Highlights
-
•
The glutamine utilization of KRAS-mutant NSCLC is higher than that of KRAS wild-type.
-
•
Targeted GLS1 and MEK inhibition enhance antitumor activity in vitro and in vivo.
-
•
The therapeutic response can be well identified by 18F-FDG PET imaging.
-
•
Dual inhibition of GLS1 and MEK induce redox and energetic stress.
-
•
Dual inhibition of GLS1 and MEK suppress the phosphorylation of AKT.
Keywords: KRAS, Glutaminase, Microenvironment, Autophagy
Abbreviations: NSCLC, non-small cell lung cancer; 18F-FDG, 18F-fluoro-2deoxyglucose; PET, positron emission tomography; 18F-Gln, 18F- (2S,4R)4-fluoroglutamine; GLS, glutaminase; IC50, the half maximal inhibitory concentration; CI, combination index; %ID/g, units of percent injected dose per gram; SUVmax, maximum standardized uptake value; PMR, partial metabolic remission
Abstract
Regulated by the tumor microenvironment, the metabolic network of the tumor is reprogrammed, driven by oncogenes and tumor suppressor genes. The metabolic phenotype of tumors of different driven-genes and different tissue types is extremely heterogeneous. KRAS-mutant non-small cell lung cancer (NSCLC) has glutamine dependence. In this study, we demonstrated that glutamine utilization of KRAS-mutant NSCLC was higher than that of KRAS wild-type. CB839, an efficient glutaminase inhibitor, synergized with the MEK inhibitor selumetinib to enhance antitumor activity in KRAS-mutant NSCLC cells and xenografts, and the therapeutic response could be well identified by 18F-FDG PET imaging. Combination therapy induced redox stress, manifesting as a decrease in mitochondrial membrane potential and an increase in ROS levels, and energetic stress manifesting as suppression of glycolysis and glutamine degradation. The phosphorylation of AKT was also suppressed. These effects combined to induce autophagy and thereby caused cancer cell death. Our results suggest that dual inhibition of the MEK-ERK pathway and glutamine metabolism activated by KRAS mutation may be an effective treatment strategy for KRAS-driven NSCLC.
Introduction
Uncontrolled uptake of glucose and amino acids is an important part of tumor metabolic reprogramming, which is a hallmark of cancer [1]. As a conditional non-essential amino acid, normal cells have no dependence on glutamine, while cancer cells, especially KRAS-mutant driven cancer cells [2], uptake glutamine frantically to meet bioenergetic and biosynthetic demands [3]. Compared with targeting glucose metabolism, targeting glutamine metabolism can inhibit cancer cell proliferation on the basis of reducing the impact on normal cells. Metabolism-based imaging such as 18F-fluoro-2deoxyglucose (18F-FDG) positron emission tomography (PET), an advanced method of evaluating tumor glucose metabolism in preclinical and clinical studies, has been used to evaluate the overall metabolic activity of tumors and the treatment response in the early stages [4], [5], [6]. Furthermore, glutamine metabolism imaging using 18F-(2S,4R)4-fluoroglutamine (18F-Gln) is under active preclinical investigation [7], [8], [9].
Beginning with glutaminase (GLS) catalyzed decomposition, glutamine provides nitrogen for the synthesis of nucleic acids, promotes the synthesis of antioxidant glutathione (GSH) to maintain redox homeostasis, and replenishes α-ketoglutarate (α-KG) for the tricarboxylic acid cycle (TCA) [3]. CB839 is an efficient GLS1 inhibitor, which can increase the radiation sensitivity of KRAS-driven NSCLC [10]. Furthermore, CB839 synergistically sensitized the BCL-2 inhibitor in leukemic cells through activation of mitochondrial apoptosis [11].
KRAS-driven cancer cells are addicted to glutamine [12,13], and in KRAS-driven NSCLC, the expression of GLS mRNA was higher than that of other mutant types [14]. Selumetinib, an inhibitor targeting the RAS-MEK-ERK pathway, significantly prolonged median progression-free survival (PFS) when combined with docetaxel in the phase 2 study [15]. Regrettably, the addition of selumetinib to docetaxel did not improve PFS compared with docetaxel alone in multinational phase 3 study [16]. There are still no specifically approved targeted therapies for this most common genetically defined subset. In preclinical study, pulsatile administration [17] or in combination with palbociclib [18], enhanced the anti-tumor effect of selumetinib in KRAS-driven NSCLC. MAPK inhibitor resistant cells showed more dependent on glutamine for growth and proliferation, selumetinib resistant melanoma cell lines had increased sensitivity to glutamine deprivation and glutaminase inhibitors than their sensitive counterparts [19].
To capitalize on the link between MEK signaling and glutamine metabolism in KRAS-mutant NSCLC, we evaluated the baseline glutamine metabolism in KRAS- mutant NSCLC, inhibited GLS1 to synergistically sensitize KRAS- mutant NSCLC to selumetinib in vitro and in vivo, and assessed therapeutic response by 18F-FDG PET imaging. Furthermore, we evaluated the redox and energetic stress induced by dual inhibition of MEK and GLS1, and preliminarily explored the mechanism of synergistic sensitivity.
Materials and methods
Cell culture
Human NSCLC KRAS-mutant cell lines A549, H23, H2122, KRAS wild-type cell line H1299, H838 and the human bronchial epithelial (HBE) cell line were obtained from the Stem Cell Bank, Chinese Academy of Sciences. A549 cells were cultured in DMEM/F12 1:1 medium (Hyclone, Logan, UT, USA), while H23, H2122,H1299, H838 and HBE cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Clark Bioscience, Houston, Texas, USA) at 37 °C and 5%CO2. In glutamine deprivation testing, NSCLC cells were cultured in RPMI-1640 without glutamine (Gibco, NY, USA) for 24 h.
Chemicals and antibodies
CB839 and selumetinib (AZD6244) were purchased from Selleckchem (Houston, TX, USA). For cellular studies, drugs were dissolved in dimethyl sulfoxide (DMSO), and for xenograft experiments, drugs were dissolved in 10% CMC—Na solution. The antibodies against GLS, Glut1, PKM2, MEK, ERK1/2, p-ERKT202+204, AKT, p-AKTs472–474were obtained from Abcam (Cambridge, UK). The antibodies against P62, p-ULK1s555 were obtained from Cell Signaling Technology (Danvers, MA, USA). The antibody against β-actin was obtained from Boster Biological Technology (Wuhan, China).
Cell viability and drug combination assay
Cells were cultured in 96-well plates (2000 cells/well) for 24 h, and were treated with the indicated concentrations (range from 10 to 105 nM)of CB839, selumetinib, or both drugs in combination for 72 h. Cell viability was measured using the Cell Counting Kit 8 (CCK8, Dojindo Laboratories, Tokyo, Japan) and microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) at the 450 nm wavelength. The half maximal inhibitory concentration (IC50) and 10% inhibitory concentration (IC10) were calculated by Probit model. Combination index (CI) data were obtained using CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA). CI values <1 indicated a synergistic drug-drug interaction, while CI values <0.3 indicated a strong synergistic effect.
Cell proliferation assay
After 72 h of drug treatment, cell proliferation was evaluated using the YF594 Click-iT EdU Imaging Kit (Everbright, California, USA) according to the manufacturer's protocol. For the colony formation assay, cells treated by different drugs were seeded into 6-well plates at a density of 400 cells per well. After culturing in the cell incubator (37 °C, 5% CO2) for 14 days, the cells were fixed in 70% ethanol and then stained with crystal violet.
Cell metabolism assay
To evaluate the changes in cell metabolism, cellular lactate, ATP, glutamate production, and glucose consumption were determined. According to the manufacturer's instructions, relative cellular ATP content was measured using an ATP assay kit (Beyotime, Shanghai, China), while the levels of lactate and glutamate in the external medium were measured using a lactic acid assay kit (KeyGEN, Nanjing, China) and a glutamate assay kit (Jiancheng, Nanjing, China). 18F-FDG cellular uptake can reflect the glucose consumption of cells. After exposure to 18F-FDG at a final concentration of 4 μCi/mL in medium at 37 °C and 5% CO2 for 60 min, the radioactivity of the whole-cell lysates was assessed by γ counter. These readouts were normalized by the amount of the corresponding protein.
Mitochondrial membrane potential and reactive oxygen species (ROS) assay
The mitochondrial membrane potential was evaluated using JC-1 Assay Kit (Beyotime, Shanghai, China) and images were analyzed by Image J. ROS levels were evaluated using ROS Assay Kit (Beyotime, Shanghai, China) and detected by a fluorescence microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). The readouts were normalized by the amount of cell numbers.
Cell transfection
The A549 cell line was transfected with small interfering RNA (siRNA) against KRAS or negative control(RiboBio, Guangzhou, China), denoted as siRNA KRAS, siRNA NC (siRNA sense: 3′-GGACGAATATGATCCAACA-5′).The H1299 cell line was transfected with the KRAS plasmid or pCMV6 Entry vector (RiboBio, Guangzhou, China), denoted as KRAS OE, KRAS NC. Transfection was performed using riboFECT™ CP Transfection Agent (RiboBio, Guangzhou, China) with a 50 nM concentration following the manufacturer's protocol.
Western blotting analysis
Total protein was extracted from cells using RIPA buffer (Beyotime, Shanghai, China) and protein concentration was measured by the BCA Protein Assay Kit (Beyotime, Shanghai, China). Protein lysates were separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were incubated with the primary antibodies at 4 °C overnight after blocking for 2 h with 5% skim milk. Following incubation with the secondary antibody (1:5000, ZSJQ Biosciences, Beijing, China), the blot signals were captured by Bio-Rad Image System using ECL Advanced Western Blot Detection Kit (Thermo Fisher Scientific, Waltham, MA, USA), and then analyzed using Image Lab software (Bio-Rad).
Xenograft experiments and PET imaging
Female BALB/c nude mice (5 week old) were injected subcutaneously into the right back with A549 cells or H1299 cells (1 × 107 cells). Tumor volume was calculated as: volume = length × (width)2/2. To evaluate the metabolic baseline levels of glutamine, 3 mice injected A549 cells and 3 mice injected H1299 cells underwent PET/MR (United imaging uPMR790, Shanghai, China) imaging with 18F-Gln and 18F-FDG on consecutive days. Coronal T1MR and static PET images were collected. To evaluate the efficacy of drugs, when tumor volumes reached approximately 200–300 mm3, the mice were randomly assigned to one of four groups (5 mice per group) and treated via oral gavage twice per day for six days as follows: control (vehicle alone, 10% CMC—Na solution), selumetinib (25 mg/kg), CB839 (200 mg/kg), selumetinib (25 mg/kg) + CB839 (200 mg/kg). Before and after therapy, mice underwent micro-PET (Madic, Shandong, China) imaging with 18F-FDG. Static PET images were collected. Mice were injected via lateral tail vein with radiolabeled probe (300 μCi per mouse), underwent 60 min uptake and image acquisition under 2% isoflurane anesthesia. Animal studies were in compliance with the Guide for the Care and Use of Laboratory Animal Resources (1996), National Research Council and approved by the China Medical University Animal Ethics Committee (IACUC Issue No.2019079). All procedures accepted the supervision and inspection by the committee and laboratory animal department.
Immunohistochemical analysis (IHC)
The following primary antibodies were used: GLS (1:100), Glut1 (1:200), MEK (1:100), ERK1/2 (1:100), p-ERKT202+204 (1:200), AKT (1:100), p-AKTs472–474 (1:100), P62 (1:1000). The mean of the integrated option density (IOD) was calculated using Image-Pro-Plus 6.0(Media Cybernetics, Inc., Bethesda, MD, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 19.0 (SPSS, Inc., Chicago, IL, USA). One-way ANOVA or the Student's t-test was used to measure differences between groups. CI were presented as median (P25, P75), other data were presented as mean±SD. A p-value<0.05 was considered statistically significant (*p<0.05, ⁎⁎p<0.01,⁎⁎⁎p<0.001).
Results
NSCLC with and without KRAS mutation showed differences in baseline glutamine metabolism
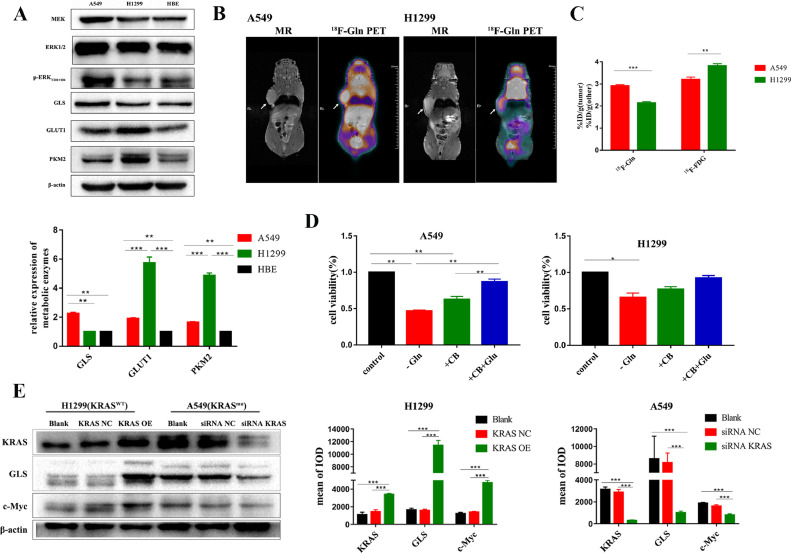
To confirm that NSCLC with KRAS mutation has glutamine addiction, we evaluated baseline glucose and glutamine metabolism in A549(KRAS-mutant), H1299(KRAS wild-type) and HBE cell lines. The expression of GLS, the key enzyme for glutamine degradation, in A549 cells was higher than that in H1299 cells, while the expression of enzymes related to glucose metabolism such as GLUT1 and PKM2 in A549 cells was lower than that in H1299 cells (Fig. 1A). We next performed coronal PET/MR imaging in A549 and H1299 mouse xenografts with 18F-FDG and 18F-Gln radiotracers on consecutive days and calculated units of percent injected dose per gram (%ID/g) (Fig. 1B-C, S1). The results demonstrated that A549 xenografts had higher glutamine and lower glucose uptake than H1299 xenografts. When glutamine was deprived (by culturing in medium without glutamine or treatment by CB839 for 24 h), cell proliferation of A549 was significantly inhibited, when glutamic acid was added, this effect could be rescued (Fig. 1D). Under identical culture conditions, KRAS was knockdown in A549 cell and overexpressed in H1299 cell. After the knockdown of KRAS in A549 cell, the expression of GLS1 was significantly decreased, while that was significantly increased in H1299 cell when KRAS was overexpression (Fig. 1E). As another crucial regulator of GLS1 expression, the change trend of c-Myc was consistent with that of GLS1 when KRAS was knockdown or overexpressed(Fig. 1E).
Fig. 1.
NSCLC with and without KRAS mutation showed differences in baseline glucose and glutamine metabolism. (A) The expression of metabolic key enzyme GLS, GLUT1, and PKM2 in A549, H1299 and HBE cells. (B) Representative PET/MR imaging of mice bearing A549 or H1299 xenografts with 18F-Gln(n = 3). Tumors are indicated by arrows. Scale bars:50 mm. (C) Quantification of 18F-FDG and 18F-Gln uptake using%ID/g. (D) Cell viability analysis following glutamine deprivation for 24 h. Cells were cultured in normal medium (control), in medium without glutamine (-Gln), or treated with CB839 (+CB,105nM), or with CB839 and glutamic acid (+CB+Glu). (E) Western blotting analysis to evaluate the expression of KRAS,GLS1 and c-Myc when KRAS was knockdown in A549 cell and overexpressed in H1299 cell. Data were presented as mean±SD from three independent experiments. A p-values<0.05 are considered statistically significant (*p<0.05, **p<0.01,***p<0.001).
CB839 synergized with selumetinib to inhibit proliferation of KRAS-mutant NSCLC cells
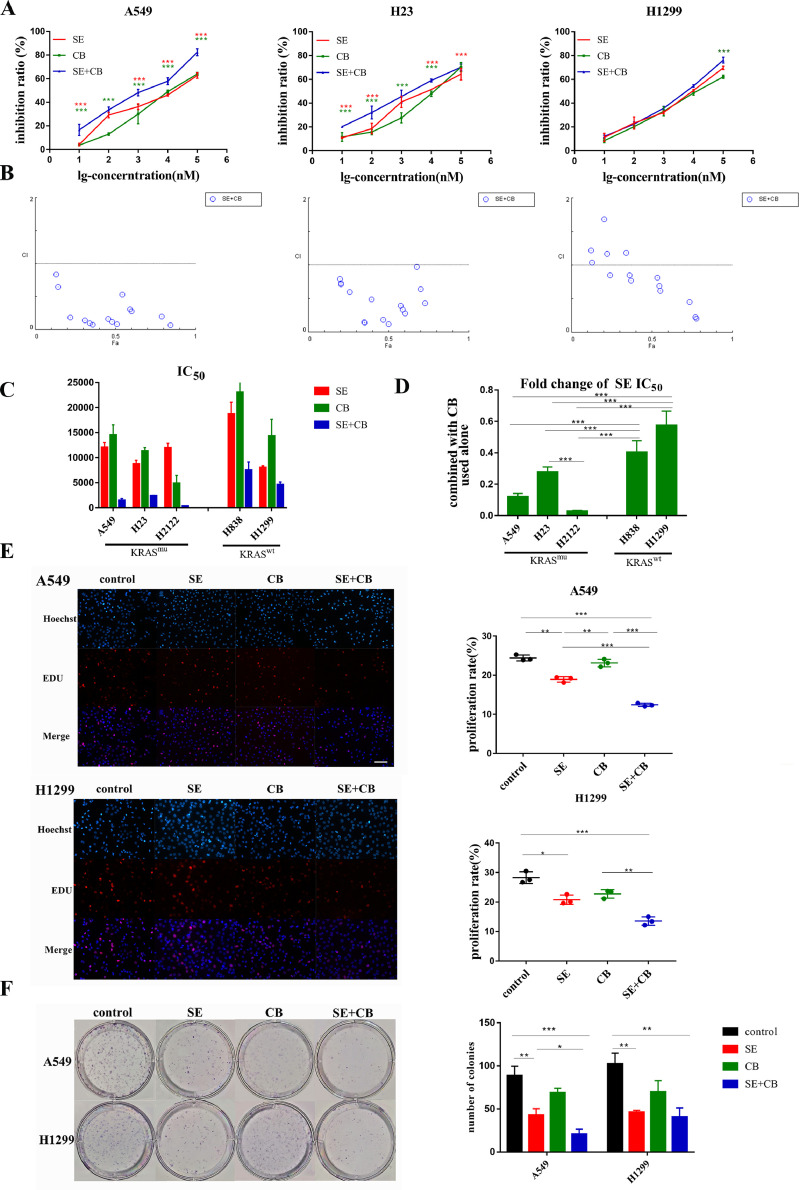
The CCK8 assay was performed to test whether CB839 would synergize with selumetinib to inhibit cell proliferation. KRAS-mutant cells A549, H23, H2122 and the KRAS wild-type cell line H1299, H838 were treated with selumetinib, CB839, or a combination of both at escalating doses that ranged from 10 to 105 nM for 72 h. The combination of selumetinib and CB839 significantly inhibited the proliferation of A549, H23 and H2122 cells compared with single drug treatment while it didn't show added benefit when compared to selumetinib treatment alone in H1299 and H838 cells (Fig. 2A,S2A). Selumetinib and CB839 showed moderate or strong synergy in the KRAS mutant NSCLC cell line A549 [CI=0.159(0.086,0.307)], H23 [CI=0.427(0.181,0.716)] and H2122[CI=0.125(0.037,0.157)], but weak synergy or only additive in the KRAS wild-type cell line H1299[CI=0.848(0.621,1.185)] and H838[CI=0.664(0.439,1.049)] (Fig. 2B,S2B). When combined with CB839, the IC50 of selumetinib was sharply decreased to 2.86%−27.6% in KRAS-mutant cells, but 40.38%−57.49% in KRAS wild-type cell (Fig. 2C-2D). Based on the results above, subsequent cell proliferation experiments were carried out according to the following combinations and drug doses: control, selumetinib (SE, ½IC50 concentration), CB839 (CB, IC10 concentration), and selumetinib+CB839 (SE ½IC50 concentration +CB IC10 concentration). After 72 h of drug treatment, the Edu assay was performed to test the short-term response to the combination therapy while colony formation assay was performed to test the long-term response in A549 and H1299 cells. The outcomes (Fig. 2E-2F) were consistent with the CCK8 assay.
Fig. 2.
CB839 synergized with selumetinib to inhibit proliferation of KRAS-mutant NSCLC cells. KRAS-mutant cells A549, H23, H2122 and the KRAS wild-type cell line H1299, H838 were treated with selumetinib, CB839, or a combination of both at escalating doses that ranged from 10 to 105 nM for 72 h. Subsequent cell proliferation experiments were carried out according to the following combinations and drug doses: control, selumetinib (SE, ½IC50 concentration), CB839 (CB, IC10 concentration), and selumetinib+CB839 (SE ½IC50 concentration +CB IC10 concentration). (A) CCK8 assay of A549,H23 and H1299 cells treated with monotherapy or a combination of selumetinib and CB839. (B) The combination index (CI) plots of selumetinib and CB839 in A549, H23, and H1299 cells. CI values <1 indicated a synergistic drug-drug interaction,CI values <0.3 indicated a strong synergistic effect. (C) IC50 of selumetinib, CB839 alone, and selumetinib combined with CB839. (D) Fold change of IC50 of selumetinib when combined with CB839. (E) Edu assay of cell proliferation of A549 and H1299 cells to test the short-term response to the combination therapy. (200x) (F) The colony formation assay of A549 and H1299 cells to test the long-term response. CI were presented as median (P25, P75), other data were presented as mean ± SD from three independent experiments. A p-value<0.05 was considered statistically significant (*p<0.05, **p<0.01,***p<0.001;* in red: compared with SE group, * in green: compared with CB group).
Combination therapy inhibited tumor growth in the KRAS-mutant NSCLC xenograft and the therapeutic response could be identified by 18F-FDG PET imaging
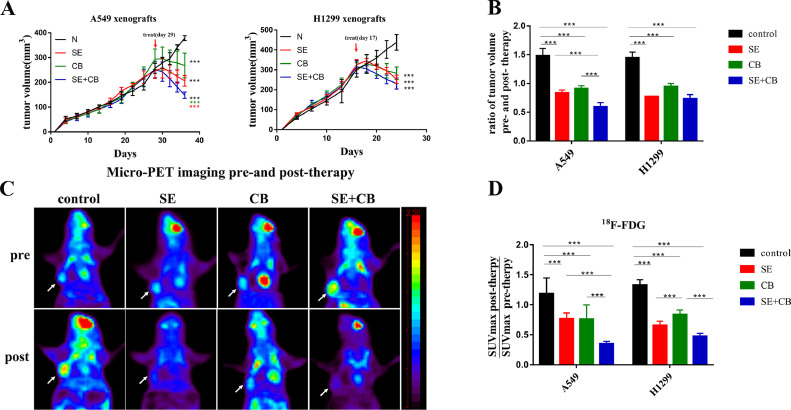
We next tested the efficacy of SE+CB combination therapy compared with monotherapy on A549 and H1299 mouse xenografts. When tumor volumes reached approximately 200–300 mm3, mice were randomly assigned to one of four groups (5 mice per group) and treated via oral gavage twice per day for six days acutely: control (vehicle alone), selumetinib (25 mg/kg) [20], CB839 (200 mg/kg) [21,22], selumetinib (25 mg/kg)+CB839 (200 mg/kg) Monotherapy and combination therapy induced significant tumor regression compared to vehicle (Fig. 3A). In A549 xenografts, combination therapy induced a significantly larger reduction in tumor volume than either selumetinib or CB839 alone, while in H1299 xenografts, combination therapy did not provide added benefit when compared with monotherapy (Fig. 3B). Before and after therapy, mice underwent micro-PET imaging with 18F-FDG to evaluate the therapeutic response from a metabolic perspective. As shown in Fig. 3C, micro-PET scans of A549 and H1299 xenografts showed efficient uptake of 18F-FDG in tumors before therapy, while monotherapy and combination therapy significantly inhibited the uptake of 18F-FDG in tumors. With reference to the European Organization for Research and Treatment of Cancer (EORTC) criteria of solid tumors [23], a decrease in maximum standardized uptake value (SUVmax) greater than 25% is defined as partial metabolic remission (PMR). Overall, 40% (2 in 5) of A549 xenografts treated by selumetinib alone achieved the PMR, while 100% (5 in 5) of A549 xenografts receiving SE+CB treatment achieved PMR (Fig. 3D). Meanwhile, in H1299 xenografts, CB839 induced less decrease in 18F-FDG uptake than selumetinib, which was equal to that of combination therapy (Fig. 3D). The ratio of SUVmax pre- and post-therapy closely mirrored the fold change in tumor volume, implying that the therapeutic response could be identified by 18F-FDG PET imaging.
Fig. 3.
Combination therapy inhibited tumor growth in the KRAS-mutant NSCLC xenograft and the therapeutic response could be identified by 18F-FDG PET imaging. When tumor volumes reached approximately 200–300 mm3, mice were randomly assigned to one of four groups (5 mice per group) and treated via oral gavage twice per day for six days acutely: control (vehicle alone), selumetinib (25 mg/kg), CB839 (200 mg/kg), selumetinib (25 mg/kg)+CB839 (200 mg/kg). (A)The tumor volume curve before and after treatment. (B) Tumor volumes represented as fold change pre- and post-treatment. (C) Representative micro-PET images of mice bearing A549 xenografts with18F-FDG pre- and post-treatment. Tumors are indicated by arrows. (D) Quantification of 18F-FDG uptake represented as fold change of SUVmax pre- and post-treatment. Data were presented as mean±SD(n = 5). A p-value <0.05 was considered statistically significant (*p<0.05, **p<0.01,***p<0.001; *in black: compared with control group, * in red: compared with SE group, * in green: compared with CB group).
Combination therapy induced redox and energetic stress
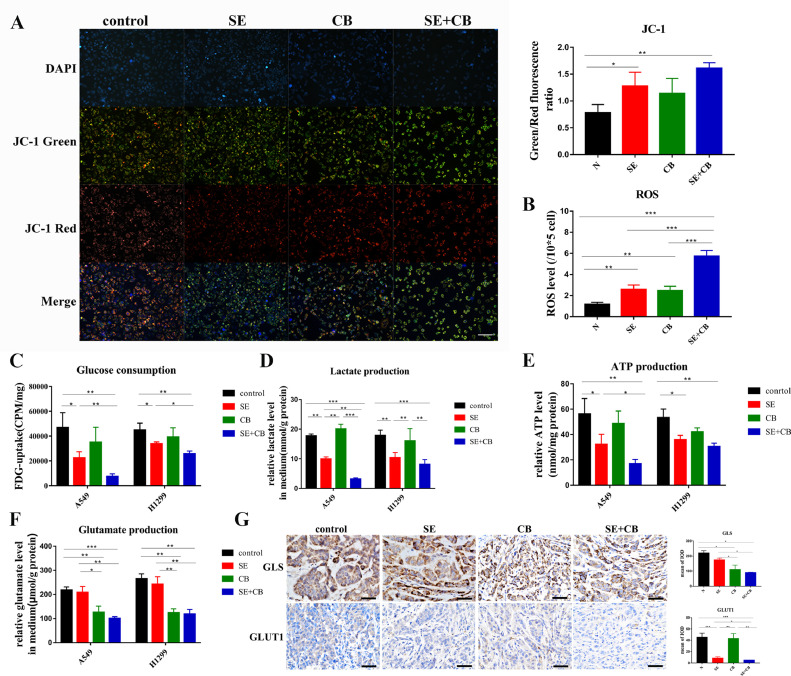
In the metabolic assay, IC10 concentration of selumetinib and CB839 were used. When the mitochondrial membrane potential decreases, the fluorescence of JC-1 staining changes from red to green. After a 72 h drug treatment, the green/red fluorescence ratio increased, demonstrating that the mitochondrial membrane potential decreased after selumetinib and combination therapy in the KRAS-mutant NSCLC (Fig. 4A). ROS levels also increased in all treatment groups, and the increase in combination therapy was significantly greater than that following monotherapy (Fig. 4B). We next examined cellular glucose consumption and lactate, ATP, and glutamate production of A549 and H1299 cells. 18F-FDG uptake was used to evaluate the glucose uptake capacity in vitro. Selumetinib alone or combined with CB839 reduced the consumption of glucose (Fig. 4C) and the production of lactate (Fig. 4D) and ATP (Fig. 4E) in both A549 and H1299 cells. Combination therapy induced a significantly sharper change compared with selumetinib alone only in A549 cells (Fig. 4C-E). The degradation of glutamine decreased when GLS1 was inhibited by CB839 (alone or with selumetinib) (Fig. 4F), implying that the nitrogen supply from glutamine was suppressed. In A549 xenografts, the expression of GLS decreased when mice were treated by CB839 (alone or in combination with selumetinib) (Fig. 4G), such a decrease in GLS1 level was probably associated with a decrease in the metabolic activity of the tumor after treatment which was reflected by 18F-FDG PET imaging. The reduced expression of GLUT1 could only be found in the SE+CB treatment group (Fig. 4G). The results of IHC were consistent with the results of micro-PET imaging. Interestingly, the expression of GLS analyzed by western blotting moderately declined following selumetinib treatment in vitro, while GLS expression was equal to the control group in vivo (Fig. 5A). Taken together, the above results suggested that combination therapy induced redox and energetic stress in the KRAS-mutant A549 cell line in vitro and in vivo.
Fig. 4.
Combination therapy induced redox and energetic stress. In the metabolic assay, IC10 concentration of selumetinib and CB839 were used. (A) JC-1 mitochondrial membrane potential staining. (200x) (B) ROS level assay. (C) 18F-FDG cellular uptake analysis to reflect the glucose consumption. (D-F) Detection of lactate, ATP, glutamate production. (G) Representative IHC images from tumors stained GLS and GLUT1. (400x) Quantification of mean of IOD(n = 3–15 fields of view). Data were presented as mean±SD. A p-value <0.05 was considered statistically significant (*p<0.05, **p<0.01,***p<0.001).
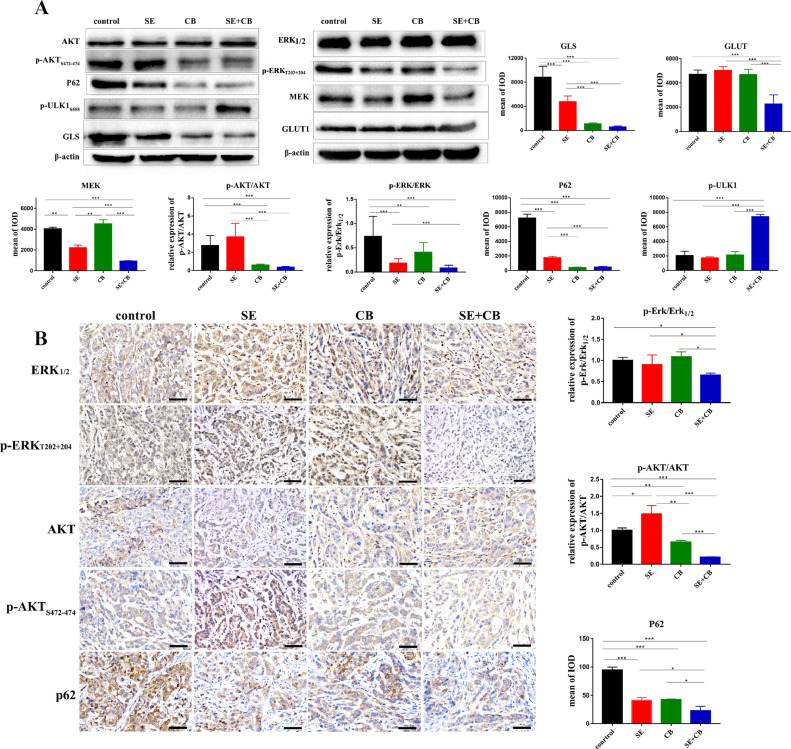
Fig. 5.
Dual inhibition of MEK and GLS1 suppressed the phosphorylation of AKT and induced autophagy in vitro and in vivo. (A) Western blotting analysis to evaluate the phosphorylation level of ERK, AKT and the expression of p-ULK1, P62, GLS, GLUT1, and MEK in A549 cell line. (B) Representative IHC images from tumors staining for ERK, p-ERK, AKT, p-AKT, and P62 in A549 xenografts (400x). Quantification of the mean IOD (n = 3–15 fields of view). Data were presented as mean±SD. A p-value <0.05 was considered statistically significant (*p<0.05, **p<0.01,***p<0.001).
Dual inhibition of MEK and GLS1 suppressed the phosphorylation of AKT and induced autophagy in vitro and in vivo
AKT activation bypassing MEK signally was one of the main reasons that KRAS- mutant NSCLC cells developed intrinsic resistance to MEK inhibition [24], and the activity of mTORC1, a negative regulator of autophagy, is regulated by AKT [25]. Considering that combination therapy had created a microenvironment characterized by redox and energetic stress, we evaluated the phosphorylation status of ERK, AKT, and the expression of autophagy biomarker p-ULK1, p62 to investigate the mechanism by which dual inhibition of MEK and GLS1 was more effective in KRAS-mutant NSCLC cells. The phosphorylation of ERK was inhibited by selumetinib and that of AKT was promoted compensatorily, while the phosphorylation of ERK and AKT was significantly suppressed by dual inhibition of MEK and GLS1 in A549 cell and xenografts (Fig. 5A-B). At the same time, the phosphorylation of autophagy positive regulator ULK increased and the level of autophagy substrate P62 decreased following combination therapy (Fig. 5A-B).Collectively, our results implied that dual inhibition of MEK and GLS1 suppressed the activation of PI3K-AKT-mTOR pathway, induced autophagy in the tumor microenvironment characterized by redox and energetic stress, and ultimately led to tumor cell death.
Discussion
In this study, we confirmed that glutamine uptake of NSCLC with KRAS mutation was higher than that of KRAS wild-type. Regulated by the tumor microenvironment, the metabolic network of the tumor is reprogrammed, driven by oncogenes and tumor suppressor genes such as KRAS [26], c-Myc [27], and TP53 [28]. The metabolic phenotype of tumors of different driven-genes and different tissue types is extremely heterogeneous. How tumors obtain nutrition is speculative, even in a specific tumor lesion, the potential contributions of glucose and non-glucose nutrients differ in poor-and well-perfused tumor areas [29].
In addition to targeting mutation sites, activation pathways and immune mechanisms, targeting cancer metabolism have emerged as a hot topic. Targeted metabolic therapy has many targets available including key enzymes (GLUT1, HK2 etc.), product (lactic acid, ROS etc.) and organelles (mitochondria). Some tumors are dependent on specific metabolic processes, and targeting the sweet spot always brings a gratifying effect. For example, JHU083, a glutamine antagonist, metabolically dismantled the immunosuppressive microenvironment and improved the efficacy of anti-PD-1 agents [30]. Currently, targeting glutamine mainly involves targeted inhibition of key enzymes such as GLS1 and ASCT2. The expression or activity of GLS1 increases in the lung, liver [31], breast cancer [32]etc. Drugs targeting GLS1 include BPTES [33], CB839 [34,35], and UPGL00004 [36]. As a potent GLS1 inhibitor, CB839 increased the sensitivity of IDH1-mutant gliomas to mTOR inhibitors [37], suppressed the proliferation of NSCLC when combined with THZ1, an inhibitor of cyclin-dependent kinase 7 (CDK7) [38], synergized with signal transduction pathway inhibitors such as erlotinib, pazopanib and everolimus in several renal cell carcinoma and NSCLC cell lines [39]. In early phase clinical trials, CB839 showed well safety and tolerability, it was absorbed rapidly (Tmax 1–2 h fasted and 2–4 h fed) and cleared with a half-life of about 4 h, plasma Cmin was > 450 nM at steady state at the recommended phase 2 dose (RP2D) of 600 mg BID [40]. Clinical trials combining CB839 with targeted therapies, such as palbociclib in advanced/metastatic NSCLC with KRAS mutation (NCT03965845), osimertinib, sapanisertib in advanced stage NSCLC (NCT03831932,NCT04250545) are ongoing. Besides, phase I/II trial on PET/CT imaging biomarkers of CB839 in combination with panitumumab and irinotecan in patients with colorectal cancer has also been started and are currently recruiting patients (NCT03263429).
KRAS-driven tumors rewire metabolic networks. KRAS4A colocalizes with HK1 on the outer mitochondrial membrane to directly regulate glucose metabolism [41]. KRAS mutation accompanied by additional mutations present different degrees of glutamine dependence. LKB1 and KEAP1/NRF2 pathways cooperatively enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma [42], it explained to a certain extent why the A549 (KRAS,LKB1,KEAP1 mutant) cell line was more sensitive to combination therapy than the H23 (KRAS,LKB1 mutant) cell line. Metabolic interfering drugs can be attempted to tackle KRAS mutant cancers with sufficient consideration of the mutation background, the tissue types and the crosstalk between the tumor and the microenvironment [43].
Selumetinib is an effective and highly selective MEK inhibitor and inhibitor of phosphorylation of ERK. Selumetinib was absorbed rapidly (Tmax 1 h) and cleared with terminal elimination half-life of about 6 h, plasma Cmin was 886 ng/ml at dose of 25 mg/m2 BID, the most common toxic effects of it included acneiform rash, gastrointestinal effects and asymptomatic creatine kinase elevation (NCT01362803) [44]. Clinical trials combining selumetinib with paclitaxel as second-line treatment (NCT02503358) and small molecule inhibitor such as vandetanib (NCT01586624), gefitinib (NCT02025114) in NSCLC are ongoing. The PI3K-AKT-mTOR pathway is a considerable pathway involved in cell proliferation and energy metabolism. AKT activation bypassing MEK is one of the main reasons that KRAS-mutant NSCLC cells developed intrinsic resistance to selumetinib [24]. In this study, the phosphorylation of AKT increased compensatorily when given selumetinib monotherapy both in vivo and in vitro and was suppressed in combination therapy. We showed that combination therapy induced a decrease in mitochondrial membrane potential and an increase in ROS levels, suppressed glycolysis, and glutamine degradation. In a microenvironment characterized by redox and energetic stress induced by combination therapy, the activity of the negative autophagy regulator mTORC1 decreased and induced autophagy, manifesting as an increase in the phosphorylation of autophagy positive regulator ULK and a decrease in the level of autophagy substrate P62, resulting in cell death.
Changes in tumor metabolism can be monitored in vivo by molecular imaging methods, so-called immunohistochemistry in vivo. PET is a representative of molecular imaging. By labeling specific molecules in tumor metabolism or pathophysiological changes with radionuclides, PET can show the molecular mechanism of the disease in vivo. 18F-FDG (18F labeled glucose analogues) is the most commonly used radiotracer in clinical practice. Based on the Warburg effect [45], means tumors tend to undergo aerobic glycolysis, tumor cells undergo vigorous and continuous glucose uptake and utilization, appearing as abnormal accumulation of 18F-FDG on the image. 18F-FDG PET imaging plays an important role in tumor diagnosis, staging, therapeutic effect evaluation, recurrence monitoring, and prognostic evaluation [46]. In this study, we used micro-PET imaging with 18F-FDG to evaluate the therapeutic response from a metabolic perspective. Before therapy, xenografts of both KRAS-mutant and wild type showed efficient uptake of 18F-FDG. Considering the requirements of animal ethics, we referred to the EORTC criteria rather than RECIST 1.1 (requiring that the diameter and length of the measurable lesion to be ≥ 20 mm) criteria [47] to perform therapeutic effect evaluation by imaging studies. According to the EORTC criteria, when the 18F-FDG uptake of lesions is same as the surrounding normal tissues, this defines a complete remission (CR), when the decrease in SUV greater than 25%, it is defined as PMR [23]. All (100%, 5 in 5) A549 xenografts receiving SE+CB treatment achieved a PMR. Furthermore, the ratio of SUVmax pre- and post-therapy closely mirrored the fold change in tumor volume and the result of IHC, implying that the therapeutic response could be well identified by 18F-FDG PET imaging.
In conclusion, targeted inhibition of GLS1 synergized with selumetinib to enhance antitumor activity in KRAS-mutant NSCLC, the therapeutic response could be well identified by 18F-FDG PET imaging. This combination therapy induced redox and energetic stress and suppressed the phosphorylation of AKT. These effects combined to induce autophagy, resulting in cell death. Targeting the pathways and addicted metabolism activated by the KRAS mutation, and using PET imaging to dynamically monitor therapeutic efficacy may be a feasible model for NSCLC tumor management.
Author contribution
Meng Xia: Experimental design, investigation, data curation, validation and writing manuscript.
Xuena Li: Experimental design.
Yao Diao: Assisted the animal imaging.
Bulin Du: Experimental design.
Yaming Li: Experimental design, Supervision and Funding acquisition.
All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare they have no competing interests.
Acknowledgments
Acknowledgments
We are grateful to Prof. Zhi Yang from Department of Nuclear Medicine, Peking University Cancer Hospital; Prof. Hank F. Kung from Department of Radiology, University of Pennsylvania School of Medicine and Prof. Lin Zhu from Key Laboratory of Radiopharmaceuticals, Ministry of Education, Beijing Normal University for their kindly assistance in synthesis of 18F-Gln.
Funding
This work was supported by the National Natural Science Foundation of China [Grant No. 81971652,81771868].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100920.
Appendix. Supplementary materials
References
- 1.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell. Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., Subbaraj L., Martinez B., Bronson R.T., Prigge J.R., Schmidt E.E., Thomas C.J., Goparaju C., Davies A., Dolgalev I., Heguy A., Allaj V., Poirier J.T., Moreira A.L., Rudin C.M., Pass H.I., Vander Heiden M.G., Jacks T., Papagiannakopoulos T. Keap1 loss promotes KRAS-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernfeld E., Foster D.A. Glutamine as an essential amino acid for KRAS-driven cancer cells. Trends Endocrinol. Metab. 2019;30:357–368. doi: 10.1016/j.tem.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Morelli M.P., Tentler J.J., Kulikowski G.N., Tan A.C., Bradshaw-Pierce E.L., Pitts T.M., Brown A.M., Nallapareddy S., Arcaroli J.J., Serkova N.J., Hidalgo M., Ciardiello F., Eckhardt S.G. Preclinical activity of the rational combination of selumetinib (AZD6244) in combination with vorinostat in KRAS-mutant colorectal cancer models. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2012;18:1051–1062. doi: 10.1158/1078-0432.CCR-11-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evangelista L., Cuppari L., Menis J., Bonanno L., Reccia P., Frega S., Pasello G. 18F-FDG PET/CT in non-small-cell lung cancer patients: a potential predictive biomarker of response to immunotherapy. Nucl. Med. Commun. 2019;40:802–807. doi: 10.1097/MNM.0000000000001025. [DOI] [PubMed] [Google Scholar]
- 6.Ito K., Teng R., Schöder H., Humm J.L., Ni A., Michaud L., Nakajima R., Yamashita R., Wolchok J.D., Weber W.A. (18)F-FDG PET/CT for monitoring of ipilimumab therapy in patients with metastatic melanoma. J. Nucl. Med. 2019;60:335–341. doi: 10.2967/jnumed.118.213652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H., Liu F., Zhang Y., Yang J., Xu X., Guo X., Liu T., Li N., Zhu L., Kung H.F., Yang Z. (2S,4R)-4-[(18)F]fluoroglutamine as a PET indicator for bone marrow metabolism dysfunctional: from animal experiments to clinical application. Mol. Imaging Biol. 2019;21:945–953. doi: 10.1007/s11307-019-01319-4. [DOI] [PubMed] [Google Scholar]
- 8.Liu F., Xu X., Zhu H., Zhang Y., Yang J., Zhang L., Li N., Zhu L., Kung H.F., Yang Z. PET imaging of (18)F-(2 S,4 R)4-fluoroglutamine accumulation in breast cancer: from xenografts to patients. Mol. Pharm. 2018;15:3448–3455. doi: 10.1021/acs.molpharmaceut.8b00430. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman B.P., Ploessl K., Wang L., Qu W., Zha Z., Wise D.R., Chodosh L.A., Belka G., Thompson C.B., Kung H.F. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J. Nucl. Med. 2011;52:1947–1955. doi: 10.2967/jnumed.111.093815. [DOI] [PubMed] [Google Scholar]
- 10.Boysen G., Jamshidi-Parsian A., Davis M.A., Siegel E.R., Simecka C.M., Kore R.A., Dings R.P.M., Griffin R.J. Glutaminase inhibitor CB839 increases radiation sensitivity of lung tumor cells and human lung tumor xenografts in mice. Int. J. Radiat. Biol. 2019;95:436–442. doi: 10.1080/09553002.2018.1558299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacque N., Ronchetti A.M., Larrue C., Meunier G., Birsen R., Willems L., Saland E., Decroocq J., Maciel T.T., Lambert M., Poulain L., Hospital M.A., Sujobert P., Joseph L., Chapuis N., Lacombe C., Moura I.C., Demo S., Sarry J.E., Recher C., Mayeux P., Tamburini J., Bouscary D. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., Sabatini D.M. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell. 2017;168:890–903.e815. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong C.C., Xu J., Bian X., Wu J.L., Kang W., Qian Y., Li W., Chen H., Gou H., Liu D., Yat Luk S.T., Zhou Q., Ji F., Chan L.S., Shirasawa S., Sung J.J., Yu J. In colorectal cancer cells with mutant KRAS, SLC25A22-mediated glutaminolysis reduces DNA demethylation to increase WNT signaling. Stemness, Drug Resist. Gastroenterol. 2020 doi: 10.1053/j.gastro.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Meijer T.W.H., Looijen-Salamon M.G., Lok J., van den Heuvel M., Tops B., Kaanders J., Span P.N., Bussink J. Glucose and glutamine metabolism in relation to mutational status in NSCLC histological subtypes. Thorac. Cancer. 2019;10:2289–2299. doi: 10.1111/1759-7714.13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jänne P.A., Shaw A.T., Pereira J.R., Jeannin G., Vansteenkiste J., Barrios C., Franke F.A., Grinsted L., Zazulina V., Smith P., Smith I., Crinò L. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 16.Jänne P.A., van den Heuvel M.M., Barlesi F., Cobo M., Mazieres J., Crinò L., Orlov S., Blackhall F., Wolf J., Garrido P., Poltoratskiy A., Mariani G., Ghiorghiu D., Kilgour E., Smith P., Kohlmann A., Carlile D.J., Lawrence D., Bowen K., Vansteenkiste J. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA. 2017;317:1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi H., Deng J., Li S., Silk T., Dong L., Brea E.J., Houghton S., Redmond D., Zhong H., Boiarsky J., Akbay E.A., Smith P.D., Merghoub T., Wong K.K., Wolchok J.D. Pulsatile MEK inhibition improves anti-tumor immunity and T cell function in murine KRAS mutant lung cancer. Cell Rep. 2019;27:806–819.e805. doi: 10.1016/j.celrep.2019.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Zhang S., Chen X., Zheng X., Yao Y., Lu G., Zhou J. Palbociclib, a selective CDK4/6 inhibitor, enhances the effect of selumetinib in RAS-driven non-small cell lung cancer. Cancer Lett. 2017;408:130–137. doi: 10.1016/j.canlet.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez-Davies J.E., Tran T.Q., Reid M.A., Rosales K.R., Lowman X.H., Pan M., Moriceau G., Yang Y., Wu J., Lo R.S., Kong M. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J. Transl. Med. 2015;13:210. doi: 10.1186/s12967-015-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon A.M., Telfer B.A., Smith P.D., Babur M., Logie A., Wilkinson R.W., Debray C., Stratford I.J., Williams K.J., Wedge S.R. The mitogen-activated protein/extracellular signal-regulated kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) enhances the radiation responsiveness of lung and colorectal tumor xenografts. Clin. Cancer Res. 2009;15:6619–6629. doi: 10.1158/1078-0432.CCR-08-2958. [DOI] [PubMed] [Google Scholar]
- 21.Momcilovic M., Bailey S.T., Lee J.T., Fishbein M.C., Magyar C., Braas D., Graeber T., Jackson N.J., Czernin J., Emberley E., Gross M., Janes J., Mackinnon A., Pan A., Rodriguez M., Works M., Zhang W., Parlati F., Demo S., Garon E., Krysan K., Walser T.C., Dubinett S.M., Sadeghi S., Christofk H.R., Shackelford D.B. Targeted inhibition of EGFR and glutaminase induces metabolic crisis in EGFR mutant lung cancer. Cell Rep. 2017;18:601–610. doi: 10.1016/j.celrep.2016.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momcilovic M., Bailey S.T., Lee J.T., Fishbein M.C., Braas D., Go J., Graeber T.G., Parlati F., Demo S., Li R., Walser T.C., Gricowski M., Shuman R., Ibarra J., Fridman D., Phelps M.E., Badran K., St John M., Bernthal N.M., Federman N., Yanagawa J., Dubinett S.M., Sadeghi S., Christofk H.R., Shackelford D.B. The GSK3 signaling axis regulates adaptive glutamine metabolism in lung squamous cell carcinoma. Cancer Cell. 2018;33:905–921.e905. doi: 10.1016/j.ccell.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young H., Baum R., Cremerius U., Herholz K., Hoekstra O., Lammertsma A.A., Pruim J., Price P. Measurement of clinical and subclinical tumour response using [18F]-fluorodeoxyglucose and positron emission tomography: review and 1999 EORTC recommendations. Eur. Organ. Res. Treat. Cancer (EORTC) PET Study Group, Eur. J. Cancer (Oxford, Engl.: 1990) 1999;35:1773–1782. doi: 10.1016/s0959-8049(99)00229-4. [DOI] [PubMed] [Google Scholar]
- 24.Kim J.Y., Welsh E.A., Fang B., Bai Y., Kinose F., Eschrich S.A., Koomen J.M., Haura E.B. Phosphoproteomics reveals MAPK inhibitors enhance MET- and EGFR-driven AKT signaling in KRAS-mutant lung cancer. Molecular cancer research: MCR. 2016;14:1019–1029. doi: 10.1158/1541-7786.MCR-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X., Yang Y., Zhong X.Z., Cao Q., Zhu X.H., Zhu X., Dong X.P. A negative feedback regulation of MTORC1 activity by the lysosomal Ca(2+) channel MCOLN1 (mucolipin 1) using a CALM (calmodulin)-dependent mechanism. Autophagy. 2018;14:38–52. doi: 10.1080/15548627.2017.1389822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J.B., Bardeesy N., Asara J.M., Haigis M.C., DePinho R.A., Cantley L.C., Kimmelman A.C. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhutia Y.D., Babu E., Ramachandran S., Ganapathy V. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 28.Hernández-Reséndiz I., Gallardo-Pérez J.C., López-Macay A., Robledo-Cadena D.X., García-Villa E., Gariglio P., Saavedra E., Moreno-Sánchez R., Rodríguez-Enríquez S. Mutant p53(R248Q) downregulates oxidative phosphorylation and upregulates glycolysis under normoxia and hypoxia in human cervix cancer cells. J. Cell. Physiol. 2019;234:5524–5536. doi: 10.1002/jcp.27354. [DOI] [PubMed] [Google Scholar]
- 29.Hensley C.T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L., Wodzak M., Klimko C., McMillan E., Butt Y., Ni M., Oliver D., Torrealba J., Malloy C.R., Kernstine K., Lenkinski R.E., DeBerardinis R.J. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone R.D., Zhao L., Englert J.M., Sun I.M., Oh M.H., Sun I.H., Arwood M.L., Bettencourt I.A., Patel C.H., Wen J., Tam A., Blosser R.L., Prchalova E., Alt J., Rais R., Slusher B.S., Powell J.D. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai W., Xu L., Yu X., Zhang G., Guo H., Liu H., Song G., Weng S., Dong L., Zhu J., Liu T., Guo C., Shen X. OGDHL silencing promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J. Hepatol. 2020;72:909–923. doi: 10.1016/j.jhep.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Gross M.I., Demo S.D., Dennison J.B., Chen L., Chernov-Rogan T., Goyal B., Janes J.R., Laidig G.J., Lewis E.R., Li J., Mackinnon A.L., Parlati F., Rodriguez M.L., Shwonek P.J., Sjogren E.B., Stanton T.F., Wang T., Yang J., Zhao F., Bennett M.K. Antitumor activity of the glutaminase inhibitor CB839 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 33.Kono M., Yoshida N., Maeda K., Suarez-Fueyo A., Kyttaris V.C., Tsokos G.C. Glutaminase 1 inhibition reduces glycolysis and ameliorates lupus-like disease in MRL/lpr mice and experimental autoimmune encephalomyelitis. Arthritis Rheumatol. (Hoboken, N.J.) 2019;71:1869–1878. doi: 10.1002/art.41019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., Li D., Xu N., Fang J., Yu Y., Hou W., Ruan H., Zhu P., Ma R., Lu S., Cao D., Wu R., Ni M., Zhang W., Su W., Ruan B.H. Novel 1, 3, 4-selenadiazole-containing kidney-type glutaminase inhibitors showed improved cellular uptake and antitumor activity. J. Med. Chem. 2019;62:589–603. doi: 10.1021/acs.jmedchem.8b01198. [DOI] [PubMed] [Google Scholar]
- 35.Xu X., Kuang Z., Han J., Meng Y., Li L., Luan H., Xu P., Wang J., Luo C., Ding H., Li Z., Bian J. Development and characterization of a fluorescent probe for GLS1 and the application for high-throughput screening of allosteric inhibitors. J. Med. Chem. 2019;62:9642–9657. doi: 10.1021/acs.jmedchem.9b01035. [DOI] [PubMed] [Google Scholar]
- 36.Huang Q., Stalnecker C., Zhang C., McDermott L.A., Iyer P., O'Neill J., Reimer S., Cerione R.A., Katt W.P. Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism. J. Biol. Chem. 2018;293:3535–3545. doi: 10.1074/jbc.M117.810101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K., Sasayama T., Irino Y., Takata K., Nagashima H., Satoh N., Kyotani K., Mizowaki T., Imahori T., Ejima Y., Masui K., Gini B., Yang H., Hosoda K., Sasaki R., Mischel P.S., Kohmura E. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J. Clin. Invest. 2015;125:1591–1602. doi: 10.1172/JCI78239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Z.J., Miao D.L., Su Q.Y., Tang X.L., Wang X.L., Deng L.B., Shi H.D., Xin H.B. THZ1 suppresses human non-small-cell lung cancer cells in vitro through interference with cancer metabolism. Acta Pharmacol. Sin. 2019;40:814–822. doi: 10.1038/s41401-018-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirna Rodriguez W.Z., Bennett Mark, Emberley Ethan, Gross Mathew, Janes Julie, MacKinnon Andrew, Pan Alison, Steggerda Susanne, Works Melissa, Parlati Francesco. Proceedings of the 106th Annual Meeting of the American Association for Cancer Research; 2015 Apr 18-22; Philadelphia, PA. Philadelphia (PA): AACR; Cancer Res. Vol. 75. 2015. CB839, a selective glutaminase inhibitor, synergizes with signal transduction pathway inhibitors to enhance anti-tumor activity.[abstract] Abstract nr 4711. [Google Scholar]
- 40.Harding J.J., Telli M.L., Munster P.N., Le M.H., Molineaux C., Bennett M.K., Mittra E., Burris H.A., Clark A.S., Dunphy M., Meric-Bernstam F., Patel M.R., DeMichele A., Infante J.R. Safety and tolerability of increasing doses of CB839, a first-in-class, orally administered small molecule inhibitor of glutaminase, in solid tumors. J. Clin. Oncol. 2015;33:2512. -2512. [Google Scholar]
- 41.Amendola C.R., Mahaffey J.P., Parker S.J., Ahearn I.M., Chen W.C., Zhou M., Court H., Shi J., Mendoza S.L., Morten M.J., Rothenberg E., Gottlieb E., Wadghiri Y.Z., Possemato R., Hubbard S.R., Balmain A., Kimmelman A.C., Philips M.R. KRAS4A directly regulates hexokinase 1. Nature. 2019;576:482–486. doi: 10.1038/s41586-019-1832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galan-Cobo A., Sitthideatphaiboon P., Qu X., Poteete A., Pisegna M.A., Tong P., Chen P.H., Boroughs L.K., Rodriguez M.L.M., Zhang W., Parlati F., Wang J., Gandhi V., Skoulidis F., DeBerardinis R.J., Minna J.D., Heymach J.V. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79:3251–3267. doi: 10.1158/0008-5472.CAN-18-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pupo E., Avanzato D., Middonti E., Bussolino F., Lanzetti L. KRAS-driven metabolic rewiring reveals novel actionable targets in cancer. Front. Oncol. 2019;9:848. doi: 10.3389/fonc.2019.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dombi E., Baldwin A., Marcus L.J., Fisher M.J., Weiss B., Kim A., Whitcomb P., Martin S., Aschbacher-Smith L.E., Rizvi T.A., Wu J., Ershler R., Wolters P., Therrien J., Glod J., Belasco J.B., Schorry E., Brofferio A., Starosta A.J., Gillespie A., Doyle A.L., Ratner N., Widemann B.C. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N. Engl. J. Med. 2016;375:2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farwell M.D., Pryma D.A., Mankoff D.A. PET/CT imaging in cancer: current applications and future directions. Cancer. 2014;120:3433–3445. doi: 10.1002/cncr.28860. [DOI] [PubMed] [Google Scholar]
- 47.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Euro. J. Cancer (Oxford, Engl.: 1990) 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.