Highlights
-
•
EEG-fMRI has been instrumental in characterizing brain networks in epilepsy.
-
•
Its value is documented in the pre-surgical assessment of drug-resistant epilepsy.
-
•
The delineation of brain areas to resect is fundamental for the post-surgical outcome.
-
•
Standard EEG-fMRI in epilepsy assesses static functional connectivity of the network.
-
•
EEG-fMRI dynamic connectivity identifies transitory features of specific connections.
-
•
We integrate dynamic fMRI connectivity and dynamic patterns of simultaneous scalp EEG.
-
•
This allows to better characterize the spatiotemporal aspects of epileptic networks.
-
•
This may help in more efficiently target the surgical intervention.
Keywords: EEG-fMRI, Epilepsy, Dynamic functional connectivity, Pre-surgical planning
Abstract
Epileptic networks, defined as brain regions involved in epileptic brain activity, have been mapped by functional connectivity in simultaneous electroencephalography and functional magnetic resonance imaging (EEG-fMRI) recordings. This technique allows to define brain hemodynamic changes, measured by the Blood Oxygen Level Dependent (BOLD) signal, associated to the interictal epileptic discharges (IED), which together with ictal events constitute a signature of epileptic disease. Given the highly time-varying nature of epileptic activity, a dynamic functional connectivity (dFC) analysis of EEG-fMRI data appears particularly suitable, having the potential to identify transitory features of specific connections in epileptic networks. In the present study, we propose a novel method, defined dFC-EEG, that integrates dFC assessed by fMRI with the information recorded by simultaneous scalp EEG, in order to identify the connections characterised by a dynamic profile correlated with the occurrence of IED, forming the dynamic epileptic subnetwork. Ten patients with drug-resistant focal epilepsy were included, with different aetiology and showing a widespread (or multilobar) BOLD activation, defined as involving at least two distinct clusters, located in two different lobes and/or extended to the hemisphere contralateral to the epileptic focus. The epileptic focus was defined from the IED-related BOLD map. Regions involved in the occurrence of interictal epileptic activity; i.e., forming the epileptic network, were identified by a general linear model considering the timecourse of the fMRI-defined focus as main regressor. dFC between these regions was assessed with a sliding-window approach. dFC timecourses were then correlated with the sliding-window variance of the IED signal (VarIED), to identify connections whose dynamics related to the epileptic activity; i.e., the dynamic epileptic subnetwork. As expected, given the very different clinical picture of each individual, the extent of this subnetwork was highly variable across patients, but was but was reduced of at least 30% with respect to the initially identified epileptic network in 9/10 patients. The connections of the dynamic subnetwork were most commonly close to the epileptic focus, as reflected by the laterality index of the subnetwork connections, reported higher than the one within the original epileptic network. Moreover, the correlation between dFC timecourses and VarIED was predominantly positive, suggesting a strengthening of the dynamic subnetwork associated to the occurrence of IED. The integration of dFC and scalp IED offers a more specific description of the epileptic network, identifying connections strongly influenced by IED. These findings could be relevant in the pre-surgical evaluation for the resection or disconnection of the epileptogenic zone and help in reaching a better post-surgical outcome. This would be particularly important for patients characterised by a widespread pathological brain activity which challenges the surgical intervention.
1. Introduction
Focal epilepsy is a network disease where the abnormal activity occurs within networks of cortical and subcortical brain structures limited to one hemisphere, either discretely localized or more widely distributed (Berg et al., 2010, Pittau et al., 2014).
The simultaneous recording of electroencephalogram (EEG) and functional magnetic resonance imaging (fMRI); i.e., EEG-fMRI, has been instrumental in revealing the large-scale nature of the networks involved in the epileptic process, by identifying widespread activation/deactivation correlated with focal epileptic activity (Gotman, 2008, Laufs, 2012, Pittau et al., 2012, Thornton et al., 2011). To investigate the functional coupling between brain regions, functional connectivity (FC) at rest is commonly assessed from fMRI data with correlational approaches (seed- or atlas-based), where the statistical interdependence between signals at different brain locations is evaluated, as well as with whole-brain decomposition techniques such as independent component analysis (ICA) (McKeown and Sejnowski, 1998). FC applied to EEG-fMRI contributed to describe alterations of connectivity patterns related to epilepsy (for an extended review, see (Centeno and Carmichael, 2014)) and to predict the outcome after surgery (Bettus et al., 2010, Morgan et al., 2017, Negishi et al., 2011).
Epilepsy is characterised by a particular type of resting state (the interictal period) with occurrences of interictal epileptiform discharges (IED), captured by scalp or intracranial EEG. The identification of FC features associated to IED is crucial to understand in which measure the brain functional coupling at rest is altered in the epileptic disease. EEG-fMRI studies have investigated the role of IED in the organization of the epileptic network, demonstrating the preservation of its spatial structure even in the absence of (detectable) IED, by comparing epochs with and without IED (Luo et al., 2014) or by regressing out the IED effect (Iannotti et al., 2016). However, these observations were obtained through the investigation of static FC; i.e., correlations computed over the whole EEG-fMRI recording time (6 to 20 min).
Recent evidence has suggested the relevance of dynamic features of FC, as FC was shown to fluctuate substantially within recording times of the duration of a run (Hutchison et al., 2013, Preti and Van De Ville, 2017). Among the different available methodologies of dynamic FC (dFC), the most commonly applied approach is the sliding-window technique (for a review, see (Preti et al., 2017)). This method assesses pairwise correlations in epochs (or windows) of generally 40–60 s and spanning the entire duration of the recording time.
If exploring FC dynamics is one of the key tools to understand brain function in healthy individuals, we can presume that this becomes even more crucial in epilepsy, which, by definition, is characterised by transitory, dynamic events, as IED likely induce substantial changes in network properties. Some connections of the epileptic network identified with static FC approaches might be specifically modulated during such dynamic events with clinical relevance. In line with this, Chiang and colleagues (Chiang et al., 2018) showed better classification performances in distinguishing patients with temporal lobe epilepsy (TLE) and healthy controls when including dFC features. Further, the inclusion of the EEG signal information in the analysis provides a proxy of the epileptic activity signature, to be associated with changes in connectivity. Few studies have explored dFC with simultaneous EEG-fMRI in epilepsy (Laufs et al., 2014, Lopes et al., 2014, Omidvarnia et al., 2017, Preti et al., 2014) and only three of them included patients with focal epilepsy not limited to the temporal lobe and showing a widespread EEG-fMRI activation, sometimes including the hemisphere contralateral to the focus (Abreu et al., 2019, Omidvarnia et al., 2017, Preti et al., 2014). These latter studies integrated the pathological EEG characteristics in the dynamic analysis; i.e., not only for IED or sleep localisation, but for direct correlation with dFC measures. In particular, Preti et al. (Preti et al., 2014) developed a new approach to allow the direct correlation of these two measures and tested it on two cases of focal epilepsy. Abreu and colleagues (Abreu et al., 2019) applied a machine learning approach (more specifically a dictionary learning) to dFC data integrated with EEG synchronization features to identify epileptic dFC states, which were concordant with IED type and ictal propagation. Following a different approach, Omidvarnia et al. (Omidvarnia et al., 2017) demonstrated the strong link between changes in connectivity and region-specific interictal EEG power, arguing that the complement of EEG information would describe additional aspects of the epileptic network, compared to the standard FC analysis alone. Studies focused on TLE (Laufs et al., 2014, Lopes et al., 2014) had already confirmed that dFC is essential to capture the pathology extension beyond the focus site. These findings motivate a better characterisation of dFC especially for patients who are candidate for resective surgery and whose EEG-fMRI examination shows a widespread activation, in order to identify the regions, even outside the focus, whose removal could improve the surgical outcome.
To this aim, we propose a novel approach that we call dFC-EEG, preliminarily introduced in (Preti et al., 2014), that combines dFC analysis with the temporal characteristics of the epileptic activity extracted from the scalp EEG to define how connections strength can be dynamically related to the occurrence of IED. We hypothesise that only a portion of the original network, i.e., the dynamic epileptic subnetwork, has connections that fluctuate with the transient occurrence of IED. This approach results in a selective definition of the regions in the epileptic network that could deserve particular attention in the pre-surgical evaluation, in order to prevent post-operative remission and/or comorbidities.
2. Methods
2.1. Subjects and acquisitions
We studied the same cohort of EEG-fMRI patients with drug-resistant epilepsy included in our previous study (Iannotti et al., 2016): 10 patients (six female; mean age at evaluation, 17 years). Criteria used for patients’ selection included: i) diagnosis of drug-resistant focal epilepsy; ii) widespread (or multilobar) IED-related BOLD map, with involvement of at least 2 lobes ispilateral or contralateral to the epileptic focus as resulting from a GLM analysis with the IED as main regressor, after applying a threshold of FWE- p-value < 0.05 and minimal size of 20 contiguous voxels. The 10 patients were recruited in the context of a multi-center study, between the Institute of Child Health in London and the University Hospital in Geneva. In our database 35% of patients had IED during the scanning period. 29% of them (8 patients) had a widespread IED-related BOLD map. From London’s database we included 2 out of 10 preselected patients who had a widespread BOLD activation. The clinical details of the recruited patients are reported in Table 1.
Table 1.
Clinical details of patients.
| Patient | Age at evaluation/ Gender/ Age at onset (y) | IED location | Number |
Inter-IED duration | MRI | EZ | Anatomical details of IED-related BOLD map (max t-value) | Concordance fMRI-seed vs estimated EZ (non-invasive) | Resection area | Post-operative outcome (time after surgery) (y) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Single IED | Burts of IED | ||||||||||
| (mean, [min, max]) (s) | (mean, [min, max]) (s) | ||||||||||
| P1 | 46/F/12 | R frontal (F4) | 10 (1.8; [0.5, 2.8]) | 66,4; [7.1, 194.5] | normal | R Fronto-parietal (pericentral) | Posterior part of R SFG (+10.7), R pre- and postcentral gyrus, R PO junction, Posterior part of L SFG, paracentral lobule, R putamen, R thalamus, pons, mesencephalon, L cerebellum | Concordant | |||
| P2 | 13/F/1 | L frontal (Fp1-F3) | 81 | 14,9 [0.2;122,9] | Tuberous sclerosis | L prefrontal (tuber) | L superior frontal sulcus (+7.2) tuber, L anterior and mid-cingulate, R TO junction, L and R insula, L thalamus | Concordant | L frontal tuber; L central tuber | I (5) | |
| P3 | 7/M/2 | L central (Fz-Fc1) | 53 (162.8; [13.7, 278,5]) | 4.5; [0.2, 23.9] | normal | L central | L mesial paracentral lobule (+8.1), L precentral gyrus, L putamen, Lcaudate, bilateral cerebellum | Concordant | |||
| P4 | 20/F/9 | R frontal (Fp2-F8) | 120 | 9.9; [0.4, 49.0] | FCD | R Frontal pole/orbital | R frontopolar (+7.5) FCD, R occipital, R cerebellum | Concordant | R Fronto-basal region | II (1.5) | |
| P5 | 18/M/8 | R frontal (F4) | 106 | 11.1; [0.1, 302.3] | Normal | R Fronto-temporal | R superior frontal sulcus (+12), L superior frontal sulcus, R thalamus, R superior frontal gyri, mesencephalon, precuneus | Concordant | |||
| P6 | 15/F/1 | L temporo-insular (T7) | 9 (1.9; [1.3, 2.8]) | 81; [19, 160] | Ischemic lesion | L large perilesional area, fronto-centrotemporal (multifocal) | L planum temporal (+6.7) (perilesional), L TPO junction, R PO junction | Concordant | |||
| P7 | 17/F/4 | R frontal (F4) | 28 | 54.2; [7.3; 201.2] | Normal | R frontal | R (+16.3) and L SFG and MFG, L/R caudate nucleus, L/R thalamus, bilateral cerebellum | Concordant | |||
| P8 | 7/M/0 | L temporoparietal (T7) | 36 (23.7;[2.8; 62]) | 2.3, [1.2; 6.1] | Abscess | L inferior frontal or temporal | L planum temporale (-15.4) (perilesional), L Thalamus, R and L caudate nucleus, R superior temporal gyrus, mesencephalon | Concordant | L temporo-parieto-occipital region | I (0.5) | |
| P9 | 12/M/7 | L parietal (C3-Cz) | 100 1.5; [0.5, 2.9] | 15.1; [1.7, 167,1] | DNET | R Parietal | Posterior part of the L SFG (+7.0), bilateral cerebellum, mid-cingulate, L putamen, R MFG | Discordant | L occipital parasagittal region | II (1.5) | |
| P10 | 15/F/1 | L temporo-occipital (T5) | 42 (353.6; [16.3, 353.6]) | 14.3; [0.3, 65.5] | Polymicrogiria | L parieto-temporal insular | L T-O region (+6.3), L IFG, L caudate nucleus, P-O junction, L superior temporal gyrus | Concordant | |||
IED: interictal epileptic discharges; EZ: epileptogenic zone; DNET: dysembryoplastic neuroepithelial tumor; F: female; FCD: focal cortical dysplasia; IFG: inferior frontal gyrus; L: left; M: male; MFG: middle frontal gyrus; O: occipital; P: parietal; R: right; SFG: superior frontal gyrus; T: temporal; (s): seconds; (y): years. Three certified epileptologists marked the epileptic events, i.e., single IED or burst of IED. Column 4 and 5 define the characteristics of the epileptic events, i.e., the total number of IED observed and the duration in case of burst of IED. Column 6 reports the temporal occurrence of the epileptic events. Column 7 reports possible abnormalities observed with structural IRM. Column 8 describes the clinical estimation of the epileptogenic zone, resulting from the multimodal non-invasive clinical information. Column 9 indicates the anatomical location of clusters in the IED-related BOLD activation map, as visually defined by a certified neurologist; in particular for the most significant cluster (or global maximum) the statistical t-value is indicated. Column 10 defines if the main seed of the IED-related BOLD map (the region in column 8 for which the max t-value is indicated) was concordant with the presumed EZ (column 9). Columns 11 and 12 defines the characteristics of the 4/10 patients who underwent surgery: the resected area and the outcome expressed according to Engel’s class.
After providing informed consent in agreement with the requirements of the Declaration of Helsinki and the local ethics committees, the patients underwent simultaneous EEG-fMRI recording. Twenty minutes of resting state EEG-fMRI data were collected for each patient, instructed to lie still in the MR scanner with eyes closed and without thinking of anything in particular.
The EEG was acquired with MR-compatible EEG systems equipped with 64 (BrainCap MR, Brain Products GmbH, Munich, Germany; London patients P3 and P10) or 256 electrodes (Electrical Geodesic, Inc., Eugene, OR, U.S.A., Geneva patients) in 1.5 Tesla (Siemens Avanto; London patients) or 3 Tesla (Siemens Trio; Geneva patients) MR-scanner. T1-weighted structural images and T2*-weighted single-shot gradient echo echo-planar functional images were acquired for all patients. Different parameters were used for the functional volumes depending on the site and the date of acquisition, as shown in Table 2.
Table 2.
Acquisition parameters.
| Patient | Number Volumes | Number Slices | TR (ms) | TE (ms) | Acquisition | Field map | Resolution (mm3) |
|---|---|---|---|---|---|---|---|
| P1 | 400 | 32 | 2880 | 30 | Sequential | Yes | 3x3x3.75 |
| P2 | 1100 | 25 | 1500 | 35 | Sequential | Yes | 3.75x3.75x5 |
| P3 | 300 | 30 | 2160 | 30 | Sequential | Yes | 3.3x3.3x4 |
| P4 | 600 | 32 | 1990 | 30 | Sequential | Yes | 3x3x3.75 |
| P5 | 600 | 32 | 1984 | 30 | Sequential | Yes | 3.75x3.75x5 |
| P6 | 600 | 32 | 1984 | 30 | Sequential | Yes | 3.75x3.75x5 |
| P7 | 1100 | 25 | 1500 | 35 | Sequential | Yes | 3.75x3.75x5 |
| P8 | 1100 | 25 | 1500 | 35 | Sequential | Yes | 3.75x3.75x5 |
| P9 | 600 | 32 | 1990 | 30 | Sequential | Yes | 3x3x3.75 |
| P10 | 300 | 30 | 2160 | 30 | Sequential | Yes | 3.3x3.3x4 |
For each patient, the details about the fMRI sequence used are reported. TR: Repetition time; TE: echo time.
2.2. Data preprocessing
EEG artifact correction was performed off-line, by using the Imaging Artifact Reduction (IAR) and the Average Template Subtraction (AAS) algorithms for correcting, consecutively, the gradient and the pulse artifact (Allen et al., 2000, Allen et al., 1998). For the 64 channels system we used the algorithm implementation of the BrainVision Analyser software (Brain Products GmbH, Munich, Germany). For the 256 channel EEG, we adopted an in-house MATLAB pipeline that applies the IAR and AAS algorithms and integrates the pulse artifact detection based on scalp topography, according to the method validated in (Iannotti et al., 2015).
Structural and functional MRI were analysed in patients’ individual space. Structural images were segmented into gray matter, white matter and cerebrospinal fluid using SPM8 software (http://www.fil.ion.ucl.ac.uk/spm) and converted into binary masks, applying a threshold of 0.5 on the probability values of tissue classes.
Functional MRI volumes were preprocessed with SPM8 software. Functional images were realigned and corrected for the susceptibility artifact, by considering the field map sequence acquired during the EEG-fMRI (SPM8 ‘realignment & unwarp’ option), corrected for slice timing, linearly co-registered to the structural image and smoothed with an isotropic Gaussian kernel with FWHM = 6 mm. Nuisance regressors (linear and quadratic trends, six motion parameters, and average white matter and cerebrospinal fluid signals) were regressed out with a GLM. A band-pass filter of the range [0.03–0.15] Hz, to focus the analysis on low frequency fluctuations, previously shown to be of interest in resting state network analysis (Biswal et al., 1995, Smith et al., 2013), and at the same time removing too low frequencies that would cause spurious fluctuations in the sliding-window procedure applied in a successive step to estimate dFC.
2.3. Epileptic network identification
The complete pipeline described in paragraphs 2.3–2.6 is illustrated in Fig. 1.
Fig. 1.
Pipeline of the dFC-EEG approach. The figure describes the steps to investigate the dependency of the epileptic network dynamic from the epileptic activity, expressed as the variance of the convolved IED (VarIED), by showing patient P2 (see Table 1 for IED type, number, and inter-IED period). The IED detected from the corrected EEG (A.a) were convolved with multiple HRF (A.b) and used to derive the IED-related BOLD map (A.c), from which the epileptic focus was identified. Average BOLD timecourses were extracted from the fROIs defined by a second GLM with the focus timecourse as main regressor (A.d). The fROIs-to-fROIs dFC was evaluated with a sliding-window approach (B.f), that was also applied to derive the variance of the convolved IED; i.e., the VarIED signal (B.e). The significant Spearman correlations between fROIs-to-fROIs dFC timecourses and the VarIED defined the connections included in the dynamic epileptic subnetwork (C.g).
First of all, for each patient we obtained the IED-related BOLD activation map by following the multi-peak GLM approach shown in (Iannotti et al., 2015): IED were identified by three expert epileptologists (F. P., S.V., and M. C.) on the scalp EEG acquired inside the MR scanner and corrected for gradient and pulse artifacts; the obtained IED signal was convolved with five HRF and used as regressor of interest in five separate general linear models (Bagshaw et al., 2004, Iannotti et al., 2015). The resulting IED-related BOLD map was obtained by considering for each voxel the highest t‐value across the single maps and applying a threshold of p = 0.05, FWE corrected. The global maximum of such map was considered as center of a 10 mm-diameter sphere, and the average BOLD signal of all voxels within that sphere was considered as the epileptic focus BOLD timecourse.
This was used as seed for a second GLM, aiming to identify voxels co-activating with the focus BOLD timecourse, and forming the epileptic network. In detail, the seed timecourse was used as regressor of interest and significant clusters in the resulting t-contrast map were considered (p = 0.05 FWE, at least 20 contiguous voxels, excluding cerebellum). Then, N Functional Regions-of-Interest (fROIs) built using 10 mm-diameter spheres at the coordinates of the t-map activation peaks and masked for grey matter, were obtained as part of the epileptic network, N being patient-specific (Fig. 1A). The static FC within the epileptic network was assessed by computing Pearson correlation coefficients between averaged timecourses of all pairs of fROIs, yielding an NxN symmetric FC matrix (see Figures S1-S10 in Supplementary Materials).
2.4. dFC-EEG: Dynamic functional connectivity of the epileptic network
In order to explore the dynamic features of the epileptic network, the dFC between fROIs was assessed by using a sliding-window approach (Chang and Glover, 2010, Hindriks et al., 2016, Hutchison et al., 2013, Tagliazucchi and Laufs, 2015). Pairwise Pearson correlations were computed in sliding-windows between the N regional timecourses of the fROIs, yielding a number of W FC matrices (one per window) of size NxN. The parameters for the sliding-window analysis were chosen according to the specific application as: window size = 30 s, in order to capture the fast connectivity changes that we can expect to arise in epilepsy, and step = 6 s, the shortest possible to be homogeneous across subjects, taking into account the different repetition time (TR) of functional sequences (Table 2). The whole pipeline was applied also with window sizes of 40 s and 50 s to check for consistency of results.
Due to the symmetry of the FC matrix, the analysis was restricted to its lower triangular part values (connections), which were vectorized and concatenated across windows, yielding a dFC matrix, including as rows the timecourses of fROIs-to-fROIs connections (Fig. 1.B).
2.5. Dependence of epileptic network dynamics on the epileptic activity
We investigated the relation between the epileptic network dynamics and the observed epileptic activity using the recorded EEG on the scalp. Following the approach introduced by Preti and colleagues (Preti et al., 2014), we used a “second-order” measure to describe the epileptic activity, consisting in the variance of the recorded IED signal in each window, named VarIED (Fig. 1 B). Indeed, this is a necessary step to obtain a variable of the same order as dFC correlation timecourses.
The correlation between each row of the matrix (representing the evolution of a connection in time) and the VarIED signal were then computed in terms of Spearman correlation coefficient, yielding an fROIs-to-fROIs matrix (Fig. 1 C.g).
2.6. Statistical identification of the dynamic epileptic sub-network
The dynamic epileptic subnetwork was defined, as formed by connections with significant Spearman correlation with VarIED; i.e., connections whose dynamics relate to the epileptic activity. Spearman correlation coefficient was chosen here instead of Pearson correlation, which is commonly adopted for FC, due to the not Gaussianly distributed nature of the VarIED signal.
To assess the statistical significance of the Spearman correlation values between dFC timecourses and the VarIED signal, we performed a non-parametric permutation test (p-value < 0.05) based on a null distribution of the correlation computed using 999 random surrogates of the VarIED signal. In particular, in order to preserve the inter-IED temporal features of the empirical VarIED in the permuted signals, for each patient: i) we evaluated the average inter-IED duration converted to the time resolution of fMRI (seconds) by the convolution with the canonical HRF; ii) we subdivided the total duration of VarIED in epochs lasting for a period (in windows) equal to the average duration (converted from seconds to windows); iii) we randomized the VarIED signal in each epoch; iv) we concatenated the randomized epochs. Connections reporting significant correlations were considered part of the dynamic epileptic subnetwork (Fig. 1 C).
2.7. Spatial organization of the dynamic epileptic subnetwork
BrainNet Viewer toolbox ((Xia et al., 2013) http://www.nitrc.org/projects/bnv/) was used to visualize dynamic epileptic subnetworks as brain graphs, where nodes (of standardized size) represent fROIs and edges represent the correlation of dFC timecourses to the epileptic activity. fROIs were labeled according to decreasing t-values resulting from the second GLM (paragraph 2.3); label 1 will therefore always identify the seed region; i.e., the epileptic focus. The size of the dynamic epileptic subnetwork was evaluated for each subject as the percentage of its connections with respect to all connections within the (static) epileptic network. The subnetwork size was correlated with the disease duration by means of Pearson correlation coefficient.
We divided the dynamic epileptic subnetwork connections in: i) ipsilateral to the epileptic focus (i.e., both connected nodes belong to the same hemisphere of the focus, number of connections = ); ii) contralateral to the epileptic focus (i.e., both connected nodes are in the hemisphere contralateral to the focus, number of connections = ); iii) inter-hemispheric connections (i.e., the connected nodes belong to two different hemispheres, number of connections = ). Building on previous studies (Negishi et al., 2011) that defined the laterality index of the nodes in the static epileptic network and associated higher values (closer to 1) to good post-surgical outcomes (Engel’s class I, (Wieser et al., 2001)), we defined the laterality index of connections :
| (Eq.1) |
This index describes the network laterality with respect to the focus as the proportion of connections involving the focus hemisphere with respect to the ones entirely lying in the opposite hemisphere. We assessed and compare the values of such index in the static epileptic network and in the dynamic epileptic subnetwork, for each patient.
3. Results
3.1. Static epileptic network identification
The static epileptic network consisted in a median across subjects of 10 fROIs. The static FC matrices for all individuals are reported in Supplementary Materials (Figures S1-S10) together with the original IED-related BOLD map.
3.2. Dynamics of connections and correlation with the epileptic activity
The spatial patterns of the dynamic epileptic subnetworks obtained with a window-size of 40 and 50 s were comparable with the ones resulting from the chosen window of 30 s (spatial correlation equal to 0.8 ± 0.1 with 40 s and 0.8 ± 0.2 with 50 s).
The distribution of sizes of the dynamic epileptic subnetwork, expressed as percentage of connections with respect to the number of connections of the full static epileptic network in each subject, is shown in Fig. 2. The median value was equal to 25%, with remarkable differences across patients (range 10%-100%), even if in the majority of cases (9/10 patients) the size of the dynamic subnetwork showed a reduction with respect to the full (static) epileptic network of at least 30% (relative subnetwork size<=70%, see Fig. 2), with a maximum reduction of 90% and median value of 80%, across the patients.
Fig. 2.
Size of the dynamic epileptic subnetwork. The bar plot in a) shows in red the percentage of connections that are part of the dynamic epileptic subnetwork, with respect to the total number of connections of the full (static) epileptic network, for each patient. In b) the total number of connections N within the epileptic network are reported for each patient. In c), the details on the acquisition parameters (see, Table 2) are shown in color-coded modality: red and orange for the MR scanner magnetic strength (3 and 1.5 Tesla, respectively); dark and light blue for the number of electrodes used for the simultaneous EEG-fMRI acquisition (64 and 256, respectively); dark, semi-dark and light green for parameters of the functional sequence used for the fMRI (combination of the repetition time (TR) expressed in seconds and the number of functional volumes). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A linear trend was found between the size of the dynamic epileptic subnetwork and the duration of the disease, across patients: longer disease duration resulted in a higher percentage of connections included in the dynamic epileptic subnetwork. The correlation value was found equal to 0.5 but did not reach the significance threshold (p-value = 0.08), likely due to the reduced sample size of our patients’ group.
3.3. Spatial organization of the dynamic epileptic sub-network
The spatial organization of the dynamic subnetwork was heterogeneous across patients, in line with the diverse characteristics of the disease, as expressed as well by the number of initial fROIs (Fig. 3).
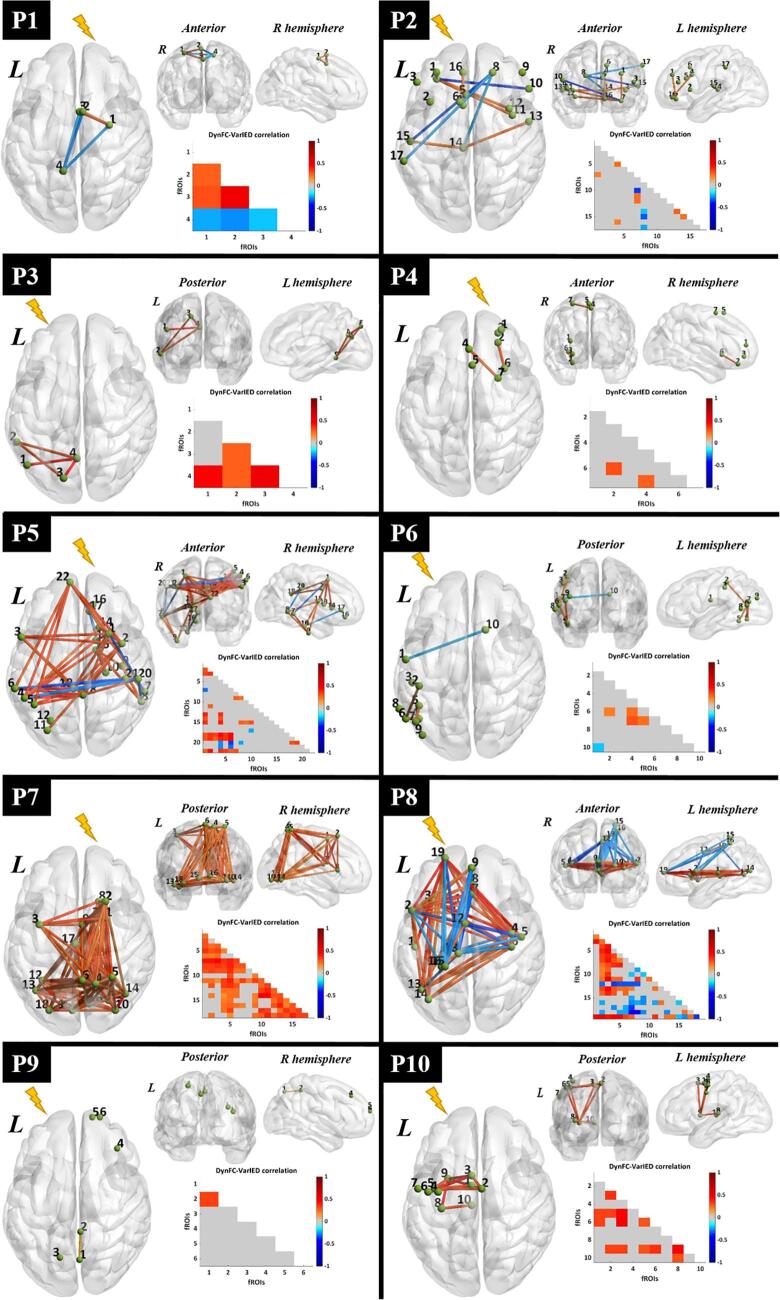
Fig. 3.
Visualization of dynamic epileptic subnetwork. For each patient, the dynamic epileptic subnetwork is shown in the form of a brain graph in axial, coronal and sagittal view. Green spheres of equal size represent fROIs, labelled with a number indicating their statistical relevance in the epileptic network (Paragraph 2.7). The strength of significant connections is color-coded according to a global color bar scaled in the range [-1, 1]. The dynamic epileptic subnetwork is also reported in the form of lower triangular correlation matrix with equivalent color-code. The lightning bolt indicates the epileptogenic hemisphere for each patient. L : left ; R : right. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The percentages of ipsilateral, contralateral and inter-hemispheric connections within the dynamic epileptic subnetwork are reported for each patient in Fig. 4. A significant larger proportion of ipsilateral and inter-hemispheric connections than contralateral ones were observed (p < 0.05, Wilcoxon ranksum test). Across patients, the average percentages of such connections, evaluated over the total amount of connections within the dynamic epileptic subnetwork, were 51%, 45% and 4%, respectively.
Fig. 4.
Summary of the dynamic epileptic subnetwork lateralization with respect to the focus. For each patient, the percentages of intra-hemispheric connections ipsilateral (in yellow) to the epileptic focus, contralateral (in light blue) to the epileptic focus as well as inter-hemispheric (in teal) are reported, normalized to the total number of connections of the dynamic epileptic subnetwork. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Further, at the group level, the number of connections (ipsilateral, contralateral and inter-hemispheric) that positively correlated with VarIED were significantly higher than the connections that negatively correlated (Wilcoxon rank sum test, p-value < 0.05).
By considering the results for the laterality index of connections (Eq.1), in respect of the static epileptic network, the dynamic subnetwork showed: i) higher lateralisation (closer to 1) in 4/10 patients; ii) equal lateralisation in 4/10 patients; iii) lower lateralisation in 2/10 patients, as shown in Fig. 5. Importantly, for patients P2 and P8, who reported a good post-surgical outcome, was found higher for the dynamic subnetwork than for the static network. Instead, in one of the patients who reported bad surgical outcome, P9, the dynamic subnetwork had inverted sign with respect to the static network, pointing to the hemisphere opposite to the one where the surgery was performed. For the other one, P4, no difference was observed.
Fig. 5.
Laterality index of the connections. For each patient, the laterality index of connections, evaluated according to the definition in (Eq.1), is shown for the static epileptic network (pink) and for the dynamic epileptic subnetwork (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
This study aimed at better characterising how focal epileptic activity modulates functional connectivity between nodes of the epileptic network, defined as brain areas that activate with the epileptic focus. We built the current work on previous findings (Iannotti et al., 2016), which described the persistent spatial organization of this network, independently from the IED occurrence. In more details, we included patients who had a widespread (or multilobar) BOLD activation, including at least two clusters located in two distinct lobes. We hypothesised that only a sub-portion of this (static) epileptic network, defined here as dynamic epileptic subnetwork, might show connectivity fluctuations related to the occurrence of IED. These connections seem to play a more relevant role in the pathophysiological processes with respect to the rest of the epileptic network. Therefore, their role in the pre-surgical evaluation and potential added value for targeting epilepsy surgery should be further evaluated. To identify the dynamic epileptic subnetwork, we used a novel approach that we define dFC-EEG, integrating dynamic functional connectivity with the EEG dynamics.
Given the high clinical heterogeneity of our patients, the connections included in the dynamic epileptic subnetwork were highly variable in number, proportion and strength across individuals. This emphasizes the importance of an individualized approach, such as the one proposed, which aims at finding the most efficient intervention for each patient. In 90% of cases the dynamic subnetwork was reduced of at least 30% of the original epileptic network. This confirms the hypothesis that a subset of the regions identified by the static analysis appears to play a particularly critical role in the epileptic network and shows an IED-related connectivity behavior. Further, this subset seems to increase in proportion with the disease duration, even if the correlation between these two variables was found to be non-significant, probably due to the small sample size considered.
Our findings suggest an increase in connectivity within the dynamic epileptic subnetwork correlated to the occurrence of IED (predominant positive correlation of dFC timecourses with the IED regressor), which can be seen as a strengthening of connections between selected regions during IED.
In the literature, increased lateralization of nodes from the epileptic network is associated to good outcomes (Negishi et al., 2011, Zhang et al., 2015). In our case, most of the dynamic subnetwork connections were found to be ipsilateral to the epileptic focus (51% involving two ipsilateral nodes vs 4% involving two contralateral nodes). This suggests a major intrahemispheric involvement ispilateral to the presumed epileptogenic zone. Moreover, switching the observation from nodes to connections, by assessing the lateralisation of connections, enhances the description of the network properties with measures that may correlate with the post-surgical outcomes. Indeed, the evaluation of this index in the subgroup of patients with subsequent surgery demonstrated a higher lateralization of the dynamic subnetwork than the static network to the epileptic hemisphere. In particular, 2/2 patients with good post-operative outcome had higher lateralisation value of the dynamic subnetwork than the static network. On the other side, in 1/2 patient with poor outcome, the dynamic network was lateralized to the hemisphere contralateral to the unsuccessful surgery. Larger studies including post-operative follow-up will show whether such laterality index of dynamic epileptic subnetworks could serve as a tool for the identification of the epileptic hemisphere and a predictive index of surgery outcome after resective or disconnective surgery.
Connectivity studies based on intracranial EEG can offer more specific and time-resolved information about the coupling between structures within or outside the different zones of the epileptic network (Lagarde et al., 2018), but their sparse and often unilateral spatial distribution may not always inform about the global lateralisation of these findings.
Recent studies have linked the dFC of epileptic networks to features extracted from the EEG in EEG-fMRI recordings. For example, Abreu et al. demonstrated the advantage of integrating information about EEG synchronization to retrieve the brain (dynamic) states in 5 epileptic patients (Abreu et al., 2019, Abreu et al., 2018). Omidvarnia et al. studied the coupling between interictal EEG power and local fMRI connectivity demonstrating that it adds complementary information to the standard IED-related BOLD in defining the extent of epileptic networks in focal epilepsy (Omidvarnia et al., 2017). Our work uses a more direct approach to model epileptic activity in the dynamic FC analysis, allowing to highlight specific connections that are directly modulated by the IED.
The results of this work on dFC-EEG compared to the static FC approach adopted previously (Iannotti et al., 2016), suggest that the network obtained by static FC analysis represents the global epileptic network, from which only selected connections (i.e., the components of the dynamic epileptic subnetwork) emerge as being modulated by the IED. Based on our preliminary observations on operated patients, we speculate that these specific connections might prove clinically useful in epileptic surgery to tailor resection/disconnection of regions connected to the epileptogenic zone.
4.1. Methodological considerations
Previous studies used a sliding-window approach to investigate the dynamic of the epileptic networks and focused mainly on temporal lobe epilepsy patients (Laufs et al., 2014). In our work, we extended the analysis to different epilepsy types.
The proposed methodology to integrate sliding-window FC and EEG was introduced in a preliminary study on two focal epilepsy patients (Preti et al., 2014). We propose here an individual-level approach to define the regions of interest (fROIs) for the FC analysis, to avoid the limitations associated to the use of an anatomical atlas. Indeed, the choice of the atlas and the associated spatial resolution (i.e., number of parcels) was shown to influence FC results and their interpretation (de Reus and van den Heuvel, 2013, Wang et al., 2009). Moreover, the reliability of an atlas-based approach becomes even more questionable in clinical populations, as it generally relies on brain sub-divisions extracted from healthy subjects and does not consider the re-organization (both in structure and function) of the diseased brains (Holmes et al., 2014, McCormick et al., 2013). Instead of using an anatomical atlas, we have therefore considered functional regions that are part of the subject-specific epileptic network, so that the analysis is adapted to each patient’s specific clinical situation. Moreover, the definition of fROIs as spheres of equal diameter centered in the local maxima of the epileptic network instead of the original clusters, allowed for an adequate coverage of the regions of interest by avoiding, at the same time, the bias due to the regional BOLD cluster size in the estimation of the fROIs timecourses. The information from the scalp EEG was initially used to identify the fROIs; i.e., regions whose activity correlates to the IED, and then to detect how the connectivity between these regions is modulated by the dynamics of IED.
The choice of limiting the dynamic analysis to the obtained static epileptic network was made under the reasonable assumption that the regions involved in the epileptic process are part of this network. This restriction does not directly affect the dFC results for the considered regions, as the dFC analysis acts connection-wise. However, some regions that only briefly synchronize with the IED may not show a significant correlation on the (static) analysis of the whole recording duration. Such regions would potentially not be revealed by static FC and therefore be excluded from the dynamic analysis.
Future developments might explore other ways to identify the dynamic epileptic network, by deriving dynamically co-activated spatial patterns (iCAPs) from periods of absence of IED (Karahanoglu and Van De Ville, 2015), or the development of a voxel-wise method allowing to cover the whole brain without the need of an atlas (e.g., Preti and Van De Ville, 2017), also overcoming the limitations deriving from the selection of the threshold used for the extraction of fROIs.
The identification of the dynamic epileptic subnetwork relies on the dynamics of IED identified on the scalp, and is therefore limited to the number of events visible on scalp EEG. Simultaneous intracranial-EEG and fMRI could improve the analysis and better identify the effect of epileptiform cortical activity on the epileptic network connectivity.
In the GLM used to define the IED-related BOLD map for the identification of the epileptic focus (global maximum), that was concordant with the clinical assessment, we did not include regressors accounting for physiological noise (namely, cardiac and respiratory signals). Other work has shown that the addition of such confounds could enhance the identification of the network activated by the IED in epileptic patients (Abreu et al., 2017, Liston et al., 2006, van Houdt et al., 2010), even if no significant difference was observed in the spatial location of the global maxima (Liston et al., 2006, van Houdt et al., 2010). These findings support our strategy for selecting the IED-correlated maximum as the epileptic seed, which is then used to define the static epileptic network and further to analyse the connections dynamically related to IED occurrence.
All patients were selected based on multifocal BOLD changes correlated with IED. Most of the patients (8/10) either had non-lesional epilepsy (MRI negative, 4/10 patients), large lesions (3/10) or multifocal lesions (1/10). Therefore, the localisation of epileptogenic zone was difficult to estimate in these difficult cases. With these limits in mind, we have performed such estimation based on the putative seizure onset zone and other multimodal non-invasive clinical information. We have confronted this estimation with the localisation of the IED-correlated BOLD change that was used as seed for the connectivity analysis.We found that the seed localisation was concordant with our estimates of the EZ in 9/10 patients. In the discordant patient, the IED localisation was discordant with the epileptogenic lesion (central midline IED anterior to parietal lesion). The IED networks may therefore show a maximum related to propagation of epileptic activity rather than to EZ.
In literature, a good reproducibility of the BOLD response, across different acquisition sessions, acquisition parameters and MR scanner strength, has been shown both in healthy subjects and in patients with epilepsy (Brown et al., 2011, Gountouna et al., 2010, Voyvodic, 2006). In particular, selected epileptic patients (5/7) with concordant frequency of IED during simultaneous EEG-fMRI evaluated at 1.5 and 3 Tesla showed similarity in the spatial extent of the IED-related BOLD map. However, the statistical power and the definition of the global maxima in the BOLD map was higher at a high magnetic field. Moreover, it remains difficult to assess the influence of the moderate differences in recording parameters on the characteristics of the IED-related BOLD map and on the number of fROIs, because a very important factor is the frequency and pattern of occurrence of sporadic IED that may vary across recording sessions. With this in mind, we favored the choice of the highest scanner strength which was available at the time of the acquisition for each patient (1.5 or 3 T), in order to profit of better signal to noise ratio. Further, we adopted one of the most advanced acquisition sequence (for TR, number of volumes, etc.) and validated processing algorithms, to optimally reduce artefacts from the different acquisition strategies.
4.2. Limitations and future perspectives
In this work, the patients’ selection was stringent, as we selected subjects with diagnosed focal (non temporal lobe) epilepsy, showing a widespread epileptic activity at the EEG-fMRI investigation. The spatial distribution of the network derived by the EEG-fMRI was including at least two distinct clusters of activation located in at least two different lobes (as visually assessed by an expert neurologist, see Column 9 of Table 1) and extended to both hemispheres in almost all the patients (i.e., except P4). Even if the proposed method allowed us to study patient-specific patterns, independently from the type and the localisation of the epilepsy, the sample size limits the generalization of our findings.
A relatively short window size (30 s) for the dFC analysis was chosen in this context to be able to capture faster-scale events related to the IED. This was done at the expense of keeping a smaller frequency range in the original timecourses (higher high-pass filter cutoff), in accordance with the recommendations by Leonardi & Van De Ville (2015), to avoid spurious fluctuations. However, we checked that the spectral content of the original IED signal was preserved, and the tests obtained with windows of 40 and 50 s were consistent in terms of dynamic epileptic subnetwork spatial distribution.
Further investigations are needed to establish the role of the identified dynamic epileptic subnetwork as diagnostic and prognostic marker, notably in the pre-surgical planning of patients with epilepsy. For instance, in cases with widespread IED-related BOLD maps, this technique could help to better delineate the areas with activity linked to the epileptic focus that should be considered for the surgical intervention. However, the small number of available post-surgical outcomes in the current study limits our capability to generalize the potential benefits of this approach to the surgical interventions.
The method shown here was applied to patients with sustained IED during the simultaneous EEG-fMRI recordings. The use of other markers of epileptic activity could be considered to define the nodes of the epileptic network to be studied by dFC, such as the patient’s specific EEG topographic map associated to the IED (Grouiller et al., 2011) or EEG-based connectivity measures (Quin, Jian et al, Neuroimage Clin 2019). Alternatively, the simultaneous acquisition of intracranial EEG and fMRI, iEEG-fMRI (Hawsawi et al., 2017, Ridley et al., 2017, Vulliemoz et al., 2011) may help to better characterize the regional functional coupling and its interaction with IED with a better electrophysiological sensitivity and regional specificity. Finally, the exploration of directed FC based on EEG analysis (Coito et al., 2016, Coito et al., 2015), or fMRI with underlying biophysical models (Friston et al., 2013, Friston, 2011) could add directionality information and better characterize the behavior of the dynamic epileptic subnetwork.
5. Conclusions
In this work, we demonstrated that dFC-EEG allows to identify a dynamic epileptic subnetwork, which involves brain areas whose connectivity is directly influenced by the IED. Beyond the methodological relevance, represented by the direct integration of dFC with the EEG information about the IED occurrence, this method could be of particular utility in clarifying the condition of specific patients, for whom IED modulate a highly widespread activation (often involving the hemisphere contralateral to the focus). By supplying a tool to identify more critical brain regions, this approach could help in future neurosurgical decisions for these patients with widespread epileptic network, where it appears fundamental to restrict the surgical resection to selected brain areas or perform limited disconnections.
CRediT authorship contribution statement
G.R. Iannotti: Conceptualization, Data curation, Formal analysis, Methodology, Writing - original draft, Writing - review & editing. M.G. Preti: Formal analysis, Methodology, Writing - original draft, Writing - review & editing. F. Grouiller: Data curation, Methodology. M. Carboni: Methodology. P. De Stefano: Methodology. F. Pittau: Conceptualization. S. Momjian: Investigation. D. Carmichael: Data curation. M. Centeno: Investigation, Resources. M. Seeck: Investigation, Resources. C.M. Korff: Investigation, Resources. K. Schaller: Investigation, Resources, Funding acquisition. D. Van De Ville: Formal analysis, Methodology, Supervision, Writing - original draft, Writing - review & editing. S. Vulliemoz: Conceptualization, Funding acquisition, Resources, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project was funded by the Swiss National Science Foundation (SNSF) under grants CRSII5_170873, 169198, 320030_182497/1, 320030_182497 and by the Gertrude von Meissner Foundation. This work was supported in part by the Center for Biomedical Imaging (CIBM) of the Geneva - Lausanne Universities and the EPFL (192749).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102467.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abreu R., Leal A., Figueiredo P. Identification of epileptic brain states by dynamic functional connectivity analysis of simultaneous EEG-fMRI: a dictionary learning approach. Sci Rep. 2019;9:638. doi: 10.1038/s41598-018-36976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu R., Leal A., Lopes da Silva F., Figueiredo P. EEG synchronization measures predict epilepsy-related BOLD-fMRI fluctuations better than commonly used univariate metrics. Clin Neurophysiol. 2018;129:618–635. doi: 10.1016/j.clinph.2017.12.038. [DOI] [PubMed] [Google Scholar]
- Abreu R., Nunes S., Leal A., Figueiredo P. Physiological noise correction using ECG-derived respiratory signals for enhanced mapping of spontaneous neuronal activity with simultaneous EEG- fMRI. Neuroimage. 2017;154:115–127. doi: 10.1016/j.neuroimage.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Allen P.J., Josephs O., Turner R. A method for removing imaging artifact from continuous EEG recorded during functional MRI. Neuroimage. 2000;12:230–239. doi: 10.1006/nimg.2000.0599. [DOI] [PubMed] [Google Scholar]
- Allen P.J., Polizzi G., Krakow K., Fish D.R., Lemieux L. Identification of EEG events in the MR scanner: the problem of pulse artifact and a method for its subtraction. Neuroimage. 1998;8:229–239. doi: 10.1006/nimg.1998.0361. [DOI] [PubMed] [Google Scholar]
- Bagshaw A.P., Aghakhani Y., Benar C.G., Kobayashi E., Hawco C., Dubeau F., Pike G.B., Gotman J. EEG-fMRI of focal epileptic spikes: analysis with multiple haemodynamic functions and comparison with gadolinium-enhanced MR angiograms. Hum Brain Mapp. 2004;22:179–192. doi: 10.1002/hbm.20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg A.T., Berkovic S.F., Brodie M.J., Buchhalter J., Cross J.H., van Emde Boas W., Engel J., French J., Glauser T.A., Mathern G.W., Moshe S.L., Nordli D., Plouin P., Scheffer I.E. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- Bettus G., Bartolomei F., Confort-Gouny S., Guedj E., Chauvel P., Cozzone P.J., Ranjeva J.P., Guye M. Role of resting state functional connectivity MRI in presurgical investigation of mesial temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 2010;81:1147–1154. doi: 10.1136/jnnp.2009.191460. [DOI] [PubMed] [Google Scholar]
- Biswal B., Yetkin F.Z., Haughton V.M., Hyde J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Brown G.G., Mathalon D.H., Stern H., Ford J., Mueller B., Greve D.N., McCarthy G., Voyvodic J., Glover G., Diaz M., Yetter E., Ozyurt I.B., Jorgensen K.W., Wible C.G., Turner J.A., Thompson W.K., Potkin S.G. Multisite reliability of cognitive BOLD data. Neuroimage. 2011;54:2163–2175. doi: 10.1016/j.neuroimage.2010.09.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno M., Carmichael D.W. Network Connectivity in Epilepsy: Resting State fMRI and EEG- fMRI Contributions. Front Neurol. 2014;5:93. doi: 10.3389/fneur.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Glover G.H. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S., Vankov E.R., Yeh H.J., Guindani M., Vannucci M., Haneef Z., Stern J.M. Temporal and spectral characteristics of dynamic functional connectivity between resting-state networks reveal information beyond static connectivity. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coito A., Genetti M., Pittau F., Iannotti G.R., Thomschewski A., Holler Y., Trinka E., Wiest R., Seeck M., Michel C.M., Plomp G., Vulliemoz S. Altered directed functional connectivity in temporal lobe epilepsy in the absence of interictal spikes: A high density EEG study. Epilepsia. 2016;57:402–411. doi: 10.1111/epi.13308. [DOI] [PubMed] [Google Scholar]
- Coito A., Plomp G., Genetti M., Abela E., Wiest R., Seeck M., Michel C.M., Vulliemoz S. Dynamic directed interictal connectivity in left and right temporal lobe epilepsy. Epilepsia. 2015;56:207–217. doi: 10.1111/epi.12904. [DOI] [PubMed] [Google Scholar]
- de Reus M.A., van den Heuvel M.P. The parcellation-based connectome: limitations and extensions. Neuroimage. 2013;80:397–404. doi: 10.1016/j.neuroimage.2013.03.053. [DOI] [PubMed] [Google Scholar]
- Friston K., Moran R., Seth A.K. Analysing connectivity with Granger causality and dynamic causal modelling. Curr Opin Neurobiol. 2013;23:172–178. doi: 10.1016/j.conb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K.J. Friston Functional and effective connectivity: a review. Brain Connect 1, 13–36. [DOI] [PubMed]
- Gotman, J., 2008. Epileptic networks studied with EEG-fMRI. Epilepsia 49 Suppl 3, 42–51. [DOI] [PMC free article] [PubMed]
- Gountouna, V.E., Job, D.E., McIntosh, A.M., Moorhead, T.W., Lymer, G.K., Whalley, H.C., Hall, J., Waiter, G.D., Brennan, D., McGonigle, D.J., Ahearn, T.S., Cavanagh, J., Condon, B., Hadley, D.M., Marshall, I., Murray, A.D., Steele, J.D., Wardlaw, J.M., Lawrie, S.M., 2010. Functional Magnetic Resonance Imaging (fMRI) reproducibility and variance components across visits and scanning sites with a finger tapping task Neuroimage 49 2011 552 560. [DOI] [PubMed]
- Grouiller F., Thornton R.C., Groening K., Spinelli L., Duncan J.S., Schaller K., Siniatchkin M., Lemieux L., Seeck M., Michel C.M., Vulliemoz S. With or without spikes: localization of focal epileptic activity by simultaneous electroencephalography and functional magnetic resonance imaging. Brain. 2011;134:2867–2886. doi: 10.1093/brain/awr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawsawi H.B., Carmichael D.W., Lemieux L. Safety of Simultaneous Scalp or Intracranial EEG during MRI: A Review. Frontiers. Physics. 2017;5 [Google Scholar]
- Hindriks, R., Adhikari, M.H., Murayama, Y., Ganzetti, M., Mantini, D., Logothetis, N.K., Deco, G., 2016. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage 127, 242-256. [DOI] [PMC free article] [PubMed]
- Holmes M., Folley B.S., Sonmezturk H.H., Gore J.C., Kang H., Abou-Khalil B., Morgan V.L. Resting state functional connectivity of the hippocampus associated with neurocognitive function in left temporal lobe epilepsy. Hum Brain Mapp. 2014;35:735–744. doi: 10.1002/hbm.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison R.M., Womelsdorf T., Allen E.A., Bandettini P.A., Calhoun V.D., Corbetta M., Della Penna S., Duyn J.H., Glover G.H., Gonzalez-Castillo J., Handwerker D.A., Keilholz S., Kiviniemi V., Leopold D.A., de Pasquale F., Sporns O., Walter M., Chang C. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti G.R., Grouiller F., Centeno M., Carmichael D.W., Abela E., Wiest R., Korff C., Seeck M., Michel C., Pittau F., Vulliemoz S. Epileptic networks are strongly connected with and without the effects of interictal discharges. Epilepsia. 2016;57:1086–1096. doi: 10.1111/epi.13400. [DOI] [PubMed] [Google Scholar]
- Iannotti G.R., Pittau F., Michel C.M., Vulliemoz S., Grouiller F. Pulse artifact detection in simultaneous EEG-fMRI recording based on EEG map topography. Brain Topogr. 2015;28:21–32. doi: 10.1007/s10548-014-0409-z. [DOI] [PubMed] [Google Scholar]
- Karahanoglu F.I., Van De Ville D. Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat Commun. 2015;6:7751. doi: 10.1038/ncomms8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H. Functional imaging of seizures and epilepsy: evolution from zones to networks. Curr Opin Neurol. 2012;25:194–200. doi: 10.1097/WCO.0b013e3283515db9. [DOI] [PubMed] [Google Scholar]
- Laufs H., Rodionov R., Thornton R., Duncan J.S., Lemieux L., Tagliazucchi E. Altered FMRI connectivity dynamics in temporal lobe epilepsy might explain seizure semiology. Front Neurol. 2014;5:175. doi: 10.3389/fneur.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A.D., Lund T.E., Salek-Haddadi A., Hamandi K., Friston K.J., Lemieux L. Modelling cardiac signal as a confound in EEG-fMRI and its application in focal epilepsy studies. Neuroimage. 2006;30:827–834. doi: 10.1016/j.neuroimage.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Lopes R., Moeller F., Besson P., Ogez F., Szurhaj W., Leclerc X., Siniatchkin M., Chipaux M., Derambure P., Tyvaert L. Study on the Relationships between Intrinsic Functional Connectivity of the Default Mode Network and Transient Epileptic Activity. Front Neurol. 2014;5:201. doi: 10.3389/fneur.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., An D., Yao D., Gotman J. Patient-specific connectivity pattern of epileptic network in frontal lobe epilepsy. Neuroimage Clin. 2014;4:668–675. doi: 10.1016/j.nicl.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C., Quraan M., Cohn M., Valiante T.A., McAndrews M.P. Default mode network connectivity indicates episodic memory capacity in mesial temporal lobe epilepsy. Epilepsia. 2013;54:809–818. doi: 10.1111/epi.12098. [DOI] [PubMed] [Google Scholar]
- McKeown M.J., Sejnowski T.J. Independent component analysis of fMRI data: examining the assumptions. Hum Brain Mapp. 1998;6:368–372. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<368::AID-HBM7>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan V.L., Englot D.J., Rogers B.P., Landman B.A., Cakir A., Abou-Khalil B.W., Anderson A.W. Magnetic resonance imaging connectivity for the prediction of seizure outcome in temporal lobe epilepsy. Epilepsia. 2017;58:1251–1260. doi: 10.1111/epi.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M., Martuzzi R., Novotny E.J., Spencer D.D., Constable R.T. Functional MRI connectivity as a predictor of the surgical outcome of epilepsy. Epilepsia. 2011;52:1733–1740. doi: 10.1111/j.1528-1167.2011.03191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidvarnia A., Pedersen M., Vaughan D.N., Walz J.M., Abbott D.F., Zalesky A., Jackson G.D. Dynamic coupling between fMRI local connectivity and interictal EEG in focal epilepsy: A wavelet analysis approach. Hum Brain Mapp. 2017;38:5356–5374. doi: 10.1002/hbm.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittau F., Dubeau F., Gotman J. Contribution of EEG/fMRI to the definition of the epileptic focus. Neurology. 2012;78:1479–1487. doi: 10.1212/WNL.0b013e3182553bf7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittau F., Megevand P., Sheybani L., Abela E., Grouiller F., Spinelli L., Michel C.M., Seeck M., Vulliemoz S. Mapping epileptic activity: sources or networks for the clinicians? Front Neurol. 2014;5:218. doi: 10.3389/fneur.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M.G., Bolton T.A., Van De Ville D. The dynamic functional connectome: State-of-the-art and perspectives. Neuroimage. 2017;160:41–54. doi: 10.1016/j.neuroimage.2016.12.061. [DOI] [PubMed] [Google Scholar]
- Preti MG, L.N., Işik Karahanŏglu F, Grouiller F, Genetti M, Seeck M, Vulliemoz S and Van De Ville D, 2014. EPILEPTIC NETWORK ACTIVITY REVEALED BY DYNAMIC FUNCTIONAL CONNECTIVITY IN SIMULTANEOUS EEG-FMRI. 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI).
- Preti M.G., Van De Ville D. Dynamics of functional connectivity at high spatial resolution reveal long-range interactions and fine-scale organization. Sci Rep. 2017;7:12773. doi: 10.1038/s41598-017-12993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, B., Wirsich, J., Bettus, G., Rodionov, R., Murta, T., Chaudhary, U., Carmichael, D., Thornton, R., Vulliemoz, S., McEvoy, A., Wendling, F., Bartolomei, F., Ranjeva, J.P., Lemieux, L., Guye, M., 2017. Simultaneous Intracranial EEG-fMRI Shows Inter-Modality Correlation in Time-Resolved Connectivity Within Normal Areas but Not Within Epileptic Regions. Brain Topogr 30, 639-655. [DOI] [PubMed]
- Smith, S.M., Beckmann, C.F., Andersson, J., Auerbach, E.J., Bijsterbosch, J., Douaud, G., Duff, E., Feinberg, D.A., Griffanti, L., Harms, M.P., Kelly, M., Laumann, T., Miller, K.L., Moeller, S., Petersen, S., Power, J., Salimi-Khorshidi, G., Snyder, A.Z., Vu, A.T., Woolrich, M.W., Xu, J., Yacoub, E., Ugurbil, K., Van Essen, D.C., Glasser, M.F., Consortium, W.U.-M.H., 2013. Resting-state fMRI in the Human Connectome Project. Neuroimage 80, 144-168. [DOI] [PMC free article] [PubMed]
- Tagliazucchi E., Laufs H. Multimodal imaging of dynamic functional connectivity. Front Neurol. 2015;6:10. doi: 10.3389/fneur.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton R., Vulliemoz S., Rodionov R., Carmichael D.W., Chaudhary U.J., Diehl B., Laufs H., Vollmar C., McEvoy A.W., Walker M.C., Bartolomei F., Guye M., Chauvel P., Duncan J.S., Lemieux L. Epileptic networks in focal cortical dysplasia revealed using electroencephalography- functional magnetic resonance imaging. Ann Neurol. 2011;70:822–837. doi: 10.1002/ana.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houdt P.J., Ossenblok P.P., Boon P.A., Leijten F.S., Velis D.N., Stam C.J., de Munck J.C. Correction for pulse height variability reduces physiological noise in functional MRI when studying spontaneous brain activity. Hum Brain Mapp. 2010;31:311–325. doi: 10.1002/hbm.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyvodic J.T. Activation mapping as a percentage of local excitation: fMRI stability within scans, between scans and across field strengths. Magn Reson Imaging. 2006;24:1249–1261. doi: 10.1016/j.mri.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Vulliemoz S., Carmichael D.W., Rosenkranz K., Diehl B., Rodionov R., Walker M.C., McEvoy A.W., Lemieux L. Simultaneous intracranial EEG and fMRI of interictal epileptic discharges in humans. Neuroimage. 2011;54:182–190. doi: 10.1016/j.neuroimage.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang L., Zang Y., Yang H., Tang H., Gong Q., Chen Z., Zhu C., He Y. Parcellation- dependent small-world brain functional networks: a resting-state fMRI study. Hum Brain Mapp. 2009;30:1511–1523. doi: 10.1002/hbm.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser H.G., Blume W.T., Fish D., Goldensohn E., Hufnagel A., King D., Sperling M.R., Luders H., Pedley T.A., Commission on Neurosurgery of the International League Against E. ILAE Commission Report. Proposal for a new classification of outcome with respect to epileptic seizures following epilepsy surgery. Epilepsia. 2001;42:282–286. [PubMed] [Google Scholar]
- Xia M., Wang J., He Y. BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.H., Lu Y., Brinkmann B., Welker K., Worrell G., He B. Lateralization and localization of epilepsy related hemodynamic foci using presurgical fMRI. Clin Neurophysiol. 2015;126:27–38. doi: 10.1016/j.clinph.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.