Abstract
Mammalian genomes encode thousands of long noncoding RNAs (lncRNAs), yet the biological functions of most of them remain unknown. A particularly rich repertoire of lncRNAs found in mammalian brain and in the early embryo. We used RNA‐seq and computational analysis to prioritize lncRNAs that may regulate commitment of pluripotent cells to a neuronal fate and perturbed their expression prior to neuronal differentiation. Knockdown by RNAi of two highly conserved and well‐expressed lncRNAs, Reno1 (2810410L24Rik) and lnc‐Nr2f1, decreased the expression of neuronal markers and led to massive changes in gene expression in the differentiated cells. We further show that the Reno1 locus forms increasing spatial contacts during neurogenesis with its adjacent protein‐coding gene Bahcc1. Loss of either Reno1 or Bahcc1 leads to an early arrest in neuronal commitment, failure to induce a neuronal gene expression program, and to global reduction in chromatin accessibility at regions that are marked by the H3K4me3 chromatin mark at the onset of differentiation. Reno1 and Bahcc1 thus form a previously uncharacterized circuit required for the early steps of neuronal commitment.
Keywords: Bahcc1, embryonic stem cells, long noncoding RNA, neuronal differentiation, Reno1
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Neuroscience; RNA Biology
Reno1 lncRNA is required for early stages of neuronal differentiation of mouse embryonic stem cells, and acts together with its adjacent protein coding gene Bahcc1.
Introduction
Genome‐wide transcriptome analyses performed over the past two decades revealed that a major proportion of the eukaryotic genome is transcribed into noncoding RNAs (Okazaki et al, 2002; Bertone et al, 2004; Carninci et al, 2005; Iyer et al, 2015). These include long noncoding RNAs (lncRNAs), a class of RNA Pol. II products that are longer than 200 nucleotides, begin with a 5′ cap, and end with a poly(A) tail. Thousands of lncRNAs have been reported in different vertebrates (Guttman et al, 2009; Cabili et al, 2011; Ulitsky et al, 2011; Derrien et al, 2012; Pauli et al, 2012; Necsulea et al, 2014; Hezroni et al, 2015), but it is unclear how many of them are functional.
Both human and mouse brains express a relatively rich repertoire of lncRNAs (Ravasi et al, 2006; Cabili et al, 2011; Derrien et al, 2012), many of which display unique temporal and spatial expression patterns within the central nervous system (CNS) (Mercer et al, 2008, 2010; Ramos et al, 2013), pointing at specific functions in different types of neurons and/or during different stages of neuronal development. Indeed, several lncRNAs have been shown to be required for proper neuronal differentiation in various in vivo and in vitro models (Hezroni et al, 2020).
lncRNA loci are enriched in proximity to genes involved in transcription (Ponjavic et al, 2009; Ulitsky et al, 2011; Hezroni et al, 2015), and some such lncRNAs were found to either regulate the transcription or affect the activity of their adjacent transcription factors (TFs). For example, Evx1as was found to promote the transcription of its neighboring TF Evx1 and regulate mesendodermal differentiation of mouse ES cells (Luo et al, 2016). An example for a lncRNA which affects the activity of an adjacent TF is Six3OS, a lncRNA transcribed from an independent promoter separated by ~ 4 kb from the promoter of Six3, and was found to be involved in retinal cell specification in neonatal mice, by binding factors known to co‐regulate Six3 target genes, possibly through recruitment of histone modifiers (Rapicavoli et al, 2011), and there are other examples of such cooperation in the nervous system (Hezroni et al, 2020).
A few large‐scale studies based on RNAi screens have enabled the identification of lncRNAs related to pluripotency and neuronal differentiation of mouse ES cells. One of these studies generated shRNAs targeting 226 different lncRNAs expressed in mouse ES cells, and found that knockdown of 137 of the 147 lncRNAs which could be efficiently targeted caused a significant impact on gene expression patterns (Guttman et al, 2011). Another study generated a genome‐scale shRNA library that enabled the identification of 20 lncRNAs involved in pluripotency (Lin et al, 2014). Knockdown during neuronal differentiation of one of these lncRNAs, Tuna [also known as Megamind (Ulitsky et al, 2011)], was shown to reduce the differentiation efficiency (Lin et al, 2014).
Taken together, these studies show that lncRNAs play important roles in diverse fundamental processes that occur during the development of the nervous system and in the regulation of gene expression patterns in ES cells. However, for the vast majority of lncRNAs expressed in the CNS and during the establishment of neural cell fate, no known function has been found yet, and the mechanisms underlying lncRNA roles during neurogenesis are poorly understood.
Results
Identification of conserved lncRNAs induced during neuronal differentiation of mouse ES cells
In order to study the roles of lncRNAs during neuronal differentiation, we used a previously described stepwise differentiation protocol (Ying et al, 2003), constituting of differentiation from mouse ES cells into neural progenitor cells (NPCs), followed by differentiation into neurons. After 4 days of differentiation in a monolayer culture the cells express NPC markers such as Nestin, and after four additional days they form a neural population, with over 70% of the cells expressing the neuronal marker Tuj1 (TUBB3) (Fig 1A). To identify lncRNAs with potential roles during neuronal differentiation, we performed strand‐specific RNA‐seq in ES cells as well as in cells after 4 or 8 days of differentiation. Protein‐coding genes upregulated during differentiation included TFs known to regulate neuronal development, such as Dbx1, Nr2f1, and Hes5, as well as neuronal genes, such as Nefm which comprises the axoskeleton of neurons (Fig EV1A), and were enriched for GO terms related to neurogenesis and development of the nervous system, as well as general developmental processes (Fig EV1B). We quantified the expression levels of mouse lncRNAs previously annotated using PLAR (Hezroni et al, 2015), and could detect the expression (FPKM > 1) of 2,294 lncRNA genes in at least one of the differentiation time points (Dataset EV1). Out of these, 1,190 were upregulated (> 1.5‐fold) either during differentiation from ES cells to NPCs or from NPCs to neurons (Dataset EV2). To narrow this list down to lncRNAs that are more likely to be functional, we focused on conserved lncRNAs. Based on clusters of orthologous lncRNAs from 17 vertebrate species (Hezroni et al, 2015), we identified 179 lncRNAs with sequence similarity to a lncRNA in at least two additional species (Dataset EV3). Finally, we included only lncRNAs highly expressed (FPKM > 5) in the CNS, either in adult mice or during embryonic development, based on public datasets. These criteria led us to focus on 121 lncRNAs (Dataset EV4), which included several with known functions in the nervous system, such as Cyrano (Ulitsky et al, 2011), Six3OS (Rapicavoli et al, 2011), Miat (Sone et al, 2007; Barry et al, 2014), and Crnde (Ellis et al, 2012), as well as lncRNAs adjacent to TFs known to be involved in the development of the CNS, such as Nr2f1 (Naka et al, 2008), Lhx1 (Zhao et al, 2007), Sox4 (Bergsland et al, 2006), and Irx2 (Matsumoto et al, 2004).
Figure 1. Expression of candidate lncRNAs during neuronal differentiation of mouse ES cells.
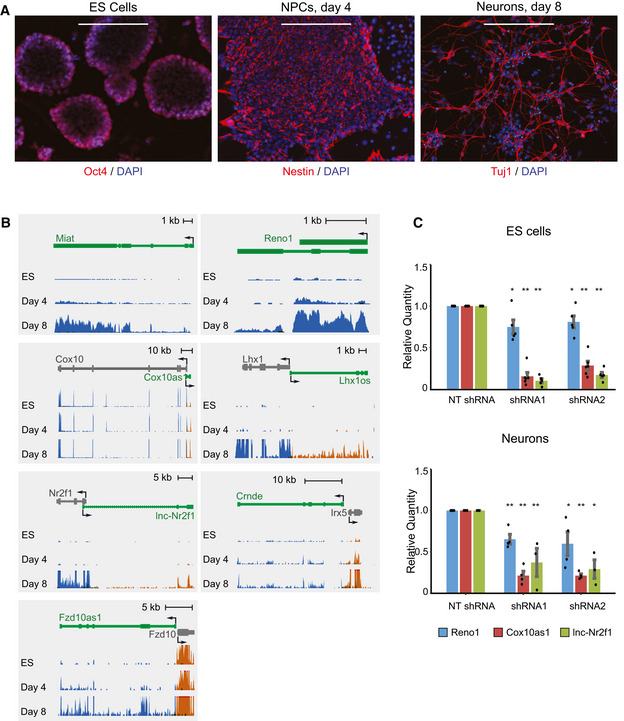
- Immunostaining of ES cells (Oct4), NPCs (day 4, Nestin), and neurons (day 8, Tuj1). Scale bar: 200 μm.
- Expression levels of candidate lncRNAs during three stages of differentiation, measured by RNA‐seq. Gene models of lncRNAs appear in green, and adjacent protein‐coding genes appear in gray. Blue depicts transcripts from the “+” strand, and orange represents transcripts from the “−” strand. All three RNA‐seq tracks were scaled separately to the maximum coverage in each locus.
- qRT–PCR of Reno1, Cox10as1, and lnc‐Nr2f1 in ES cells (top) and following 8 days of neuronal differentiation (bottom), following knockdown using two different shRNAs, normalized to sh‐NT. Mean ± SEM is shown for 3–6 independent experiments, *P < 0.05, **P < 0.01 (unpaired two‐sample t‐test).
Figure EV1. Induction of neuronal and developmental genes during neuronal differentiation of ES cells.
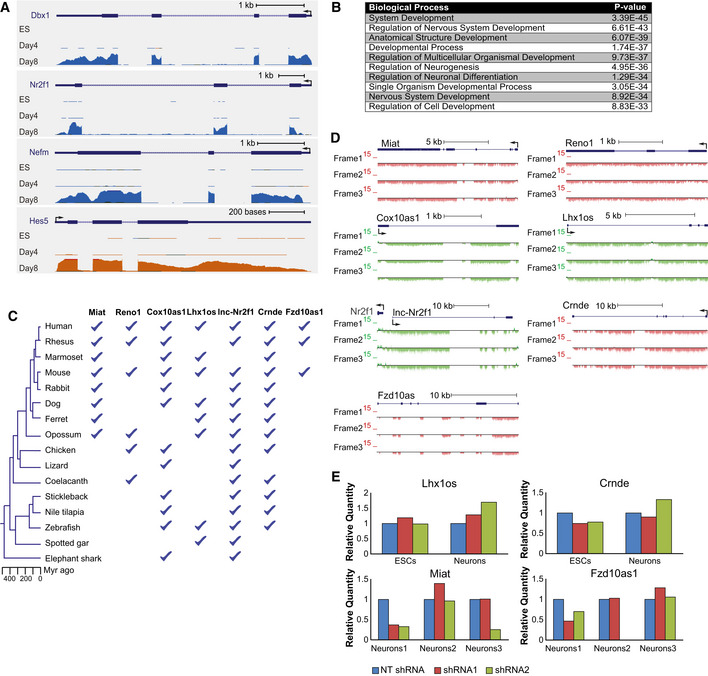
- RNA‐seq read coverage at four of the protein‐coding genes which were the most upregulated during differentiation. Colors and scaling are as described in Fig 1B.
- Top Biological Process Gene Ontology (GO) categories enriched in genes upregulated during neuronal differentiation, as identified using GOrilla (Eden et al, 2009).
- Species represented in clusters of sequence‐similar lncRNAs containing each of the candidate lncRNAs, from Hezroni et al (2015).
- PhyloCSF scores (Lin et al, 2011) throughout the candidate lncRNAs loci in the three possible frames.
- qRT–PCR of Lhx1os, Crnde, Miat, and Fzd10as1 following infection with two targeting shRNAs for knockdown in ES cells and following 8 days of neuronal differentiation (top), or three biological repeats of cells following 8 days of differentiation (bottom), all normalized to the non‐targeting shRNA (sh‐NT).
Knockdown of Reno1 and lnc‐Nr2f1 inhibits neuronal differentiation
We focused on seven lncRNAs for functional studies: 2810410L24Rik (which we named Regulator of Early Neurogenesis 1, or Reno1), lnc‐Nr2f1 (A830082K12Rik), Cox10as1 (2810001G20Rik), Miat, Lhx1os, Fzd10as1 (5930412G12Rik), and Crnde. All seven lncRNAs have sequence‐conserved orthologs in several other species (Fig EV1C), are predicted to be noncoding by the three coding predictors in PLAR (Hezroni et al, 2015) and by PhyloCSF (Lin et al, 2011; Fig EV1D), which is not part of PLAR, and their expression levels are gradually upregulated during neuronal differentiation of mouse ES cells (Fig 1B). In order to interrogate the functions of the candidate lncRNAs during neuronal differentiation, we carried out RNAi knockdown (KD) experiments, using lentiviral delivery of pLKO.1 shRNA plasmids. ES cells were infected with two shRNAs targeting each of the candidate lncRNAs, or with a non‐targeting shRNA (sh‐NT). The cells were then induced to differentiate, and lncRNA expression levels were measured in ES cells and in differentiated neurons using qRT–PCR. We were able to obtain efficient and reproducible KD for three out of the seven candidate lncRNAs: Reno1, Cox10as1, and lnc‐Nr2f1. For Cox10as1 and lnc‐Nr2f1 we obtained > 70% reduction in ES cells and > 60% reduction in differentiated neurons, while for Reno1 we obtained ~ 20% reduction in ES cells and ~ 30% reduction in neurons (Fig 1C). The other four lncRNAs could not be efficiently targeted using shRNAs: the shRNAs either did not have any effect on the expression levels of the lncRNAs at any stage (Lhx1os), or no effect following differentiation (Crnde), or the effect was not reproducible (Miat, Fzd10as1) (Fig EV1E).
The efficiency of neuronal differentiation following KD of Reno1, lnc‐Nr2f1, or Cox10as1 was estimated by immunostaining of the neuronal marker Tuj1. The total number of cells, number of Tuj1‐positive cells, and the total neurite length were quantified in ten non‐overlapping fields for each shRNA. Reno1 KD led to a reduced number of cells that survived the differentiation process, as well as a lower percentage of Tuj1‐positive cells and shorter neurites (Fig 2A and B), indicating that depletion of Reno1 inhibits neuronal differentiation. KD of lnc‐Nr2f1 led to an increased cell number, a decreased percentage of Tuj1‐positive cells, and a decreased neurite length (Fig EV2A and B), indicating that it is required for proper neuronal differentiation as well. Unlike Reno1 and lnc‐Nr2f1, for which targeting by two different shRNAs resulted in highly similar effects on the differentiated cells, only one of the two shRNAs targeting Cox10as1 led to a significantly reduced number of cells that survived following differentiation, despite comparable KD efficiencies (Fig EV2C and D), suggesting that the reduced cell number observed with the first shRNA is probably due to off‐target effects. Even with this shRNA, the percentage of Tuj1‐positive cells and the neurite length per neuron were similar to that of sh‐NT infected cells, indicating that this shRNA probably affected cell survival rather than differentiation efficiency.
Figure 2. Knockdown of Reno1 inhibits neuronal differentiation.
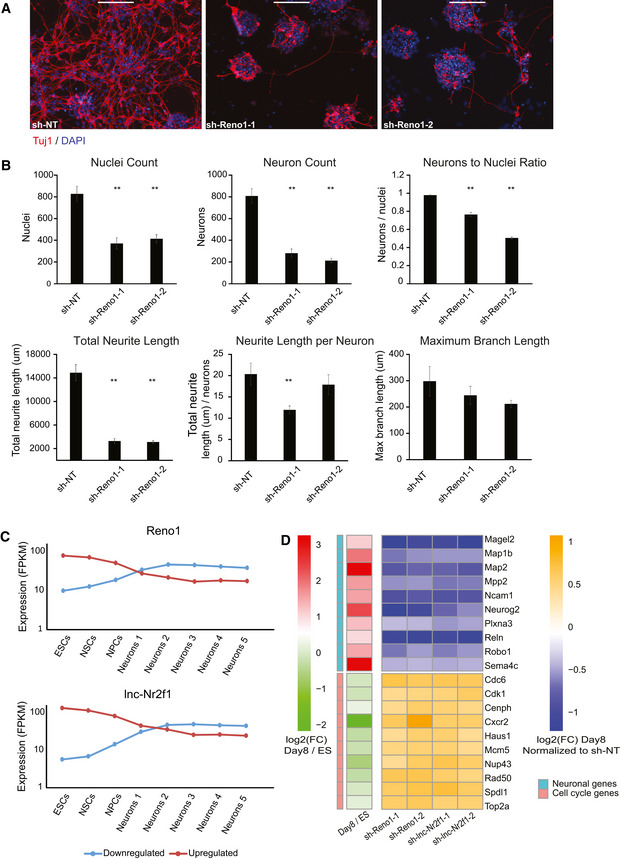
- Immunostaining using anti‐TUJ1 antibody of ES‐cell-derived neurons following infection with either non‐targeting shRNA (sh‐NT), or two different shRNAs targeting Reno1. Scale bar: 200 μm.
- Quantification of cell numbers and neurite lengths of ten images of non‐overlapping fields for each shRNA. Mean ± SEM is shown, **P < 0.01 (unpaired two‐sample t‐test).
- Mean expression levels of genes which were down‐ or upregulated following knockdown (KD) of Reno1 or lnc‐Nr2f1 during an eight time point neuronal differentiation and maturation of mouse ES cells: Neurons1 corresponds to day in vitro (DIV)1; Neurons2—DIV7; Neurons3—DIV16; Neurons4—DIV21; and Neurons5—DIV28 (Hubbard et al, 2013).
- Transcriptional response of cell cycle and neuron‐related genes. Heatmaps show log2FC of cells infected with shRNAs targeting Reno1 or lnc‐Nr2f1 compared to cells infected with sh‐NT, following 8 days of neuronal differentiation, and of day 8 of differentiation compared to ES cells during differentiation of WT cells.
Figure EV2. Effects of Reno1, lnc‐Nr2f1, and Cox10as1 KD on neuronal differentiation.
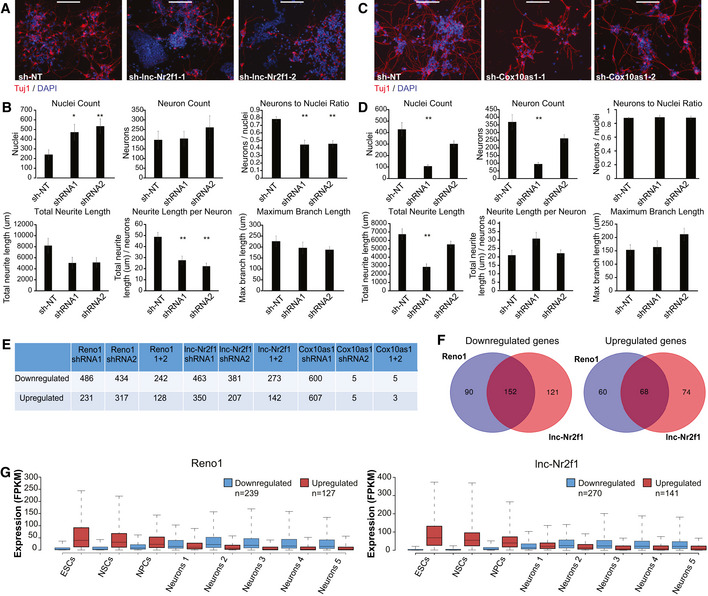
- Immunostaining using anti‐Tuj1 antibody of ES‐cell-derived neurons following infection with either sh‐NT, or two different shRNAs targeting lnc‐Nr2f1. Scale bar: 200 μm.
- Quantification of cell numbers and neurite lengths of ten images of non‐overlapping fields for each shRNA targeting lnc‐Nr2f1 or sh‐NT. Mean ± SEM is shown, *P < 0.05, **P < 0.01 (unpaired two‐sample t‐test).
- Immunostaining using anti‐Tuj1 antibody of ES‐cell-derived neurons following infection with either sh‐NT, or two different shRNAs targeting Cox10as1. Scale bar: 200 μm.
- Quantification of cell numbers and neurite lengths of 10 images of non‐overlapping fields for each shRNA targeting Cox10as1 or sh‐NT. Mean ± SEM is shown, **P < 0.01 (unpaired two‐sample t‐test).
- Number of genes which were significantly (P < 0.01, DESeq2) down‐ or upregulated following KD of Reno1, lnc‐Nr2f1, or Cox10as1.
- Overlap between genes that were significantly down‐ (left) or upregulated (right) following Reno1 and lnc‐Nr2f1 KD.
- Boxplots indicating the median, quartiles, and 5th and 95th percentiles of expression levels of genes which were down‐ or upregulated following KD of Reno1 or lnc‐Nr2f1 during the same neuronal differentiation time points described in Fig 2C. The numbers of genes in each group are indicated in the legend.
Reno1 or lnc‐Nr2f1 KD during neuronal differentiation leads to major dysregulation of gene expression
In order to further characterize the effect of KD of lncRNAs on neuronal differentiation, we performed RNA‐seq at the end of the differentiation process (day 8) after knocking down Reno1, Cox10as1, or lnc‐Nr2f1. Hundreds of genes were significantly down‐ or upregulated (P < 0.05, DESeq2) following infection of shRNAs targeting either Reno1 or lnc‐Nr2f1, while only one of the shRNAs targeting Cox10as1 (the same one that affected cell viability) significantly affected gene expression (Fig EV2E). Due to the inconsistent effect of shRNAs targeting Cox10as1, we focused on Reno1 and lnc‐Nr2f1 for further analyses. Many genes were significantly dysregulated following KD of both Reno1 and lnc‐Nr2f1 (Fig EV2F). In order to characterize the expression patterns of the genes which were affected by Reno1 or lnc‐Nr2f1 KD, we examined the expression levels of these genes in a previously published RNA‐seq dataset of eight time points throughout a 29‐day protocol of neuronal differentiation and maturation of mouse ES cells (Hubbard et al, 2013). Genes which were downregulated in our data following KD of either Reno1 or lnc‐Nr2f1 were induced during neuronal differentiation in this dataset, while genes that were upregulated were reduced (Figs 2C and EV2G). More specifically, neuronal genes such as genes encoding for proteins involved in microtubule assembly, TFs that regulate neuronal development, and proteins involved in axon guidance and other processes that occur during development of the nervous system, were all downregulated following KD Reno1 or lnc‐Nr2f1, while genes related to cell cycle progression and proliferation were upregulated (Fig 2D).
Taken together, our results indicate that Reno1 and lnc‐Nr2f1 are required for proper neuronal differentiation of mouse ES cells. While each lncRNA may employ a unique mode of action, the highly similar effect of KD of these two lncRNAs on neuronal differentiation suggests that depletion of either lncRNA does not allow the cells to acquire neuronal identity and activate the proper molecular pathways.
Reno1 is an intergenic, deeply conserved lncRNA found in close spatial proximity to Bahcc1
We then focused on Reno1, a novel lncRNA which has not been studied previously in any context. Reno1 is an intergenic, highly conserved lncRNA, with sequence‐similar homologs expressed in human, sheep, opossum, chicken, and coelacanth fish (Fig 3A), and a syntenic lncRNA expressed in zebrafish (see Discussion). In the RefSeq database it is annotated as a three‐exon transcript, but based on RNA‐seq data and 3′ RACE in ES‐derived neurons, we propose that there are two main Reno1 transcript variants: a 1.8 kb single‐exon variant, which is typically more abundant, and a spliced three‐exon variant, which is expressed at lower levels and whose relative abundance increases in more mature neurons (Figs 3B and EV3A). Based on RNA‐seq data, Reno1 is predominantly localized to the nucleus, both in ES cells and in motor neurons (Fig 3B). The human homolog RENO1 is also expressed mainly in the fetal brain, and also found mainly in the nucleus (Fig 3C). Single‐molecule fluorescence in situ hybridization (smFISH) in neurons differentiated from WT and Reno1 m/m ES cells (see below) and in mouse intestine confirmed the nuclear localization of Reno1 and suggested that 5–10 copies of Reno1 are found per cell on average, with substantial variability (Figs 3D and EV3B). Reno1 copy number per cell was also estimated using qRT–PCR in ES cells and in cells differentiated for 4 or 8 days. This analysis showed that Reno1 is not detectably expressed in ES cells and five to eight copies of it are found in cells following 4 or 8 days of differentiation, in agreement with the smFISH results (Fig EV3C).
Figure 3. Reno1 is a deeply conserved lncRNA, found in close spatial proximity to Bahcc1 .
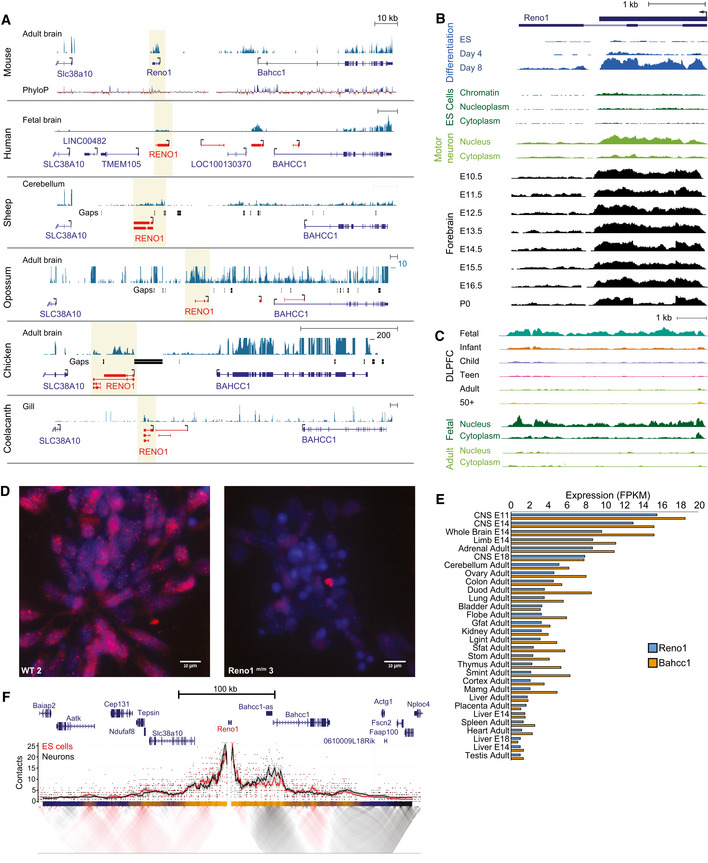
- Reno1 locus in six vertebrate species. Shaded region indicates the Reno1 locus. Blue gene models are from RefSeq or Ensembl, and red gene models are based on PLAR transcript reconstructions (Hezroni et al, 2015). RNA‐seq datasets from indicated tissues were taken from publicly available datasets: BodyMap (Li et al, 2017a) (mouse), Reference Epigenome (human), FAANG (sheep), SRP011985 (opossum), SRP016501 (chicken), and DRP000627 (coelacanth). RNA‐seq data were scaled separately in each species to the highest coverage in the window, except for opossum and chicken, where a custom upper threshold was set as indicated.
- The mouse Reno1 locus. Expression levels during neuronal differentiation of ES cells, in different cellular fractions of ES cells (data from Engreitz et al, 2016) and motor neurons (GSE90913), and in forebrain during different stages of embryonic development (data from the ENCODE project). Each set of tracks was normalized separately.
- RENO1 locus in human. Expression levels in dorsolateral prefrontal cortex across six age groups, and in different cellular fractions in fetal and adult cells. Each set of tracks was normalized separately.
- Reno1 smFISH signal (red) and DAPI staining (blue) in WT and Reno1 m/m cells at day 8 of neuronal differentiation, imaged using 100× objective. Group sizes indicated in Fig EV2G.
- Expression levels of Reno1 and Bahcc1 in 30 adult and embryonic tissues. Data from the ENCODE project, and quantification was done using RSEM (Li & Dewey, 2011).
- Targeted Chromosome Conformation Capture (4C) using the Reno1 promoter as bait. Top, smoothed trend lines and raw counts of the contact profile in ES cells (red) or differentiated neurons (black); bottom, domainogram showing mean contact per fragment end for a series of window sizes.
Figure EV3. Expression levels of Reno1 and Bahcc1 in various tissues and cells.
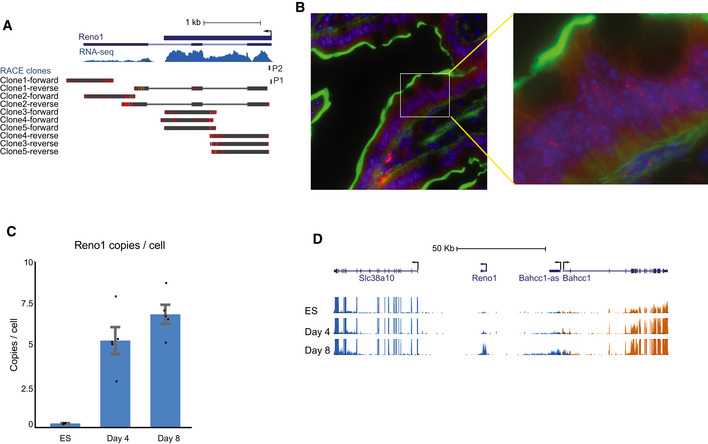
- RNA‐seq read coverage and 3′ RACE on RNA extracted from cells following 8 days of differentiation. BLAT alignments of Sanger sequencing results of five independent 3′ RACE clones and the position of the primer used for 3′RACE (P1) are shown.
- Reno1 smFISH signal (red), DAPI staining (blue) and phall (green) in mouse intestine, imaged using 100× objective.
- Absolute quantification of Reno1 RNA copy number per cell, estimated by qRT–PCR. Mean ± SEM is shown for 3–4 independent experiments.
- RNA‐seq read coverage at the Reno1, Bahcc1, and Slc38a10 loci. Blue depicts transcripts from the “−” strand, and orange represents transcripts from the “+” strand. All tracks are scaled to the same level.
The genes flanking Reno1 throughout vertebrates are Bahcc1 and Slc38a1, which are located ~ 40 kb upstream or downstream of mouse Reno1, respectively (Fig 3A). In mouse, Bahcc1 is gradually upregulated during neuronal differentiation, in a similar manner to Reno1, while Slc38a1 is highly expressed throughout the differentiation process (Fig EV3D). The expression levels of Reno1 and Bahcc1 are highly correlated in various adult and embryonic mouse tissues (Spearman's r = 0.89, Fig 3E), with high expression levels observed mainly during embryonic development of the CNS and differentiation of ES cells to neurons (Fig 3E and Appendix Fig S1).
Since Reno1 is not found in close proximity to any gene on the linear genome, we examined the landscape of its spatial contacts using targeted chromosome conformation capture (4C) (Schwartzman et al, 2016) with Reno1 promoter as the viewpoint. The most dominant association was with the 5′ region of Bahcc1, and this association became stronger following neuronal differentiation (Fig 3F). Together with the highly correlated expression patterns of Reno1 and Bahcc1, these results indicate that Reno1 and Bahcc1 are likely related to each other, in that they are either co‐regulated or regulate each other's expression.
Generation of Reno1 m/m and Bahcc1 m/+ ES cell lines
To further study the functions of Reno1, we transfected ES cells with Cas9 and with two gRNAs, one targeted upstream of the Reno1 promoter and the other in the first exon, leading to a ~ 600 nt deletion (Fig 4A). Using PCR with primers flanking the deleted region, we identified one colony in which one allele contains the desired deletion and the other allele has a shorter deletion (Reno1 m/m 1), and two colonies in which both alleles contain the desired deletion (Reno1 m/m 2 and 3, Fig EV4A), with which we continued for further analysis. Deletions in clones 2 and 3 were validated by Sanger sequencing (Fig EV4B). qRT–PCR analysis showed a strong depletion of Reno1 transcript in both Reno1 m/m 2 and Reno1 m/m 3 lines (Fig 4B). RNA‐seq of two WT lines and Reno1 m/m 2 and 3 confirmed that there are no reads coming from the deleted region, and while there is residual RNA‐seq read coverage downstream of the deleted region, the entire Reno1 transcript is expressed at substantially lower levels compared to WT clones (Fig EV4B). This suggests that a weaker cryptic promoter is likely found downstream of the deleted region, which enables some level of transcription when the main Reno1 promoter is removed.
Figure 4. Reno1 m/m and Bahcc1 m/+ ES cells proliferate normally but fail to survive neuronal differentiation.
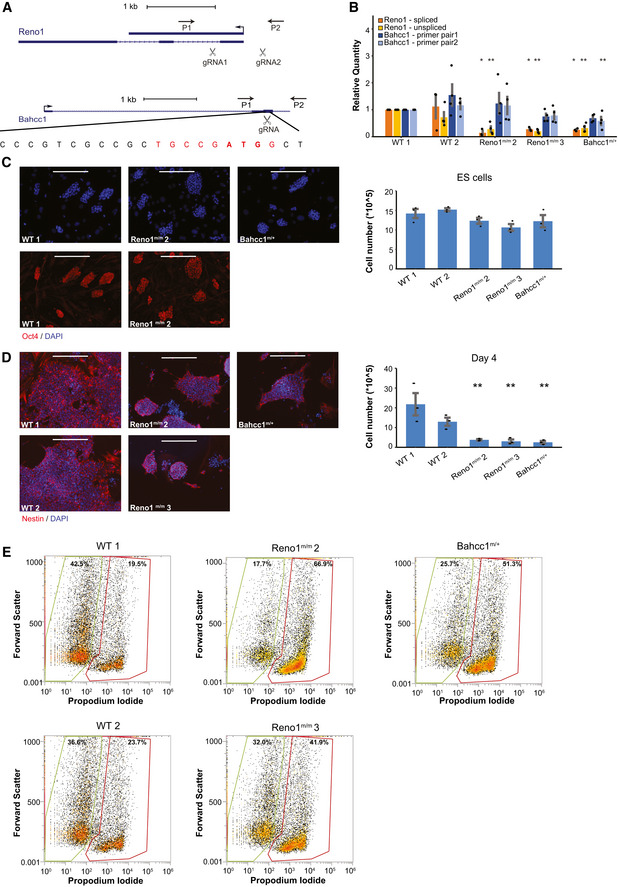
- Generation of Reno1 m/m and Bahcc1 m/+ ES cells using CRISPR‐Cas9. Primers used for colony screening marked by arrows.
- Expression levels of Reno1 (both the spliced and unspliced isoforms) and Bahcc1 following 4 days of differentiation of the indicated cells. Mean ± SEM is shown for 3–4 independent experiments, *P < 0.05, **P < 0.01 (unpaired two‐sample t‐test).
- Left: Oct4 staining of the indicated cells. Scale bar: 200 μm. Right: Cell count of the indicated ES cells grown in ES medium. Cells were harvested from one well of a 6‐well plate and counted using Orflo MOXI Z Mini Automated Cell Counter. Mean ± SEM is shown for three independent experiments.
- Left: Nestin staining of the indicated cells following 4 days of differentiation. Scale bar: 200 μm. Right: Cell count of the indicated cells following 4 days of differentiation. Cells were harvested from one well of a 6‐well plate and counted using Orflo MOXI Z Mini Automated Cell Counter. Mean ± SEM is shown for three independent experiments. **P < 0.01 (unpaired two‐sample t‐test).
- Flow cytometry analyses of dead and live cells in the indicated ES cells following 4 days of differentiation. Y axis: forward scatter; X axis: PI fluorescence intensity.
Figure EV4. Depletion of Reno1 or Bahcc1 inhibits neuronal differentiation.
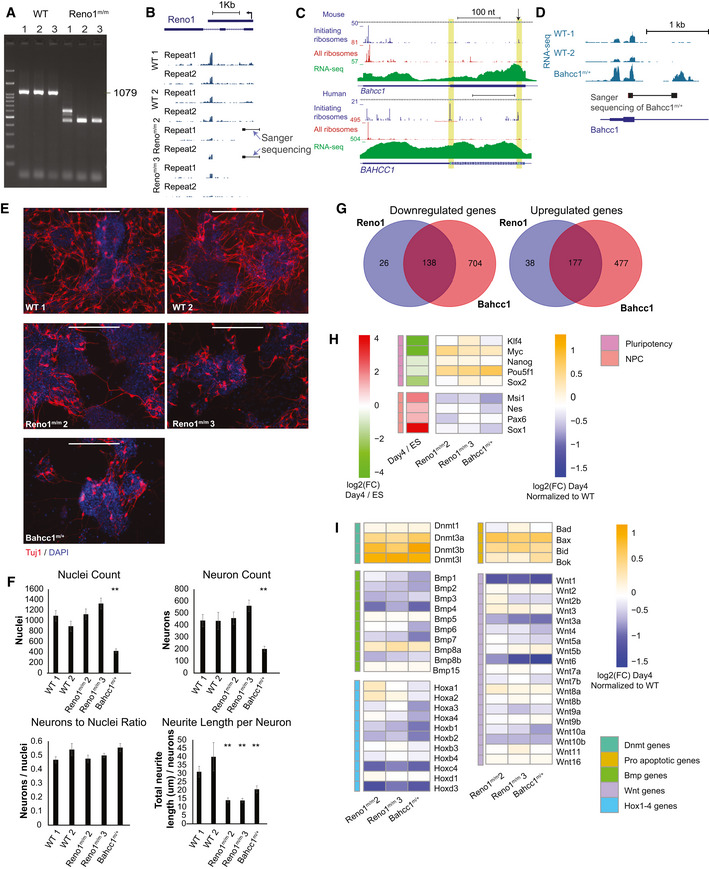
-
AGenotyping of Reno1 m/m ES cells using primers flanking the deleted region.
-
BRNA‐seq read coverage in the Reno1 locus in the RNA‐seq data of differentiated cells in the indicated clones. Deletions validated by Sanger sequencing are shown—the thick boxes indicate Sanger‐sequenced regions and the thin line the gap in the sequence.
-
CRibo‐seq and RNA‐seq read coverage from multiple studies accumulated in the GWISP‐vis database (Michel et al, 2014) for the first coding exon of Bahcc1 in human and mouse. Regions of the two possible start codons in frame with the main coding sequence are highlighted. An arrow indicates that position targeted by the gRNA used to generate Bahcc1 m/+ cells.
-
DRNA‐seq read coverage around the first coding exon of Bahcc1 in the RNA‐seq data of differentiated cells in the indicated clones. Deletion validated by Sanger sequencing in the first clone is shown—the thick boxes indicate Sanger‐sequenced regions and the thin line the gap in the sequence.
-
EImmunostaining using anti‐Tuj1 antibody following 8 day differentiation of WT and Reno1 m/m ES cells.
-
FQuantification of cell numbers and neurite lengths of 16 images of non‐overlapping fields for each shRNA. Mean ± SEM is shown, *P < 0.05, **P < 0.01 (unpaired two‐sample t‐test).
-
GOverlap between genes that were significantly down‐ (left) or upregulated (right) in Reno1 m/m or Bahcc1 m/+ cells at day 4 of differentiation.
-
H, IChanges in gene expression of pluripotency‐ and NPC‐related genes (H) and genes from the indicated gene families (I) after 4 days of differentiation. Heatmaps show log2‐transformed fold changes of mutant clones compared to WT cells following 4 days of neuronal differentiation, and of day 4 of differentiation compared to undifferentiated ES cells.
In order to generate a Bahcc1 mutant, we examined Ribo‐seq data from human and mouse cells and found that Bahcc1 translation initiates from either the annotated ATG codon, or from an alternative ATG found ~ 50 codons downstream in the same exon (Fig EV4C). To deplete Bahcc1 levels, we therefore transfected ES cells with Cas9 and with a gRNA targeting the downstream ATG codon (Fig 4A). We selected colonies where PCR products of primers flanking the start codon were not digested by BccI, which in WT cuts the region of the start codon (Fig 4A). Genotyping of these colonies by Sanger sequencing and RNA‐seq results showed one clone that was heterozygous for a 622 nt deletion surrounding the gRNA cleavage site [akin to such deletions recently described in (Kosicki et al, 2018)] (Fig EV4D). Despite screening > 140 additional clones from ES cells transfected with the same gRNA or with another gRNA targeting exon 5 of Bahccc1, we did not identify any homozygous clones, and so we continued to analyze the heterozygous deletion clone which we refer to as Bahcc1 m/+. The expression level of Bahcc1 mRNA was significantly reduced in this clone (Fig 4B, tested after 4 days of differentiation as Bahcc1 levels are very low in mouse ES cells). Interestingly, Reno1 was also significantly reduced (Fig 4B), suggesting BAHCC1 may regulate Reno1 expression (see Discussion). Reno1 m/m and Bahcc1 m/+ lines grown on MEFs in medium containing serum and LIF formed Oct4 positive colonies similar to WT lines, and when colonies were harvested and counted from one well containing each line, cell numbers were similar (Fig 4C), indicating that depletion of Reno1 or Bahcc1 does not affect the pluripotency of the cells or their proliferation rates.
Low survival rates of Reno1 m/m and Bahcc1 m/+ cell lines when induced to differentiate into neurons
Next, we wanted to examine the effect of Reno1 and Bahcc1 depletion on neuronal differentiation. We induced two WT, two Reno1 m/m, and one Bahcc1 m/+ lines to differentiate. Major cell death was observed in the Reno1 m/m and Bahcc1 m/+ lines, and only few cells survived the first 4 days of differentiation (Fig 4D and E). When surviving cells were harvested and seeded for the second step of differentiation, Reno1 m/m and Bahcc1 m/+ lines formed neurons with shorter neurites compared to WT cells (Fig EV4E and F). These results indicate that even the Reno1 m/m and Bahcc1 m/+ cells that did survive the first step of differentiation, did not form NPCs that were capable of proper differentiation into neurons.
To further characterize the effect of Reno1 and Bahcc1 depletion on the differentiation process, we performed RNA‐seq following 4 days of differentiation. Hundreds of genes were dysregulated in Reno1 m/m and Bahcc1 m/+ cells compared to WT cells, and many genes were up‐ or downregulated following depletion of either Reno1 or Bahcc1 (Fig EV4G). Notably > 80% of the genes significantly affected by Reno1 depletion were also affected by Bahcc1 depletion, suggesting that Reno1 mainly functions in the Bahcc1 pathway, whereas Bahcc1 may have additional, Reno1‐independent functions. Pluripotency genes such as Pou5f1 (Oct4), Nanog, Klf4, Sox2, and Myc were upregulated in Reno1 m/m and Bahcc1 m/+ cells compared to WT cells, while NPC‐related genes such as Nestin, Pax6, Sox1, and Msi1 were downregulated (Fig EV4H). Moreover, specific gene families that have important functions during development were also dysregulated in the Reno1‐ and Bahcc1‐depleted cells including Dnmt3 genes (Dnmt3a, Dnmt3b, Dnmt3l), which are essential for de novo methylation and play important roles in mammalian development (Okano et al, 1999; Chedin et al, 2002), pro‐apoptotic gene members of the Bcl family: Bax, Bid, and Bok, and most members of the Bmp, Wnt, and Hox1–4 families (Fig EV4I). Taken together, our results indicate that when ES cells depleted of Reno1 or Bahcc1 are induced to differentiate into neurons, most of them undergo apoptosis at early differentiation stage. The cells that do survive after 4 days fail to acquire the transcriptional programs that allow proper neuronal differentiation.
Significant effect of Reno1 depletion on gene expression at an early stage of differentiation
We next wanted to characterize the effect of Reno1 perturbation on gene expression following just 2 days of differentiation, prior to the massive cell death observed at day 4. We used RNA‐seq to characterize cells that experienced Reno1 knockdown by shRNAs (Appendix Fig S2A), Reno1 depletion by promoter deletion, or Reno1 knockdown by GapmeRs (Appendix Fig S2B). Transfection of GapmeRs targeting Reno1 led to a significant reduction in number of viable cells at an early stage of differentiation which resembled the outcome of Reno1 depletion by promoter deletion (Appendix Fig S2C). Reno1 depletion did not have a significant effect on Bahcc1 mRNA expression level in any of the perturbation methods (Appendix Fig S2D). Hundreds of genes were dysregulated in Reno1‐depleted cells at day 2, and the gene expression pattern was highly concordant for the different perturbation methods (Fig 5A and B, and Appendix Fig S2E). The 168 genes which were downregulated in at least two of the three perturbations were enriched for GO terms related to cell migration and motility, and to developmental process, while the 94 genes which were upregulated in at least two of the three perturbations methods were enriched for GO terms related to metabolic processes related to translation (Appendix Fig S2F). The upregulated genes are highly expressed in ES cells and their expression is reduced in early stages of neuronal differentiation, while downregulated genes are expressed at low levels in ES cells, and they are induced in early stages of differentiation (Fig 5C and Appendix Fig S2G). These results suggest that appropriate levels of Reno1 transcript at an early stage of neuronal differentiation are required in order to acquire the transcriptional programs that allow proper differentiation.
Figure 5. Transcriptional response to Reno1 depletion and overexpression at day 2 of differentiation.
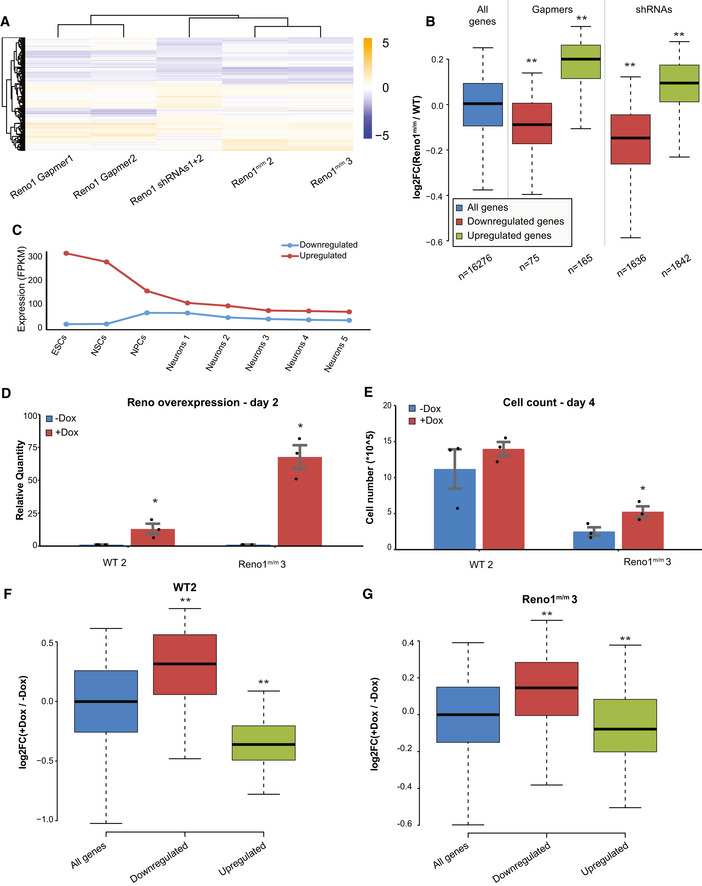
-
AHeatmap showing log2‐transformed fold changes of Reno1‐depleted cells compared to their respective controls. All genes which were significantly differentially expressed (P < 0.05) in any of the treatments are shown.
-
BBoxplots indicating the median, quartiles, and 5th and 95th percentiles of changes in expression levels of Reno1 m/m cells compared to WT cells, for genes with significant (P < 0.05) change in cells where Reno1 was perturbed with the indicated method. Number of genes in each group is indicated in the x axis. **P < 0.01 (two‐sided Wilcoxon rank sum test).
-
CMean expression levels of genes in genes which were significantly (P < 0.05) dysregulated by at least two out of three Reno1 perturbations at day 2 in Fig 5A (n = 168 downregulated and 94 upregulated), during an eight time point neuronal differentiation and maturation of mouse ES cells: Neurons1 corresponds to day in vitro (DIV)1; Neurons2—DIV7; Neurons3—DIV16; Neurons4—DIV21; and Neurons5—DIV28 (Hubbard et al, 2013).
-
DExpression levels of Reno1 following Dox addition in WT and Reno1 m/m cells. Mean ± SEM is shown for three independent experiments, *P < 0.05 (unpaired two‐sample t‐test).
-
ECell count of WT and Reno1 m/m cells following 4 days of differentiation, with or without Dox addition. Cells were harvested from one well of a 6‐well plate and counted using Orflo MOXI Z Mini Automated Cell Counter. Mean ± SEM is shown for three independent experiments. *P < 0.05 (unpaired two‐sample t‐test).
-
F, GBoxplots indicating the median, quartiles, and 5th and 95th percentiles of changes in expression levels of WT (F) or Reno1 m/m (G) cell treated with Dox for the first 2 days of differentiation compared to untreated cells, in genes which were significantly (P < 0.05) downregulated (n = 252) or upregulated (n = 223) by at least two out of three Reno1 perturbations at day 2 in Fig 5A, compared to all genes (n = 16,511). **P < 0.01 (two‐sided Wilcoxon rank sum test).
Expression of Reno1 RNA in Reno1 m/m ES cells leads to reduced cell death at the onset of differentiation and affects expression levels of genes dysregulated in Reno1‐depleted cells
In order to test whether expression of Reno1 produced in trans can rescue the phenotype observed in Reno1‐depleted cells, we infected one WT and one Reno1 m/m line with lentivirus carrying a doxycycline (Dox)‐inducible cDNA of the unspliced short isoform of Reno1. We added Dox at the onset of differentiation and measured Reno1 levels at day 2. Reno1 was overexpressed by ~ 10‐fold in the WT cells and ~ 70‐fold in the Reno1 m/m cells following Dox addition (Fig 5D). Dox addition increased the number of live Reno1 m/m cells at day 4 of differentiation (Fig 5E). We also used RNA‐seq to characterize gene expression at day 2 of differentiation, and found that Reno1 induction through Dox addition to either WT or Reno1 m/m cells upregulated the 168 gene signature that was significantly downregulated in Reno1‐depleted cells, and downregulated the 94 gene signature that was significantly upregulated upon Reno1 loss‐of‐function (Fig 5F and G). In Reno1 m/m cells, overexpression of Reno1 did not have a significant effect on Bahcc1 expression level (Appendix Fig S2H). These results demonstrate that ectopic expression of Reno1 can at least partially rescue the effect of Reno1 depletion on cell death and gene expression at early stages of differentiation and suggest that Reno1 might be able to carry its function when produced in trans to its regular site of transcription.
Global reduction in chromatin accessibility of H3K4me3‐marked regions in Reno1‐ or Bahcc1‐deficient cells during early stages of differentiation
In order to examine the effect of loss of Reno1 on the chromatin landscape, we used ATAC‐seq (Buenrostro et al, 2013) to map accessible chromatin in two WT clones, two Reno1 m/m clones, and the Bahcc1 m/+ clone, as well as WT cells transfected with shRNAs targeting Reno1, Bahcc1, or a non‐targeting control (two shRNAs per target and two replicates per treatment, Figs EV4A and EV5A, and Appendix Fig S2A). In order to obtain sufficient and clean material, we analyzed cells at day 2 following the beginning of differentiation, before the onset of massive cell death. Combining the ATAC‐seq datasets, we obtained and quantified 49,722 robust peaks, and for each peak computed the average ratio of read coverage between the perturbed and the control cells. We then divided the peaks into those that fell in “core promoter” [< 300 nt from transcription start site (TSS)], “extended promoter” (< 2 kb from TSS), “gene body”, or “intergenic” regions relative to RefSeq annotations. Within each of these categories, we also further divided the peaks into those found within 1 Mb of Reno1 (and thus might be regulated in cis) and those elsewhere in the genome (trans peaks). Strikingly, peaks found within core or extended promoters exhibited globally reduced accessibility relative to the intergenic peaks in both Reno1 m/m and Bahcc1 m/+ cells, regardless of their location relative to Reno1, and similar effects were seen in Reno1 or Bahcc1 KD cells (Fig 6A). There was also significantly reduced accessibility in the cis peaks compared to the other peaks (P < 0.05 for each comparison of Reno1‐ or Bahcc1‐perturbed cells vs. controls), which could be partially explained by the increased prevalence of promoter peaks in the cis group compared to the rest of the genome (58.8% of cis peaks were in promoters compared to 28.9% of the trans peaks). When we directly compared cis and trans peaks within each group, there was significant reduction in some cases (Fig 6A), which was less pronounced than the genome‐wide trans effect on promoter accessibility (Fig 6A). Overall we observed substantial and reproducible changes in promoter accessibility in hundreds of genes: 837 promoter peaks were reduced by > 30% by Reno1 KO and KD, 564 were reduced by > 30% in Bahcc1 KO and KD, 128 of these were reduced in all conditions. Differences in promoter accessibility in shRNA‐infected cells were correlated with changes in gene expression in these cells, as evaluated by RNA‐seq at the same stage (Fig 6B). These changes in gene expression at day 2 in Reno1 KD cells were substantial (1,135/735 genes up/downregulated by at least 30% with adjusted P < 0.05), and correlated with those observed at day 8 of differentiation (Spearman R = 0.245, P < 10−15), suggesting that the neuronal differentiation defects observed at the late stages in differentiation could be traced back to chromatin and transcriptional dysregulation already at day 2.
Figure EV5. Characterization of Reno1‐ and Bahcc1‐perturbed cells.
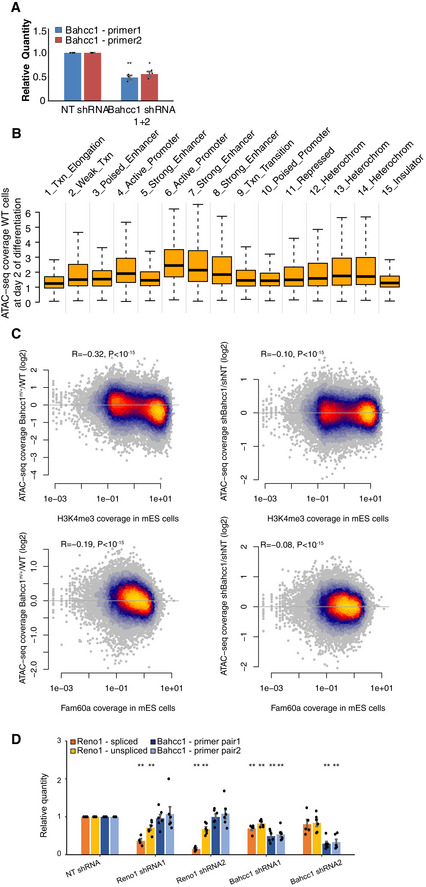
- qRT–PCR using the indicated primer pairs (see Materials and Materials and Methods) in cells at day 2 of differentiation following Bahcc1 shRNA knockdown. Mean ± SEM is shown for three independent experiments. *P < 0.05; **P < 0.01 (unpaired two‐sample t‐test).
- ATAC‐seq accessibility in WT cells following 2 days of differentiation in peaks that overlap the indicated chromatin states in mouse ES cells.
- Correlation between changes in ATAC‐seq signal following the indicated treatment and H3K4me3 ChIP‐seq signal (top) or Fam60a ChIP‐seq signal (bottom) in mouse ES cells (bottom).
- qRT–PCR of Reno1 and Bahcc1 following knockdown in N2a cells using two targeting shRNAs, normalized to a non‐targeting shRNA (sh‐NT Mean ± SEM is shown for 5–6 independent experiments. **P < 0.01 (unpaired two‐sample t‐test).
Figure 6. Global reduction in chromatin accessibility of H3K4me3‐marked regions in Reno1‐ or Bahcc1‐deficient cells.
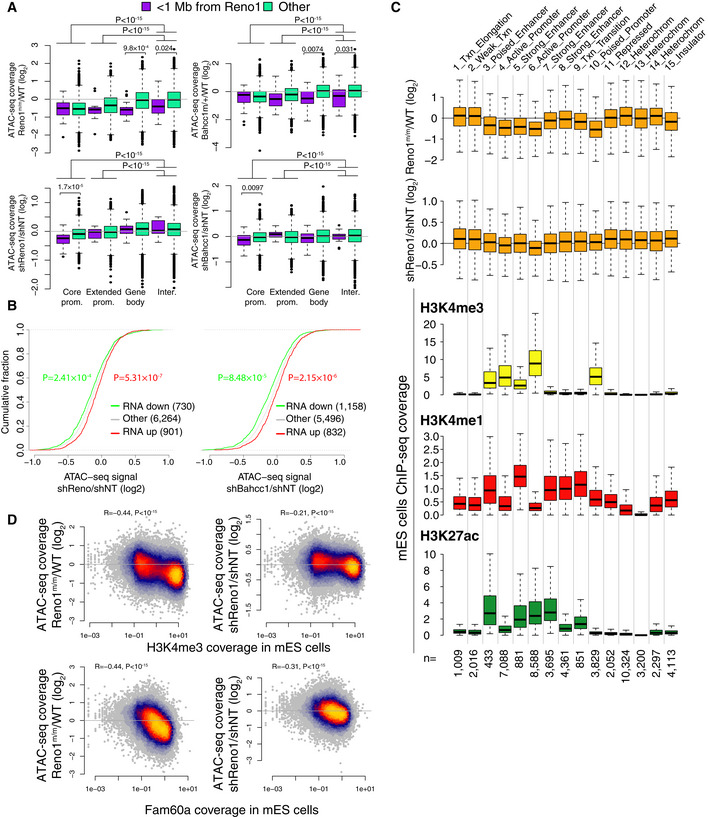
- Boxplots indicating the median, quartiles and 1.5 time the interquartile range of ratios of ATAC‐seq signal for the indicated group of peaks, comparing the indicated genotypes or treatments. Core promoter peaks are < 300 nt from a TSS; extended promoter is more than 300 nt but < 2 Kb for a TSS; gene body peaks overlap transcription units, and intergenic do not. Each group of peaks was compared to the intergenic peaks. In addition, for each group of peaks, the peaks within 1 Mb of Reno1 were compared to peaks further away. P‐values computed using two‐sided Wilcoxon rank sum test and shown only for cases where P < 0.05. Number of data points (from left to right): 39; 10,777; 11; 3,550; 17; 14,380; 18; 20,930.
- Changes in ATAC‐seq signal for peaks assigned as falling within core or extended promoters, of genes in the indicated group (up/downregulated by at least 30% and with adjusted P < 0.05) in cells with Reno1 (left) or Bahcc1 (right) KD. P‐values computed using two‐sided Wilcoxon rank sum test.
- Top: Boxplots indicating the median, quartiles and 1.5 time the interquartile range of changes in ATAC‐seq signal on day 2 of neuronal differentiation of peaks from indicated Reno1 perturbation, classified according to the underlying chromatin state in mouse ES cells; bottom: boxplots indicating the median, quartiles, and 1.5 time the interquartile range of ChIP‐seq coverage of the indicated chromatin marks in ENCODE mouse ES data in the peak regions. Number of peaks in each group is indicated at the bottom.
- Correlation between changes in chromatin accessibility on day 2 of neuronal differentiation in the indicated perturbations and H3K4me3 or Fam60a ChIP coverage from mouse ES cells (H3K4me3 is from ENCODE project and Fam60a from Streubel et al, 2017). Coefficient and P‐value computed using Spearman's correlation. n = 49,722 peaks.
In order to explore any common characteristics of the regions affected by loss of Reno1, we intersected the ATAC‐seq peaks with chromatin state annotations in mouse ES cells (Bogu et al, 2015). Reno1‐deficient cells exhibited a substantial reduction of accessibility of peaks in five of the 14 observed states (states 3–6 and 10, Fig 6C), which correspond to all three of the “promoter” states and two of the four “enhancer” states. In contrast, there were no obvious changes in accessibility in the other two enhancer states (8 and 9), where the baseline chromatin accessibility in WT cells was similar to the affected “enhancer” states (Fig EV5B). The main distinguishing factor between the affected and the unaffected states was the level of the H3K4me3 modification (Fig 6C). Overall, there was a striking negative correlation between change in ATAC‐seq signal at day 2 of differentiation and H3K4me3 levels in ENCODE mouse ES data (Spearman R = −0.44, Figs 6D and EV5C). As the Sin3a complex with which BAHCC1 is associated was reported to be enriched at H3K4me3‐marked regions, we also tested the association between occupancy of FAM60A, a member of the same complex, and chromatin accessibility, and again observed a striking correlation (Figs 6D and EV5C), especially considering that the compared data are coming from different studies and different differentiation states (ES cells vs. day 2 of differentiation). The main observed effect of loss of Reno1 or Bahcc1 at the chromatin level is thus reduction in chromatin accessibility in regions bearing H3K4me3, which mostly correspond to promoters, possibly explaining the inability of mouse ES cells to execute the proper transcriptional program required for neurogenic commitment.
Discussion
Of the thousands of lncRNAs that are expressed in different vertebrates, many are restricted to the nervous system, and some to very specific cell types in the brain or to specific time points during neuronal development. However, it is not clear how many of these transcripts carry out any function or exert any fitness advantage. Here, by combining sequence conservation and expression patterns, we compiled a list of 121 lncRNAs with potential functional importance in neurogenesis, tested seven of them, and identified among them two lncRNAs required for neuronal differentiation of mouse ES cells: Reno1 and lnc‐Nr2f1. KD of either of these lncRNAs during neuronal differentiation of mouse ES cells led to reduced differentiation efficiency, which we characterized both by immunostaining for the neuronal marker Tuj1 and by transcriptome analyses. Notably, Reno1 and lnc‐Nr2f1 are also some of the most conserved lncRNAs in vertebrates—sequence‐similar homologs of Reno1 are found in tetrapods and coelacanth (Figs 3A and EV1C, and Appendix Fig S3A). lnc‐Nr2f1 belongs to a cluster of sequence‐similar and syntenic lncRNAs that are found throughout vertebrates (Fig EV1C) and regions alignable to the human lnc‐Nr2f1 are found in amniotes (Appendix Fig S3B). A lncRNA syntenic to Reno1 with similar embryonic expression patterns and locus architecture, but no detectable sequence similarity is also found in zebrafish upstream of bahcc1b (Appendix Fig S4).
Lnc‐Nr2f1 (A830082K12Rik, known as NR2F1AS1 in human), a lncRNA which is transcribed in the opposite direction from the nearby neurogenic TF Nr2f1, was reported to be upregulated, together with Nr2f1, in a Waardenburg syndrome type 4 mouse model (Bergeron et al, 2016). KD of this lncRNA was recently reported to suppress oxaliplatin resistance of hepatocellular carcinoma cells (Huang et al, 2018). A recent study found that lnc‐Nr2f1 is mutated in human patients with autism spectrum disorder and intellectual disability, and its expression promotes the conversion from MEFs to induced neurons, when co‐expressed with Ascl1 (Ang et al, 2019). In agreement with our findings, when lnc‐Nr2f1 KO mouse ES cells were differentiated into neurons, hundreds of genes were differentially expressed in the differentiated neurons compared to WT cells, with downregulated genes being enriched with neuronal pathfinding and axon guidance genes (Ang et al, 2019).
The second lncRNA which we found to affect the efficiency of neuronal differentiation was Reno1. Our results indicate that the function of Reno1 during neuronal differentiation is related to the function of the nearby Bahcc1 gene (also known as KIAA1447). BAHCC1 is a poorly characterized protein, whose precise function is ¬ known yet. However, Bahcc1 KO mice have been previously generated, and Bahcc1 −/− mice usually died immediately after birth, which can be consistent with a role in the nervous system (Nakayama et al, 2006). Only one mouse survived, and it displayed a hind leg motor dysfunction, which was attributed to a potential defect in the motor neurons controlling the muscle (Nakayama et al, 2006). Supporting a role for BAHCC1 in neuronal differentiation, KD of Bahcc1 in primary NPCs using RNAi led to reduced neurogenesis (Benayoun et al, 2014). BAHCC1 is a very large protein (280 kDa), containing a BAH domain at its C‐terminal end and a tandem pair of Tudor domains (Faure & Callebaut, 2013). Lack of an effective commercially available antibody for BAHCC1 currently restricts the ability to study the function of the protein. However, both BAH and Tudor domains are usually found in proteins with functions related to chromatin modification, such as DNMTs and ARID4A (Lasko, 2010; Yang & Xu, 2013). According to two databases of experimentally recovered protein–protein interactions, STRING (Szklarczyk et al, 2017) and Bio‐GRID (Chatr‐Aryamontri et al, 2017), BAHCC1 interacts with members of the Sin3a histone deacetylase complex, including HDAC1, SAP30, FAM60A, RBBP4, RBBP7, and ING2 (Streubel et al, 2017). This suggests that BAHCC1 might associate with the Sin3a complex and modulate its activity under certain conditions. Hdac1 and Hdac2 have been shown to be important for proper neuronal development, as well as to ES cells differentiation (Hsieh et al, 2004; Montgomery et al, 2009; Dovey et al, 2010; Jang & Jeong, 2018), and the Sin3a complex has been recently found to repress the expression of a subset of somatic genes during reprogramming of somatic cells to pluripotent stem cells, including genes related to brain development and dendrite neurogenesis (Li et al, 2017b).
Reno1 and Bahcc1 functions appear to be related to the H3K4me3 modification, as regions marked with H3K4me3 preferentially lose accessibility in Reno1 m/m or Bahcc1 m/+ cells and in cells after KD of either Reno1 or Bahcc1 during early stages of differentiation. Interestingly, loss of Mll1, an H3K4me3 “writer”, is associated with impairment of neurogenesis (Lim et al, 2009). Further, a variant of the Sin3a complex in mouse ES cells, which includes BAHCC1, is associated specifically with H3K4me3‐marked regions (Streubel et al, 2017), and depletion of Fam60a, the defining member of this variant Sin3a complex that is enriched at regions that are affected by Reno1 or Bahcc1 loss (Fig 6D), leads to reduction in neuroectodermal lineage markers and increase in mesodermal markers. Lastly, Bahcc1 gene has been previously identified as one of the genes with the broadest H3K4me3 domains in the mouse genome (Benayoun et al, 2014), suggesting it may be involved in genome‐wide H3K4me3 regulation, as transcriptional regulators are often regulated by the same epigenetic/chromatin processes they regulate, so as to form feedback loops (Crews & Pearson, 2009).
lncRNAs found in proximity to transcriptional regulators may act in cis to regulate their neighbors, or potentially collaborate with them, e.g., by binding and stabilizing the protein or scaffolding its interactions with other proteins; examples of both scenarios can be found in the literature. The increase in physical interactions between the Reno1 and the Bahcc1 loci observed upon neuronal differentiation (Fig 3F) suggests that similarly, Reno1 may regulate Bahcc1; however, we observed no change in Bahcc1 mRNA levels during differentiation of Reno1 m/m cells (Fig 4B), and exogenous expression of Reno does not change Bahcc1 level (Appendix Fig S2H). It is possible that loss of Reno1 causes a transient reduction in Bahcc1 levels during differentiation, perhaps through changes in chromatin accessibility we observe in some peaks found < 1 Mb from Reno1, Fig 6A. It is further possible that cells where Bahcc1 levels are reduced undergo rapid cell death precluding us from observing a reduction in Bahcc1 mRNA levels. KD of Reno1 using RNAi in a “static” Neuro2a (N2a) cell line (Fig EV5D) did not affect Bahcc1 levels either, suggesting that if Reno1 regulates Bahcc1 expression it does so in a condition‐specific manner. Interestingly, reduction of Bahcc1 in N2a cells or in Bahcc1 m/+ ES cells led to a substantial decrease in Reno1 levels (Figs 4B and EV5D. These suggest that Bahcc1 and Reno1 may form part of the same pathway, but not in the direction that has been typically observed, in which the lncRNA regulates the expression of the protein‐coding gene, but rather through co‐regulation (supported by the high co‐expression between Reno1 and Bahcc1, Fig 3E), and potential cooperation, reminiscent of that observed for the Six3os and Paupar lncRNAs and the TFs SIX3 and PAX6, respectively (Rapicavoli et al, 2011; Vance et al, 2014; Pavlaki et al, 2018), and the Pnky and POU3F2 lncRNA/TF pair that are co‐located in the genome but appear to act independently in the developing brain (Andersen et al, 2019). In the Reno1‐BAHCC1 case, such cooperation could occasionally be reinforced by regulation of Reno1 by Bahcc1. We speculate that Reno1 may bind the BAHCC1 protein, potentially affecting its participation in the Sin3a complex. If such activity, that can potentiate the complex activity at H3K4me3‐decorated loci, occurs near the Reno1/Bahcc1 site of transcription, it can explain the preferential effect of Reno1 and Bahcc1 depletion on chromatin accessibility in the 1 Mb region flanking the locus (Fig 6A). Future studies into the biology of BAHCC1 and Reno1, which will be facilitated by the generation of effective antibodies for BAHCC1, will uncover the mechanism through which Reno1 and BAHCC1 act, and determine whether their activity is required solely for neuronal commitment or for other early developmental events.
Materials and Methods
Cell culture and neuronal differentiation
R1 mouse ES cells were grown on a feeder layer of irradiated mouse embryonic fibroblasts (MEFs) on 0.1% gelatin‐coated cell culture dishes. Culture medium consisted of DMEM (Gibco), 15% ES‐grade fetal calf serum (FCS, Biological Industries), 1× Glutamax (Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM nonessential amino acids (Gibco), 0.1 mM β‐mercaptoethanol (Sigma), 100 U/ml penicillin and 0.1 mg/ml streptomycin (Biological Industries), and 1,000 U/ml LIF. Neuronal differentiation was performed as previously described (Ying et al, 2003). Pluripotent stem cells were first grown in the absence of MEFs for two passages and then seeded on gelatin‐coated plates at a density of 2.1*104/cm2 in N2B27 medium: 1:1 mixture of DMEM/F12 (Sigma) supplemented with N2 (Gibco), and Neurobasal medium (Gibco) supplemented with B27 (Gibco), 1× Glutamax (Gibco), 0.1 mM β‐mercaptoethanol (Sigma), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Biological industries). After 4 days under these conditions, the cells were dissociated and re‐plated on plates coated with Poly‐d‐Lysine (Sigma) and Laminin (Sigma), at a density of 3.7*104/cm2 in N2B27 medium supplemented with 20 ng/ml FGF2 (Peprotech). After 24 h, FGF2 was removed from medium and cells were grown for three additional days.
RNA interference using shRNAs
Short sequences (21nt) complementary to target genes (Table 1) were synthetized as 58nt single‐stranded oligos as forward oligo: 5′‐CCGG—21nt sense—CTCGAG—21nt antisense—TTTTTG‐3′, and reverse oligo: 5′‐AATTCAAAAA—21nt sense—CTCGAG—21nt antisense‐3′. Forward and reverse oligos were phosphorylated and annealed to each other by the addition of T4 ligation buffer (NEB) and T4 PNK (NEB) and incubation at 37°C for 30 min followed by incubation at 95°C for 5 min and gradual cooling to room temperature at a rate of 5°C/min. The annealed oligo pairs were ligated to pLKO.1 vector (Sigma # SHC001) which was digested by AgeI and EcoRI. Ligated plasmids were transformed into NEB stable competent bacteria. A plasmid containing an shRNA not targeting a mammalian sequence (Sigma #SHC002) was used as negative control.
Table 1.
Sequences of shRNAs targeting lncRNAs
| Gene | shRNA1 sequence | shRNA2 sequence |
|---|---|---|
| Reno | ACGGATCCCATTTCCAGAAAG | GTCTCTTTGAGGAAGATTGCA |
| Cox10as1 | GAAGAACCTGTTGCTTAAGTT | ACCTGTAAGGCTTTAAGTAAC |
| Lnc‐Nr2f1 | GGACCTTAGCTATAATTTAGC | AGGAGCTTTGGCAATAGAAGA |
| Miat | GCTCCTTGTTCGGTTTATATC | GAGCCTCAAAGTGCTAATGGG |
| Fzd10as1 | ACCTCTCTGATCTGATTTCTG | GTGCTGAGATTACAAACTGGA |
| Lhx1os | GACCGCGTTTAGAGGATCAGT | GCTACTTAGCTGGTACTAATT |
| Crnde | GGTGATTTAGAAGACAGTGAG | AGCTTCGCCACTGTAAAGTCA |
| Bahcc1 | CGCATCCAGAAGAAGCTATCT | GAAGCGAAGCAAACTGGGAAA |
Lentiviral particles were generated as previously described (Tiscornia et al, 2006): HEK293T cells were transfected with a mixture of shRNA plasmid: pMDL: pVSVG: pRev at a ratio of 1:0.65:0.35:0.25 respectively, using PEI. Medium was collected from plates 48 and 72 h after transfection, filtered, and frozen at −80°C.
Mouse ES cells were infected by the addition of growth medium containing lentiviral particles and 8 μg/ml Polybren (Sigma) to attached cells. Selection was done by the addition of Puromycin (InvivoGen) at a concentration of 0.8 μg/ml. Pools of infected cells that survived the selection were used for neuronal differentiation and analysis of RNA.
Reno1 knockdown using LNA GapmeRs
Mouse ES cells were transfected with Reno1 GapmeR1 (TACGACGAAGATGGAT) or Reno1 GapmeR2 (GGATCAGCAGAGAGTG), ordered from Qiagen, using a Nucleofector instrument (Lonza), using P3 solution and CG‐104 program, with 50 nM of each GapmeR. Transfected cells were then seeded for neuronal differentiation, and RNA was analyzed following 2 days.
RNA‐seq library preparation and data analysis
RNA was extracted from cells using TRI reagent (MRC) according to manufacturer's protocol. Strand‐specific RNA‐seq libraries were prepared from 500 to 2,000 ng total RNA using either the TruSeq Stranded mRNA Library Prep Kit (Illumina), or the SENSE‐mRNA‐Seq‐V2 (Lexogen), according to the manufacturers’ protocol, and sequenced on a NextSeq 500 machine to obtain 75 nt or 150 nt single‐ or paired‐end reads (Table EV1). RNA‐seq reads were mapped to the mouse genome using STAR (Dobin et al, 2013). Expression levels, both of RefSeq genes and of lncRNAs previously annotated using PLAR (Hezroni et al, 2015), were quantified using RSEM (Li & Dewey, 2011). Differential expression analysis following shRNA KD or genome editing was done using DESeq2 (Love et al, 2014). Genes which were up‐ or downregulated by two different shRNAs, or in two different cell lines, in two biological repeats with P value < 0.05 were considered differentially expressed. Enriched gene ontology (GO) terms within were identified using GOrilla (Eden et al, 2009). Heatmaps of expression levels of differentially expressed genes were generated with pheatmap R package.
Immunofluorescence staining
Cultured cells were fixed with 4% paraformaldehyde for 10 min. Blocking and permeabilization were done with 5% normal goat serum, and 0.1% Triton X‐100 in PBS for 30 min in room temperature. Primary antibodies were diluted in permeabilization buffer and incubated with fixed cells for 1 h at room temperature or 4°C overnight. Secondary antibodies were diluted in permeabilization buffer and incubated with fixed cells for 30 min at room temperature. Antibodies used: rabbit anti‐Oct4 (Santa Cruz sc‐9081), mouse anti‐Nestin (Abcam ab6142), rabbit anti‐beta III Tubulin (Tuj1, Abcam ab18207), Donkey anti Mouse Alexa 594 (Molecular Probes A21203), Goat anti Rabbit Alexa 594 (Abcam ab150080). Imaging was done using EVOS FL Cell Imaging System.
Image analysis of Tuj1 (TUBB3) immunostaining
Ten to 16 non‐overlapping fields were imaged for each condition. Quantification of total cell number, number of Tuj1 stained cells and total length of neurites in each field were quantified using the Fiji software (Schindelin et al, 2012). Background was subtracted by means of a rolling ball method, from the DAPI channel as well as the 594 channel used for Tuj1 staining. Then, nuclei were identified and counted by thresholding of the DAPI channel using a fixed value. Cells positive for Tuj1 were defined as nuclei with high fluorescence in the 594 channel. Finally, the Tubeness plugin was used to identify the total neurite length in each field. An estimation of the average neurite length was obtained by dividing the total neurite length per field by the number of Tuj1 stained cells.
Quantitative reverse‐transcription PCR (qRT–PCR)
RNA was extracted from cells using TRI reagent (MRC) according to manufacturer's protocol and treated with DNAse (Quanta Biosciences) for 30 min at 37°C. Reverse transcription was done using qScript Flex cDNA synthesis kit (Quanta Biosciences), using random primers. Quantitative PCR was performed in a ViiA 7 Real‐Time PCR System (Thermo) in a 10 μl reaction mixture containing 0.1 μM forward and reverse gene‐specific primers (Table 2), fast SYBR master mix (Applied Biosystems), and template cDNA. A reaction containing DDW instead of cDNA was used as a no‐template control and was amplified for each primer pair. Only samples free of DNA contamination were further analyzed. Five‐fold serial dilutions of the template cDNA (ranging from 1,000 ng to 1.6 ng) were used to generate a standard curve in order to determine the linear range for each primer pair. After determining the linear range for each primer pair, a dilution within that range was used to determine the initial quantity of each RNA in the different cDNA samples. To measure Reno1 copies per cell, the unspliced variant of Reno1 was cloned into pLIX_402 (Addgene #41394). This plasmid was then used to obtain a standard curve for absolute quantification of Reno1 abundance in defined numbers of cells.
Table 2.
Gene‐specific primer pairs used for mRNA and lncRNA expression level analysis
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Ppib | TGATCCAGGGTGGAGACTTC | ATTGGTGTCTTTGCCTGCAT |
| Sdha | GCTCCTGCCTCTGTGGTTGA | AGCAACACCGATGAGCCTG |
| Reno—spliced | CTGGCTGGTGGGGTAGAAAC | TAGGGAAAGATCCGCAGAAA |
| Reno—unspliced | AGGGTGGCTTTGTGAGAAGA | CTTCCTCAAAGAGACTGGGAGT |
| Cox10as1 | GAGCCATCCCTCCTGTCAT | GGGAATTGCCAGGAGTACAG |
| Lnc‐Nr2f1 | TTCCACTGAACTCTGCTTGC | CCAAAGCTCCTTCCTCAACA |
| Miat | TGCATCACTACAGCTCAGCA | CCAGTTCCAGGAGGTCAGAA |
| Fzd10as1 | CATGGGAGCCAGACTTCTCT | CGTCTGCTCTCTTCCCTCAC |
| Lhx1os | GCTGATGGAGAGAGGCTGAA | TCGGGGCTACAGAGAGAAAC |
| Crnde | CACTGTGTGGGGAACATCAG | CTGAGTGACCTGTGGCCTTC |
| Bahcc1—primer1 | GGGCAACATCGTTTCTATGG | TTGGCGGGTAGTATGGTAGG |
| Bahcc1—primer2 | AGCTCTGGAAGTGGTCAGGA | GTCTCCTTGCCTCGGACAAT |
3′RACE
3′RACE was performed with RNA from ES‐derived neurons using SMARTerRACE 5′/3′kit (Clontech, #634859). Briefly, RACE products were amplified using nested primers GATTACGCCAAGCTT CTTCTCTTCCCGGAGGGCTCAGAGTT and GATTACGCCAAGCTT CCCCTTTTCGCGATTGGGAACGTGGA. PCR products were then purified from 1% agarose gel using NucleoSpin (#740609.50), cloned into the pRACE vector (provided with the kit), and transformed into Stellar competent cells. RACE products were Sanger sequenced with M13F primer: TGTAAAACGACGGCCAGT and M13R primer: AACAGCTATGACCATGATTA and aligned to the mouse genome.
Single‐molecule FISH
Cells at day 8 of differentiation were fixed with 4% paraformaldehyde. Intestine tissue was frozen in Tissue‐Tek O.C.T compound (Sakura 4583) blocks and sectioned using a Leica cryostat (CM3050) at 10 μm thickness. A library of 48 probes (Table EV2) was designed to target the unspliced isoform of the mouse Reno1 RNA sequence (Stellaris RNA FISH probes, Biosearch Technologies). Hybridization conditions and imaging were as described previously (Lyubimova et al, 2013). smFISH imaging was performed on a Nikon‐Ti‐E inverted fluorescence microscope with a 100× oil‐immersion objective and a Photometrics Pixis 1,024 CCD camera using MetaMorph software as previously described (Bahar Halpern & Itzkovitz, 2016).
Targeted chromosome conformation capture (4C) analysis
ES cells were trypsinized and depleted from MEFs by 20 min incubation on gelatin‐coated plates. 3C was carried out on 5 × 106 ES cells/neurons, essentially as described (Olivares‐Chauvet et al, 2016), with the following slight modification: permeabilization buffer constitution was 10 mM Tris–HCl pH 8, 10 mM NaCl, 0.5% NP‐40, supplemented with protease inhibitors. 4C libraries were prepared as described (Schwartzman et al, 2016), with primers directed to the promoter region of Reno1 (upstream primer sequence: CCACCAACAAAAAACCACAACTCTG, downstream primer sequence: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTATCTCCTAACCCCGCAAATCTGG). Libraries were sequenced in an Illumina NextSeq 500 instrument, and analyzed as described (Schwartzman et al, 2016).
Generation of Reno1 m/m and Bahcc1 m/+ ES cells using CRISPR‐Cas9
To generate Reno1 m/m ES cells we deleted the promoter and first exon of Reno1 (a total of 631 bp) by transfection of pCas9_GFP plasmid (Addgene #44719) and two gRNA plasmids. To generate Bahcc1 m/+ cells we targeted the start codon of Bahcc1 by transfection of pCas9_GFP plasmid (Addgene #44719) and one gRNA plasmid targeting Bahcc1 start codon. Transfections were done using a Nucleofector instrument (Lonza), using P3 solution and CG‐104 program, with 2 μg Cas9 plasmid and 250 ng of each gRNA plasmid. 106 cells were transfected with the plasmid mix, and 7*105 cells were seeded on one well of a six well plate, in the absence of MEFs, and in medium supplemented with CHIR 99021 and PD 0325901 (2i, Axon). 24 h after transfection, 1 μg/μl Puromycin was added to the medium, and selection was carried out for 48 h. The cells were then seeded on a 10 cm plate for recovery, and 8 days after the transfection, cells were seeded in low density for colony picking. Colonies were genotyped by PCR using primers flanking the deletion region (for Reno1) or by PCR with primers flanking the start codon (Table 3), followed by restriction reaction using BccI enzyme, which recognized a restriction site within the amplicon which was abolished when the start codon was cleaved by Cas9.
Table 3.
Sequences of primers used for cloning of gRNA plasmids and for genotyping of colonies
| Name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Reno gRNA1 | CACCGAAGCTACTCCCTATCCGGCGT | TAAAACGCCGGATAGGGAGTAGCTTC |
| Reno gRNA2 | CACCGCAAACCAATGGCATCGCGTGT | TAAAACACGCGATGCCATTGGTTTGC |
| Bahcc1 gRNA | CACCGTGTGCGAAGCCATCGGCAGGT | TAAAACCTGCCGATGGCTTCGCACAC |
| Reno genotyping | GGTTGGGGGATAACTGTATGAA | CTTCTCGGGGAAATAGACCCT |
| Bahcc1 genotyping | GCATTTGCTATCGGAGCG | CATTGCACAGGGAAATGAAAC |
gRNAs sequences were designed using CHOPCHOP (Labun et al, 2016) and cloned into a pKLV‐U6gRNA vector containing Puromycin resistance gene (Addgene #50946) as previously described (Cong et al, 2013).
Analysis of cell viability
ES cells following 4 days of neuronal differentiation were washed in 1× PBS and dissociated into single cells using 0.05% trypsin for 5 min at 37°C. Simultaneously, the supernatant containing dead cells was collected, treated with trypsin to dissociate the clumps of dead cells and mixed back with the corresponding sample containing the live cell fraction. Cells were centrifuged for 5 min at 300 g and cell pellets were resuspended in 1 ml of PBS containing 10 μg/ml propidium iodide and incubated at room temperature for 15 min. Samples were then analyzed with Attune NxT Flow Cytometer.
ATAC‐seq
ATAC‐seq was performed as previously described (Buenrostro et al, 2013) with minor adjustment for application to ES cells. To harvest the cells, we used 200 μl of a buffer containing 1× PBS, 2 mM EDTA, and 1% BSA and spun the cells at 500 × g, 4°C for 20 min. 25,000 cells per sample after 2 days of neuronal differentiation were used. Libraries were sequenced with paired‐end sequencing on Illumina NextSeq 500. Reads were aligned to the mm9 genome assembly using Bowtie2 (Langmead et al, 2009). Normalized read coverage files were computed by MACS2 (Feng et al, 2012). Peaks were called using MACS2 combining all the reads from all the samples, identifying peak summits and padding them with 70 bases on each side. Read coverage in the peaks was computed using bigWigAverageOver bed UCSC utility (Karolchik et al, 2003) and normalized by the mean number of reads per peak in that sample after excluding the peaks in the top and bottom 5% of values. We then averaged the coverage in the samples from the same genotype or treatment and excluded peaks where the maximum average was below 1 or above 20, or where the coefficient of variation (standard deviation divided by the mean) among the samples from the same genotype was above 2. Fold changes between read coverages were computed after adding a pseudocount of 0.25. Peaks were assigned to the closest RefSeq gene and annotated as “Core promoter” if they fell within 300 nt of a TSS; “Extended promoter” if they fell within 2,000 nt of a TSS; “Gene body” if they overlapped a transcription unit; or “Intergenic” otherwise.
Data availability
The datasets produced in this study are available in the following database: RNA‐seq and ATAC‐seq data: Gene Expression Omnibus GSE124517 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124517).
Author contributions
HH and IU conceptualized the study; HH and RB‐TP designed the methodology; HH, RB‐TP, NG, and ND investigated the study; HH, NG, and IU involved in formal analysis; HH and IU wrote the original draft of the manuscript; HH and IU wrote, reviewed, and edited the manuscript; IU involved in funding acquisition; and IU supervised the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Review Process File
Acknowledgements
We would like to thank Ulitsky Lab members for helpful discussions and comments on the manuscript, Elena Ainbinder for assistance with stem cell work, and Liat Alyagor for assistance with single‐molecule FISH. This work was supported by research grants from the European Research Council project lincSAFARI; the I‐CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation (grant no 1796/12); Nella and Leon Benoziyo Center for Neurological Disease; The Helen and Martin Kimmel Institute for Stem Cell Research; Lapon Raymond; and the Abramson Family Center for Young Scientists. I.U. is incumbent of the Sygnet Career Development Chair for Bioinformatics.
EMBO Reports (2020) 21: e51264
References
- Andersen RE, Hong SJ, Lim JJ, Cui M, Harpur BA, Hwang E, Delgado RN, Ramos AD, Liu SJ, Blencowe BJ et al (2019) The long noncoding RNA pnky is a trans‐acting regulator of cortical development in vivo . Dev Cell 49: 632–642 e637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang CE, Ma Q, Wapinski OL, Fan S, Flynn RA, Lee QY, Coe B, Onoguchi M, Olmos VH, Do BT et al (2019) The novel lncRNA lnc‐NR2F1 is pro‐neurogenic and mutated in human neurodevelopmental disorders. Elife 8: e41770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar Halpern K, Itzkovitz S (2016) Single molecule approaches for quantifying transcription and degradation rates in intact mammalian tissues. Methods 98: 134–142 [DOI] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW et al (2014) The long non‐coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia‐associated alternative splicing. Mol Psychiatry 19: 486–494 [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E et al (2014) H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron KF, Nguyen CM, Cardinal T, Charrier B, Silversides DW, Pilon N (2016) Upregulation of the Nr2f1‐A830082K12Rik gene pair in murine neural crest cells results in a complex phenotype reminiscent of Waardenburg syndrome type 4. Dis Model Mech 9: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J (2006) The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev 20: 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S et al (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Bogu GK, Vizan P, Stanton LW, Beato M, Di Croce L, Marti‐Renom MA (2015) Chromatin and RNA maps reveal regulatory long noncoding RNAs in mouse. Mol Cell Biol 36: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA‐binding proteins and nucleosome position. Nat Methods 10: 1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C et al (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chatr‐Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A et al (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res 45: D369–D379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Lieber MR, Hsieh CL (2002) The DNA methyltransferase‐like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA 99: 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Pearson JC (2009) Transcriptional autoregulation in development. Curr Biol 19: R241–R246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Cowley SM (2010) Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA 107: 8242–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis BC, Molloy PL, Graham LD (2012) CRNDE: a long non‐coding RNA involved in CanceR, Neurobiology, and DEvelopment. Front Genet 3: 270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure G, Callebaut I (2013) Identification of hidden relationships from the coupling of hydrophobic cluster analysis and domain architecture information. Bioinformatics 29: 1726–1733 [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS (2012) Identifying ChIP‐seq enrichment using MACS. Nat Protoc 7: 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP et al (2009) Chromatin signature reveals over a thousand highly conserved large non‐coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Perry RBT, Ulitsky I (2020) Long noncoding RNAs in development and regeneration of the neural lineage. Cold Spring Harb Symp Quant Biol 84: 165–177 [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH (2004) Histone deacetylase inhibition‐mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA 101: 16659–16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J (2018) LncRNA NR2F1‐AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR‐363. J Cell Mol Med 22: 3238–3245 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hubbard KS, Gut IM, Lyman ME, McNutt PM (2013) Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000Res 2: 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Jeong HS (2018) Histone deacetylase inhibition‐mediated neuronal differentiation via the Wnt signaling pathway in human adipose tissue‐derived mesenchymal stem cells. Neurosci Lett 668: 24–30 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ et al (2003) The UCSC genome browser database. Nucleic Acids Res 31: 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A (2018) Repair of double‐strand breaks induced by CRISPR‐Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44: W272–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P (2010) Tudor domain. Curr Biol 20: R666–R667 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12: 323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L (2017a) A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA‐seq. Sci Rep 7: 4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S et al (2017b) Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell 21: 819–833 e816 [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia‐Verdugo JM, Wysocka J, Ernst P, Alvarez‐Buylla A (2009) Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M (2011) PhyloCSF: a comparative genomics method to distinguish protein coding and non‐coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ et al (2014) An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53: 1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P et al (2016) Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652 [DOI] [PubMed] [Google Scholar]
- Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, van Oudenaarden A (2013) Single‐molecule mRNA detection and counting in mammalian tissue. Nat Protoc 8: 1743–1758 [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Nishihara S, Kamimura M, Shiraishi T, Otoguro T, Uehara M, Maeda Y, Ogura K, Lumsden A, Ogura T (2004) The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci 7: 605–612 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF (2010) Long noncoding RNAs in neuronal‐glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, Kiran AM, De Bo C, Oconnor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV (2014) GWIPS‐viz: development of a ribo‐seq genome browser. Nucleic Acids Res 42: D859–D864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN (2009) Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA 106: 7876–7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H (2008) Requirement for COUP‐TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci 11: 1014–1023 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Iida M, Koseki H, Ohara O (2006) A gene‐targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J 20: 1718–1720 [DOI] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H et al (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full‐length cDNAs. Nature 420: 563–573 [DOI] [PubMed] [Google Scholar]
- Olivares‐Chauvet P, Mukamel Z, Lifshitz A, Schwartzman O, Elkayam NO, Lubling Y, Deikus G, Sebra RP, Tanay A (2016) Capturing pairwise and multi‐way chromosomal conformations using chromosomal walks. Nature 540: 296–300 [DOI] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A et al (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, Woodcock DJ, Ponting CP, Szele FG, Vance KW (2018) The long non‐coding RNA Paupar promotes KAP1‐dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J 37: e98219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP (2009) Genomic and transcriptional co‐localization of protein‐coding and long non‐coding RNA pairs in the developing brain. PLoS Genet 5: e1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales‐Roybal G, Oldham MC, Song JS, Lim DA (2013) Integration of genome‐wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo . Cell Stem Cell 12: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S (2011) The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev 6: 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM et al (2006) Experimental validation of the regulated expression of large numbers of non‐coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman O, Mukamel Z, Oded‐Elkayam N, Olivares‐Chauvet P, Lubling Y, Landan G, Izraeli S, Tanay A (2016) UMI‐4C for quantitative and targeted chromosomal contact profiling. Nat Methods 13: 685–691 [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S (2007) The mRNA‐like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Streubel G, Fitzpatrick DJ, Oliviero G, Scelfo A, Moran B, Das S, Munawar N, Watson A, Wynne K, Negri GL et al (2017) Fam60a defines a variant Sin3a‐Hdac complex in embryonic stem cells required for self‐renewal. EMBO J 36: 2216–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P et al (2017) The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res 45: D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Verma IM (2006) Production and purification of lentiviral vectors. Nat Protoc 1: 241–245 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP (2014) The long non‐coding RNA Paupar regulates the expression of both local and distal genes. EMBO J 33: 296–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Xu RM (2013) Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol 48: 211–221 [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H (2007) LIM‐homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA 104: 13182–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Review Process File
Data Availability Statement
The datasets produced in this study are available in the following database: RNA‐seq and ATAC‐seq data: Gene Expression Omnibus GSE124517 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124517).


