Summary
Photoreceptors, especially cones, which are enriched in the human macula, have high energy demands, making them vulnerable to metabolic stress. Metabolic dysfunction of photoreceptors and their supporting retinal pigment epithelium (RPE) is an important underlying cause of degenerative retinal diseases. However, how cones and the macula support their exorbitant metabolic demand and communicate with RPE is unclear. By profiling metabolite uptake and release and analyzing metabolic genes, we have found cone-rich retinas and human macula share specific metabolic features with upregulated pathways in pyruvate metabolism, mitochondrial TCA cycle, and lipid synthesis. Human neural retina and RPE have distinct but complementary metabolic features. Retinal metabolism centers on NADH production and neurotransmitter biosynthesis. The retina needs aspartate to sustain its aerobic glycolysis and mitochondrial metabolism. RPE metabolism is directed toward NADPH production and biosynthesis of acetyl-rich metabolites, serine, and others. RPE consumes multiple nutrients, including proline, to produce metabolites for the retina.
Subject Areas: Biochemistry, Sensory Neuroscience, Metabolomics
Graphical Abstract
Highlights
-
•
Besides glucose, mouse and human retinas heavily consume aspartate and glutamate
-
•
Mature retinal organoids robustly consume aspartate and glutamate
-
•
Cone-rich retinas and the neural macula use more pyruvate
-
•
Neural retinas and RPE differ in their metabolite uptake and release
Biochemistry; Sensory Neuroscience; Metabolomics
Introduction
Retinal photoreceptors need an enormous supply of ATP, reducing equivalents and other metabolites to maintain visual transduction and renew their daily-shed outer segments (Hurley et al., 2015; Leveillard and Sahel, 2017; Okawa et al., 2008). Mutations of metabolic genes or defects in metabolic processes have a role in many common degenerative retinal diseases such as inherited retinal degeneration, diabetic retinopathy, and age-related macular degeneration (AMD) (Ferrington et al., 2020; Leveillard and Sahel, 2017; Narayan et al., 2017).
Like tumors, retinas are known for aerobic glycolysis, also called the Warburg effect (Hurley et al., 2015). Aerobic glycolysis can produce large amounts of lactate from glucose, even in the presence of O2. It is estimated that 80%–96% of glucose in isolated retinas is converted into lactate and only a small fraction is oxidized in the mitochondria (Hurley et al., 2015). Recent studies confirm that mammalian retinas strongly express enzymes associated with the Warburg effect such as the M2 isoform of pyruvate kinase, hexokinase 2, and lactate dehydrogenase A (Casson et al., 2016; Lindsay et al., 2014; Petit et al., 2018; Rajala et al., 2013; Rueda et al., 2016). However, retinas are also densely packed with mitochondria, which consume oxygen rapidly (Hoang et al., 2002; Narayan et al., 2017). Since glucose is the putative primary nutrient for the retina, it remains unclear how the retina supports its active mitochondrial metabolism while converting most of its glucose to lactate.
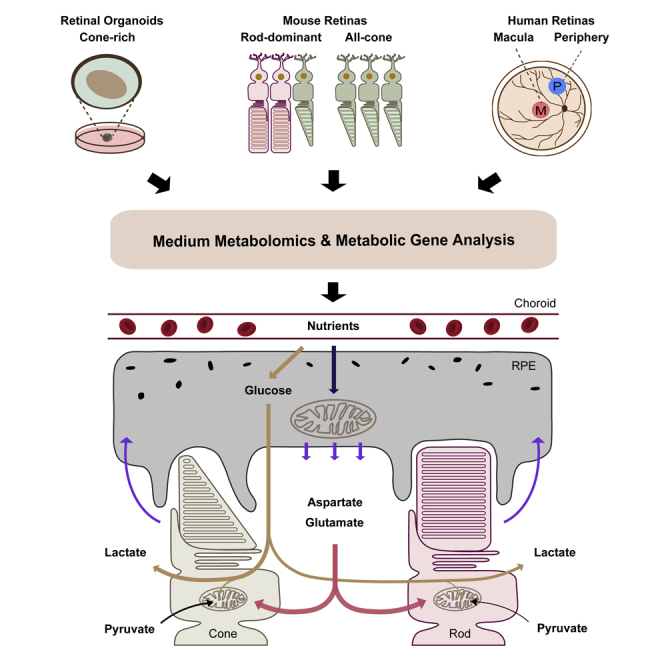
Rods and cones are the two major types of photoreceptors in the retina. Rods are responsible for dimly lit vision, whereas cones are for high acuity, central vision, and color vision. Cones are highly enriched in the center of the retina, called the macula, in humans and some birds. Cone photoreceptor degeneration is the ultimate cause of clinically significant vision loss in conditions such as AMD (Hoang et al., 2002; Narayan et al., 2017). Cones expend more energy than rods (Johnson et al., 2007; Perkins et al., 2003), but we still know very little about their metabolism compared with rods. The study of cone metabolism is challenging, since cones account for only 2%–3% of photoreceptors in mice and rodents do not have the macula. The recent development of all-cone mice by deletion of neural retina leucine zipper (Nrl), a gene required for rod development (Daniele et al., 2005; Mears et al., 2001), and cone-rich human retinal organoids (Kim et al., 2019) facilitate cone research. To investigate cone photoreceptor metabolism, we used all-cone mice, cone-rich retinal organoids, and human macula from postmortem donors in this study.
Photoreceptors rely on metabolic support from their neighboring retinal pigment epithelium (RPE). Disruption of RPE metabolism contributes to photoreceptor degeneration (Ferrington et al., 2020; Fisher and Ferrington, 2018). Inhibition of RPE mitochondrial metabolism is sufficient to cause AMD-like retinal degeneration in mice (Han et al., 2020; Kurihara et al., 2016; Zhao et al., 2011). Conversely, impairment of photoreceptor metabolism also influences RPE health (Brown et al., 2019). Accumulating evidence supports the concept that photoreceptors and RPE form an interdependent metabolic ecosystem (Bisbach et al., 2020; Brown et al., 2019; Kanow et al., 2017; Reyes-Reveles et al., 2017; Wang et al., 2019). Photoreceptors shed lipid-rich outer segment discs and export lactate due to the Warburg effect; the RPE converts the outer segment discs into ketone bodies to fuel photoreceptors and it utilizes lactate to preserve glucose for photoreceptors(Adijanto et al., 2014; Reyes-Reveles et al., 2017). We have also reported that reductive carboxylation (reversed TCA cycle) is a very active metabolic pathway in RPE, which it prefers to fuel with proline (Chao et al., 2017; Du et al., 2016). Proline-derived intermediates, including citrate, glutamate, and aspartate, can be exported by RPE to fuel retinal mitochondria (Yam et al., 2019). However, most of these studies come from cultured RPE cells and mice. Native human RPE are more dense and multinucleated in the macula than in the periphery (Rashid et al., 2016; Starnes et al., 2016). The macular RPE is where AMD starts, but the metabolic differences between the RPE of the macula and the peripheral retina, and their symbiotic relations with the corresponding neural retinal regions, are still unknown.
We used a targeted metabolomics approach in this study to analyze consumed and released metabolites in the media from cone-rich human retinal organoids, rod-dominant versus all-cone mouse retinas, human macular versus peripheral neural retinas, and human macular versus peripheral RPE/choroid. By integrating our results with the analysis of the expression of metabolic genes, we have identified key metabolic features of retinal organoid development, rods versus cones, macula versus periphery, and neural retina versus RPE. These features may elucidate the different metabolic needs of retinal mitochondria, cones, and human macula and reveal the metabolic communications between human retina and RPE at the macula and peripheral retina.
Results
Metabolite Consumption and Metabolic Gene Expression in Human Cone-Rich Retinal Organoids
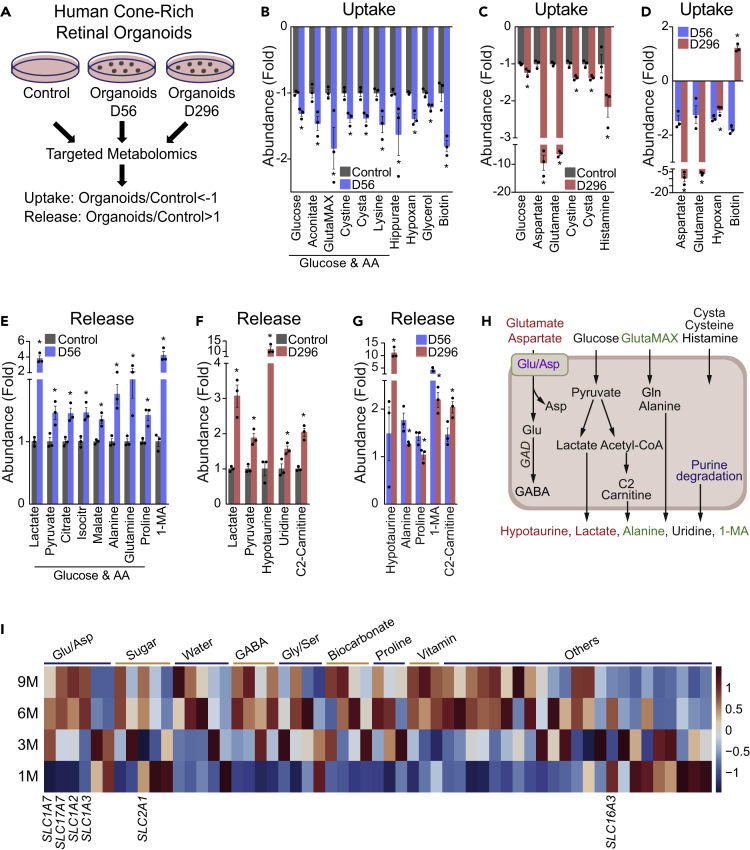
Cell differentiation in retinal organoids largely recapitulated transcriptomic and electrophysiological properties of retinogenesis in vivo (Kim et al., 2019). Retinal organoids at D56 were mainly composed of retinal progenitor cells and early differentiated retinal cells, whereas retinal organoids at D296 were mostly composed of maturing cones, rods, and Müller glial cells (Kim et al., 2019). To study the nutrient needs during the development of the human retina, we collected spent media 48 h after the medium change from human retinal organoids on 56 days (D56) and 296 days (D296) (Figure 1A). Blank media without cells in the same culture plate for 48 h served as baseline control. We targeted 219 metabolites covering major nutrients and pathways (Table S1) and detected 89 metabolites from these media. To determine metabolite uptake and release, we divided the ion abundance from organoids by those from the control media (see details in Transparent Methods). Retinal organoids at both D56 and D296 consumed glucose, cystine, and cystathionine (Figures 1B and 1C). Retinal organoids at D56 used L-Alanyl-Glutamine (GlutaMAX), hypoxanthine, lysine, hippurate, glycerol, and biotin, but D296 did not (Figures 1B–1D). Surprisingly, retinal organoids at D296 consumed 6- to 8-fold more aspartate and glutamate from the media, but organoids at D56 did not use them at all (Figures 1B and 1C). These results indicate that the consumption of aspartate and glutamate is specific for mature retinal cells. Additionally, retinal organoids at D296 also consumed histamine. Glucose can be metabolized through glycolysis and the mitochondrial TCA cycle into lactate, pyruvate, and other mitochondrial intermediates. GlutaMAX could be hydrolyzed into glutamine and alanine. As expected, the products from glucose and GlutaMAX, including lactate, pyruvate, alanine, glutamine, proline, citrate, and other metabolites, were produced significantly by D56 organoids (Figure 1E). D296 organoids generated similar amounts of lactate and pyruvate as D56 organoids, but they produced ∼11-fold of hypotaurine and 2-fold of acetyl-carnitine (C2-carnitine) (Figures 1F and 1G).
Figure 1.
Metabolite Consumption and Metabolic Gene Expression in Human Cone-Rich Retinal Organoids
(A) Schematic for profiling medium metabolites in retinal organoid culture. Retinal organoids on Day 56 (D56) and D296 were cultured in a medium (see Transparent Methods) for 48 h. The spent media were analyzed for metabolite consumption or release, and the medium in culture wells without retinal organoids was used as a control.
(B–D) Metabolite consumption in retinal organoids. Metabolite uptake in retinal organoids at (B) 56 days and (C) 296 days. (D) The comparison of metabolite uptake between D56 and D296 retinal organoids. Data were relative ion abundance over the control. N = 3, ∗p < 0.05 versus Control or D56 organoids. Data are represented as mean ± SEM. t test.
(E–G) Metabolite release from organoids. Metabolite release in retinal organoids at (E) 56 days and (F) 296 days. (G) The comparison of metabolite release between D56 and D296 retinal organoids. Data were relative ion abundance over the control. N = 3, ∗p < 0.05 versus Control or D56 organoids. Data are represented as mean ± SEM. t test.
(H) A schematic of nutrient uptake and release in retinal organoids. Increased metabolites in mature organoids at D296 are colored in red and increased metabolites in D56 organoids are colored in green.
(I) Heatmap of gene expression of top-ranked small molecule transporters in the human retinal organoids cultured at 1-, 3-, 6-, and 9-month stages. FPKM values of bulk RNA sequencing (Kim et al., 2019) was used for plotting.
Small molecule transporters and metabolic enzymes contribute to specific profiles in metabolite uptake and release. To compare the expression of transporters and enzymes, we curated 2,764 classic metabolism genes from KEGG pathways (Table S2) and analyzed their expression in the transcriptome of retinal organoids at stages between 1 and 9 months (Kim et al., 2019). Differentially expressed genes along the time course of human retinal organoids identified previously (Kim et al., 2019) were intersected with the genes in metabolic pathways, resulting in 378 metabolic genes that were differentially expressed during the development of retinal organoids. More than half of the differentially expressed genes were ion and small molecule transporters (Figure S1A, Table S3). Remarkably, glutamate and aspartate transporters including solute carrier 1A3 (SLC1A3), SLC17A7, SLC1A7, and SLC1A2 were the most highly enriched of the upregulated small molecule transporters in 9-month organoids (Figure 1H and Table S3). These results indicate that mature retinal organoids need more aspartate and glutamate. Other amino acids, bicarbonate, and water transporters were also upregulated in the maturing organoids. Glucose transporter 1 (SLC2A1), the major glucose transporter, was expressed similarly during organoid development except for a decrease at 3 months. Monocarboxylate transporter (MCT) is the major lactate/pyruvate transporter in the retina, and MCT4 or SLC16A3 was expressed similarly between 3- and 9-month organoids (Figure 1H). These results were consistent with our metabolite consumption data on glucose and lactate. Glutamate decarboxylase 1 and 2 (GAD1/2) convert glutamate into 4-aminobutyrate (GABA) (Figure 1I). They were highly upregulated in D296 organoids (Figure S1B), providing further evidence of the need for glutamate. GABA transporters were also increased in the mature organoids (Table S3). Genes in cysteine, cystathionine, glutathione, and histidine/histamine metabolism were highly enriched in the mature organoids (Figure S1B). This may implicate in the changed consumption of cysteine derivatives such as cystine, hypotaurine, and histamine. Apart from aspartate and glutamate metabolism, most upregulated genes in mature retinal organoids were involved in glycan, glycolysis, lipids, ATP-binding cassette (ABC) transporter (lipid transporter), and nucleotides, especially cyclic nucleotides and purine degradation (Figures S1C–S1E, Table S3). Increased purine degradation and recycling may be the cause of the reduced consumption of hypoxanthine that we observed (Figures 1D and 1I). To sum up, retinal organoids showed distinctive features of nutrient consumption and developmentally regulated expression of metabolic genes. Mature retinal organoids may import and utilize aspartate and glutamate through their upregulated transporters for neurotransmitter synthesis and energy metabolism (Figure 1I).
Metabolite Consumption and Metabolic Gene Expression of Rod-Dominant and All-Cone Mouse Retinas
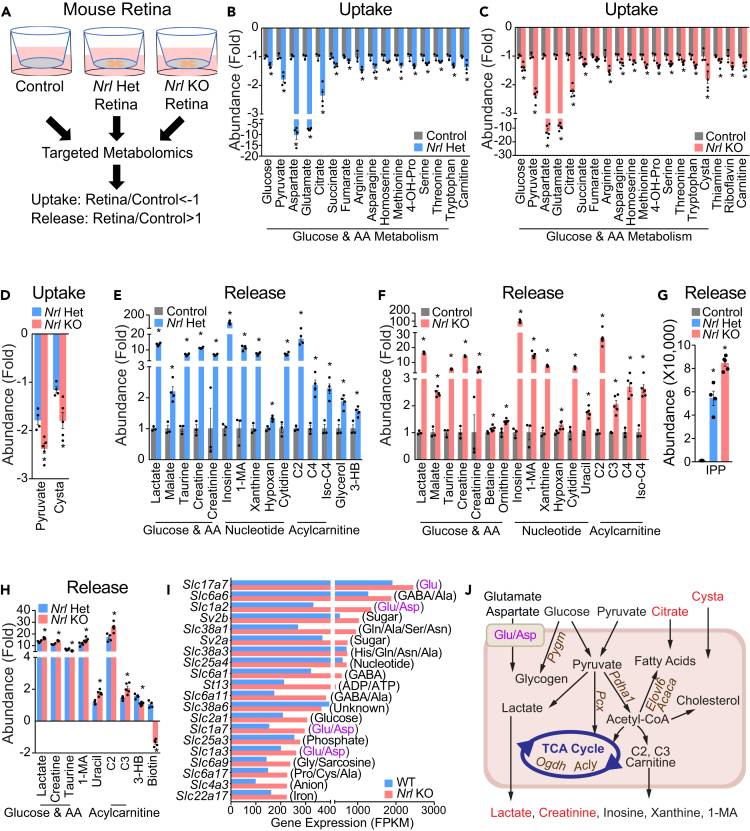
Rods comprise ∼97% of photoreceptors in mouse retinas. Knockout (KO) of Nrl converts all rods into cone-like photoreceptors (Mears et al., 2001) to make all-cone retinas. We cultured retinas from ∼1-month-old Nrl-heterozygous (Het) (rod-dominant similar to wild type [WT]) mice and Nrl KO mice in transwells and analyzed metabolites in the media after culturing for 24 h (Figure 2A). Compared with the control media, 32 metabolites from Nrl Het and 37 metabolites from Nrl KO media were significantly different (Figures 2B–2H). Most changed metabolites were in a similar pattern in the Het and KO retinas. Interestingly, aspartate and glutamate were among the most consumed metabolites in both Het and KO retinas, with a decrease of 8- to 11-fold compared with baseline media (Figures 2B and 2C). Additionally, both Het and KO retinas consumed glucose, amino acids, carnitine, and intermediates in mitochondrial metabolism including citrate, succinate, and fumarate (Figures 2B and 2C). Like retinal organoids, mouse retinas excreted large amounts of lactate, acetylcarnitine, and 1-methyladenosine (Figures 2E and 2F). However, mouse retinas also released nucleosides, other acyl-carnitines, amino acids, and derivatives from amino acids such as creatine and creatinine (Figures 2E and 2F). Isopentenyl pyrophosphate (IPP), an intermediate of the mevalonate pathway, was not detected in the control media but was significantly increased in the spent media of mouse retinas, especially the Nrl KO group (Figure 2G). These results indicate that mouse retinas have very active mitochondrial, lipid, and nucleotide metabolism.
Figure 2.
Metabolite Consumption and Metabolic Gene Expression in Nrl Het (Rod-Dominant) and Nrl KO (all-cone) Mouse Retina
(A) Schematic for profiling medium metabolites in mouse retinal explants. Retinas from Nrl Het and Nrl KO mice were dissected, placed in transwells, and maintained in culture for 24 h. The culture media without retinas were the baseline control. All media metabolites were analyzed by targeted metabolomics.
(B–D) Metabolite consumption from mouse retinal explants. Metabolite uptake in (B) Nrl Het and (C) Nrl KO retinas. (D) The comparison of metabolite uptake between Nrl Het and Nrl KO retinas. Data were relative ion abundance over the control or Nrl Het. N = 6, ∗P < 0.05 versus Control or Nrl Het retinas. Data are represented as mean ± SEM. t test.
(E–H) Metabolite release from mouse retinal explants. Metabolite release in (E) Nrl Het and (F) Nrl KO retinas. (G and H) The comparison of metabolite uptake between Nrl Het and Nrl KO retinas. Data were relative ion abundance over the control or Nrl Het or absolute ion abundance when those metabolites were not detected in the baseline control. N = 6, ∗P < 0.05 versus Control or Nrl Het retinas. Data are represented as mean ± SEM. t test.
(I) Gene expression of the top 20 abundant small molecule transporters in the Nrl KO and Het mouse retinas.
(J) Schematic of nutrient uptake and release in Nrl KO retinas. Metabolites with enhanced uptake or release in Nrl KO retinas are colored red. Upregulated genes in Nrl KO are colored brown. Glu/Asp represents transporters for glutamate and aspartate.
See also Figures S2–S3 and Tables S2, S4, S7, and S10.
Twelve metabolites were different between Het and Nrl KO retinas (Figures 2D and 2G–2H). Nrl KO consumed more pyruvate and cystathionine and produced more lactate, acylcarnitines, creatine, and 1-methyladenosine (Figures 2D, 2G–2H). In contrast to retinal organoids, neither Nrl Het nor KO retinas excreted hypotaurine; however, mouse retinas exported more taurine, particularly the Het retinas (Figure 2H). Ketone bodies are important alternative fuels for neurons. Intriguingly, only rod-dominant retinas produced the ketone body, 3-HB (Figures 2E–2H), suggesting that there might be interdependent ketone body utilization between rods and cones.
We then analyzed the expression of metabolic genes from a transcriptome database of WT and Nrl KO retinas (Brooks et al., 2011). Among the most abundant small molecule transporters ranked by either WT or KO retinas, transporters for glutamate and aspartate were among the most enriched (Figures 2I and S2). In particular, Slc17a7 had more abundant transcripts than the other small molecule transporters (Figures 2I and S2). Transporters for glucose, GABA, glutamine, lactate, pyruvate, and ketone bodies were also among the top transporters. Compared with WT, 800 metabolic genes were changed, with 401 upregulated in Nrl KO retinas (Figure S3, Table S4). Ion transport, lipid metabolism, small molecule transport, glycan, amino acid, nucleotide, and sugar metabolism were the major changed pathways (Figure S3). Key enzymes in glycogen metabolism (Pygm, Gaa), pyruvate metabolism (Pcx, Pdha1, Me1), TCA cycle (Cs, Acly, Ogdh, Idh3a), cysteine/cystathionine metabolism (Suox, Ado, Cdo1, Gclm), and fatty acid synthesis (Acaca, Acacb, Oxsm, Elovl6) were significantly upregulated in Nrl KO retinas (Figures 2J and Table S4). These data support the nutrient uptake and release results, indicating that cone photoreceptors require more pyruvate and other nutrients for their robust mitochondrial TCA cycle, lipid synthesis, and amino acid metabolism (Figure 2J).
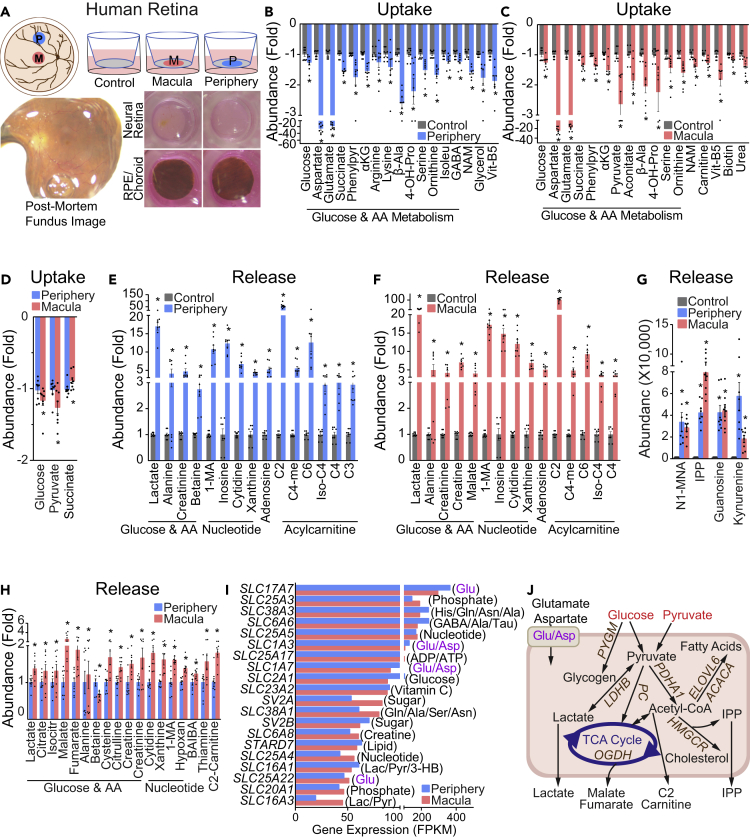
Metabolite Consumption and Metabolic Gene Expression of Human Peripheral and Macular Retina
To investigate the nutrient utilization of the human macula and peripheral retina, we cultured human neural retinal explants punched from the macular and peripheral regions from the same donor (Figure 3A). Human donors had no known retinal diseases, and postmortem fundus photographs showed no abnormalities (Figures 3A and Table S5). After culturing for 24 h, media levels of 52 metabolites in the peripheral retina and 51 metabolites in the macula were significantly different (Figures 3B–3H and S4). Strikingly, aspartate and glutamate were almost completely depleted in spent media from both peripheral retina and macular punches (Figures 3B and 3C). Aspartate and glutamate were supplemented at ∼0.15 mM in all the media of retinal cultures, and they were depleted in mature retinal organoids, mouse retinas, and human retinas (Tables S6–S8). These findings demonstrate that retinas need substantial amounts of aspartate and glutamate. As well as glucose, carnitine, and amino acids, human retinas also actively consumed vitamins such as pantothenic acid, nicotinamide, and biotin (Figures 3B–3D and S4). β-Alanine, which can be metabolized into acetyl-CoA in mitochondria, was also reduced 2- to 3-fold in the media. Similar to mouse retinas, both macular and peripheral retinas released lactate, nucleosides, and acylcarnitines, confirming that human retinas are very active in metabolizing glucose, amino acids, and lipids (Figures 3E–3G and S4). Creatine, creatinine, and citrulline increased 3- to 7-fold in the media with retinal cultures (Figures 3E–3G). These metabolites, synthesized from arginine, are involved in the synthesis and degradation of the high-energy metabolite phosphocreatine. IPP, guanosine, kynurenine, and N1-methylnicotinamide were not detected in the control media but significantly increased in spent media from both macular and peripheral neural retinas (Figure 3G). These results confirm that the mevalonate pathway, purine catabolism, and NAD metabolism are active in the retina.
Figure 3.
Metabolite Consumption and Metabolic Gene Expression in the Human Macular and Peripheral Retina
(A) Schematic diagram of human macular and peripheral explant culture. Macula and Peripheral punches from neural retinas or their corresponding RPE/choroid punches were cultured on inserts in transwell plates for 24 h. Representative fundus images from one donor showed healthy retinas. Representative images of cultured macular and peripheral neural retinas and RPE/choroids in transwell plates.
(B–D) Metabolite consumption from human neural retinal explant culture. Metabolite uptake from (B) peripheral and (C) macular retinal explants. (D) The comparison of metabolite uptake between macular and peripheral retinal explants. Data were relative ion abundance over the control or peripheral retinas. N = 8, ∗P < 0.05 versus Control or peripheral retinas. Data are represented as mean ± SEM. t test.
(E–H) Metabolite release from human neural retinal explant culture. Metabolite release from (E) peripheral and (F) macular retinal explants. (G and H) The comparison of metabolite release between macular and peripheral retinal explants. Data were relative ion abundance over the control or peripheral retinas, or absolute abundance when detected in the baseline control. N = 8, ∗P < 0.05 versus Control or peripheral retinas. Data are represented as mean ± SEM. t test.
(I) Gene expression of the top 20 abundant small molecule transporters in human peripheral and macular retinas.
(J) Schematic of nutrient uptake and release in macular retinas. Metabolites with enhanced uptake or release in macular retinas are colored red. Upregulated genes in macular retinas are colored brown. Glu/Asp represents transporters for glutamate and aspartate.
Twenty-five metabolites in the media were significantly different between the macular and the peripheral retina (Figures 3D and 3G–3H). In general, the neural macula consumed and released more metabolites than the peripheral retina except for succinate, betaine, and kynurenine (Figures 3D and 3G–3H). Like cone-dominant Nrl KO retinas, the neural macula consumed more pyruvate from the media and released more lactate, acetylcarnitine, 1-methyladenosine, and creatine into the media. These data suggest that the neural macula has more active energy metabolism than the peripheral retina. In support of this, the neural macula released more nucleosides and several TCA cycle intermediates than the peripheral retina (Figure 3H).
To further understand the genetic basis for this differential nutrient consumption, we analyzed the expression of metabolic genes in a human transcriptome database (Whitmore et al., 2014). As in the mouse retinas, aspartate and glutamate transporters were highly enriched among the most abundant small molecular transporters in both the macula and peripheral retina, with SLC17A7 being the most abundant of all (Figures 3I and S5). Transporters for glucose, glutamine, taurine, creatine, lactate, and pyruvate were also among the most abundant transporters (Figures 3I and S5). Metabolic gene analysis found 307 upregulated and 192 downregulated genes in neural macula, compared with peripheral retina (Table S9). Similar to mouse Nrl KO retinas, the most changed pathways were in ion transport, small molecule transport, lipid, glycan, amino acid, nucleotide, and sugar metabolism (Figure S6). Changed small molecule transporters were enriched for the transport of sugar, glutathione, glycine, serine, arginine, citrulline, and glutamate (Figure S6). Glycan, a polysaccharide that conjugates with lipid and protein to make the cell membrane, is synthesized mostly from glucose and fructose. Strikingly, most genes in glycan synthesis were upregulated in the macula, but genes responsible for its degradation were downregulated, indicating the neural macula requires more exogenous glucose to meet the high demand for glycans.
Key enzymes in several major metabolic pathways were significantly upregulated in the macula compared with the peripheral retina such as those responsible for glycogen metabolism (PYGM, PYGB, GBE1, UGP2), pyruvate metabolism (PC, ME2, LDHB, LDHD), TCA cycle (OGDH, ACLY, MDH1, ACO1, SDHA), fatty acid synthesis (ACACA, ACSS2, FASN, ELOVL6), mevalonate and cholesterol metabolism (IDI1, HMGCR, HMGCS1), ketone body metabolism (OXCT2, HMGCLL1), and NAD synthesis (QPRT, NMNAT2) (Table S9, Figure 3J). These upregulated pathways in human neural macula were similar to all-cone Nrl KO retinas. By comparing the changed metabolic genes in Nrl KO with the human macula, we found 119 of 177 genes were changed in the same way, including those in the major metabolic pathways (Table S10). These results indicate that all-cone mouse retinas have some metabolic similarities with the cone-enriched human macula. Notably, pyruvate was the only nutrient that was consumed more by both Nrl KO retinas and human macula. Pyruvate carboxylase (PC in human and Pcx in mouse), which utilizes pyruvate for TCA cycle and gluconeogenesis, was also increased in both Nrl KO retinas and macular retinas (Figure 3J). These data demonstrate that the neural macula meets its higher metabolic requirements by utilizing more glucose and pyruvate for the TCA cycle and synthesis of glycan, glycogen, and lipids.
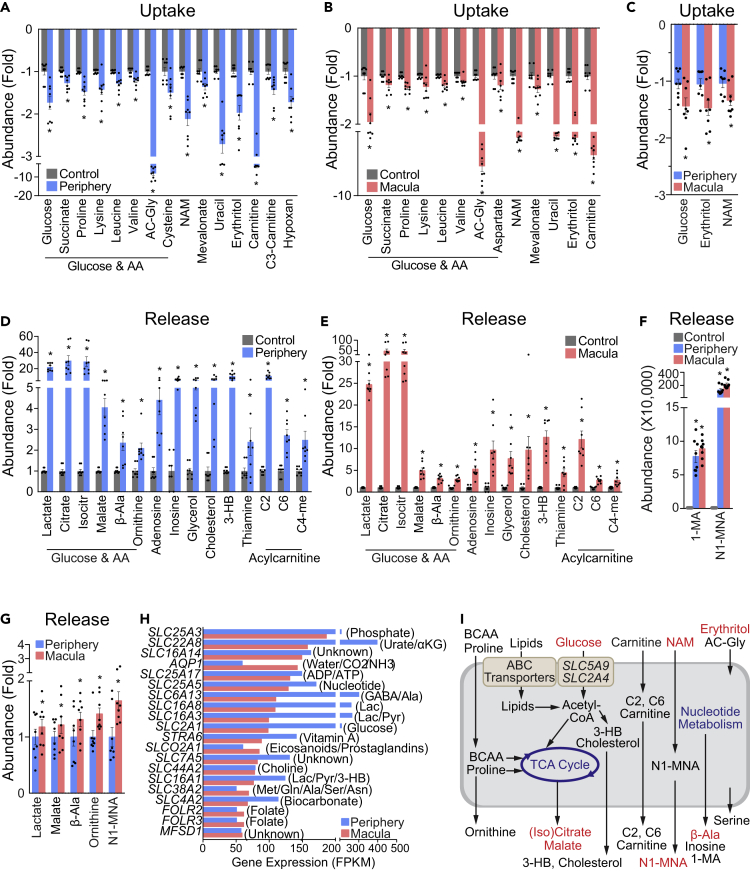
Metabolite Consumption and Metabolic Gene Expression in Human Peripheral and Macular RPE/Choroid
The RPE, tightly bound to the choroid, is critical to transport metabolites to and from the retinas. To understand the metabolic communications between retina and RPE, we studied the nutrient uptake and release of punches of human RPE/choroid using the same donor eyes from the neural retinal experiments (Figure 3A). The profiles of consumption and release by human RPE/choroid punches from both macula and periphery were similar to those of our previously reported studies of human RPE in vitro (Chao et al., 2017) but quite different from those in human retinas (Figures 4A–4G and S7). As well as specific nutrients in the media, such as carnitine, nicotinamide, cysteine, and proline, human RPE/choroid also consumed nutrients not specified in the medium formula such as N-acetylglycine, erythritol, and succinate (Figures 4A and 4B). These metabolites may come from serum and other supplements in the media. In contrast to neural retinas, RPE/choroid did not consume glutamate, and only the macular RPE used a small amount of aspartate (<1.5-fold), confirming that the consumption of glutamate and aspartate is specific to the neural retina. In contrast to human neural retinas, human RPE/choroid consumed, rather than excreted, taurine and betaine (Figures 4D–4G and S7), suggesting that the neural retina and RPE might exchange these metabolites.
Figure 4.
Metabolite Consumption and Metabolic Gene Expression in Human Macular and Peripheral RPE/Choroids
(A–C) Metabolite consumption from human RPE/choroid explant culture. Metabolite uptake from (A) peripheral and (B) macular RPE/choroid explants. (C) The comparison of metabolite uptake between macular and peripheral RPE/choroid explants. Data were relative ion abundance over the control or peripheral RPE/choroid explants. N = 8, ∗P < 0.05 versus Control or peripheral RPE/choroid explants. Data are represented as mean ± SEM. t test.
(D–G) Metabolite release from the macular and peripheral RPE/choroid explant culture. Metabolite release from (D) peripheral and macular (E) RPE/choroid explants. (F–G) The comparison of metabolite uptake between macular and peripheral RPE/choroid explants. Data were relative ion abundance over the control or peripheral RPE/choroid explants, or absolute abundance when detected in the baseline control. N = 8, ∗P < 0.05 versus Control or peripheral RPE/choroid explants. Data are represented as mean ± SEM. t test.
(H) Gene expression of the top 20 abundant small molecule transporters in human peripheral and macular RPE/choroid.
(I) Schematic of nutrient uptake and release in macular RPE/choroid. Red-colored metabolites are those with increased uptake or release in macular RPE/choroid. ABC transporters and glucose transporters (SLC5A9 and SLC2A4) are upregulated in macular RPE/choroid.
See also Figures S7–S9 and Tables S2, S5, S11, and S16.
As well as lactate, nucleosides, and acylcarnitines, RPE/choroid from both macula and periphery exported considerable amounts of citrate, isocitrate, 3-HB, and cholesterol (Figures 4D and 4E), supporting previous reports (Adijanto et al., 2014; Reyes-Reveles et al., 2017) that RPE actively metabolizes lipids. Compared with their corresponding neural retinas, RPE/choroid exported ∼40 times more N1-methylnicotinamide (methylated from nicotinamide), indicating that RPE/choroid has highly active nicotinamide catabolism (Figure 4F). Additionally, RPE/choroid released serine, β-alanine, and glycerol, which were consumed by the human neural retinas (Figures 4D–4E and S7), providing further evidence for metabolic communication between the neural retina and the RPE.
The peripheral RPE/choroid punches consumed and released significantly more metabolites than the macular punches (Figures 4 and S7). However, the macular punches generally consumed or released more of the changed metabolites that they had in common with the peripheral RPE/choroid punches (Figures 4C and 4G). The macular RPE/choroid punches utilized more nicotinamide, erythritol, and glucose than their peripheral counterparts (Figure 4C). Erythritol comes from either additives or endogenous synthesis from glucose. It can be metabolized into erythronic acid, which is abundant in human aqueous humor (Harding et al., 1999). RPE/choroid punches from the macular region also released more N1-methylnicotinamide, malate, β-alanine, and ornithine, which may have been derived from the methylation of nicotinamide and glucose catabolism (Figures 4F, 4G, and 4I).
Unlike neural retinas, no aspartate/glutamate transporters were found in the top 20 most abundant small molecule transporters in the RPE/choroid (Figures 4H and S8). The abundance of small molecule transporters was generally higher in the peripheral RPE than the macula, supporting our data that there were more changed metabolites in the periphery. MCTs for lactate, pyruvate, and ketone bodies and transporters for α-ketoglutarate, sugar, leucine, proline, and choline were abundant in peripheral RPE/choroid (Figures 4H and S8), consistent with the altered uptake and release of these metabolites. The analysis of expression of metabolic genes found that there were 127 genes that were differentially expressed between macular and peripheral RPE/choroid (Table S11). The major altered pathways included lipid metabolism, transporters, and amino acid metabolism (Figure S9). Interestingly, genes involved in glycolysis and the TCA cycle were either decreased or unchanged in the macular RPE/choroid, whereas glucose transporters (SLC2A4, SLC5A9) were significantly increased (Table S11, Figures 4I and S9). Overall, genes in both lipid synthesis and lipid transporters were upregulated in the macular RPE/choroid punches compared with those from the peripheral RPE/choroid retina (Figure 4I).
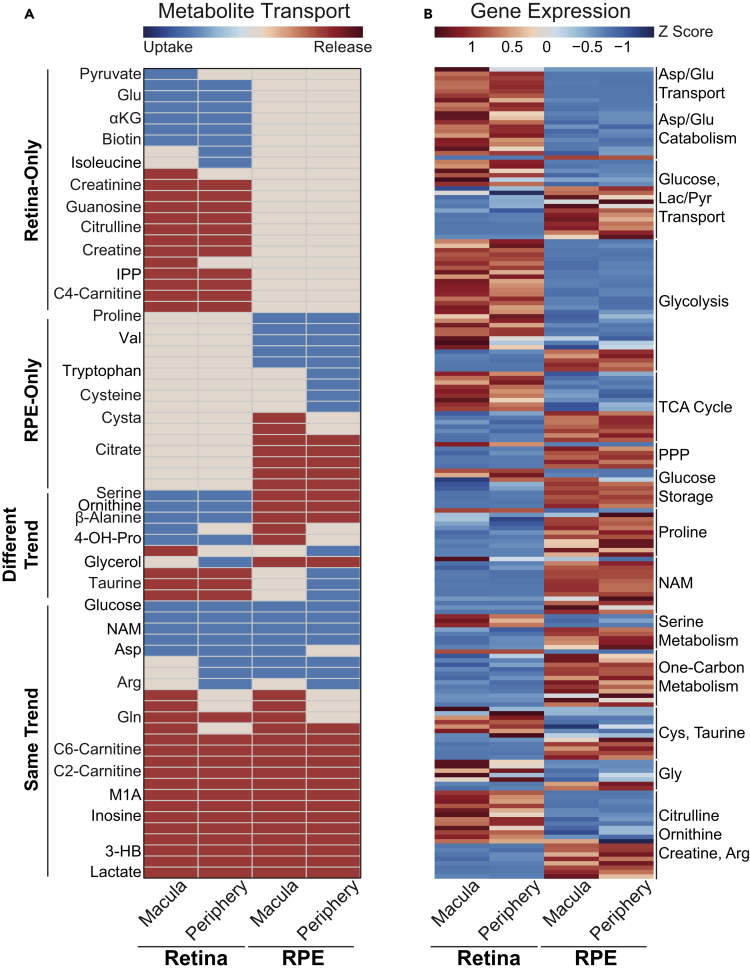
Human Retina and RPE/Choroid Have Distinctive but Symbiotic Metabolic Features
To compare the metabolic features of the human retina and RPE/choroid, we studied the uptake and release of metabolites in the media from their cultures (Figures 3, 4 and 5A). We found 25 metabolites were consumed and released in the same pattern between retinas and RPE/choroid such as glucose, nicotinamide, carnitine, lactate, and inosine. Twenty-two metabolites were consumed or released only by the neural retina, indicating these metabolites are neural retina specific. Sixteen metabolites were consumed or released only by RPE/choroid, indicating that they are RPE specific (Figure 5A). Interestingly, ten metabolites were consumed or released in opposite patterns, such as serine, ornithine, and β-alanine (Figure 5A), which were released by RPE/choroid but consumed by the neural retinas, suggesting that retinas and RPE are metabolically interdependent. Overall, we found neural retinas and RPE/choroid not only share some common pathways but also have their distinctive preferences in nutrient consumption and production.
Figure 5.
Comparison of Metabolite Consumption and Metabolic Genes between Human Retinas and RPE/Choroids
(A) Differential metabolite consumption and release between human retinas and RPE/choroids. Metabolites are colored for changes over their baseline controls. Blue, metabolite consumption; red, metabolite release; and yellow, unchanged.
(B) Comparing gene expression in major metabolic pathways between human retinas and RPE/choroids.
See also Figures S10–S11 and Tables S12, S13, and S14.
About half of the 2,764 metabolic genes we studied were expressed differentially in human neural retinas versus RPE/choroids: 1,473 in the macula and 1,770 in the periphery (Tables S12 and S13). Transporters, lipids, glycan, amino acid, nucleotide, and sugar metabolism were again the major altered pathways between retinas and RPE/choroids (Figure S4). All genes involved in aspartate/glutamate transport and catabolism were upregulated in the neural retinas except aspartoacylase, which regenerates aspartate from N-acetyl-aspartate (Figures 5B and Table S13). This strongly suggests that retinas have a specific requirement for aspartate and glutamate, as summarized in Figure 6. More transporters for glucose, lactate, and pyruvate were found in RPE/choroid but they were also enriched in the retinas, indicating that these metabolites are essential nutrients for both tissues. As expected, neural retinas predominantly expressed genes involved in glycolysis to support their robust aerobic glycolysis (Figures 5B and S10, Table S14). Interestingly, many mitochondrial TCA cycle genes expressed different isoforms in neural retinas versus RPE/choroid. For example, NAD+-dependent IDH3A/B, ADP-dependent SUCLA2, ACO2, and L2HGDH were predominantly expressed in the neural retinas, whereas NADP+-dependent IDH1/2, GDP-dependent SUCLG2, ACO1, and D2HGDH were mainly expressed in the RPE/choroid (Figure 6, Table S14). Notably, upregulated TCA cycle genes in the neural retinas were mostly NAD+ dependent. This differential expression suggests neural retinas and RPE have different roles in mitochondrial bioenergetics and biosynthesis. Strikingly, NADP(H)-associated pathways, including the pentose phosphate pathway (PPP) and one-carbon metabolism, were highly upregulated in the RPE/choroid compared with neural retinas (Figure 5B). Genes in serine synthesis were upregulated in RPE/choroid, but serine transporters were upregulated in the neural retinas. This was consistent with our findings that serine was released by RPE/choroid and consumed by retinas (Figures 5B and Table S14). Remarkably, genes involved in proline metabolism and nicotinamide/NAD metabolism were significantly upregulated in RPE/choroid, consistent with our findings that RPE/choroid consumed proline and nicotinamide. Many genes in amino acid metabolism pathways, such as those for cysteine, taurine, and citrulline, were partly upregulated in neural retinas or RPE/choroids, further indicating that there are symbiotic metabolic relationships between them (Figure 6).
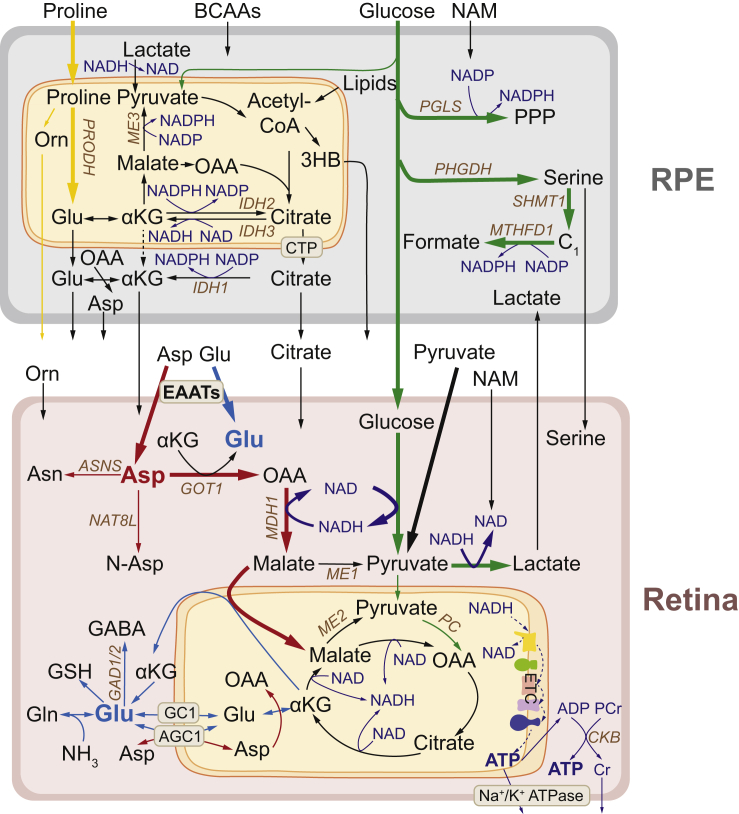
Figure 6.
A Proposed Model of Metabolic Communications between Human Retina and RPE/Choroid
Glucose transports through RPE from the choroid to the retina (bold green line). RPE strongly expresses genes in PPP, serine de novo synthesis, and one-carbon metabolism, which can use glucose to generate NADPH. The retina strongly expresses glycolic genes to produce lactate and NADH. The retina imports aspartate and glutamate through upregulated transporters (bold red line). Aspartate can be converted into OAA and malate to recycle NADH into NAD+ and bring electrons into mitochondria to produce ATP. Aspartate can also be used for the synthesis of glutamate, asparagine (ASN), N-acetylaspartate (Asp). The retina needs a large amount of glutamate for neurotransmission, the mitochondrial TCA cycle, and biosynthesis of GABA, glutathione (GSH), and glutamine (Gln) (bold blue line). RPE mitochondria use different substrates such as proline (yellow line), BCAAs, and lipids to synthesize NADPH and other metabolites that can be released to be used by the retina. Cone-rich retinas or human macula consumes more pyruvate (bold black line) for mitochondrial metabolism. Brown colored genes are upregulated genes in the retinas or RPE.
See also Figure S11.
Discussion
We have compiled an atlas of nutrient consumption and utilization in human retinal organoids, mouse retinas, all-cone mouse retinas of Nrl KO, and human macular and peripheral retinas. By combining nutrient consumption with metabolic gene analysis, we have identified the following key features of neural retinal and RPE metabolism (Figure 6).
-
•
Neural retinal metabolism centers on NADH-related metabolism for ATP production and the synthesis of neurotransmitters such as glutamate and GABA. The neural retina consumes glucose, aspartate, glutamate, citrate, nicotinamide, and other nutrients to sustain its active glycolysis, mitochondrial oxidative metabolism, and neurotransmitter synthesis.
-
•
Cone-rich retinas and the neural macula have more active glycolysis and mitochondrial metabolism, through which they consume more pyruvate, than the rod-rich peripheral neural retina.
-
•
Mature retinal organoids have aspartate/glutamate metabolism that is similar to the human neural retina.
-
•
RPE/choroid metabolism centers on NADPH-related and acetyl-CoA metabolism for biosynthesis. RPE/choroid consumes glucose, proline, nicotinamide, BCAAs, and other nutrients for its activate PPP, reductive carboxylation, serine synthesis, one-carbon metabolism, citrate synthesis, ketone body synthesis, and cholesterol synthesis.
-
•
Neural retinas and RPE/choroid have specific and complementary profiles in nutrient consumption and production that support their tight metabolic communications.
Uptake of aspartate and glutamate was a common feature in cone-rich retinal organoids and mouse and human retinas. Transporters and enzymes responsible for aspartate and glutamate metabolism were also highly enriched in the retina. Why do retinas actively consume aspartate and glutamate? Aspartate and glutamate are interconvertible by transamination with their ketoacids through GOT1/2 (Figure 6). We recently also reported that aspartate transamination is the primary pathway for the catabolism of aspartate and glutamate in mouse neural retinas (Xu et al., 2020). As a major neurotransmitter, glutamate is the most abundant amino acid in the retina (Kalloniatis et al., 1996; Marc et al., 1995). Retinal cells with the highest glutamate levels also have the highest aspartate levels (Kalloniatis et al., 1996). We have previously reported that aspartate is utilized robustly to provide both carbons and nitrogen groups for glutamate synthesis (Du et al., 2013; Lindsay et al., 2014; Xu et al., 2020). Notably, aspartate transamination also generates oxaloacetate (OAA), which can be converted to malate by cytosolic malate dehydrogenase (MDH1) to carry electrons into mitochondria through the malate-aspartate shuttle (Figure 6). This is important because it may help address the paradox of how the retina supports its active mitochondrial metabolism, whereas its metabolism relies on the Warburg effect that allows only a small proportion of pyruvate to be oxidized. MDH1 and GOT1 transcripts are more abundant than any other TCA cycle enzymes in both human and mouse neural retinas (Figure S11), supporting that aspartate catabolism is enhanced in the retina. Aspartate catabolism may (1) recycle NADH into NAD+ to sustain the highly active glycolysis, (2) provide cytosolic electrons for ATP production, (3) replenish mitochondrial OAA for biosynthesis, and (4) produce cytosolic and mitochondrial pyruvate through malic enzymes (Figure 6). Interestingly, low glucose or inhibition of glucose oxidation could cause massive accumulation of aspartate in the neural retina with severe depletion of glutamate (Du et al., 2013; Grenell et al., 2019; Zeevalk and Nicklas, 2000). Therefore, aspartate utilization supports active glycolysis, mitochondrial metabolism, and glutamate synthesis. Additionally, aspartate is an immediate precursor for asparagine and N-acetylaspartate (NAA), both of which are important for neuronal function (Ramos et al., 2011; Ruzzo et al., 2013) (Figure 6). Deficiency of the mitochondrial aspartate/glutamate carrier 1(AGC1) reduces aspartate and NAA, resulting in brain defects and impaired visual function (Contreras et al., 2016; Jalil et al., 2005; Ramos et al., 2011).
Besides being a neurotransmitter, glutamate is an important substrate for the TCA cycle and the synthesis of glutamine and glutathione (Figure 6). Decreased glutamate availability substantially reduces the levels of glutamine and glutathione (Grenell et al., 2019). Glutamate also serves as a major nitrogen donor to synthesize non-essential amino acids by transaminases and is a precursor for GABA by decarboxylation (Figure 6). Consistently, glutamate alanine transaminase and glutamate decarboxylases are strongly expressed in the neural retina (Whitmore et al., 2014). Blood levels of aspartate and glutamate are quite low (Dasarathy et al., 2010; Hui et al., 2017); it remains unclear how they are transported across retina-blood barriers. RPE may be a source of aspartate and glutamate for the retina. We have reported that RPE can metabolize glucose and proline into glutamate and aspartate, which are exported to be used by the retina (Chao et al., 2017; Yam et al., 2019). Notably, glutamate content decreases in cone photoreceptors overlying RPE defects in atrophic AMD (Jones et al., 2016). There is a moderate expression of multiple transporters for aspartate and glutamate in the RPE (Whitmore et al., 2014).
Cones are known to have much higher energy needs than rods (Korenbrot, 2012). It is postulated that mitochondrial metabolism may support the extra energy demand, as cones have 2–10 times more mitochondria and 3 times more cristae membrane surface than rods (Hoang et al., 2002; Perkins et al., 2003, 2004). The generation of higher lactate, acetylcarnitine, and IPP (synthesized from acetyl-CoA) in both all-cone mouse retinas and the neural macula, which is cone-rich, provides strong evidence that cones are more metabolically active than rods. We also found all-cone neural retinas and macula consume more pyruvate than the peripheral neural retina, which is rod dominant. This may explain how cones fuel their abundant mitochondria even when they have active aerobic glycolysis. The SLC16A3 (MCT4), which has low affinity for pyruvate in order to preserve intracellular pyruvate levels (Bergersen, 2015), is strongly expressed in the neural macula (Table S9). Inhibition of pyruvate utilization causes visual impairment in mice and fish due to photoreceptor degeneration (Grenell et al., 2019; Taylor et al., 2004). Pyruvate provides metabolic advantages that enhance both glycolysis and mitochondrial metabolism. Pyruvate can regenerate NAD+ through lactate dehydrogenase to stimulate glycolysis. Mitochondria need a constant supply of acetyl groups from acetyl-CoA and OAA to synthesize citrate to sustain the TCA cycle for oxidative and biosynthetic metabolism (Figure 6). Pyruvate provides both acetyl groups and OAA through pyruvate dehydrogenase and PC. The neural retina can also use fatty acids and ketone bodies, but these nutrients only provide acetyl groups (Joyal et al., 2018; Reyes-Reveles et al., 2017). Consistently, pyruvate carboxylase expression was upregulated in both all-cone retinas and neural macula. Hepatic deletion of PC in mice diminishes TCA cycle intermediates, especially aspartate, and depletes NADPH and glutathione (Cappel et al., 2019). PPP is the classic pathway to generate NADPH for the regeneration of visual pigment, but its activity is very low in the neural retina and photoreceptors (Adler et al., 2014; Futterman and Kinoshita, 1959; Winkler et al., 1986). The regeneration of visual pigment in cones is at least 100 times faster than rods (Chen et al., 2009). Pyruvate could stimulate PPP activity 15 times in isolated neural retinas (Futterman and Kinoshita, 1959) and may contribute to the high demand for NADPH in cones. Pyruvate also increases the availability of glutamate and glutamine in vitro (Gamberino et al., 1997; Lieth et al., 2001). We have reported that retinal deletion of the mitochondrial pyruvate carrier leads to the accumulation of pyruvate and aspartate but the depletion of glutamate, glutamine, and glutathione results in retinal degeneration (Grenell et al., 2019). Interestingly, pyruvate supplementation prevents light-induced retinal degeneration (Natoli et al., 2016; Ren et al., 2011; Sato et al., 2020). Further research is warranted to determine whether increasing pyruvate utilization by supplementation or genetic approaches might have a neuroprotective effect on the macula.
The RPE is closely coupled with photoreceptors in nutrient utilization (Han et al., 2020; Kanow et al., 2017; Reyes-Reveles et al., 2017; Xu et al., 2020). In agreement with other studies of RPE cells (Chao et al., 2017; Xu et al., 2020; Yam et al., 2019), we found that human RPE/choroid explants consumed glucose, proline, nicotinamide, and BCAAs and released plenty of 3-HB, citrate, and cholesterol. All these released metabolites are made from acetyl-CoA, indicating that RPE mitochondria function like mitochondria in the liver that oxidize nutrients to support other tissues. Interestingly, retinas could readily utilize the released 3-HB, citrate, and cholesterol for bioenergetics and biosynthesis (Chao et al., 2017; Futterman and Kinoshita, 1959; Grenell et al., 2019; Reyes-Reveles et al., 2017; Tserentsoodol et al., 2006). The different consumption of metabolites by the neural retina and RPE/choroid suggests an important metabolic coupling between them. For example, RPE released serine, but the retina consumed serine. Serine is an essential precursor for phospholipid synthesis and one-carbon metabolism. Photoreceptors require active phospholipid biosynthesis to renew their daily shedding of outer segments, but enzymes for serine synthesis are highly expressed in RPE (Figure 5B) (Sinha et al., 2020). The uptake of serine by our neural retinal cultures and upregulated serine transporters sideroflexin 1/3 (SFXN1/3) in the neural retina further suggest that retinas need exogenous serine. Mutations of SFXN3 leads to progressive outer retinal degeneration (Chen et al., 2020). Remarkably, low serine from the RPE may account for the synthesis of toxic deoxysphingolipids in the degenerative macular condition, macular telangiectasia type 2 (Gantner et al., 2019).
In summary, profiling nutrient uptake and release by tissue explants provides a useful platform to investigate tissue-specific and inter-tissue metabolism and to functionally annotate the ever-increasing tissue-specific metabolic transcriptome. By profiling mouse and human retinal explants, we reveal distinctive metabolic features in progenitor versus differentiated retinal organoids, mouse rod versus cone-retinas, and human peripheral neural retina versus the neural macula. These findings provide a resource of basic information for future studies on the retina and RPE metabolism.
Limitations of the Study
Traditional culture media were used for our organoid and retinal explant culture. These media may not reflect the microenvironment in vivo. For example, no lactate was included in these media, but RPE in an eye may be exposed to high amounts of lactate from the neural retina. A recent study showed that RPE can uptake lactate to preserve glucose for photoreceptors (Kanow et al., 2017). Future investigation of metabolite concentrations in choroid and vortex veins will be important for developing a medium formulation that more accurately mimics in vivo conditions. Additionally, the glucose concentrations in our culture were higher than physiological conditions owing to the limitation of traditional static culture. A microfluidic perifusion with a physiological concentration of glucose is under exploration to optimize human retinal explant culture. Our human RPE explant culture provides information for metabolite uptake and release, but it cannot be used to study metabolite transport because the RPE explant only covers part of the transwell membrane and there is no biological barrier between the upper and lower chamber. Human neural retinal explants cultured together with human RPE cells can be investigated with stable-isotope tracers to understand metabolite transport and communication between neural retina and RPE. Cone-like photoreceptors in Nrl KO eyes may still be different from mature cones in wild animals. Future studies on cone-rich animals such as squirrels, fish, and chicken will help to validate our findings on cone-specific metabolism. A limitation of our targeted metabolomics analysis is that we reported the ion abundance, not absolute abundance. Evaluation of absolute abundance will require labeled internal standard for each metabolite to avoid matrix effect in the mass spectrometry analyses. Future technical improvements in our analysis procedure will focus on determining the absolute concentration of many of the key metabolites in retinas and RPE. Finally, the human retinas from our explant culture underwent postmortem delay, which can influence metabolism in these tissues.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianhai Du (jianhai.du@wvumedicine.org).
Materials Availability
Materials are available upon request from Dr. Jianhai Du.
Data and Code Availability
The Nrl mouse transcriptome datasets are available at NEI Commons: https://neicommons.nei.nih.gov/#/dataDetail/442, https://neicommons.nei.nih.gov/#/dataDetail/445. All data used in this manuscript are available upon request from the lead author. No custom code was used in the analysis of the data.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Anand Swaroop and Dr. Zachary Batz from the National Eye Institute for sharing Nrl KO animals and their transcriptome datasets. We appreciate the comments from Dr. James Hurley from the University of Washington. We acknowledge the NSW lion tissue bank's support for this research. This work was supported by NIH Grants EY026030 (J.D.), EY029806 (W.L.), EY022645 (W.L.), the Retina Research Foundation (J.D.), the BrightFocus Foundation (J.D.), the Orphan Disease Center, University of Pennsylvania (MDBR-19-130-CRB1 W.L.), funds for Core facilities P20 GM103434 (WV INBRE grant), WVCTSI grant GM104942, funds from Australian NHMRC project grant (APP1145121 M.C.G.) and the Lowy Medical Research Institute. M.C.G. is a Sydney Medical School Fellow and supported by an Australian NHMRC practitioner fellowship.
Author Contributions
Conceptualization, J.D.; Investigation, B.L., T.Z., W.L., Y.W., R.X., S. Zeng, R.Z., S. Zhu, L.Z.; Writing, B.L., T.Z., W.L., L.Z., M.C.G., and J.D.; Funding Acquisition, J.D.; Supervision, L.Z., M.C.G., and J.D.
Declaration of Interests
The authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101672.
Supplemental Information
References
- Adijanto J., Du J., Moffat C., Seifert E.L., Hurle J.B., Philp N.J. The retinal pigment epithelium utilizes fatty acids for ketogenesis. J. Biol. Chem. 2014;289:20570–20582. doi: 10.1074/jbc.M114.565457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler L.t., Chen C., Koutalos Y. Mitochondria contribute to NADPH generation in mouse rod photoreceptors. J. Biol. Chem. 2014;289:1519–1528. doi: 10.1074/jbc.M113.511295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergersen L.H. Lactate transport and signaling in the brain: potential therapeutic targets and roles in body-brain interaction. J. Cereb. Blood Flow Metab. 2015;35:176–185. doi: 10.1038/jcbfm.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbach C.M., Hass D.T., Robbings B.M., Rountree A.M., Sadilek M., Sweet I.R., Hurley J.B. Succinate can shuttle reducing power from the hypoxic retina to the O2-rich pigment epithelium. Cell Rep. 2020;31:107606. doi: 10.1016/j.celrep.2020.107606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M.J., Rajasimha H.K., Roger J.E., Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes. Mol. Vis. 2011;17:3034–3054. [PMC free article] [PubMed] [Google Scholar]
- Brown E.E., DeWeerd A.J., Ildefonso C.J., Lewin A.S., Ash J.D. Mitochondrial oxidative stress in the retinal pigment epithelium (RPE) led to metabolic dysfunction in both the RPE and retinal photoreceptors. Redox Biol. 2019;24:101201. doi: 10.1016/j.redox.2019.101201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappel D.A., Deja S., Duarte J.A.G., Kucejova B., Inigo M., Fletcher J.A., Fu X., Berglund E.D., Liu T., Elmquist J.K. Pyruvate-carboxylase-Mediated anaplerosis promotes antioxidant capacity by sustaining TCA cycle and redox metabolism in liver. Cell Metab. 2019;29:1291–1305.e8. doi: 10.1016/j.cmet.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson R.J., Wood J.P., Han G., Kittipassorn T., Peet D.J., Chidlow G. M-Type pyruvate kinase isoforms and lactate dehydrogenase A in the mammalian retina: metabolic implications. Invest. Ophthalmol. Vis. Sci. 2016;57:66–80. doi: 10.1167/iovs.15-17962. [DOI] [PubMed] [Google Scholar]
- Chao J.R., Knight K., Engel A.L., Jankowski C., Wang Y., Manson M.A., Gu H., Djukovic D., Raftery D., Hurley J.B. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J. Biol. Chem. 2017;292:12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Aredo B., Ding Y., Zhong X., Zhu Y., Zhao C.X., Kumar A., Xing C., Gautron L., Lyon S. Forward genetic analysis using OCT screening identifies Sfxn3 mutations leading to progressive outer retinal degeneration in mice. Proc. Natl. Acad. Sci. U S A. 2020;117:12931–12942. doi: 10.1073/pnas.1921224117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Blakeley L.R., Koutalos Y. Formation of all-trans retinol after visual pigment bleaching in mouse photoreceptors. Invest. Ophthalmol. Vis. Sci. 2009;50:3589–3595. doi: 10.1167/iovs.08-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras L., Ramirez L., Du J., Hurley J.B., Satrustegui J., de la Villa P. Deficient glucose and glutamine metabolism in Aralar/AGC1/Slc25a12 knockout mice contributes to altered visual function. Mol. Vis. 2016;22:1198–1212. [PMC free article] [PubMed] [Google Scholar]
- Daniele L.L., Lillo C., Lyubarsky A.L., Nikonov S.S., Philp N., Mears A.J., Swaroop A., Williams D.S., Pugh E.N., Jr. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasarathy J., Gruca L.L., Bennett C., Parimi P.S., Duenas C., Marczewski S., Fierro J.L., Kalhan S.C. Methionine metabolism in human pregnancy. Am. J. Clin. Nutr. 2010;91:357–365. doi: 10.3945/ajcn.2009.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Cleghorn W.M., Contreras L., Lindsay K., Rountree A.M., Chertov A.O., Turner S.J., Sahaboglu A., Linton J., Sadilek M. Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. J. Biol. Chem. 2013;288:36129–36140. doi: 10.1074/jbc.M113.507285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Yanagida A., Knight K., Engel A.L., Vo A.H., Jankowski C., Sadilek M., Tran V.T., Manson M.A., Ramakrishnan A. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. U S A. 2016;113:14710–14715. doi: 10.1073/pnas.1604572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrington D.A., Fisher C.R., Kowluru R.A. Mitochondrial defects drive degenerative retinal diseases. Trends Mol. Med. 2020;26:105–118. doi: 10.1016/j.molmed.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.R., Ferrington D.A. Perspective on AMD pathobiology: a bioenergetic crisis in the RPE. Invest. Ophthalmol. Vis. Sci. 2018;59:AMD41–AMD47. doi: 10.1167/iovs.18-24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterman S., Kinoshita J.H. Metabolism of the retina. I. Respiration of cattle retina. J. Biol. Chem. 1959;234:723–726. [PubMed] [Google Scholar]
- Gamberino W.C., Berkich D.A., Lynch C.J., Xu B., LaNoue K.F. Role of pyruvate carboxylase in facilitation of synthesis of glutamate and glutamine in cultured astrocytes. J. Neurochem. 1997;69:2312–2325. doi: 10.1046/j.1471-4159.1997.69062312.x. [DOI] [PubMed] [Google Scholar]
- Gantner M.L., Eade K., Wallace M., Handzlik M.K., Fallon R., Trombley J., Bonelli R., Giles S., Harkins-Perry S., Heeren T.F.C. Serine and lipid metabolism in macular disease and peripheral neuropathy. N. Engl. J. Med. 2019;381:1422–1433. doi: 10.1056/NEJMoa1815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenell A., Wang Y., Yam M., Swarup A., Dilan T.L., Hauer A., Linton J.D., Philp N.J., Gregor E., Zhu S. Loss of MPC1 reprograms retinal metabolism to impair visual function. Proc. Natl. Acad. Sci. U S A. 2019;116:3530–3535. doi: 10.1073/pnas.1812941116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.Y.S., Kinoshita J., Bisetto S., Bell B.A., Nowak R.A., Peachey N.S., Philp N.J. Role of monocarboxylate transporters in regulating metabolic homeostasis in the outer retina: insight gained from cell-specific Bsg deletion. FASEB J. 2020;34:5401–5419. doi: 10.1096/fj.201902961R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding J.J., Hassett P.C., Rixon K.C., Bron A.J., Harvey D.J. Sugars including erythronic and threonic acids in human aqueous humour. Curr. Eye Res. 1999;19:131–136. doi: 10.1076/ceyr.19.2.131.5334. [DOI] [PubMed] [Google Scholar]
- Hoang Q.V., Linsenmeier R.A., Chung C.K., Curcio C.A. Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis. Neurosci. 2002;19:395–407. doi: 10.1017/s0952523802194028. [DOI] [PubMed] [Google Scholar]
- Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Le Z., Yanxiang Guo J. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.B., Lindsay K.J., Du J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015;93:1079–1092. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalil M.A., Begum L., Contreras L., Pardo B., Iijima M., Li M.X., Ramos M., Marmol P., Horiuchi M., Shimotsu K. Reduced N-acetylaspartate levels in mice lacking aralar, a brain- and muscle-type mitochondrial aspartate-glutamate carrier. J. Biol. Chem. 2005;280:31333–31339. doi: 10.1074/jbc.M505286200. [DOI] [PubMed] [Google Scholar]
- Johnson J.E., Jr., Perkins G.A., Giddabasappa A., Chaney S., Xiao W., White A.D., Brown J.M., Waggoner J., Ellisman M.H., Fox D.A. Spatiotemporal regulation of ATP and Ca2+ dynamics in vertebrate rod and cone ribbon synapses. Mol. Vis. 2007;13:887–919. [PMC free article] [PubMed] [Google Scholar]
- Jones B.W., Pfeiffer R.L., Ferrell W.D., Watt C.B., Tucker J., Marc R.E. Retinal remodeling and metabolic alterations in human AMD. Front. Cell. Neurosci. 2016;10:103. doi: 10.3389/fncel.2016.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyal J.S., Gantner M.L., Smith L.E.H. Retinal energy demands control vascular supply of the retina in development and disease: the role of neuronal lipid and glucose metabolism. Prog. Retin. Eye Res. 2018;64:131–156. doi: 10.1016/j.preteyeres.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalloniatis M., Marc R.E., Murry R.F. Amino acid signatures in the primate retina. J. Neurosci. 1996;16:6807–6829. doi: 10.1523/JNEUROSCI.16-21-06807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., Linton J.D., Farnsworth C.C., Sloat S.R., Rountree A. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6:e28899. doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lowe A., Dharmat R., Lee S., Owen L.A., Wang J., Shakoor A., Li Y., Morgan D.J., Hejazi A.A. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. U S A. 2019;116:10824–10833. doi: 10.1073/pnas.1901572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbrot J.I. Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: facts and models. Prog. Retin. Eye Res. 2012;31:442–466. doi: 10.1016/j.preteyeres.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Westenskow P.D., Gantner M.L., Usui Y., Schultz A., Bravo S., Aguilar E., Wittgrove C., Friedlander M., Paris L.P. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife. 2016;5:e14319. doi: 10.7554/eLife.14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveillard T., Sahel J.A. Metabolic and redox signaling in the retina. Cell. Mol. Life Sci. 2017;74:3649–3665. doi: 10.1007/s00018-016-2318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieth E., LaNoue K.F., Berkich D.A., Xu B., Ratz M., Taylor C., Hutson S.M. Nitrogen shuttling between neurons and glial cells during glutamate synthesis. J. Neurochem. 2001;76:1712–1723. doi: 10.1046/j.1471-4159.2001.00156.x. [DOI] [PubMed] [Google Scholar]
- Lindsay K.J., Du J., Sloat S.R., Contreras L., Linton J.D., Turner S.J., Sadilek M., Satrustegui J., Hurley J.B. Pyruvate kinase and aspartate-glutamate carrier distributions reveal key metabolic links between neurons and glia in retina. Proc. Natl. Acad. Sci. U S A. 2014;111:15579–15584. doi: 10.1073/pnas.1412441111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc R.E., Murry R.F., Basinger S.F. Pattern recognition of amino acid signatures in retinal neurons. J. Neurosci. 1995;15:5106–5129. doi: 10.1523/JNEUROSCI.15-07-05106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Narayan D.S., Chidlow G., Wood J.P., Casson R.J. Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin. Exp. Ophthalmol. 2017;45:730–741. doi: 10.1111/ceo.12952. [DOI] [PubMed] [Google Scholar]
- Natoli R., Rutar M., Lu Y.Z., Chu-Tan J.A., Chen Y., Saxena K., Madigan M., Valter K., Provis J.M. The role of pyruvate in protecting 661W photoreceptor-like cells against light-induced cell death. Curr. Eye Res. 2016;41:1473–1481. doi: 10.3109/02713683.2016.1139725. [DOI] [PubMed] [Google Scholar]
- Okawa H., Sampath A.P., Laughlin S.B., Fain G.L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008;18:1917–1921. doi: 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G.A., Ellisman M.H., Fox D.A. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol. Vis. 2003;9:60–73. [PubMed] [Google Scholar]
- Perkins G.A., Ellisman M.H., Fox D.A. The structure-function correlates of mammalian rod and cone photoreceptor mitochondria: observations and unanswered questions. Mitochondrion. 2004;4:695–703. doi: 10.1016/j.mito.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Petit L., Ma S., Cipi J., Cheng S.Y., Zieger M., Hay N., Punzo C. Aerobic glycolysis is essential for normal rod function and controls secondary cone death in retinitis pigmentosa. Cell Rep. 2018;23:2629–2642. doi: 10.1016/j.celrep.2018.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala A., Gupta V.K., Anderson R.E., Rajala R.V. Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion. 2013;13:566–576. doi: 10.1016/j.mito.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M., Pardo B., Llorente-Folch I., Saheki T., Del Arco A., Satrustegui J. Deficiency of the mitochondrial transporter of aspartate/glutamate aralar/AGC1 causes hypomyelination and neuronal defects unrelated to myelin deficits in mouse brain. J. Neurosci. Res. 2011;89:2008–2017. doi: 10.1002/jnr.22639. [DOI] [PubMed] [Google Scholar]
- Rashid A., Bhatia S.K., Mazzitello K.I., Chrenek M.A., Zhang Q., Boatright J.H., Grossniklaus H.E., Jiang Y., Nickerson J.M. RPE cell and sheet properties in normal and diseased eyes. Adv. Exp. Med. Biol. 2016;854:757–763. doi: 10.1007/978-3-319-17121-0_101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Liu N.Y., Song X.F., Ma Y.S., Zhai X.Y. A novel specific application of pyruvate protects the mouse retina against white light damage: differential stabilization of HIF-1alpha and HIF-2alpha. Invest. Ophthalmol. Vis. Sci. 2011;52:3112–3118. doi: 10.1167/iovs.10-5605. [DOI] [PubMed] [Google Scholar]
- Reyes-Reveles J., Dhingra A., Alexander D., Bragin A., Philp N.J., Boesze-Battaglia K. Phagocytosis-dependent ketogenesis in retinal pigment epithelium. J. Biol. Chem. 2017;292:8038–8047. doi: 10.1074/jbc.M116.770784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda E.M., Johnson J.E., Jr., Giddabasappa A., Swaroop A., Brooks M.J., Sigel I., Chaney S.Y., Fox D.A. The cellular and compartmental profile of mouse retinal glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and ∼P transferring kinases. Mol. Vis. 2016;22:847–885. [PMC free article] [PubMed] [Google Scholar]
- Ruzzo E.K., Capo-Chichi J.M., Ben-Zeev B., Chitayat D., Mao H., Pappas A.L., Hitomi Y., Lu Y.F., Yao X., Hamdan F.F. Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron. 2013;80:429–441. doi: 10.1016/j.neuron.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Mochida S., Tomimoto D., Konuma T., Kiyota N., Tsuda S., Shiga Y., Omodaka K., Nakazawa T. A pyruvate dehydrogenase kinase inhibitor prevents retinal cell death and improves energy metabolism in rat retinas after ischemia/reperfusion injury. Exp. Eye Res. 2020;193:107997. doi: 10.1016/j.exer.2020.107997. [DOI] [PubMed] [Google Scholar]
- Sinha T., Naash M.I., Al-Ubaidi M.R. The symbiotic relationship between the neural retina and retinal pigment epithelium is supported by utilizing differential metabolic pathways. iScience. 2020;23:101004. doi: 10.1016/j.isci.2020.101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnes A.C., Huisingh C., McGwin G., Jr., Sloan K.R., Ablonczy Z., Smith R.T., Curcio C.A., Ach T. Multi-nucleate retinal pigment epithelium cells of the human macula exhibit a characteristic and highly specific distribution. Vis. Neurosci. 2016;33:e001. doi: 10.1017/S0952523815000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.R., Hurley J.B., Van Epps H.A., Brockerhoff S.E. A zebrafish model for pyruvate dehydrogenase deficiency: rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc. Natl. Acad. Sci. U S A. 2004;101:4584–4589. doi: 10.1073/pnas.0307074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tserentsoodol N., Sztein J., Campos M., Gordiyenko N.V., Fariss R.N., Lee J.W., Fliesler S.J., Rodriguez I.R. Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol. Vis. 2006;12:1306–1318. [PubMed] [Google Scholar]
- Wang W., Kini A., Wang Y., Liu T., Chen Y., Vukmanic E., Emery D., Liu Y., Lu X., Jin L. Metabolic deregulation of the blood-outer retinal barrier in retinitis pigmentosa. Cell Rep. 2019;28:1323–1334.e4. doi: 10.1016/j.celrep.2019.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S.S., Wagner A.H., DeLuca A.P., Drack A.V., Stone E.M., Tucker B.A., Zeng S., Braun T.A., Mullins R.F., Scheetz T.E. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res. 2014;129:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler B.S., DeSantis N., Solomon F. Multiple NADPH-producing pathways control glutathione (GSH) content in retina. Exp. Eye Res. 1986;43:829–847. doi: 10.1016/s0014-4835(86)80013-6. [DOI] [PubMed] [Google Scholar]
- Xu R., Ritz B.K., Wang Y., Huang J., Zhao C., Gong K., Liu X., Du J. The retina and retinal pigment epithelium differ in nitrogen metabolism and are metabolically connected. J. Biol. Chem. 2020;295:2324–2335. doi: 10.1074/jbc.RA119.011727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam M., Engel A.L., Wang Y., Zhu S., Hauer A., Zhang R., Lohner D., Huang J., Dinterman M., Zhao C. Proline mediates metabolic communication between retinal pigment epithelial cells and the retina. J. Biol. Chem. 2019;294:10278–10289. doi: 10.1074/jbc.RA119.007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevalk G.D., Nicklas W.J. Lactate prevents the alterations in tissue amino acids, decline in ATP, and cell damage due to aglycemia in retina. J. Neurochem. 2000;75:1027–1034. doi: 10.1046/j.1471-4159.2000.0751027.x. [DOI] [PubMed] [Google Scholar]
- Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J.L. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Nrl mouse transcriptome datasets are available at NEI Commons: https://neicommons.nei.nih.gov/#/dataDetail/442, https://neicommons.nei.nih.gov/#/dataDetail/445. All data used in this manuscript are available upon request from the lead author. No custom code was used in the analysis of the data.