YA genes in Arabidopsis. (A) Sequence alignment of GmNFYA13 and ten NF-YA genes in Arabidopsis with conserved domains highlighted in black. The linker and two subdomains are underlined. Critical amino acids were indicated by asterisks. (B) Phylogenetic analysis of GmNFYA13 and ten NF-YA genes in Arabidopsis. MEGA 7.0 was used to construct the neighbor joining tree.
Germination rate of WT and 35S:GmNFYA13 transgenic Arabidopsis lines under treatment of control, salt, drought and ABA. (A) Germination rates on 1/2 MS medium. (B–C) Germination rates on 1/2 MS medium with 50 and 80 mM NaCl. (D–E) Germination rates on 1/2 MS medium with 8 and 10% PEG6000. (F–G) Germination rates on 1/2 MS medium with 0.5 and 0.8 μM ABA. Asterisks represent significant differences at P < 0.05 in comparison to the corresponding controls. Each experiment had three biological replicates.
Stomatal apertures of the leaves in WT and 35S:GmNFYA13 transgenic Arabidopsis lines were measured under control and ABA treatments. (A) Stomatal apertures subjected to 0 and 5 μM ABA solution. (B) The ruler tool developed in Adobe Photoshop CS5 was used to measure width/length of the stomatal aperture with 0, 5, and 10 μM ABA. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05.
The transcript levels of GmNFYA13 in WT and 35S:GmNFYA13 lines were evaluated with RT-PCR. AtTub8 was the internal control.
The primer sets and gene accessions for qRT-PCR used in this study.
References
- Alam M. M., Tanaka T., Nakamura H., Ichikawa H., Kobayashi K., Yaeno T., et al. (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 13 85–96. 10.1111/pbi.12239 [DOI] [PubMed] [Google Scholar]
- Amir Hossain M., Lee Y., Cho J. I., Ahn C. H., Lee S. K., Jeon J. S., et al. (2010). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72 557–566. 10.1007/s11103-009-9592-9 [DOI] [PubMed] [Google Scholar]
- Ballif J., Endo S., Kotani M., Macadam J., Wu Y. (2011). Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. Biochem. 49 579–583. 10.1016/j.plaphy.2011.01.013 [DOI] [PubMed] [Google Scholar]
- Barrero J. M., Rodriguez P. L., Quesada V., Piqueras P., Ponce M. R., Micol J. L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29 2000–2008. 10.1111/j.1365-3040.2006.01576.x [DOI] [PubMed] [Google Scholar]
- Bihmidine S., Lin J., Stone J. M., Awada T., Specht J. E., Clemente T. E. (2013). Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237 55–64. 10.1007/s00425-012-1740-9 [DOI] [PubMed] [Google Scholar]
- Cai X., Ballif J., Endo S., Davis E., Liang M., Chen D., et al. (2007). A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 145 98–105. 10.1104/pp.107.102079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang Q. Y., Cheng X. G., Xu Z. S., Li L. C., Ye X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353 299–305. 10.1016/j.bbrc.2006.12.027 [DOI] [PubMed] [Google Scholar]
- Chen M., Zhao Y., Zhuo C., Lu S., Guo Z. (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13 482–491. 10.1111/pbi.12270 [DOI] [PubMed] [Google Scholar]
- Cui X. Y., Gao Y., Guo J., Yu T. F., Zheng W. J., Liu Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180 605–620. 10.1104/pp.19.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. T., Zhao M. J., Wang C. T., Gao Y., Wang Y. X., Liu Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18:320. 10.1186/s12870-018-1551-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. 10.3389/fpls.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Luking I., Yang O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30 1383–1391. 10.1007/s00299-011-1068-0 [DOI] [PubMed] [Google Scholar]
- Guan R. X., Qu Y., Guo Y., Yu L. L., Liu Y., Jiang J. H., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80 937–950. 10.1111/tpj.12695 [DOI] [PubMed] [Google Scholar]
- Hackenberg D., Keetman U., Grimm B. (2012). Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 13 3458–3477. 10.3390/ijms13033458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G. P., Zhang X. H., Wang Y. Q., Wu Z. Y., Huang C. L. (2009). Nucleotide variation in the NCED3 region of Arabidopsis thaliana and its association study with abscisic acid content under drought stress. J. Integr. Plant Biol. 51 175–183. 10.1111/j.1744-7909.2008.00786.x [DOI] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- Hsieh T. H., Li C. W., Su R. C., Cheng C. P., Sanjaya, Tsai Y. C., et al. (2010). A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231 1459–1473. 10.1007/s00425-010-1147-4 [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imahori Y., Bai J. H., Baldwin E. (2016). Antioxidative responses of ripe tomato fruit to postharvest chilling and heating treatments. Sci. Hortic. 198 398–406. 10.1016/j.scienta.2015.12.006 [DOI] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. 10.1002/j.1460-2075.1987.tb02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong R. W., Bui A. Q., Lee H., Kwong L. W., Fischer R. L., Goldberg R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18. 10.1105/tpc.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor D. W. (2013). Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J. Exp. Bot. 64 83–108. 10.1093/jxb/ers326 [DOI] [PubMed] [Google Scholar]
- Lee D. K., Kim H. I., Jang G., Chung P. J., Jeong J. S., Kim Y. S., et al. (2015). The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 241 199–210. 10.1016/j.plantsci.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Li B., Liu Y., Cui X. Y., Fu J. D., Zhou Y. B., Zheng W. J., et al. (2019). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 10:549. 10.3389/fpls.2019.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Mallano A. I., Bo L., Wang T., Nisa Z., Li Y. (2017). Soybean transcription factor GmNFYB1 confers abiotic stress tolerance to transgenic Arabidopsis plants. Can. J. Plant Sci. 97 501–515. 10.1139/CJPS-2016-0158 [DOI] [Google Scholar]
- Li W. X., Oono Y., Zhu J., He X. J., Wu J. M., Iida K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251. 10.1105/tpc.108.059444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F., Sanchez D. H., Musialak M., Schlereth A., Scheible W. R., Hincha D. K., et al. (2009). AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 149 1761–1772. 10.1104/pp.108.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Xu Z. S., Pan-Pan L., Hu D., Chen M., Li L. C., et al. (2013). A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J. Exp. Bot. 64 2915–2927. 10.1093/jxb/ert133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Bai X., Sun X., Zhu D., Liu B., Ji W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64 2155–2169. 10.1093/jxb/ert073 [DOI] [PubMed] [Google Scholar]
- Ma X. J., Yu T. F., Li X. H., Cao X. Y., Ma J., Chen J., et al. (2020). Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 20:123. 10.1186/s12870-020-02337-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Mittal A., Gampala S. S., Ritchie G. L., Payton P., Burke J. J., Rock C. D. (2014). Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 12 578–589. 10.1111/pbi.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Ito Y., Serizawa A., Kurata N. (2003). OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 36 532–540. 10.1046/j.1365-313X.2003.01897.x [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 1042–1054. 10.1104/pp.108.126342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers Z. A., Kumimoto R. W., Siriwardana C. L., Gayler K. K., Risinger J. R., Pezzetta D., et al. (2016). NUCLEAR FACTOR Y, Subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 12:e1006333. 10.1371/journal.pgen.1006333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Shinwari Z. K., Sakuma Y., Seki M., Miura S., Shinozaki K., et al. (2000). Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 42 657–665. 10.1023/A:1006321900483 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32 959–970. 10.1007/s00299-013-1418-1 [DOI] [PubMed] [Google Scholar]
- Ni Z., Hu Z., Jiang Q., Zhang H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82 113–129. 10.1007/s11103-013-0040-5 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Cheng H. Y., Chen C., Yu X. W., Yang J. N., Gao W. R., et al. (2009). A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 166 1934–1945. 10.1016/j.jplph.2009.05.013 [DOI] [PubMed] [Google Scholar]
- Qiu Q. S., Guo Y., Dietrich M. A., Schumaker K. S., Zhu J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A 99 8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Suzuki T., Takahashi F., Shinozaki K., Yamaguchi-Shinozaki K. (2019). NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 180 1677–1690. 10.1104/pp.19.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson T. J., Mcintyre C. L., Collet C., Xue G. P. (2011). TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct. Integr. Genomics 11 327–340. 10.1007/s10142-011-0212-9 [DOI] [PubMed] [Google Scholar]
- Sussmilch F. C., Brodribb T. J., Mcadam S. A. M. (2017). Up-regulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. J. Exp. Bot. 68 2913–2918. 10.1093/jxb/erx124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao N. P., Tran L. S. (2012). Potentials toward genetic engineering of drought-tolerant soybean. Crit. Rev. Biotechnol. 32 349–362. 10.3109/07388551.2011.643463 [DOI] [PubMed] [Google Scholar]
- Tian W., Hou C. C., Ren Z. J., Pan Y. J., Jia J. J., Zhang H. W., et al. (2015). A molecular pathway for CO2 response in Arabidopsis guard cells. Nat. Commun. 6:6057. 10.1038/ncomms7057 [DOI] [PubMed] [Google Scholar]
- Uchida A., Jagendorf A. T., Hibino T., Takabe T., Takabe T. (2002). Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 163 515–523. 10.1016/S0168-9452(02)00159-0 [DOI] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21 3170–3184. 10.1105/tpc.109.069179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhang W. X., Qin M. Y., Li S., Qiao M., Liu Z. H., et al. (2017). Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a Novel R2R3-MYB transcription factor. Plant Cell Physiol. 58 1764–1776. 10.1093/pcp/pcx111 [DOI] [PubMed] [Google Scholar]
- Wang X. Y., Huang W. L., Liu J., Yang Z. M., Huang B. R. (2017). Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 15 237–248. 10.1111/pbi.12609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li T., John S. J., Chen M., Chang J., Yang G., et al. (2018). A CBL-interacting protein kinase TaCIPK27 confers drought tolerance and exogenous ABA sensitivity in transgenic Arabidopsis. Plant Physiol. Biochem. 123 103–113. 10.1016/j.plaphy.2017.11.019 [DOI] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153 1747–1758. 10.1104/pp.110.156943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18 2971–2984. 10.1105/tpc.106.043299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. L., Jiang J. F., Ge L., Xu Y. Y., Chen H., Zhao Y., et al. (2005). FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep. 24 79–85. 10.1007/s00299-004-0906-8 [DOI] [PubMed] [Google Scholar]
- Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60 3781–3796. 10.1093/jxb/erp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liting Wang J. X., Si L., Zheng S., Yang Y., Yang H., et al. (2020). Wheat TabZIP8, 9, 13 participate in ABA biosynthesis in NaCl-stressed roots regulated by TaCDPK9-1. Plant Physiol. Biochem. 151 650–658. 10.1016/j.plaphy.2020.03.039 [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhuang K., Wang S., Lv J., Ma N., Meng Q. (2017). A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco. J. Integr. Plant Biol. 59 102–117. 10.1111/jipb.12514 [DOI] [PubMed] [Google Scholar]
- Zhang W., Liao X. L., Cui Y. M., Ma W. Y., Zhang X. N., Du H. Y., et al. (2019). A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 15:e1007798. 10.1371/journal.pgen.1007798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Z., Zheng W. J., Cao X. Y., Cui X. Y., Zhao S. P., Yu T. F., et al. (2019). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and Soybean. Front. Plant Sci. 10:1453. 10.3389/fpls.2019.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. Y., Tian A. G., Zou H. F., Xie Z. M., Lei G., Huang J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6 486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- Zhou Y. B., Liu C., Tang D. Y., Yan L., Wang D., Yang Y. Z., et al. (2018). The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30 1100–1118. 10.1105/tpc.17.01000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The synthetic fragment RNAi-GmNFYA13. The hairpin structure contained three parts: the positive sequence in blue, the reverse complementary sequence in green and the intron of GmNFYA13 in purple.
The sequence alignment and phylogenetic analysis of GmNFYA13 and ten NF-YA genes in Arabidopsis. (A) Sequence alignment of GmNFYA13 and ten NF-YA genes in Arabidopsis with conserved domains highlighted in black. The linker and two subdomains are underlined. Critical amino acids were indicated by asterisks. (B) Phylogenetic analysis of GmNFYA13 and ten NF-YA genes in Arabidopsis. MEGA 7.0 was used to construct the neighbor joining tree.
Germination rate of WT and 35S:GmNFYA13 transgenic Arabidopsis lines under treatment of control, salt, drought and ABA. (A) Germination rates on 1/2 MS medium. (B–C) Germination rates on 1/2 MS medium with 50 and 80 mM NaCl. (D–E) Germination rates on 1/2 MS medium with 8 and 10% PEG6000. (F–G) Germination rates on 1/2 MS medium with 0.5 and 0.8 μM ABA. Asterisks represent significant differences at P < 0.05 in comparison to the corresponding controls. Each experiment had three biological replicates.
Stomatal apertures of the leaves in WT and 35S:GmNFYA13 transgenic Arabidopsis lines were measured under control and ABA treatments. (A) Stomatal apertures subjected to 0 and 5 μM ABA solution. (B) The ruler tool developed in Adobe Photoshop CS5 was used to measure width/length of the stomatal aperture with 0, 5, and 10 μM ABA. Data indicate three biological replicates ± SD. Different letters above the columns represent significant differences at P < 0.05.
The transcript levels of GmNFYA13 in WT and 35S:GmNFYA13 lines were evaluated with RT-PCR. AtTub8 was the internal control.
The primer sets and gene accessions for qRT-PCR used in this study.
Data Availability Statement
Soybean gene sequences in this study were downloaded from the plant genomics resource (https://phytozome.jgi.doe.gov/pz/portal.html). Arabidopsis gene sequences in this study were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). The gene accessions are listed in Supplementary Table 1.