Abstract
Understanding how severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) highjacks epithelial cells and infiltrates the lung, as well as other organs and tissues, is essential for developing treatment strategies and vaccines against this highly contagious virus. Another major goal is to fully elucidate the mechanisms by which SARS-CoV- 2 bypasses the innate immune system and induces a cytokine storm, and its effects on mortality. Currently, SARS- CoV-2 is thought to evade innate antiviral immunity, undergo endocytosis, and fuse with the host cell membrane by exploiting ACE2 receptors and the protease TMMPRSS2, with cathepsin B/L as alternative protease, for entry into the epithelial cells of tissues vulnerable to developing coronavirus disease 2019 (COVID-19) symptoms. However, the incorporation of new and unique binding sites, i.e., O-linked glycans, and the preservation and augmentation of effective binding sites (N-linked glycans) on the outer membrane of SARS-CoV-2 may represent other strategies of infecting the human host. Here, I will rationalize the possibility that other host molecules—i.e., sugar molecules and the sialic acidsN-glycolylneuraminic acid, N-acetylneuraminic acid, and their derivates could be viable candidates for the use as virus receptors by SARS-CoV-2 and/or serve as determinants for the adherence on ACE2 of SARS-CoV-2.
Keywords: SARS-COV-2, COVID-19, Sialic acids, Glycan shield, Neu5Ac, Neu5Gc, Antibodies, Vaccine, Pandemic
Introduction
On the day of writing, more than 40 million people worldwide have been infected with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2 shows a human transmission rate in the range of 1 person infecting 2 or 3 others, at least in the beginning of the pandemic [1], and, although big differences between countries and regions are evident, the overall infection/fatality rate is approximately 1,0% [2], [3]
Coronaviruses (CoVs) infect humans and animals alike and cause a variety of illnesses, including respiratory, enteric, renal, and neurological diseases [4]. CoVs are classified into four genera that affect different animals. The genera alpha-CoV and beta-CoV only affect mammals [5] and mostly result in respiratory and gastrointestinal disorders, whereas gamma-CoV and delta-CoV infect birds and some mammals, including dolphins and white beluga whales [6]. The first known case of a CoV infecting a human occurred in the 1960 s when the virus was isolated from a patient with a cold [7]. Since then, other CoVs have emerged, including SARS-CoV, a lineage B beta-CoV originating from bats and palm civets in 2002–2003 that infected more than 8000 people and caused approximately 800 deaths [5]. In 2012, Middle East respiratory syndrome coronavirus (MERS-CoV), a lineage C beta-CoV, emerged in Saudi Arabia [5] and is currently responsible for 2519 confirmed cases and 866 deaths (World Health Organization, 2020). MERS is still active in the Middle East and shows the highest mortality rate (34.2%) of all CoVs affecting humans. The dromedary camel is considered the zoonotic host of MERS-CoV, although other possible intermediary hosts have not been excluded [8].
The actual data related with the structure of the virus, indicates that the virus probably evolved natural [9] and developed new mechanisms to (1) avoid direct innate immune system surveillance [10], (2) enter host cells through different membrane receptors [11], (3) increase transmission capacity between humans [12], and (4) induce cell-virus membrane or cell–cell fusion [13]. Several reports suggest that the novel infective mechanisms employed by SARS-CoV-2 arose through mutation and assimilation of certain molecules in a hitherto unknown intermediate host [14], although it is also possible that an intermediate host is absent, because of the finding that the firstly identified horseshoe bat virus is able to adhere to the human ACE2 receptor [15], so direct infection of bat to humans could have be possible.
The purpose of this review is to attract attention to the need of identifying the host sugar molecules used by SARS- CoV-2 as a result of its glycan shield. The role of glycans are very often overlooked when scientists search for remedies against viral infections. Viral glycans are much more difficult to identify than immunogenic proteins, because of their tendency to move around and their compositional flexibility and we hypothesize that different host and non-host glycans and sialic acids could be used as alternate receptors of SARS-COV-2. Therefore, it is extremely important to include glycans into the research of the overall picture of viral transmission and the development of vaccines against all virus and especially SARS-CoV-2 which is producing devastating effects not only on human health but also on the global economy. The data of two recent papers in science and nature have clarified part of the composition of the glycan shield of SARS-CoV-2 and the capacity of adhering to at least ten different sugar molecules, c.q. Glycoforms [16], [17].
Evasion of host innate antiviral immunity by SARS-CoV-2
SARS-CoV-2 has a median incubation time of approximately 5.1 days (95% CI, 4.5 to 5.8 days; [18]), indicating viral evasion of the innate antiviral immune system, which leads to a delayed immune response [19], [20]. The innate immune system is normally alerted to viral entry by pattern-recognition receptors (PRRs), including cell membrane-localized Toll-like receptors (TLR), intracellular Nod-like receptors (NLR), and DNA sensors that are triggered by nucleic acids and foreign proteins, in response to pathogen-associated molecular patterns (PAMPs) [21]. When PAMPs are sensed by PRRs, a signaling cascade is initiated that massively recruits leukocytes via type I and III interferon production, eventually limiting viral replication and preventing viral spread to other cells [21]. In the never-ending coevolution between hosts and viruses, both competitors continuously evolve and develop new survival strategies [22]. Accordingly, RNA viruses, including this new RNA coronavirus [10], have evolved multiple strategies of evading the innate antiviral immunity of the host [23].
The SARS-CoV-2 S protein consists of two subunits, S1 and S2. S1 is responsible for attachment to host molecules on the cell membrane, while S2 facilitates fusion between the cell and virus membrane and/or between neighboring cells resulting in cell–cell fusion, forming a syncytium [24]. S1-glycoprotein binding to the ACE2 receptor on the surface of human cells [25], [26] and S- protein cleavage by TMMPRSS2 are critical steps in the crosstalk between the virus and host. SARS-CoV and MERS-CoV utilize several host cell membrane sugars as viral receptors and part of their glycan shield [27]. These sugar molecules are part of sialic acid-binding immunoglobulin-like lectins (SIGLECs), which are responsible for self-recognition by innate immune cells and protection against cytotoxic immune activity [28]. SARS-CoV can bind to Neu5,7,9Ac3 and Neu4,5,Ac2, both derivates of N-acetylneuraminic acid (Neu5Ac) [29], through a host galectin [30] together with the N-linked glycans [31], [32], [33].
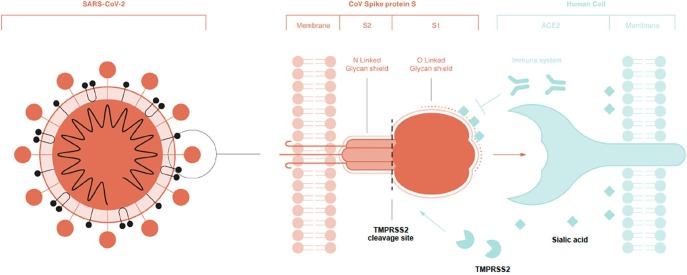
The glycan structure of the SARS-CoV-2 spike glycoprotein (S) is similar to that of SARS-CoV in terms of the N- linked glycans [29]; however, it also possesses unique O-linked glycans and a polybasic cleavage site ([9]; Fig. 1 ). The abundant presence of N- and O-linked glycans covering most of the immunogenic peptides and RNA of SARS-CoV-2 gives the virus the capacity to evade the antiviral innate immune system. Most of the N- and O-linked are linked themselves with multiple sugar molecules and some of them are heavily sialylated [17], masking the immunogenic structures of the virus [34], [16], [35].
Fig. 1.
A schematic overview of the spike glycoprotein S and its subunits S1 and S2. Note the Olinked glycans on S1 in the vicinity of the TMMPRSS2 cleavage site (dashed line). TMMPRSS2 cleaves S2 from S1, facilitating the connection between S1 and hACE2. N-linked glycans (continuous line) cover both the S1 and S2 subunit as part of the masking glycan shield. Different sialic acids serve as part of the glycan shield and could be determinants for viral entry into epithelial cells. The innate antiviral immune system is evaded. and tricked through the possible use of Neu5Gc, Neu5Ac, and their derivates (given as sialic acids) by SARSCoV-2.
Sialic acids and other host sugar molecules and their possible involvement in different SARS-CoV-2 mechanisms
Sialic acids and other host sugar molecules, which are often used as receptors by a wide range of viruses [32], [36], show the highest density on epithelial cells [37], including those in the lungs and more importantly, the oral cavity [38]. The major function of sialic acids and glycans on different cell types is to help the innate immune system discriminate between self and non-self; asa result, different tissues express tens to hundreds of millions of glycan chains per cell that are capped by sialic acids [39]. It is now known that sialic acids comprise a family of more than 50 naturally occurring derivatives of the nine-carbon sugar neuraminic acid (5-amino-3,5-dideoxy-D-glycero-D-galactononulsonic acid, [40]). One branch of the sialic acid family is N-acetylated to form N-acetylneuraminic acids (Neu5Ac, NANA, Sia), which are the most widespread form of sialic acid and almost the only form found in humans. The other branch is based on N-glycolylneuraminic acids (Neu5Gc) which are common in many animal species [41]. Humans lost the capacity to produce Neu5Gc from precursors such as Neu5Ac more than 2 million years ago, due to a mutation of the gene encoding for the cytidine monophosphate N-acetylneuraminic acid hydroxylase (CMAH) enzyme; this mutation possibly arose from a bottleneck event that occurred in response to a type of malaria [28]. Nevertheless, when humans consume Neu5Gc-rich nutrients, this sialic acid can be incorporated into different cell types and expressed on the cell membrane of multiple human tissues or in fluids such as saliva and airway surface liquid [38], [42]. All humans have circulating IgM, IgA and IgG antibodies against Neu5Gc-glycans of different levels [43]. These anti-Neu5Gc antibodies tend to interact with food derived Neu5Gc, metabolically incorporated in host glycans, producing a chronic xenosialitis that can contribute to vascular inflammation [44], diabetes type 2 [45] and cancer progression [46]. SARS-CoV-2 is heavily sialylated and both sialic acids groups (Neu5Ac and Neu5Gc) are candidates for capping the glycan shield of its spike protein S [47].
The nasopharyngeal cavity is constantly exposed to viruses, bacteria, and fungi and contains the entrance of both the gastrointestinal and respiratory tracts. The oral cavity is protected by multiple components of the innate immune system, including immune cells, defensins and—important in the relationship with SARS-CoV-2—a wide range of glycans [38]. Pathogens entering the body through the oral cavity will first interact with host surfaces under a saliva-rich environment. The abundance of glycans in saliva protect the host against pathogenic invaders and nourish symbiotic microbes that make up the oral microbiome [48]. However, several viruses can exploit these glycans as a port of entry, with many of these interactions remaining unknown. SARS-CoV-2 glycan shield is heavily sialylated and almost all N- and O-linked glycans are capped by multiple saccharides [17] and both, saccharides and sialic acids, are abundantly present in saliva. Indeed, SARS-CoV-2 infection can be reliably measured in saliva, and several studies have shown that the virus is present in the saliva of both asymptomatic and symptomatic individuals [49], [50]. The replication rate of the virus in the nasopharyngeal compartments is also unusually high, which may be responsible for its high transmission rates between humans [14]. Moreover, Shang et al. (2020b) [51] demonstrated that SARS- CoV-2 has a 10–20-fold higher capacity than SARS-CoV of adhering to hACE2, which was confirmed by Wrapp et al. (2020) [52]. Given that ACE2 is expressedin epithelial cells of the tongue, this may explain the highly transmissible nature of the virus through saliva [12].
A more recent study [53] found that the SARS-CoV-2 entry receptor ACE2 and viral entry- associated protease TMPRSS2 are highly expressed in nasal goblet and ciliated cells. This finding implicates these cells as loci of original infection and possible reservoirs for dissemination within and between individuals. They also showed that other barrier surface tissues could also suggest further investigation into alternative transmission routes. For example, the co-expression in esophagus, ileum and colon could explain viral fecal shedding observed clinically, with implications for potential fecal–oral transmission [53]. Again, these data demand further research on the role of the glycan shield and host sugar molecules in the pathogenesis of COVID-19, because of the fact that glycans and sialic acids are most abundant on surface barriers and an important part of their integrity under stress circumstances [39], [54].
N- and O-linked glycans, cellular entry
The ongoing vaccine development efforts have primarily focused on the CoV transmembrane S glycoprotein, which extends from the viral surface and mediates host cell entry. Recent studieshave described how SARS-CoV-2 binds to hACE2 and TMPRSS2 [25], [55], [56].
Both the S glycoprotein and ACE2 receptor are known to be extensively glycosylated by complexN- linked-glycans [52]. The glycans on the spike glycoprotein serve multiple purposes, of which the glycan shield is the most well documented [11], [16], [35]. Research on the site-specific, N- linked glycosylation of MERS and SARS S glycoproteins showed that each of these glycosylation sites can be occupied by up to ten different glycans, which greatly increases epitope diversity [16], [35]. Although SARS-CoV-2 and SARS-CoV show very similar N-linked glycans on their spike glycoproteins, SARS- CoV-2 is the first CoV to also incorporate O-linked glycans (Fig. 1). Different reports predicted the presence of new O-linked glycans owing to the incorporation of a proline amino acid, and all were in agreement that this incorporation resulted from immunological pressure in some intermediate host [9]. The existence of these O-linked glycans was confirmed unambiguously in a recent study [17].
N-linked and O-linked glycans show high affinity to sialic acids through the formation of covalent bonds and their amino acid sequences. MERS-CoV shows high affinity to Neu5Ac, with no affinity to Neu5Gc; moreover, no O- linked glycans have been identified for this virus, although it could be that nobody ever looked for them. Both N- and O-linked in SARS-CoV-2 are heavily sialylated [17] and it could be that they act as a determinant factor in viral binding with hACE2 receptors [25], [47], [29].
The presence and characteristics of the O-linked glycans in SARS-CoV-2 appear highly “sophisticated” for a virus affecting humans. If O-linked glycans are essential for the life cycle of SARS-CoV-2, which has made the jump to humans as the preferred vector, why and how has it incorporated these O-linked glycans in the vicinity of the S1 and S2 subunits [57]? O- linked glycation is normally used to mask the cleavage site of proteins and inhibit cleavage ([58]; Fig. 1). Thus, the O-glycans would also impede the necessary cleavage of the SARS-CoV-2 S glycoprotein and its capacity to enter the host cell, fuse, and replicate. The most logical explanation is that the virus needed to mask its S glycoprotein from the immune system of the host and therefore incorporated the glycan shield composed of both N-linked and O-linked glycans capped by host monosaccharides and sialic acids [17].
Discussion
Glycan research in viral structures is difficult but essential to develop treatment options and vaccines against SARS- CoV-2. All the mass spec data point to the SARS-CoV-2 spike protein’s being heavily glycosylated, but less than HIV. HIV is so densely glycosylated that the enzymes that process the sugars on its surface can’t easily reach them. SARS-CoV-2′s sparser glycosylation means that the sugars are more naturally processed than the ones in HIV. But it also suggests that the coronavirus’s glycan shield may not be as effective as that of HIV [16], [35]. Further studies have to focus on the type of sialic acid that is preferentially used by the N- and O-glycans in SARS-CoV-2 and becoming part of its glycan shield.
Establishing whether the SARS-CoV-2 receptors utilize the abundant membrane sugar molecules of the host and identifying the intermediate host would help us expand our knowledge of how SARS-CoV-2 achieved its virulence and was transmitted to humans. This can aid the development of therapeutic strategies or vaccines against the virus as well as highlight lifestyle changes that can be implemented to prevent further infection. If sialic acids and especially Neu5Gc emerge as important players in the pathophysiology of SARS-CoV-2, it may also partially explain the difference in susceptibility between males, females, children, and the elderly. Given that Neu5Gc is an important sialic acid of mammalian meat and dairy products, both part of the human diet, Neu5Gc incorporation in humancell membranes would depend on the amount and frequency of consumption of Neu5Gc-rich nutrients.
Meat consumption is significantly higher by men than women [59], [60] and children are less exposed to meat than are those older than 60 years of age. If Neu5Gc is part of the possible virus receptor reservoir of SARS-CoV-2 it could explain the susceptibility of children for SARS-CoV-2 provoked Kawasaki disease. Several reports inform about an explosive outbreak of children suffering from Kawasaki since the SARS-CoV-2 pandemic. One study, based on a retrospective time-series analysis of the last 15 year, shows an 497% increase of children suffering from Kawasaki since SARS-CoV-2 and this increased incidence is similar to the peak of Kawasaki disease that occurred after the 2009 influenza A H1N1 pandemic, providing evidence of the role of viral infections in triggering Kawasaki disease. [61]. Another study found a 30-fold increased incidence of Kawasaki-like disease. Children diagnosed after the SARS-CoV-2 epidemic began showed evidence of immune response to the virus, were older, had a higher rate of cardiac involvement, and features of macrophage activation syndrome. The SARS-CoV-2 epidemic was associated with high incidence of a severe form of Kawasaki disease [62]. A similar association was found for the level of IgG and IgA antibodies against Neu5Gc and severity of Kawasaki disease in a study with 10 patients and compared with a control group of 6 healthy and 6 febrile patients [63]. In the acute phase antibodies against Neu5Gc raised significantly, stayed high during the subacute phase and decreased at convalescence in the Kawasaki patient group whereas no increase was detected in the control group at any moment [63]. The exact cause of Kawasaki disease has never been established, although different types of virus seem responsible for disease initiation [64]. It could be possible that SARS-CoV-2 uses Neu5Gc to enter children with increased metabolically incorporated Neu5Gc and this could explain the COVID-19 like Kawasaki disease symptoms. Anyway, the fact that the glycan shield of SARS-CoV-2 is heavily sialylated, supports the hypotheses that Neu5Ac and/or Neu5Gc and their derivates are possible virus receptors and/or determinants for cell entrance of the virus. If so, then it could be of interest to investigate the use of anti-sialic acid-glycan antibodies as a possible vaccine to prevent COVID-19. Nevertheless, it could still take months till definite data are available; it took years till it was shown that MERS has the capacity to adhere to Neu5Gc [65].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
I would like to thank Editage (www.editage.com) for English language editing and Leonard Haase for editing the references in this paper.
References
- 1.Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491):eabb6936. doi: 10.1126/science:abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H., Dighe A., Griffin J.T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C.A., Ghani A.C., Ferguson N.M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20(6):669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell, T. W., Hellewell, J., Jarvis, C. I., Zandvoort, K. Van, Abbott, S., Ratnayake, R., Flasche, S., Eggo, R. M., Edmunds, W. J., & Kucharski, A. J. (2020). Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Eurosurveillance, 25(12), 6–10. https://doi.org/10.2807/1560-7917.ES.2020.25.12.2000256. [DOI] [PMC free article] [PubMed]
- 4.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L.i., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q.i., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Tsang A.K.L., Hui S.-W., Fan R.Y.Y., Martelli P., Yuen K.-Y. Discovery of a Novel Bottlenose Dolphin Coronavirus Reveals a Distinct Species of Marine Mammal Coronavirus in Gammacoronavirus. J Virol. 2014;88(2):1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Kahn J.S., McIntosh K. History and Recent Advances in Coronavirus Discovery: Pediatr Infect Dis J. 2005;24(Supplement):S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 8.Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. The Lancet. 2020;395(10229):1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikkert, M. (2020). Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 12, 4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed]
- 11.Li R., Qiao S., Zhang G. Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J Infect. 2020;80(4):469–496. doi: 10.1016/j.jinf.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, H., Zhong, L., Deng, J., Peng, J., Dan, H., Zeng, X., et al. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12, 8. doi: 10.1038/s41368-020-0074-x Xu, J., Zhao, S., Teng, T., Abdalla, A.E., Zhu, W., Xie, L., et al. (2020). Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 12, E244. doi:10.3390/v12020244 Yu, C., Gao, K., Zhu, L., Wang, W., Wang, L., Zhang, F., et al. (2016). At least two Fc Neu5Gc residues of monoclonal antibodies are required for binding to anti-Neu5Gc antibody. Sci. Rep. 7, 20029. doi: 10.1038/srep20029.
- 13.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang, J., Wan, Y., Liu, C., Yount, B., Gully, K., Yang, Y., et al. (2020a). Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 16, e1008392. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed]
- 15.Lau S.K.P., Luk H.K.H., Wong A.C.P., Li K.S.M., Zhu L., He Z., Fung J., Chan T.T.Y., Fung K.S.C., Woo P.C.Y. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020;26(7):1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020 doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelemans, T., and Kikkert, M. (2019). Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses 11, E961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed]
- 20.Prompetchara, E., Ketloy, C., and Palaga, T. (2020). Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 38, 4401– 9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed]
- 21.Sparrer, K. M. J., and Gack, M. U. (2015). Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 26, 1–9. doi: 10.1016/j.mib.2015.03.001. Sun, Z., Thilakavathy, K., Kumar, S. S., He, G., and Liu, S. V. (2020). Potential factors influencing repeated SARS outbreaks in China. Int. J. Environ. Res. Public Health 17, E1633. doi: 10.3390/ijerph17051633. [DOI] [PMC free article] [PubMed]
- 22.Retel, C., Märkle, H., Becks, L., and Feulner, P. G. D. (2019). Ecological and evolutionary processes shaping viral genetic diversity. Viruses 11, 220. doi: 10.3390/v11030220. [DOI] [PMC free article] [PubMed]
- 23.Beachboard D.C., Horner S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr Opin Microbiol. 2016;32:113–119. doi: 10.1016/j.mib.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belouzard, S., Millet, J. K., Licitra, B. N., and Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4, 1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed]
- 25.Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280. doi: 10.1016/j.cell.2020.02.052. World Health Organization. (2020). Middle East respiratory syndrome coronavirus (MERS-CoV)– The Kingdom of Saudi Arabia. https://www.who.int/csr/don/24-february-2020-mers-saudi- arabia/en/ [Accessed April 2020]. [DOI] [PMC free article] [PubMed]
- 26.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, Z., Yu, C., Wang, W., Yu, G., Zhang, T., Zhang, L., et al. (2018). Aloe polysaccharides inhibit influenza A virus infection—a promising natural anti-flu drug. Front. Microbiol. 9, 2338. doi: 10.3389/fmicb.2018.02338. [DOI] [PMC free article] [PubMed]
- 28.Varki A. Uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci. 2010;107(Supplement_2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F., Goff S.P. Receptor Recognition Mechanisms of Coronaviruses: a Decade of Structural Studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.-J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasik B.R., Barnard K.N., Parrish C.R. Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol. 2016;24(12):991–1001. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr Top Microbiol Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;9983:eabb9983. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banda, K., Gregg, C. J., Chow, R., Varki, N. M., and Varki, A. (2012). Metabolism of vertebrate amino sugars with N-glycolyl groups: Mechanisms underlying gastrointestinal incorporation of the non-human sialic acid xeno-autoantigen N-glycolylneuraminic acid. J. Biol. Chem. 287, 31528852– 28864. doi: 10.1074/jbc.M112.364182, Bardor, M., Nguyen, D. H., Diaz, S., and Varki, A. (2005). Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. J. Biol. Chem. 280, 3184228– 4237. doi: 10.1074/jbc.M412040200. [DOI] [PMC free article] [PubMed]
- 38.Cross B.W., Ruhl S. Glycan recognition at the saliva – oral microbiome interface. Cell Immunol. 2018;333:19–33. doi: 10.1016/j.cellimm.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altman, M. O., and Gagneux, P. (2019). Absence of Neu5Gc and presence of anti-Neu5Gc antibodies in humans—an evolutionary perspective. Front. Immunol. 10, 789. doi: 30410.3389/fimmu.2019.00789. [DOI] [PMC free article] [PubMed]
- 40.Koehler, M., Delguste, M., Sieben, C., Gillet, L., & Alsteens, D. (2020). Initial Step of Virus Entry: Virion Binding to Cell-Surface Glycans. Annual Review of Virology, 7(1), 1–23. https://doi.org/10.1146/annurev- virology-122019-070025. [DOI] [PubMed]
- 41.Wang B., Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr. 2003;57(11):1351–1369. doi: 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 42.Martins M.d.F., Honório-Ferreira A., Martins P., Gonçalves C.A. Presence of sialic acids in bronchioloalveolar cells and identification and quantification of N-acetylneuraminic and N-glycolylneuraminic acids in the lung. Acta Histochem. 2019;121(6):712–717. doi: 10.1016/j.acthis.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Dhar C., Sasmal A., Varki A. From “Serum Sickness” to “Xenosialitis”: Past, Present, and Future Significance of the Non-human Sialic Acid Neu5Gc. Front Immunol. 2019;10(April):807. doi: 10.3389/fimmu.2019.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham, T., Gregg, C. J., Karp, F., Chow, R., Padler-Karavani, V., Cao, H., Chen, X., Witztum, J. L., Varki, N. M., & Varki, A. (2009). Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood, 114(25), 5225–5235. https://doi.org/10.1182/blood-2009-05-220400. [DOI] [PMC free article] [PubMed]
- 45.Kuipers R.S., Pruimboom L. Short comment on “A review of potential metabolic etiologies of the observed association between red meat consumption and development of type 2 diabetes mellitus”, by Yoona Kim, Jennifer Keogh, Peter Clifton. Metabolism. 2016;65(1):e3–e4. doi: 10.1016/j.metabol.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Samraj A.N., Pearce O.M.T., Läubli H., Crittenden A.N., Bergfeld A.K., Banda K., Gregg C.J., Bingman A.E., Secrest P., Diaz S.L., Varki N.M., Varki A. A red meat-derived glycan promotes inflammation and cancer progression. PNAS. 2015;112(2):542–547. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.-J., Bosch B.-J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26(6):481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koropatkin N.M., Cameron E.A., Martens E.C. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azzi, L., Carcano, G., Gianfagna, F., Grossi, P., Gasperina, D. D., Genoni, A., et al. (2020). Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. S0163–4453, 30213–30219. doi: 30910.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed]
- 50.To K.-W., Tsang O.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.-Y., Cai J.-P., Chan J.-C., Chik T.-H., Lau D.-L., Choi C.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.-K., Poon R.-S., Luo C.-T., Cheng V.-C., Chan J.-W., Hung I.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang J., Ye G., Shi K.e., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo- EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.aax0902. Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed]
- 53.Sungnak, W., Huang, N., Bécavin, C., Berg, M., Queen, R., Litvinukova, M., Talavera-López, C., Maatz, H., Reichart, D., Sampaziotis, F., Worlock, K. B., Yoshida, M., Barnes, J. L., Banovich, N. E., Barbry, P., Brazma, A., Collin, J., Desai, T. J., Duong, T. E., … Figueiredo, F. (2020). SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine, 26(5), 681–687. . Thaysen-Andersen, M., Larsen, M. R., Packer, N. H., and Palmisano, G. (2013). Structural analysis of glycoprotein sialylation-Part I: Pre-LC-MS analytical strategies. RSC Adv. 3, 22683–22705. doi: 10.1039/c3ra42960a. [DOI] [PMC free article] [PubMed]
- 54.Cioffi D.L., Pandey S., Alvarez D.F., Cioffi E.A. Terminal sialic acids are an important determinant of pulmonary endothelial barrier integrity. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2012;302(10):L1067–L1077. doi: 10.1152/ajplung.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng J. SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J. Identification of common deletions in the spike protein of SARS-CoV-2. J Virol, June. 2020 doi: 10.1128/jvi.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachert C., Linstedt A.D. A Sensor of Protein O-Glycosylation Based on Sequential Processing in the Golgi Apparatus: O-Glycosylation Sensor. Traffic. 2013;14(1):47–56. doi: 10.1111/tra.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love H.J., Sulikowski D. Of meat and men: Sex differences in implicit and explicit attitudes toward meat. Front Psychol. 2018;9:559. doi: 10.3389/fpsyg.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa S., Hart C. Where’s the beef? How masculinity exacerbates gender disparities in health behaviors. Socius. 2019;5:1–12. doi: 10.1177/2378023119831801. [DOI] [Google Scholar]
- 61.Ouldali N., Pouletty M., Mariani P., Beyler C., Blachier A., Bonacorsi S., Danis K., Chomton M., Maurice L., Le Bourgeois F., Caseris M., Gaschignard J., Poline J., Cohen R., Titomanlio L., Faye A., Melki I., Meinzer U. Emergence of Kawasaki disease related to SARS-CoV-2 infection in an epicentre of the French COVID-19 epidemic: a time-series analysis. The Lancet Child & Adolescent Health. 2020;4(9):662–668. doi: 10.1016/S2352-4642(20)30175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. The Lancet. 2020;395(10239):1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padler-Karavani V., Tremoulet A.H., Yu H., Chen X., Burns J.C. A Simple Method for Assessment of Human Anti-Neu5Gc Antibodies Applied to Kawasaki Disease. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0058443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toubiana J., Poirault C., Corsia A., Bajolle F., Fourgeaud J., Angoulvant F., Debray A., Basmaci R., Salvador E., Biscardi S., Frange P., Chalumeau M., Casanova J.L., Cohen J.F., Allali S. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ (Clinical Research Ed.) 2020;369 doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci USA. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]