Graphical Abstract
We report here the first instance of nucleic acid nanostructures that contain 2′−5′ linkages and characterize structures of different complexities ranging from a simple duplex to a 4-arm junction, a double crossover (DX) motif and a tensegrity triangle motif.
In the Marvel superhero universe, Tony Stark builds the Iron Man suit and becomes a superhero. The power of the suit comes not from a single function but from its many tools and functionalities, which Stark continuously upgrades to increase his power. We can draw analogy to this in DNA nanotechnology, where a growing list of molecular tools and biomedical functionalities continue to add power to this emerging field. So far, scientists have created a wide variety of nanostructures using DNA,1–3 with applications in biosensing,4 molecular computation,5 macromolecular scaffolding,6 biomolecular analysis,7 and in drug delivery.8 While many different assembly strategies have been optimized over the years to construct nucleic acid nanostructures, there is still ongoing work to functionalize these structures to make them better suited for real life applications. For example, modification of strand terminus with chemical groups (for example, biotin, thiol or click chemistry groups) allow the attachment of nanoparticles,9 or proteins.10 These functionalities are useful in biosensing (for optical readout in the case of nanoparticles)9 or in the creation of enzyme cascades11 and synthetic vaccines (for antigens and proteins).12 In drug delivery, incorporation of chemical groups to attach peptides provide cell targeting13 or drug encapsulation14 while photocleavable linkers are useful in triggered release.15 Modified DNA or RNA nucleotides or backbones have also been reported,16 including the use of L-DNA,17 locked nucleic acids (LNA),18 peptide nucleic acids (PNA)19 and incorporation of unnatural base pairs such as 5-Me-isoC, isoG and 2-thioT.20 A prevalent use of such modifications is to enhance the stability of nucleic acid nanostructures21 and provide additional nuclease resistance20 so that the structures do not degrade easily in physiological conditions. This increasing library of available modifications is a useful resource for researchers in the field to create and functionalize nucleic acid nanostructures for specific applications.22 Adding to this list, we report here for the first time, the incorporation of 2′−5′ linkages in nucleic acid nanostructures. We characterized the tolerance of backbone heterogeneity in the assembled structures and discuss potential use of 2′−5′ linkages in enhancing the stability of such structures against ribonucleases.
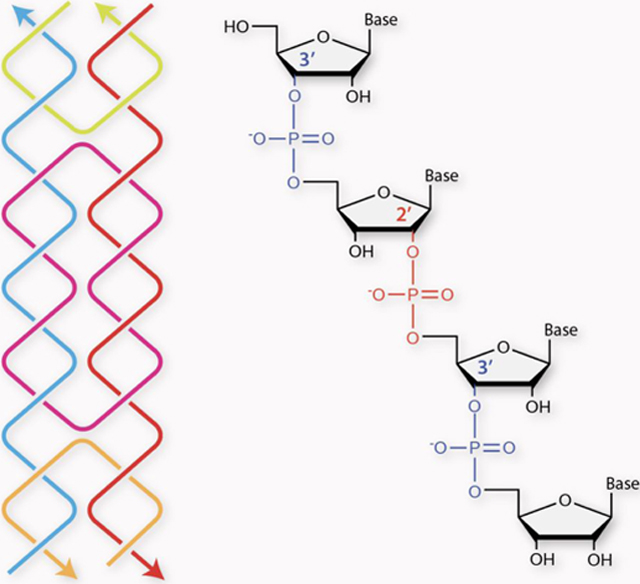
Although most RNAs have 3′−5′ linked phosphate backbone, the 2′−5′ phosphodiester bond also exists in nature and plays critical roles in many biological systems such as mRNA processing,23 neurodegenerative diseases24 and in viral replication25 (Figure 1). Further, 2′−5′ linked oligonucleotides have potential use in antisense therapeutics due to their resistance to enzymatic hydrolysis,26 diminished immune-stimulatory response27 and reduced nonspecific binding to plasma and cellular proteins.28 Other than the therapeutic applications, 2′−5′ linked RNAs have also emerged as a plausible prebiotic RNA analogue and been regarded as a well-conserved chemical relic from evolution.29 In addition, RNA aptamers and ribozymes with mixtures of 3′−5′ and 2′−5′ linkages retain their ligand binding ability and catalytic activity,29 indicating that these systems are very flexible to accommodate certain numbers of 2′−5′ linkages.
Figure 1. 3′−5′-linked vs 2′−5′-linked RNA.

Structures of single stranded (a) 3′−5′-linked “regular” RNA, (b) 2′−5′-linked RNA and (c) RNA with mixed linkages (3′−5′ linkages shown in blue and 2′−5′ linkages shown in red).
Tools in structural biology have provided insights into the conformation of 2′−5′-linked RNA within the context of RNA duplexes. In previous work, we solved the first two crystal structures for a self-complementary RNA 10-mer duplex containing two and six 2′−5′ linkages respectively.30 These data, along with molecular simulation work, provided structural insights into the conformation of 2′−5′-linked nucleotides and how RNA duplexes adjust their global and local structures to accommodate these mixed backbones through flexible sugar puckers and the flanking nucleotides compensation effect.30 Following this work, we also reported two additional high-resolution structures of the same RNA duplex containing four and eight 2′−5′ linkages at different positions, providing a dynamic view of RNA structural changes with increased numbers of 2′−5′ linkages.31 Despite this research progress, incorporation of 2′−5′-linked ribonucleotides in a DNA/RNA hybrid duplex has not been studied so far, and their structural and functional compatibility in the context of nucleic acid nanostructures have not been explored. In this study, we investigated the effect of 2′−5′ linkages on hybrid DNA/RNA assemblies using four different structures of increasing complexity: a simple duplex, a four-arm junction (one branch point), a double crossover (DX) motif (two branch points) and a tensegrity triangle motif (three branch points) (Figure 2a). We analyzed the assembly of these structures through non-denaturing polyacrylamide gel electrophoresis (PAGE), characterized the assembled motifs using UV melting studies and circular dichroism (CD), and revealed their dynamic conformational changes through molecular dynamics simulations.
Figure 2. Analysis of 2′−5′ linkages in DNA/RNA hybrid structures.

(a) Nucleic acid structures used in this study. A duplex (no branch points), a 4-arm junction (single branch point), a double crossover (DX) motif (two branch points) and a tensegrity triangle motif (each vertex of the triangle is a junction, thus three branch points). Each column shows results from (b) duplex, (c) 4-arm junction, (d) DX motif and (e) tensegrity triangle. In each case, the figure shows (i) Scheme of the structures with the RNA strand in red and the four 2′−5′ linkage positions indicated as yellow circles, (ii) results from native PAGE, and (iii) CD spectra of the assembled structures. 1L to 4L = one to four 2′−5′ linkages.
We first synthesized RNA strands with different numbers of 2′−5′ linkages through solid phase synthesis using commercially available phosphoramidites and purified them using denaturing PAGE (see supporting information and Table S1). We first tested DNA/RNA hybrid duplexes where the RNA strand contains zero to four 2′−5′ linkages (Figure 2b-i, RNA strand shown in red). We validated their formation on native PAGE (Figure 2b-ii, full gels in Figure S1) and characterized conformational changes in the hybrid duplexes using CD (Figure 2b-iii). The CD spectra showed clear transition from B-form duplex for the DNA/DNA to A-form for the DNA/RNA (evident by a dominant positive band at 260 nm and a negative band at 210 nm) and showed similar spectra for the hybrid duplex containing RNA with one to four 2′−5′ linkages. We then analyzed the thermal stability of the hybrid duplexes using UV melting studies (Figure S2). Thermal melting profiles showed that the melting temperatures of the hybrid duplexes slightly decreased as the number of 2′−5′ linkages in the component RNA strand increased. These results collectively indicate that formation of the hybrid DNA/RNA duplex did not seem to be affected by the heterogeneity of 2′−5′ and 3′−5′ linkages in the RNA strand.
Moving to motifs related to nanotechnology, we tested DNA structures containing strand crossovers, starting with a 4-arm junction and a double crossover (DX) motif. We characterized formation of the 4-arm junction containing 2′−5′-linked RNA using non-denaturing PAGE and CD, showing that the structure tolerated incorporation of 2′−5′ linkages (Figure 2c and Figure S1, S3). Our third system was a DX motif that contains 5 strands folded into two adjacent double helical domains connected by two crossovers (Figure 2d-i and Figure S4).32 DNA/RNA hybrid DX molecules33 and all-RNA DX molecules34 have previously been reported, and DX-based two dimensional arrays have been used to study the helicity of duplex domains with modified nucleotides.35,36 We modified one of the non-crossover strands to be RNA with 2′−5′ linkages (red strand in Figure 2d-i) and tested proper assembly of the motif using native PAGE (Figure 2d-ii, full gel in Figure S1). The formation of many such DNA motifs is based on strict stoichiometry, and in some cases the purity of the component strands. Similar motifs have been characterized using non-denaturing PAGE and other methods in many other studies15,37–41 and the presence of higher and lower molecular weight bands in some instances are known to indicate the formation of dimers and improperly folded products, respectively. The presence of the target band (corresponding to the structure) in high yield is indicative of proper assembly of these structures. We then characterized the hybrid assemblies using CD (Figure 2d-iii) and tested the thermal stability of the motif with increasing numbers of 2′−5′ linkages (Figure S2). The melting temperature of the DX motif containing an RNA strand (~51 °C) was lower than an all-DNA complex (~57 °C), possibly due to the change in helical parameters around the two crossover points. The melting temperature of the motif decreased slightly with the incorporation of 2′−5′ linkages but did not exhibit much variation for the complexes containing one to four 2′−5′ linkages (ranging from ~50 °C to ~54 °C).
Our final model structure was the tensegrity triangle motif, previously used in assembly of 2D lattices42 and designed 3D crystals.43 The motif consists of three duplex edges connected pairwise by strand crossovers (Figure 2e-i). The edges of the motif are in an over-and-under fashion, allowing it to be used in 3D construction (model shown in Figure 2a). In construction of 3D crystals, symmetric versions of these triangle motifs yielded higher resolution;43–45 thus, we first tested incorporation of 2′−5′-linked RNA into a symmetric 3-turn tensegrity triangle motif (with each edge containing 31 base pairs). The symmetric version is constructed using 3 unique strands in the ratio 3:3:1 where the three duplex edges all have the same sequence identity (Figure S5). We first confirmed formation of the symmetric tensegrity triangle on a non-denaturing PAGE (Figure S6). We then modified the non-crossover strand to be RNA with 2′−5′ linkages. Since the motif is symmetric, modifications will occur on all three edges when this strand is modified. Non-denaturing PAGE showed that the motifs do not form with an RNA strand containing multiple 2′−5′ linkages (Figure S7). To test the influence of symmetry, we then tested an asymmetric design of the tensegrity triangle motif. In this version, the motif is constructed using 7 unique strands where all the edges have different sequences (Figure S5). We modified the sequences of one of the edges of a triangle reported previously46 so that the sequence of the modified RNA strand is the same in both the symmetric and asymmetric designs. We confirmed successful assembly of this altered design on a non-denaturing PAGE (Figure S6). We then incorporated an RNA strand on one of the edges (shown in red in Figure 2e-i, sequences in Figure S5). We checked assembly of the motif with the RNA strand as well as RNA containing one to four 2′−5′ linkages, and non-denaturing PAGE showed proper assembly of all the motifs (Figure 2e-ii, full gels in Figure S1). The asymmetric motif is modified only on one edge (i.e. one out of seven strands is modified) and thus tolerates the 2′−5′ linkages better than the symmetric motif (three out of seven strands modified). CD spectra showed that the positive band at 260 nm is not as dominant as in other structures containing 2′−5′ linkages, indicating a predominantly B-form structure, an expected result since only one out of seven strands is changed to RNA (Figure 2e-iii).
The effects of incorporating 2′−5′ linkages in a DNA/RNA hybrid structure has not been studied before. To understand the effect of these modifications on structural and dynamics of DNA/RNA hybrid duplexes, we simulated the DNA/DNA and hybrid DNA/RNA duplexes containing zero to four 2′−5′ linkages (Figure 3). We performed simulations of length >36 ns, each consisting of 420061 atoms on average (418127, 424681, 418017, 418041, 419981, 421521 for DNA/DNA and DNA/RNA with zero to four 2′−5′ linkages, respectively) including explicit solvent and K+/Cl− ions. All the DNA/RNA duplexes were observed to be stable on the simulated timescales and maintained their overall helical structures with well-defined base pairing and base stacking. Root-mean-square deviations (RMSD) of the trajectories with respect to their initial starting structures show that the initial DNA/RNA duplex structures, even containing 2′−5′ linkages, remain rigidly folded throughout the simulations (Figure S8). RMSD values plateau after 10 ns indicating a rapid equilibration of local bending fluctuations at the modification site. Each trajectory was then structurally clustered excluding the initial 10 ns equilibration period; the centroid of each simulation’s highest populated cluster is shown in Figure 3a. In all cases, a single, highly populated cluster containing >70% of all frames was observed (Figure 3b and Figure S9).
Figure 3. Molecular dynamics simulations of 2’−5’-linked DNA/RNA hybrid duplexes.

(a) Superposition of centroids of all the structures analyzed. (b) Simulated structures of each variation, with the DNA strand shown in grey (constant in all the structures). RNA strands with none to four 2′−5′ linkages are shown in different colors for clarity. (c) Interhelical bending angle distribution of the simulated structures, color coded as in (b). 1L to 4L = one to four 2′−5′ linkages.
To further understand how these modifications affect the structural dynamics of duplexes, we analyzed the helical bending angle distribution within the main cluster of each duplex over the course of the trajectory (Figure 3c). This analysis showed a steady increase in bending dynamics when one strand of DNA is replaced with RNA and further increases with the addition of 2′−5′ modifications (increase in standard deviation of bending angle values in Figure 3c). Superposing all six centroid structures illustrates the gradually increasing amounts of helical bending as a function of the 2′−5′ linkage numbers (Figure 3a). We then used the 3DNA structural analysis package47 to measure how 2′−5′ linkages affect helix structural parameters of these DNA/RNA duplexes. This analysis revealed a transition from predominantly B-form helices in the DNA/DNA duplex to partially A-form in the DNA/RNA duplex irrespective of the presence of 2′−5′ linkages (Figure S10), in agreement with the experimentally observed CD spectra (Figure 2b-iii). This data is also in agreement with our experiments where the addition of more 2′−5′ linkages in the structure cause a monotonic decrease in melting temperatures (Figure S2). Increased availability of structural parameters and crystal studies of DNA motifs might provide additional functions to simulate more complex DNA nanostructures than just the duplexes studied here.
Since RNA strands with 2′−5′ linkages are known to resist digestion by ribonucleases (RNases),26,28,48 we tested whether these modifications confer RNase protection to DNA/RNA hybrid duplex structures (Figure 4a). We incubated the native and 2′−5′-linked hybrid duplexes with different amounts of RNase H and analyzed the samples using non-denaturing PAGE.49 When incubated with 0.5 units RNase H for 16 minutes, the modified structure showed lower levels of degradation compared to the native structure (Figure 4b and Figure S11). At a lower enzyme concentration (0.1 unit), the duplex with RNA containing four 2′−5′-linkages was almost fully intact while the duplex with unmodified RNA showed higher levels of degradation. We also monitored the digestion of native and 2′−5′-linked structures over a time course, showing that the modified structure is more resistant to RNase digestion than its native counterpart (Figure 4c).
Figure 4. Testing ribonuclease activity on 2′−5′-linked structures.

(a) Native and modified duplexes with four 2′−5′ linkages were tested for resistance against RNase H. (b) Percent structure intact after 16-min incubation with different amounts of RNase H. (c) Digestion trend of native and modified duplexes with 0.5 units RNase H.
This study is the first report on incorporating RNA strands with 2′−5′ linkages into nucleic acid nanostructures. We have shown that 2′−5′ linkages have minimal influence on the assembly of hybrid duplexes, a 4-arm branched junction and a DX motif. In a larger tensegrity triangle motif, incorporation of one modified strand does not influence the structure. However, incorporation of 2′−5′ linkages in a symmetric design where all three edges are modified affects self-assembly of the motif. Future studies on incorporation of such linkages in crossover strands (compared to the non-crossover strands presented here) will also provide useful information on structural effects. Further, the number of 2′−5′ linkages incorporated into the structures could also play a role in the assembly process, depending on the sequence contexts.27,31,50 Previous studies have shown that RNA/RNA or DNA/DNA duplexes are more stable than hybrid RNA/2′−5′ linked RNA or DNA/2′−5′ linked RNA.48 Further, a fully 2′−5′-linked RNA strand preferably binds with RNA than DNA strands.51 Thus, we think the use of a fully 2′−5′-linked strand in nucleic acid nanostructures might interfere with the assembly of the motifs to certain extent, but can be overcome by design optimization. Here, we have shown here that hybrid DNA/RNA duplexes tolerate the incorporation of quite a few 2′−5′ linked nucleotides, which would be sufficient for ideal functions of nanostructures such as enhanced RNase resistance and binding of small molecules ligands. From synthesis costs and function points of view, our current design of ~10% of 2′−5′ links in a strand is representative of the future applications.
Addition of such unusual linkages into nucleic acid nanostructures might provide additional features for certain applications. For example, mixture of 3′−5′/2′−5′ linkages have previously been tested in RNA aptamers and ribozymes,29 thus opening up the possibility of using such functionalities within nucleic acid nanostructures. As mentioned above, 2′−5′-linked RNA oligonucleotides have special binding specificity to single stranded RNA over single stranded DNA, and can discriminate between DNA and RNA.51 This feature could be exploited in future nucleic acid nanomachines where DNA or RNA-preferred strand displacement reactions can drive chemical reactions or trigger release of compounds. In addition, it is known that single stranded RNA with 2′−5′ linkages, phosphorothioate or phosphate chimeras resist enzymatic hydrolysis.26 Preliminary ribonuclease assays in this study show that 2′−5′-linked oligonucleotides can potentially be used in DNA/RNA objects that can withstand the effect of RNases better than native structures. Construction of such modified nanostructures could provide new platforms for drug delivery with better stability and functionality. For the tensegrity triangle motif tested here, one of the main applications (and the original notion of the field of DNA nanotechnology) is in the creation of designed DNA crystals that can act as a framework for the crystallization of other molecules.52 These 3D crystals have previously been used to host triplex forming oligonucleotides53 and polyaniline,54 and used to study the crystal structure and effect of torsional strain induced by base pair addition or deletion.55 In that context, understanding the tolerance of 2′−5′ linkages in this motif could lead to designed DNA crystals that can act as scaffolds for structural analysis of such unusual linkages and modifications.
Supplementary Material
FUNDING
Research reported in this publication was supported by the National Institutes of Health (NIH) through NIGMS under award R35GM124720 to K.H; R35GM133469 to AAC; R15GM124627 to J.S. and by the National Science Foundation (NSF) under award CHE1845486 to J.S. and MCB1651877 to AAC.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to declare.
REFERENCES
- 1.Seeman NC, DNA in a material world, Nature, 2003, 421, 427–431. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekaran AR, Anderson N, Kizer M, Halvorsen K and Wang X, Beyond the Fold: Emerging Biological Applications of DNA Origami, ChemBioChem, 2016, 17, 1081–1089. [DOI] [PubMed] [Google Scholar]
- 3.Xavier PL and Chandrasekaran AR, DNA-based construction at the nanoscale: emerging trends and applications, Nanotechnology, 2018, 29, 062001. [DOI] [PubMed] [Google Scholar]
- 4.Xiao M, Lai W, Man T, Chang B, Li L, Chandrasekaran AR and Pei H, Rationally Engineered Nucleic Acid Architectures for Biosensing Applications, Chem. Rev, 2019, 119, 11631–11717. [DOI] [PubMed] [Google Scholar]
- 5.Rangnekar A and LaBean TH, Building DNA Nanostructures for Molecular Computation, Templated Assembly, and Biological Applications, Acc. Chem. Res, 2014, 47, 1778–1788. [DOI] [PubMed] [Google Scholar]
- 6.Yang YR, Liu Y and Yan H, DNA Nanostructures as Programmable Biomolecular Scaffolds, Bioconjugate Chem, 2015, 26, 1381–1395. [DOI] [PubMed] [Google Scholar]
- 7.Rajendran A, Endo M and Sugiyama H, Single-Molecule Analysis Using DNA Origami, Angewandte Chemie International Edition, 2012, 51, 874–890. [DOI] [PubMed] [Google Scholar]
- 8.Madhanagopal BR, Zhang S, Demirel E, Wady H and Chandrasekaran AR, DNA Nanocarriers: Programmed to Deliver, Trends in Biochemical Sciences, 2018, 43, 997–1013. [DOI] [PubMed] [Google Scholar]
- 9.Bai S, Xu B, Guo Y, Qiu J, Yu W and Xie G, High-Discrimination Factor Nanosensor Based on Tetrahedral DNA Nanostructures and Gold Nanoparticles for Detection of MiRNA-21 in Live Cells, Theranostics, 2018, 8, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh AS, Yin H, Erben CM, Wood MJA and Turberfield AJ, DNA Cage Delivery to Mammalian Cells, ACS Nano, 2011, 5, 5427–5432. [DOI] [PubMed] [Google Scholar]
- 11.Fu J, Liu M, Liu Y, Woodbury NW and Yan H, Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures, J. Am. Chem. Soc, 2012, 134, 5516–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H and Chang Y, A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines, Nano Lett, 2012, 12, 4254–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Wu L, Wang L and Jiang W, A dual-targeting DNA tetrahedron nanocarrier for breast cancer cell imaging and drug delivery, Talanta, 2018, 179, 356–363. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Tian C, Guo F, Liu Z, Jiang W and Mao C, DNA-Directed Three-Dimensional Protein Organization, Angew. Chem. Intl. Ed, 2012, 51, 3382–3385. [DOI] [PubMed] [Google Scholar]
- 15.Valsangkar VA, Chandrasekaran AR, Zhuo L, Mao S, Lee GW, Kizer M, Wang X, Halvorsen K and Sheng J, Click and photo-release dual-functional nucleic acid nanostructures, Chem. Commun, 2019, 55, 9709–9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinheiro VB and Holliger P, Towards XNA nanotechnology: new materials from synthetic genetic polymers, Trends in Biotechnology, 2014, 32, 321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K-R, Kim HY, Lee Y-D, Ha JS, Kang JH, Jeong H, Bang D, Ko YT, Kim S, Lee H and Ahn D-R, Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs, Journal of Controlled Release, 2016, 243, 121–131. [DOI] [PubMed] [Google Scholar]
- 18.Olson X, Kotani S, Yurke B, Graugnard E and Hughes WL, Kinetics of DNA Strand Displacement Systems with Locked Nucleic Acids, J. Phys. Chem. B, 2017, 121, 2594–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flory JD, Shinde S, Lin S, Liu Y, Yan H, Ghirlanda G and Fromme P, PNA-Peptide Assembly in a 3D DNA Nanocage at Room Temperature, J. Am. Chem. Soc, 2013, 135, 6985–6993. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Liu G, Wang T, Fu J, Li R, Song L, Wang Z-G, Ding B and Chen F, Enhanced Stability of DNA Nanostructures by Incorporation of Unnatural Base Pairs, ChemPhysChem, 2017, 18, 2977–2980. [DOI] [PubMed] [Google Scholar]
- 21.Conway JW, McLaughlin CK, Castor KJ and Sleiman H, DNA nanostructure serum stability: greater than the sum of its parts, Chem. Commun, 2013, 49, 1172–1174. [DOI] [PubMed] [Google Scholar]
- 22.Madsen M and Gothelf KV, Chemistries for DNA Nanotechnology, Chem. Rev, 2019, 119, 6384–6458. [DOI] [PubMed] [Google Scholar]
- 23.Murphy WJ, Watkins KP and Agabian N, Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: Evidence for Trans splicing, Cell, 1986, 47, 517–525. [DOI] [PubMed] [Google Scholar]
- 24.Kwong LK, Neumann M, Sampathu DM, Lee VM-Y and Trojanowski JQ, TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease, Acta Neuropathol, 2007, 114, 63–70. [DOI] [PubMed] [Google Scholar]
- 25.Côté F, Lévesque D and Perreault J-P, Natural 2′,5′-Phosphodiester Bonds Found at the Ligation Sites of Peach Latent Mosaic Viroid, Journal of Virology, 2001, 75, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhan P, Bhan A, Hong M, Hartwell JG, Saunders JM and Hoke GD, 2′,5′-Linked oligo-3′-deoxyribonucleoside phosphorothioate chimeras: thermal stability and antisense inhibition of gene expression, Nucleic Acids Res, 1997, 25, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habibian M, Harikrishna S, Fakhoury J, Barton M, Ageely EA, Cencic R, Fakih HH, Katolik A, Takahashi M, Rossi J, Pelletier J, Gagnon KT, Pradeepkumar PI and Damha MJ, Effect of 2′−5′/3′−5′ phosphodiester linkage heterogeneity on RNA interference, Nucleic Acids Res, 2020, 48, 4643–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kandimalla ER, Manning A, Zhao Q, Shaw DR, Byrn RA, Sasisekharan V and Agrawal S, Mixed backbone antisense oligonucleotides: design, biochemical and biological properties of oligonucleotides containing 2′–5′-ribo- and 3′–5′-deoxyribonucleotide segments, Nucleic Acids Res, 1997, 25, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelhart AE, Powner MW and Szostak JW, Functional RNAs exhibit tolerance for non-heritable 2′–5′ versus 3′–5′ backbone heterogeneity, Nature Chemistry, 2013, 5, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng J, Li L, Engelhart AE, Gan J, Wang J and Szostak JW, Structural insights into the effects of 2′−5′ linkages on the RNA duplex, PNAS, 2014, 111, 3050–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen F, Luo Z, Liu H, Wang R, Zhang S, Gan J and Sheng J, Structural insights into RNA duplexes with multiple 2΄−5΄-linkages, Nucleic Acids Res, 2017, 45, 3537–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu TJ and Seeman NC, DNA double-crossover molecules, Biochemistry, 1993, 32, 3211–3220. [DOI] [PubMed] [Google Scholar]
- 33.Ko SH, Su M, Zhang C, Ribbe AE, Jiang W and Mao C, Synergistic self-assembly of RNA and DNA molecules, Nature Chemistry, 2010, 2, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart JM, Subramanian HKK and Franco E, Self-assembly of multi-stranded RNA motifs into lattices and tubular structures, Nucleic Acids Res, 2017, 45, 5449–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukeman PS, Mittal AC and Seeman NC, Two dimensional PNA/DNA arrays: estimating the helicity of unusual nucleic acid polymers, Chem. Commun, 2004, 1694–1695. [DOI] [PubMed] [Google Scholar]
- 36.Rinker S, Liu Y and Yan H, Two-dimensional LNA/DNA arrays: estimating the helicity of LNA/DNA hybrid duplex, Chem. Commun, 2006, 2675–2677. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W and Mao C, Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra, Nature, 2008, 452, 198–201. [DOI] [PubMed] [Google Scholar]
- 38.Huang K, Yang D, Tan Z, Chen S, Xiang Y, Mi Y, Mao C and Wei B, Self-Assembly of Wireframe DNA Nanostructures from Junction Motifs, Angew. Chem. Intl. Ed, 2019, 58, 12123–12127. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Chen Y, Liu H, Ribbe AE and Mao C, Self-Assembly of Hexagonal DNA Two-Dimensional (2D) Arrays, J. Am. Chem. Soc, 2005, 127, 12202–12203. [DOI] [PubMed] [Google Scholar]
- 40.Chandrasekaran AR and Halvorsen K, Controlled disassembly of a DNA tetrahedron using strand displacement, Nanoscale Adv., 2019, 1, 969–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valsangkar V, Chandrasekaran AR, Wang R, Haruehanroengra P, Levchenko O, Halvorsen K and Sheng J, Click-based functionalization of a 2′-O-propargyl-modified branched DNA nanostructure, J. Mater. Chem. B, 2017, 5, 2074–2077. [DOI] [PubMed] [Google Scholar]
- 42.Liu D, Wang M, Deng Z, Walulu R and Mao C, Tensegrity: Construction of Rigid DNA Triangles with Flexible Four-Arm DNA Junctions, J. Am. Chem. Soc, 2004, 126, 2324–2325. [DOI] [PubMed] [Google Scholar]
- 43.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C and Seeman NC, From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal, Nature, 2009, 461, 74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sha R, Birktoft JJ, Nguyen N, Chandrasekaran AR, Zheng J, Zhao X, Mao C and Seeman NC, Self-Assembled DNA Crystals: The Impact on Resolution of 5′-Phosphates and the DNA Source, Nano Lett, 2013, 13, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohayon YP, Hernandez C, Chandrasekaran AR, Wang X, Abdallah HO, Jong MA, Mohsen MG, Sha R, Birktoft JJ, Lukeman PS, Chaikin PM, Ginell SL, Mao C and Seeman NC, Designing Higher Resolution Self-Assembled 3D DNA Crystals via Strand Terminus Modifications, ACS Nano, 2019, 13, 7957–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen N, Birktoft JJ, Sha R, Wang T, Zheng J, Constantinou PE, Ginell SL, Chen Y, Mao C and Seeman NC, The absence of tertiary interactions in a self-assembled DNA crystal structure, Journal of Molecular Recognition, 2012, 25, 494–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Olson WK and Lu X-J, Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures, Nucleic Acids Res, 2019, 47, W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasner M, Arion D, Borkow G, Noronha A, Uddin AH, Parniak MA and Damha MJ, Physicochemical and Biochemical Properties of 2’,5’-Linked RNA and 2’,5’-RNA:3’,5’-RNA “Hybrid” Duplexes, Biochemistry, 1998, 37, 7478–7486. [DOI] [PubMed] [Google Scholar]
- 49.Chandrasekaran AR and Halvorsen K, Nuclease degradation analysis of DNA nanostructures using gel electrophoresis, Current Protocols in Nucleic Acid Chemistry, 2020, 82, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kierzek R, He L and Turner DH, Association of 2’ - 5’ oligoribonucleotides, Nucleic Acids Res, 1992, 20, 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giannaris PA and Damha MJ, Oligoribonucleotides containing 2′,5′-phosphodiester linkages exhibit binding selectivity for 3′,5′-RNA over 3′,5′-ssDNA, Nucleic Acids Res, 1993, 21, 4742–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeman NC, Nucleic acid junctions and lattices, Journal of Theoretical Biology, 1982, 99, 237–247. [DOI] [PubMed] [Google Scholar]
- 53.Rusling DA, Chandrasekaran AR, Ohayon YP, Brown T, Fox KR, Sha R, Mao C and Seeman NC, Functionalizing Designer DNA Crystals with a Triple-Helical Veneer, Angew. Chem. Intl. Ed, 2014, 53, 3979–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Sha R, Kristiansen M, Hernandez C, Hao Y, Mao C, Canary JW and Seeman NC, An Organic Semiconductor Organized into 3D DNA Arrays by “Bottom-up” Rational Design, Angew. Chem. Intl. Ed, 2017, 56, 6445–6448. [DOI] [PubMed] [Google Scholar]
- 55.Hernandez C, Birktoft JJ, Ohayon YP, Chandrasekaran AR, Abdallah H, Sha R, Stojanoff V, Mao C and Seeman NC, Self-Assembly of 3D DNA Crystals Containing a Torsionally Stressed Component, Cell Chemical Biology, 2017, 24, 1401–1406.e2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


