Abstract
Areca is a genus comprising about 50 species endemic to the humid tropics. Arecanut (Areca catechu L.) is a commercially and economically important crop in South and Southeast Asia. In addition to its contribution to the agricultural economies of countries where the crop is grown, arecanut holds an important place in the religious, cultural, and social milieu of the rural folks. The nuts have been used since time immemorial in traditional Indian (Unani and Ayurveda) and Chinese herbal systems of medicine for the treatment of various disorders like rheumatism, parasitic infection, diseases of gastrointestinal tracts, and depression. Here, we report the complete chloroplast (cp) genome sequence of arecanut. The cp genome of A. catechu was a typical circular DNA molecule with a size of 158,689 bp in length. The genome possessed a typical quadripartite structure composed of a pair of inverted repeats (IRa and IRb) of 27,137 bp separated by a large single-copy (LSC) region of 86,814 bp and a small single-copy (SSC) region of 17,601 bp and a GC content of 37.3%. The cp genome of arecanut encodes a set of 133 genes, comprising 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes; among these, 21 contained introns. A total of 70 SSR loci were detected, the majority being in inter-genic regions. Phylogenetic analysis revealed that A. catechu was closely related to A. vestiaria.
Keywords: Areca catechu, Arecaceae, Chloroplast genome, Phylogenetic analysis
Specifications Table
| Subject | Agriculture and Biological Sciences |
| Specific subject area | Plastome genomics |
| Type of data | Shallow DNA sequencing data |
| How data were acquired | Novaseq 6000 sequencing platform |
| Data format | Raw sequencing data (fastq) and analyzed data (fasta) |
| Parameters for data collection | Spindle leaves (i.e. the first unopened leaves) were collected from the South Kanara Local cultivar, and genomic DNA was extracted based on SDS protocol [1]. A quality check of the extracted DNA was carried out using Qubit 2.0 Fluorometer and Agilent 2100 Bioanalyzer. Paired-end sequencing was carried out on Novaseq 6000 platform (2 × 150 bp run configuration) (Illumina, San Diego, CA, USA). |
| Description of data collection | High-quality reads were assembled by using NOVOPlasty. The assembled scaffold was annotated using PGA and GeSeq, and the circular chloroplast genome map was drawn using OGDRAW. Alignment of complete chloroplast genome sequences of Areca catechu and other members of Arecaceae was undertaken using MAFFT version 7.467, and the phylogenetic tree was constructed using MEGA7. |
| Data source location | Vittal, Karnataka State, India (12°46′20.1"N 75°06′58.2"E) |
| Data accessibility | Repository name: NCBI A. catechu chloroplast genome- data identification number: MT559306 Direct URL to A. catechu chloroplast genome: https://www.ncbi.nlm.nih.gov/nuccore/MT559306 Raw data have been deposited under BioProject: PRJNA667176 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA667176) and SRR12777938 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR12777938) |
Value of the Data
-
•
The complete cp genome represents a useful sequence-based resource for A. catechu.
-
•
The data allows further scrutiny of the mechanisms which are involved in transcriptional regulation and translational modification of the arecanut cp genome.
-
•
The cp genome sequence presented here provides a basis for researchers for additional studies on taxonomy, population structure, and evolution of Areca spp.
-
•
The cp genome data could be useful for comparative studies of RNA editing sites in Areca spp.
1. Data Description
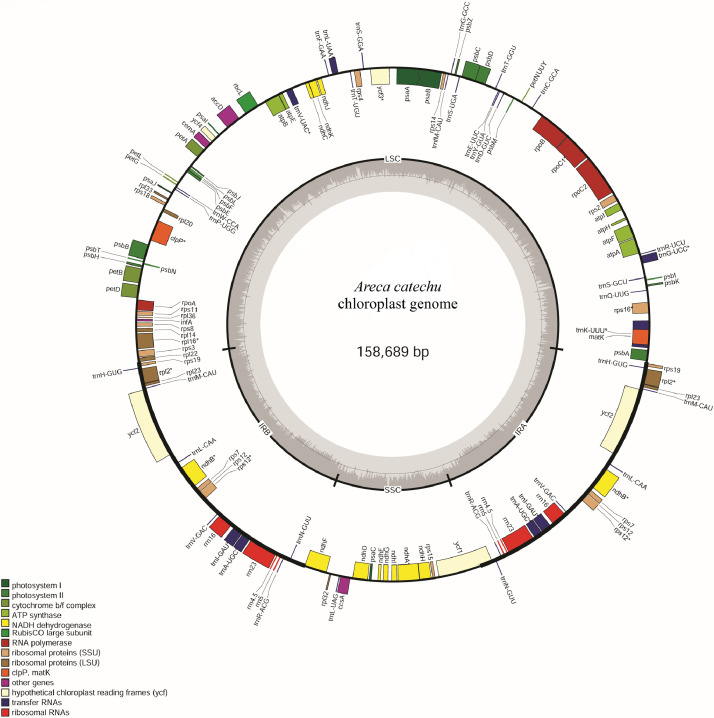
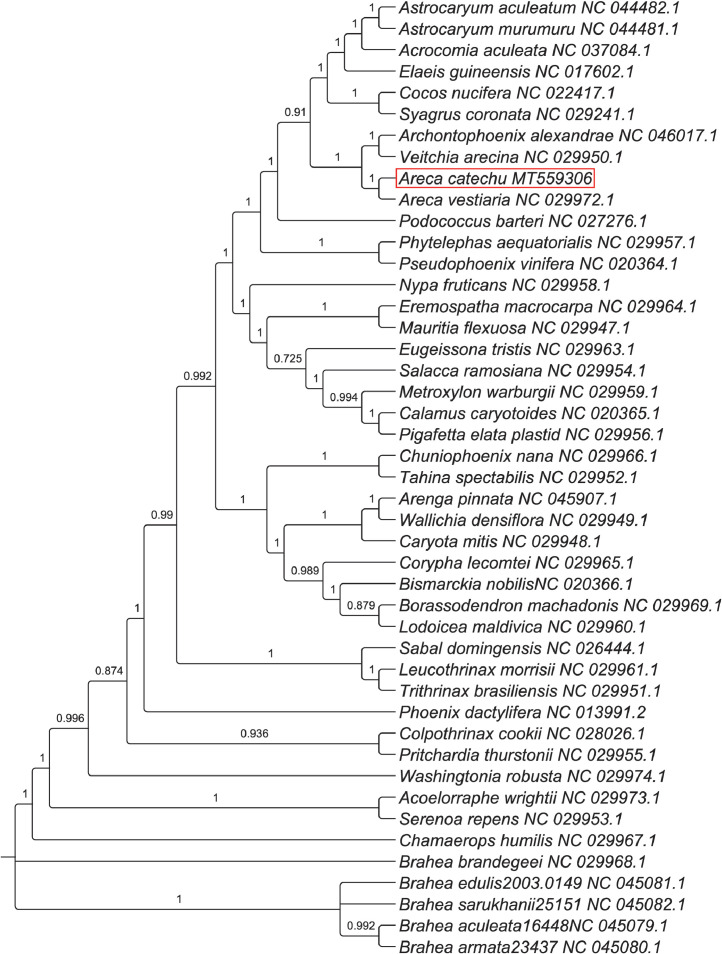
The circular map of the chloroplast (cp) genome of Areca catechu is given in Fig. 1. Table 1 lists the genes encoded by the A. catechu plastome. The list of simple sequence repeats (SSR) loci in A. catechu plastome is given in Table 2. The maximum likelihood phylogenetic tree for A. catechu based on 44 other complete cp genomes of Arecaceae is given as Fig. 2.
Fig. 1.
Circular gene map of the chloroplast genome of A. catechu. Genes drawn inside the circle are transcribed clockwise, and those outside the circle are transcribed counter-clockwise. Small single copy (SSC), large single copy (LSC), and inverted repeats (IRa, IRb) are indicated. The darker grey in the inner circle represents the GC content; conversely, the lighter one represents the AT content. Gene function or gene identifiers are displayed using colors indicated by the inner legend. The symbol ‘*' indicates genes with introns.
Table 1.
List of genes in the chloroplast genome of A. catechu. Hypothetical conserved chloroplast reading frames are shown as ‘ycf’. Numbers of copies are shown in parenthesis for genes with multiple copies. The symbol '*' indicates genes with one intron, while '**' indicates genes with two introns.
| Category | Group | Genes | |
|---|---|---|---|
| Photosynthesis related genes | Rubisco | rbcL | |
| Photosystem I | psaA, psaB | ||
| Photosystem II | psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | ||
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | ||
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN | ||
| Cytochrome C synthesis | ccsA | ||
| NADPH dehydrogenase |
ndhA*, ndhB (× 2) *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
||
| RNA genes | Ribosomal RNA | rrn16 (× 2), rrn23 (× 2), rrn4.5 (× 2), rrn5 (× 2) | |
| Transfer RNA | trnA-UGC (× 2)*,trnC-GCA, trnD-GUC,trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*, trnH-GUG (× 2),trnI-GAU (× 2)*,trnK-UUU*, trnL-CAA (× 2),trnL-UAA*, trnL-UAG, trnM-CAU (× 2),trnN-GUU (× 2), trnP-UGG, trnQ-UUG, trnR-ACG (× 2),trnR-UCU,trnS-GCU,trnS-GGA,trnS-UGA,trnT-GGU,trnT-UGU,trnV-GAC (× 2), trnV-UAC*, trnW-CCA, trnY-GUA | ||
| Transcription and translation-related genes | Transcription | rpoA, rpoB, rpoC1*, rpoC2 | |
| Ribosomal proteins | Small sub-unit | rps11, rps12 (× 2)*, rps12 (× 2), rps14, rps15, rps16*, rps18, rps19 (× 2), rps2, rps3, rps4, rps7 (× 2), rps8 | |
| Large sub-unit | rpl14, rpl16*, rpl2 (× 2)*, rpl20, rpl22, rpl23 (× 2), rpl32, rpl33, rpl36 | ||
| Translation initiation factor | infA | ||
| Other genes | RNA processing | matK | |
| Carbon metabolism | cemA | ||
| Fatty acid synthesis | accD | ||
| Proteolysis | clpP** | ||
| Genes of unknown function | Conserved ORFs | ycf1, ycf2 (× 2), ycf3**, ycf4 | |
Table 2.
A list of simple sequence repeats in the chloroplast genome of A. catechu. The SSR-containing coding regions are indicated in parentheses.
| Repeat unit | Length (No. of units) | Number | Start position |
|---|---|---|---|
| ATGTA | 4 | 1 | 27855 |
| TATTT | 3 | 1 | 43151 |
| TTTCA | 3 | 1 | 67458 |
| TTTAT | 3 | 1 | 71217 |
| ATAAT | 3 | 1 | 84531 (rpl16-intron I) |
| TCTA | 4 | 1 | 6054 |
| AATG | 3 | 1 | 63995 (cemA) |
| ATAA | 3 | 1 | 73633 (clpP) |
| AATA | 3 | 3 | 7197, 84785 (rpl16-intron I), 118973 (ndhD) |
| TTTA | 3 | 1 | 121721 |
| CAG | 4 | 1 | 716 (psbA) |
| AAT | 4 | 1 | 3924 |
| TAT | 6 | 1 | 47868 |
| ATA | 4 | 1 | 129311(ycf1) |
| AT | 8 | 1 | 8862 |
| AT | 5 | 1 | 20751 (rpoC2) |
| TA | 5 | 1 | 30383 |
| AT | 7 | 1 | 49130 |
| AT | 9 | 1 | 50075 |
| AT | 6 | 1 | 70535 |
| TC | 5 | 1 | 126168 |
| A | 10 | 7 | 3595, 3796, 7370, 8039, 9468, 10097, 12389 |
| A | 11 | 6 | 13007 (atpF-intron I), 13265 (atpF-intron I), 13942, 15278, 17177, 23506 (rpoC1) |
| A | 12 | 2 | 23827 (rpoC1-intron I), 29614 |
| A | 13 | 1 | 30258 |
| A | 14 | 1 | 33820 |
| A | 15 | 1 | 38310 (rps14) |
| T | 10 | 6 | 44492 (ycf3-intron I), 54542, 56663, 61122, 61338, 67870 |
| T | 11 | 10 | 67983, 68681, 69413, 69702, 71097, 73166 (clpP-intron I), 73434 (clpP-intron I), 73971 (clpP-intron II), 77773 (petB-intron I), 82487 |
| T | 12 | 3 | 83362, 85455 (rpl16-intron I), 86844, 87232 |
| T | 13 | 6 | 116155, 118714, 126666, 129962 (ycf1), 130141 (ycf1), 130511 (ycf1) |
| T | 14 | 1 | 130645 (ycf1) |
Fig. 2.
Maximum likelihood phylogenetic tree for A. catechu based on complete chloroplast genomes of Arecaceae. The bootstrap value is given at each node.
Around 41.34 Gb data was generated comprising of 273,784,506 reads, with a GC content of 42.28% and Q30 of 91.12%. The complete cp genome of A. catechu genome was assembled with a size of 158,689 bp in length (Fig. 1). The circular genome included two copies of inverted repeats (IRa and IRb: 27,137 bp) separated by two regions: the large single-copy region (LSC: 86,814 bp) and the small single-copy region (SSC: 17,601 bp). GC content of the whole genome, IRs, LSC, and SSC regions are 37.30, 42.48, 35.32 and 31.06 %, respectively.
The cp genome of A. catechu encoded a set of 133 genes, comprising of 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes (Table 1). Twenty-one genes contained introns.
A total of 24 forward repeats, 26 palindromic repeats, and 27 tandem repeats were identified in the A. catechu cp genome. Out of the 70 SSR loci detected, more than half (67.14%) were A and T mononucleotide repeats, followed by dinucleotide (10%), trinucleotide (5.72%), tetranucleotide repeats (10 %) and pentanucleotide (7.14%) repeats. Most of the SSRs were located in intergenic regions; some of them were also found in coding regions such as cemA, clpP, ndhD, psbA, psbA, ycf1, rpoC1, rpoC2 and rps14 (Table 2).
To examine the phylogenetic position of A. catechu, the cp genome sequences of A. catechu and 44 members of Arecaceae, for which complete cp genome sequences were available in NCBI, were aligned and a phylogenetic tree was constructed. Phylogenetic analysis revealed that A. catechu is very closely related to A. vestiaria (Fig. 2).
2. Experimental Design, Materials and Methods
2.1. Experimental material, sampling and DNA extraction
Spindle leaves (i.e. the first unopened leaves) were collected from South Kanara Local cultivar maintained at the National Arecanut Gene Bank, Vittal, Karnataka State, India (12°46′20.1"N 75°06′58.2"E). The genomic DNA was extracted based on the SDS protocol standardized earlier [1]. The quality check of the extracted DNA was carried out using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and 2100 Bioanalyzer (Agilent).
2.2. Library preparation, sequencing and sequence analysis
The genomic DNA was fragmented and size-selected through agarose gel electrophoresis. Selected DNA fragments were blunted and ligated to sequencing adapters. DNA library was constructed using the TruSeq Nano DNA kit (Illumina, USA) following the standard Illumina operating procedure and shallow sequencing (∼20x coverage) was carried out on a Novaseq 6000 platform (Illumina, USA) using the run configuration of 2 × 150 bp. High-quality reads were assembled by using NOVOPlasty [2]. The assembled scaffold was annotated using PGA [3] and GeSeq [4].
2.3. Analysis of repeat sequences
Dispersed and palindromic repeats of A. catechu plastome were identified using REPuter [5] with default parameters. Tandem repeat sequences were searched using the Tandem Repeats Finder program [6] with the following parameters: ‘2’ for alignment parameters match, ‘7’ for mismatch and indels, and ‘80’ for minimum alignment score to report repeat respectively. Simple sequence repeats (SSRs) were analyzed using MISA (http://pgrc.ipk-gatersleben.de/misa/) with the parameters of ‘10’ for mono, ‘5’ for di-, ‘4’ for tri-, and ‘3’ for tetra- and penta- nucleotide motifs.
2.4. Phylogenetic analysis
To examine the phylogenetic position of A. catechu, the cp genome sequences of A. catechu and members of Arecaceae, for which complete cp genome sequences are available in NCBI (sequences are given in Supplementary Table S1), were aligned by MAFFT version 7.467 [7]. The phylogenetic tree was constructed using MEGA7 [8], with bootstrap set to 1000, using the maximum likelihood method.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2020.106444.
CRediT Author Statement
M.K. Rajesh: Conceptualization, Methodology, Supervision, Data curation, Writing – original draft. K.P. Gangaraj: Data curation, Writing – original draft. Sudheesh K. Prabhudas: Data curation, Writing – review & editing. T.S. Keshava Prasad: Conceptualization, Methodology, Supervision, Data curation, Writing – review & editing.
Appendix. Supplementary materials
Supplementary Table S1. Chloroplast genome sequences of members of Arecaceae.
References
- 1.Rajesh M.K., Bharathi M., Nagarajan P. Optimization of DNA isolation and RAPD technique in arecanut (Areca catechu L.) Agrotropica. 2007;19:31–34. [Google Scholar]
- 2.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X.J., Moore M.J., Li D.Z., Yi T.S. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Chloroplast genome sequences of members of Arecaceae.